Summary
Type 4a pili (T4aP) are long, thin and dynamic fibres displayed on the surface of diverse bacteria promoting adherence, motility and transport functions. Genomes of many Enterobacteriaceae contain conserved gene clusters encoding putative T4aP assembly systems. However, their expression has been observed only in few strains including Enterohaemorrhagic Escherichia coli (EHEC) and their inducers remain unknown. Here we used EHEC genomic DNA as template to amplify and assemble an artificial operon composed of four gene clusters encoding 13 pilus assembly proteins. Controlled expression of this operon in non-pathogenic E. coli strains led to efficient assembly of T4aP composed of the major pilin PpdD, as shown by shearing assays and immuno-fluorescence microscopy. When compared with PpdD pili assembled in a heterologous Klebsiella T2SS type 2 secretion system (T2SS) by using cryo-electron microscopy (cryoEM), these pili showed indistinguishable helical parameters, emphasizing that major pilins are the principal determinants of fibre structure. Bacterial two-hybrid analysis identified several interactions of PpdD with T4aP assembly proteins, and with components of the T2SS that allow for heterologous fibre assembly. These studies lay ground for further characterization of T4aP structure, function and biogenesis in enterobacteria.
Keywords: type 4 pili, EHEC O157:H7, electron microscopy, type 2 secretion system, Gibson assembly, bacterial two-hybrid, Sxy, type 4 filaments
Graphical abstract
Introduction
Pili or fimbriae are hair-like appendages that decorate the surface of many bacteria and allow them to adhere to biotic or abiotic substrates. Among the prokaryotic surface structures, T4P are arguably the most widespread and ancient, sharing common evolutionary origins with archaeal flagella and pili (Makarova et al., 2016). Together with bacterial T2SSs, they are members of the type 4 filament (Tff) superfamily of structurally and functionally related prokaryotic nanomachines (Berry & Pelicic, 2015). T4P are present in Gram-positive and Gram-negative bacteria (Berry & Pelicic, 2015, Melville & Craig, 2013, Hospenthal et al., 2017). They can be grouped into three main subtypes, a, b and c (or Tad pili), which share common assembly pathway while showing differences in subunit primary sequence, assembly components, and gene organization (Tomich et al., 2007, Pelicic, 2008, Ellison et al., 2017). While the genes encoding T4b and T4c pili are typically clustered in a single operon, those encoding T4aP are split in several gene clusters localized in conserved positions around the bacterial chromosome (Pelicic, 2008).
T4P are mainly built as helical homo-polymers of a single subunit called the major pilin. In addition, one or several minor pilin subunits might participate in assembly, priming the initiation, modulating fibre dynamics or surface epitopes (Helaine et al., 2007, Berry et al., 2016, Cehovin et al., 2013, Nguyen et al., 2015, Ng et al., 2016). Pilins are initially inserted in the plasma membrane as precursors containing an N-terminal positively charged cytoplasmic leader peptide acting as a membrane anchor. A dedicated prepilin peptidase PilD removes this peptide and N-methylates mature pilins to allow for their membrane extraction and incorporation at the base of the growing pilus (Strom & Lory, 1991, Strom et al., 1993, Santos-Moreno et al., 2017).
Composition and function of T4aP assembly machinery have been extensively characterized in several model organisms, including Pseudomonas and Neisseria (Leighton et al., 2015a, Berry & Pelicic, 2015). The T4a pilus systems (T4aPS), depicted schematically in Fig. 1A, comprise four sub-complexes: the cytoplasmic ATPase motor complex, the inner-membrane (IM)- anchored assembly platform (AP) complex, the outer-membrane (OM) secretin complex (exclusively in Gram-negative bacteria), and the filament (Chang et al., 2016, Gold et al., 2015). The ATPase motor PilB (according to the P. aeruginosa nomenclature) localized at the base of the system, powers pilus assembly (Karuppiah et al., 2013, Tammam et al., 2013, McCallum et al., 2017). PilB interacts with the platform protein PilC and with the PilM component of the IM complex that also includes PilN and PilO (Ayers et al., 2009, Georgiadou et al., 2012). The IM AP complex is connected via PilP to the OM secretin channel formed by the PilQ multimer (Koo et al., 2016) that allows pilus exposure on the bacterial surface (Wolfgang et al., 1998). The major pilins in P. aeruginosa (Tammam et al., 2013) and Neisseria (Georgiadou et al., 2012) interact with the AP, which presumably transmits conformational changes in PilB to promote pilin polymerization. AP components also orchestrate pilus retraction, powered by the PilT ATPase (Misic et al., 2010, Leighton et al., 2015b). Recently, cryo-electron microscopy brought more detailed insights into several T4aP structures (Kolappan et al., 2016, Wang et al., 2017), while advances in cryo-electron tomography revealed global organization of T4aP assembly systems in situ, in the envelope of Pseudomonas (Gold et al., 2015) and Myxococcus (Chang et al., 2016).
Figure 1.
The T4aP assembly systems. A, Schematic representation of the T4aP assembly system in the Gram-negative bacterial envelope. Major (D, dark green) and minor pilins (in light green) are processed by the prepilin peptidase (O, yellow). Cytoplasmic ATPase (B) transmits conformational changes to the inner membrane (IM) platform protein (C) and the assembly complex M-N-O-P (light purple), which is connected with the secretin (Q, dark purple) in the outer membrane (OM). B, Organization of T4aP gene clusters in EDL933 genome (EHEC) and in the representative species of the order of Enterobacteriaceae: Dickeya dadantii, Serratia marcescens, Salmonella enterica, Klebsiella oxytoca and Proteus mirabilis. The ppd-hof genes are indicated as arrows whose length represents relative gene size, colour-coded as in Fig. 1A. Genes adjacent to T4P clusters are represented as black and grey arrows reflecting relative degree of conservation. For EHEC, annotation numbers are indicated above each T4P gene in blue.
The Escherichia coli chromosome harbours genes that encode all components of a putative T4aP assembly system, similar to the well-studied systems in P. aeruginosa and Neisseria spp. The promoter regions of predicted T4aP operons in E. coli contain binding motifs called CRP-S, reminiscent of the catabolic repressor protein (CRP) binding sites (Cameron & Redfield, 2006). These CRP-S motifs are low-affinity binding sites for the catalytic repressor protein (CRP) that require cAMP and the natural competence activator Sxy (or TfoX) to allow for transcriptional activation of downstream genes (Cameron & Redfield, 2006). The presence of these sites upstream of T4aP and DNA uptake genes suggests that, like Pasteurellaceae and Vibrionaceae, E. coli and other enterobacteria have the genetic potential for natural competence (Cameron & Redfield, 2006). Indeed, Sxy overproduction in E. coli activates transcription of ppd/hof operons encoding T4P, but also that of genes encoding the DNA uptake machinery (Sinha et al., 2009). Moreover, sxy expression can promote uptake of external DNA (Sinha & Redfield, 2012), supporting the role of T4aP in natural competence. In E. coli K-12 sxy expression is auto-regulated in a CRP-cAMP dependent manner and activated by the stress sigma factor RpoS (Jaskolska & Gerdes, 2015). However transcriptional analyses show that T4P genes are silent in E. coli K-12 under a variety of conditions tested (Sauvonnet et al., 2000a, Sinha et al., 2009) and specific signals and conditions that induce Sxy production in E. coli remain unknown.
The first indication that ppd/hof genes may encode a functional system has come from studies of enterohaemorrhagic E. coli (EHEC), which produce pili composed of the major subunit PpdD in minimal casein medium (Xicohtencatl-Cortes et al., 2007). EHEC is a major intestinal pathogen causing haemorrhagic colitis and haemolytic uremic syndrome (HUS), associated with significant mortality and morbidity (Monteiro et al., 2016). Sera of patients recovering from HUS contain PpdD-reactive antibodies (Xicohtencatl-Cortes et al., 2007). Ectopic expression of the plasmid-borne ppdD-hofBC operon conferred to an E. coli strain HB101 several T4P-linked phenotypes, including adherence and twitching motility (Xicohtencatl-Cortes et al., 2009). Importantly, PpdD pili (also designated HCP, for hemorrhagic coli pili) contribute to EHEC adherence to intestinal tissues (Xicohtencatl-Cortes et al., 2007) and induce pro-inflammatory signalling (Ledesma et al., 2010).
In view of this functional evidence, here we used the ppd/hof genes from the EHEC strain EDL933 as building blocks to reconstitute an artificial T4P assembly operon in non-pathogenic E. coli. Successful reconstitution of pilus assembly in this system allowed us to compare PpdD fibres polymerized by the cognate T4aPS to those produced when PpdD is assembled by the heterologous T2SS of Klebsiella oxytoca (Sauvonnet et al., 2000b, Cisneros et al., 2012b). Despite different PpdD interactions with the assembly components of these systems, the resulting pili are highly similar, consistent with the role of major pilins as key determinants of the fibre symmetry.
Results
Enterobacterial T4aP assembly systems share common features
Genes that encode T4aP assembly systems in E. coli and other enterobacterial genera are found in conserved chromosomal loci (Fig. 1), as highlighted previously (Pelicic, 2008). The ppdD-hofBC operon encoding the major pilin, the ATPase HofB and the platform protein HofC is localised between guaC and ampD genes at 3 min of the E. coli map (genes Z0116–0118 in EHEC strain EDL933) (Perna et al., 2001). At min 61, between the thyA and recC genes, the ppdAB-ygdB-ppdC operon encodes the four minor pilins. The hofMNOPQ operon encodes the assembly platform complex connecting HofB ATPase with the secretin HofQ and is located at min 75 between aroK and mrcA genes (Fig. 1B). The yggR gene encoding a PilT homologue is localized at min 66 (not depicted) and corresponds to Z4295 gene in EHEC. In Pseudomonas and Vibrio genomes the essential prepilin peptidase gene pilD is linked to the major pilin operon pilABCD (Fullner & Mekalanos, 1999). This is not the case in Enterobacteriaceae, where pilD homologues are linked to operons encoding T2SSs. The gene gspO, one of the two prepilin peptidase genes in E. coli K-12, is the last gene of the gsp operon encoding the chitinase-specific T2SS (Francetic et al., 1998, Francetic et al., 2000). In EHEC, almost all gsp genes have been lost during evolution, leaving only the gspO fragment encoding the prepilin peptidase catalytic domain (gene Z4693) (Perna et al., 2001). Additionally, EHEC strains have the gspO homologue etpO within a plasmid-encoded T2SS cluster (Schmidt et al., 1997).
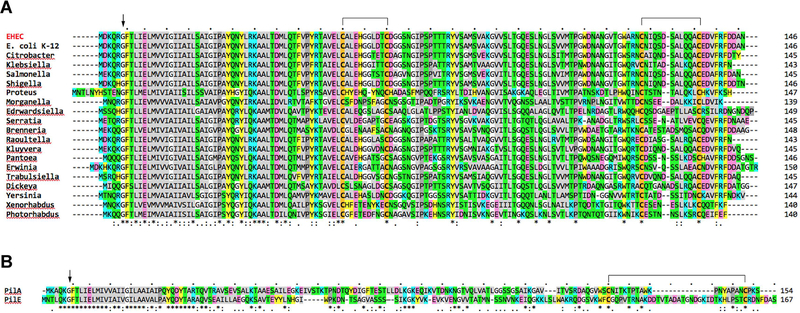
The genes encoding the major T4a pilins are remarkably conserved in all E. coli strains sequenced to date (Fig. 2). A single amino acid difference is found between the primary sequence of the major pilin PpdD from E. coli K-12 and EHEC. Furthermore, there is a high sequence similarity between the EHEC PpdD and major pilins from diverse genera of the Enterobacteriaceae family, including Salmonella (88.4% of identical residues), Yersinia (58.2%), Shigella (97.3%), Raoultella (82.9%), Dickeya (60.1%), Serratia (59.2%), Proteus (42.1%), Citrobacter (89.7%), Klebsiella (86.3%), or Pantoea (60.1%) (Fig. 2A and Fig. S1). Compared to well-studied major pilins, PilE of Neisseria and PilA of Pseudomonas, which contain two cysteine residues near their C-terminus forming a disulphide bond (Fig. 2B) (Parge et al., 1995, Craig et al., 2003), PpdD and its homologues contain an additional pair of Cys residues at positions 50 and 60. In PpdD, NMR and chemical shift analysis showed that these residues form disulphide bonds, one at the junction between the long α-helical stem and the globular domain, and the second close to the C-terminus defining the variable D loop typically found in T4a pilins (de Amorim et al., 2014). These residues are fully conserved in all PpdD homologues (Fig. 2A), suggesting that major pilins in Enterobacteraceae share a similar structure. Like PpdD, the minor pilins PpdA, PpdB, YgdB and PpdC and their homologues in the family contain two conserved Cys pairs at similar positions.
Figure 2.
Protein sequence alignments of major pilin subunits. A, Multiple alignment of major T4a pilin sequences from diverse Enterobacterial strains: EHEC (WP_000360900.1), E. coli K-12 (WP_000360904.1), Citrobacter werkmanii (WP_042312306.1), Klebsiella oxytoca (WP_064398298.1), Salmonella enterica (WP_061382948.1), Shigella dysenteriae (WP_000360691.1), Proteus mirabilis (WP_060558290.1), Morganella morganii (WP_064483293.1), Edwardsiella tarda (WP_005282704.1), Serratia marcescens (WP_060440768.1), Brenneria sp. (WP_009111461.1), Raoultella terrigena (WP_045859871.1), Kluyvera ascorbata (WP_035893064.1), Pantoea theicola (WP_103059883.1), Erwinia toletana (WP_017800450.1), Trabulsiella odontotermitis (WP_049856566.1), Dickeya dadantii (WP_038923864.1), Yersinia enterocolitica (WP_046695439.1), Xenorhabdus nematophila (WP_010845542.1) and Photorhabdus luminescens (WP_011147801.1). The arrowhead indicates the prepilin peptidase cleavage site upstream of the N-terminal F1 residue of mature pilins. B, Pairwise alignment of P. aeruginosa PilA and N. gonorrhoeae PilE. Alignments showing identical (*), conserved (:) and similar (.) residues were performed with Clustal omega (Sievers et al., 2011) and the EMBL-EBI tools (Li et al., 2015). Conserved Cys residues are highlighted in bold with orange background; putative disulphide bridges are indicated above. Hydrophobic (light grey), Pro (dark grey), Gly (white), polar (green), aromatic (yellow), negatively (pink) and positively charged (blue) residues are indicated.
Assembly of the artificial ppd-hof operon
Since the EHEC strain EDL933 assembles functional pili (Xicohtencatl-Cortes et al., 2007), we used its genomic DNA as template to PCR-amplify the ppdD-hofB-hofC, ppdAB-ygdB-ppdC and hofMNOPQ clusters, flanked with ~100 bp of noncoding DNA. We amplified a pBR322-derived vector fragment carrying the replication origin and the ApR selection marker and used the Gibson assembly approach (Gibson et al., 2009) to join these PCR products into a plasmid named pMS10 (Table 2). To complete the artificial operon, we introduced at its distal end the gspO gene, PCR-amplified from E. coli K-12 strain MG1655, to give plasmid pMS43 (Table 2). The choice of gspO was motivated by its position downstream of the Sxy-activated gene gspM in the chitinase T2SS operon in E. coli K-12 (Sinha et al., 2009), suggesting a functional link of this secretion system with T4PS. The only other T4aP-related chromosomal gene yggR encoding the putative retraction ATPase, was not included in our construct, with the aim to reconstitute efficient T4aP assembly, independent on putative retraction.
Table 2.
Plasmids used in this study
| Name | Origin/resistancea | Relevant markers | Reference |
|---|---|---|---|
| pCHAP8565 | p15A/CmR | placZ- ppdD | This study |
| pSU18 | p15A/CmR | multicloning site in lacZ’ | (Bartolome et al., 1991) |
| pBR322 | ColE1/ ApR, TcR | cloning vector | (Bolivar et al., 1977) |
| pCHAP8746 | p15A/CmR | pSU18 sxy | This study |
| pCHAP8184 | ColE1 / ApR | pulAsol all pul genes ΔpulG | (Campos et al., 2010) |
| pCHAP6116 | pSC101, ZeR | ppdAB-ygdB-ppdC | (Cisneros et al., 2012b) |
| pCHAP8889 | p15A/CmR | pSU18 hofC | This study |
| pMS10 | ColE1 / ApR | pBR322-cloned ppdD-hofB-hofC, hofMNOPQ and ppdAB-ygdB-ppdC operons flanked by 100 bp of noncoding DNA | This study |
| pMS37 | ColE1 / ApR | ppdD-hofB-hofC, hofMNOPQ and ppdAB-ygdB-ppdC | This study |
| pMS38 | ColE1 / ApR | ppdD-hofB-hofC, hofMNOPQ, ppdAB-ygdB-ppdC and gspO | This study |
| pMS39 | ColE1 / ApR | placZ ppdD-hofB-hofC, hofMNOPQ, ppdAB-ygdB-ppdC and gspO | This study |
| pMS40 | ColE1 / ApR | ppulC ppdD-hofB-hofC, hofMNOPQ, ppdAB-ygdB-ppdC and gspO | This study |
| pMS41 | ColE1 / ApR | pMS39 ΔppdD | This study |
| pMS43 | ColE1 / ApR | pMS10 carrying gspO | This study |
| pMS45 | ColE1 / ApR | pMS39 ΔppdAB-ygdB-ppdC | This study |
| pMS47 | ColE1 / ApR | pMS39 ΔhofC | This study |
| pUT18c | ColE1 / ApR | placUV5-cyaA T18 fragment in pUC18 | (Karimova et al., 1998) |
| pKT25 | P15A/KmR | placUV5-cyaA T25 fragment in pSU38 | (Karimova et al., 1998) |
| pCHAP8501 | ColE1 / ApR | pUT18c-ppdD | This study |
| pCHAP8504 | p15A/KmR | pKT25-ppdD | This study |
| pCHAP8897 | ColE1/ ApR | pUT18c-hofB | This study |
| pCHAP8945 | p15A/KmR | pKT25-hofB | This study |
| pCHAP8898 | ColE1/ ApR | pUT18c-hofC | This study |
| pCHAP8899 | p15A/KmR | pKT25-hofC | This study |
| pCHAP8907 | ColE1/ ApR | pUT18c-hofN | This study |
| pCHAP8910 | p15A/KmR | pKT25-hofN | This study |
| pCHAP8908 | ColE1/ ApR | pUT18c-hofO | This study |
| pCHAP8911 | p15A/KmR | pKT25-hofO | This study |
| pCHAP8909 | ColE1/ ApR | pUT18c-hofP | This study |
| pCHAP8912 | p15A/KmR | pKT25-hofP | This study |
| pMS30 | ColE1/ ApR | pUT18c-ppdB | This study |
| pMS26 | p15A/KmR | pKT25-ppdB | This study |
| pCHAP8246 | ColE1/ApR | pUT18c-pulJ | (Cisneros et al., 2012a) |
| pCHAP8364 | ColE1/ApR | pUT18c-pulF | (Nivaskumar et al., 2016) |
| pCHAP7330 | ColE1/ApR | pUT18c-pulG | (Nivaskumar et al., 2014) |
| pCHAP7332 | P15A/KmR | pKT25- pulG | (Nivaskumar et al., 2014) |
| pCHAP8154 | ColE1/ApR | pUT18c- pulM | (Nivaskumar et al., 2016) |
| pCHAP8113 | ColE1/ApR | pUT18c-pulC | (Nivaskumar et al., 2016) |
| pCHAP8256 | ColE1/ApR | pUT18c-pulH | (Nivaskumar et al., 2016) |
| pCHAP8472 | ColE1/ApR | pUT18c-pulL | (Nivaskumar et al., 2016) |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Ze, zeocin.
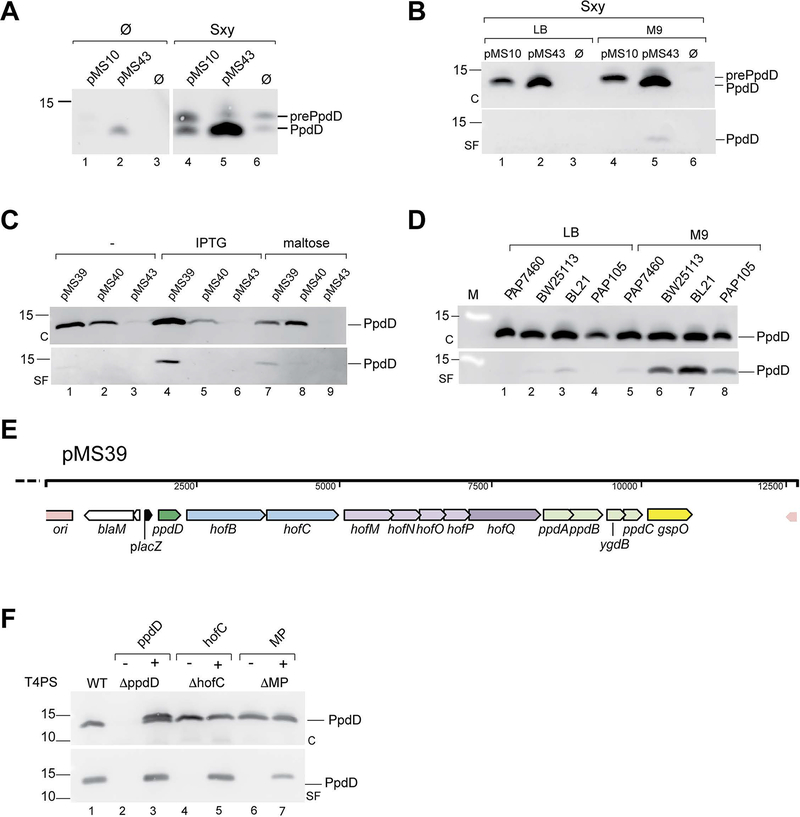
The pMS10 and pMS43 constructs were introduced in E. coli PAP7460 strain to examine production of and assembly of PpdD. In bacteria grown of LB, the expression of ppdD, the first gene in the artificial operon, from plasmid pMS10 was almost indistinguishable from the negative control containing vector alone, as revealed with anti-PpdD antibodies (Fig. 3A, lanes 1 and 3). A faint band corresponding to mature (GspO-processed) PpdD was detected in the strain with plasmid pMS43 (Fig. 3A, lane 2). To test whether Sxy would activate expression of the artificial ppd-hof operon, we cloned the E. coli sxy gene with its 197-bp upstream region under control of the placZ promoter on plasmid pCHAP8746 (Table 2). Co-expression of sxy in strain PAP7460 in the presence of isopropyl β-D-1-thiogalactopyranoside (IPTG) induced both plasmid- and chromosome-encoded PpdD production (Fig. 3A, 4–6). Partial PpdD processing was observed in the presence of pMS10 and vector, showing limiting activity of E. coli prepilin peptidases. The increase of PpdD levels required Sxy auto-regulation and was only observed with constructs containing the CRP-S site upstream of the sxy gene (Fig. S2), confirming previous findings (Jaskolska & Gerdes, 2015). We next tested pilus assembly on bacteria grown on LB or minimal glycerol plates. Pili were sheared from the bacterial surface as described (Luna Rico et al., 2018) (Experimental procedures), followed by analysis of cell and sheared fractions by immunodetection with anti-PpdD antibodies. Only a low amount of PpdD pili was found in the sheared fraction of bacteria producing mature PpdD from plasmid pMS43 cultured on minimal medium (Fig. 3B), suggesting that factors or conditions inducing pilus assembly might be missing. We hypothesized that the DNA flanking the T4aPS-coding genes might have caused inefficient translation of one or more components of the system or, alternatively, that Sxy overexpression might have activated the chromosomal retraction ATPase gene yggR to reduce piliation.
Figure 3.
Construction, optimization and complementation analysis of the synthetic ppd-hof operon. A, The Sxy-mediated induction of ppdD expression in E. coli strain PAP7460 carrying pMS10, pMS43 and pBR322, without (Ø) or with sxy gene on plasmid pCHAP8746. Total extracts from 0.15 OD600nm of bacteria cultured on LB plates containing IPTG for 48 hours were analysed by SDS-PAGE and immunodetection using anti-PpdD antibodies (1:1000); B, Bacteria of strain PAP7460 containing plasmid pCHAP8746 and either pMS10, pMS43 or vector pBR322 (Ø) cultured on LB or M9-glycerol media for 48 hours. Pilus assembly was assayed as described in Experimental procedures. C, cell fraction; SF, sheared fraction; M, molecular weight marker. C, Construct optimization. Pilus assembly in strain PAP7460 containing plasmid pMS43, compared with optimized constructs pMS39 and pMS40 cultured in LB (−) or LB containing IPTG or maltose as inducers. D, Pilus assembly in indicated E. coli host strains transformed with plasmid pMS39 and cultured on selective LB or M9 glycerol plates containing IPTG. E, Linear map of the pMS39 construct with an artificial ppd-hof operon under placZ control. Gene names are indicated and colour-coded corresponding to the scheme in Fig. 1A. F, PpdD pilus assembly in pMS39 (WT) and its deletion derivatives lacking ppdD (pMS41), hofC (pMS47) and minor pilin (MP) ppdAB-ygdB-ppdC genes (pMS45). Strain BW25113 F’lacIQ containing complementing plasmids (+) of empty vector (−) were grown on selective M9 glycerol plates containing IPTG. In Panels B, C and F samples of cell (C) and sheared fractions (SF) from 0.05 OD600nm equivalents were analysed by SDS-PAGE and immunoblot with anti-PpdD antibodies diluted 1:2000.
Figure 4.
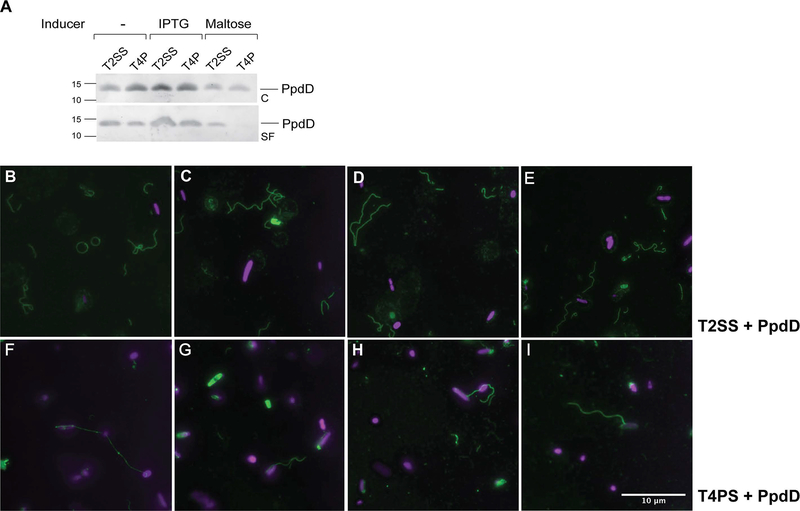
Comparison of PpdD pilus assembly via the T2SS and T4PS. A, E. coli strain BW25113 F’lacIQ containing plasmid pCHAP8565 and pCHAP8184 (T2SS) or pMS41 (T4PS) was grown on LB plates containing AP and Cm at 30°C for 48 hours, without or with indicated inducers. Fractionation and Western blot with antibodies raised against MalE-PpdD fusion protein were performed as described in Experimental procedures. Migration of molecular weight markers is indicated on the left. B-E, Immunofluorescence microscopy analysis of PpdD pili assembled by the T2SS or by the T4PS (F-I). Samples were processed as indicated in Experimental procedures. Pili are labelled in green (GFP) and bacterial nucleoid in magenta (DAPI). Scale bar indicates 10 μM.
Figure 6.
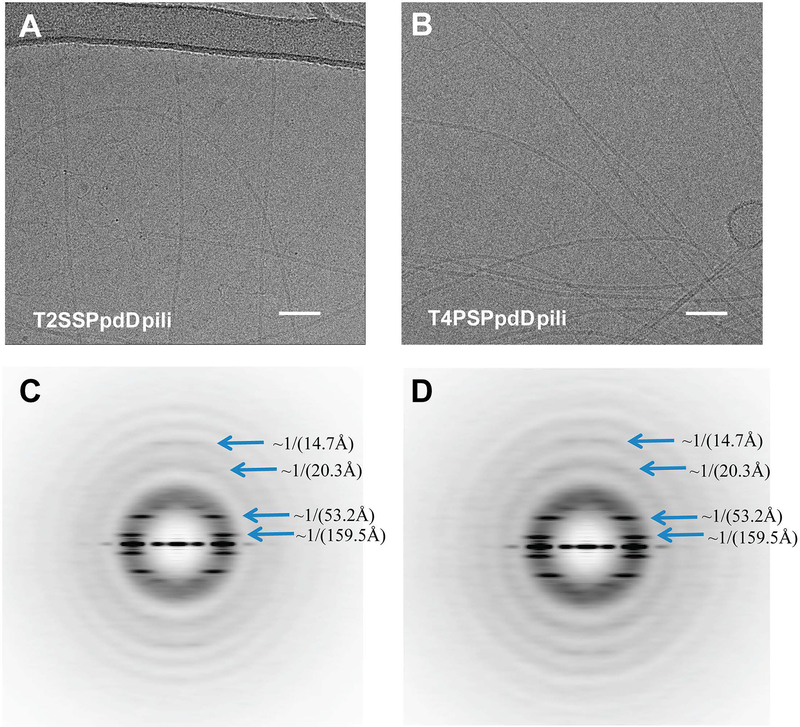
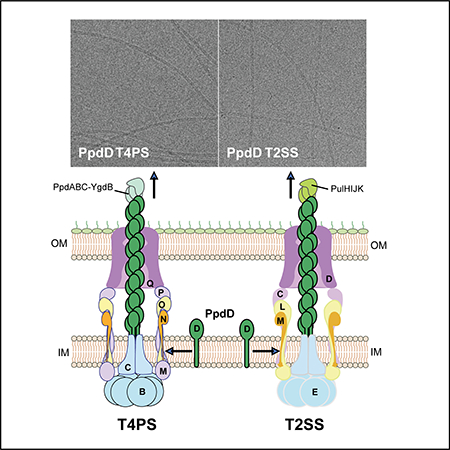
PpdD pili assembled by the Klebsiella Pul T2SS and EHEC T4SP show similar architecture and helical symmetry. Representative cryoEM images of PpdD pili from T2SS (A) and T4PS (B). Scale bar, 50 nm. The helical symmetry of PpdD pili from T2SS (C) and T4PS (D) were determined by the averaged power spectra generated from 384-pixel long segments. Due to the variability of both twist and axial rise in the PpdD filaments, sorting by helical symmetry generates improved power spectra. The power spectra in (C) and (D) were from the ~25% of segments classified as having a twist near 96° and a rise near 10.8 Å. The blue arrows indicate the dominant layer lines.
Optimization of the construct, bacterial hosts and culture conditions
To improve piliation efficiency and to render the expression of T4PS genes independent on Sxy, we sequentially eliminated noncoding DNA from individual ppd/hof gene clusters in the artificial operon, taking advantage of unique restriction sites that we had introduced in the pMS10 and pMS43 constructs (Experimental procedures). We further placed the resulting operon under control of inducible promoters, placZ in plasmid pMS39 or ppulC (a maltose-inducible promoter controlling expression of the Klebsiella T2SS pul gene cluster) in pMS40.
These plasmids were introduced in strain PAP7460 and bacteria were cultured on LB selective plates with or without inducers, as indicated. Piliation assays showed improved ppdD expression, in the absence of Sxy. Compared to the pMS43 plasmid, ppdD expression was improved in both “next generation” constructs. However, while inducible piliation was observed in strains carrying pMS39 (Fig. 3C), the pMS40 construct did not promote efficient pilus assembly, even when PpdD levels were induced by maltose (Fig. 3C lane 8). Further improvement of piliation efficiency for plasmid pMS39 was achieved by varying the E. coli expression hosts (Fig. 3D), and by culture on minimal media supplemented with 0.5 % glycerol (Fig. 3D, lanes 6, 7). For further assays, we chose the parental strain of the Keio gene knockout collection, BW25113 (Baba et al., 2006) and bacterial culture for 72 hours on M9 glycerol plates containing the IPTG at 30°C. Linear map of the ppd-hof artificial operon on plasmid pMS39 is shown in Fig. 3E.
To probe the specificity and requirements for pilus assembly, we tested the effects of several gene deletions in plasmid pMS39. As expected, no PpdD was detected in strains with plasmid pMS41 (Table 2) lacking the major pilin gene ppdD (Fig. 3F). Complementation with ppdD cloned on a compatible plasmid pCHAP8565 restored piliation, although overproduction of PpdD led to accumulation of its unprocessed form. Deletions of the gene encoding the polytopic IM platform protein (ΔhofC) and of the minor pilin operon ppdAB-ygdB-ppdC (ΔMP) also abolished pilus assembly, and piliation was restored upon expression of the corresponding genes in trans (Fig. 3F).
Observation of pili by immunofluorescence microscopy
Previous studies showed that PpdD pili could be assembled by the related heterologous machinery, the K. oxytoca pullulanase T2SS (Sauvonnet et al., 2000b, Cisneros et al., 2012b). To compare PpdD pilus assembly in the heterologous T2SS and in the reconstituted T4P system, we transformed strain BW25113 F’ lacIQ with pMS41 (encoding the T4aPS ΔppdD) or pCHAP8184 (encoding the T2SS lacking the major pseudopilin PulG) (Table 2). We complemented these strains with ppdD cloned on plasmid pCHAP8565 and probed for conditions that would allow comparable pilus assembly in both systems in bacteria cultured on minimal glycerol plates. As shown in Fig. 4A, lanes 1 and 2, pili were assembled in both strains in the absence of induction. Maltose, the inducer of T2SS gene expression, lowered PpdD levels and pilus assembly efficiency (Fig. 4A, lanes 5, 6). Induction of the hof-ppd operon and ppdD by IPTG allowed for more efficient piliation in both systems (Fig. 4A, lanes 3 and 4).
The possibility to use different Tff assembly systems for PpdD polymerization provides a unique opportunity to study the relative influence of pilin subunits and the assembly machinery on fibre structure and properties. We used immunofluorescence microscopy to compare pilus assembly via the two systems in bacteria cultured in M9 glycerol plates in the presence of IPTG (Fig. 4A, lanes 3,4). Similar numbers of PpdD pili were assembled by the T2SS (top panels, B-E) and T4PS (bottom panels, F-I). Both fibres appeared highly similar in terms of length and apparent thickness, although PpdD pili assembled by the heterologous T2SS appeared somewhat more curved and flexible. We noted that T4PS- assembled pili were more often attached to the bacterial cells when compared to those assembled by the T2SS.
Analysis of PpdD pili by electron microscopy
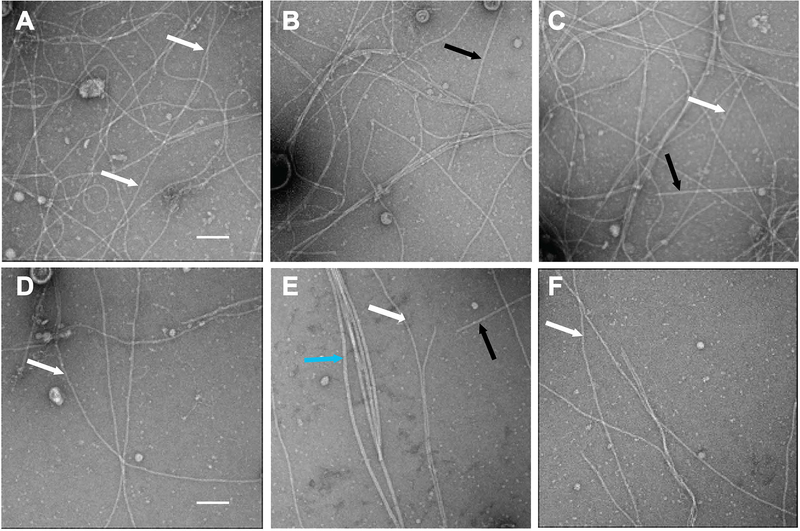
To better compare how the two systems, T2SS and T4PS, affect the PpdD pilus assembly, we imaged the two types of PpdD pili by using both negative stain electron microscopy (EM) and cryo-EM. The analysis of negatively stained pili by EM showed somewhat higher conformational heterogeneity and flexibility for the T2SS-assembled pili (Fig. 5), consistent with the IF results.
Figure 5.
Negative staining electron microscopy PpdD pili assembled by T4PS and T2SS. Crude sheared fractions of strains BW25113 F’ expressing ppdD (from pCHAP8565) and either the T2SS genes (from pCHAP8184) (panels A-C) or T4PS genes (from pMS41) (panels D-F) were analysed. Black arrows indicate Type 1 pili (in panels B and C) as shown previously in (Spaulding et al., 2018), the blue arrow indicates flagella (in panel E) and white arrows indicate PpdD T4P. The scale bar (A) is 50 nm.
We determined the helical symmetry of the pili assembled by the two different systems based on the averaged power spectra of segments imaged by cryo-EM (Fig. 6C&D). Within both of the two averaged power spectra we found dominant layer lines at 1/(14.7Å), 1/(20.3Å), 1/(53.2Å) and 1(159.5Å), indicating that they have an indistinguishable helical symmetry. Each subunit is related to an adjacent subunit by, on average, a ~11.2 Å axial rise and ~ 96.0° azimuthal rotation, forming a 1-start right-handed helix, although these parameters are quite variable in PpdD. This symmetry is very typical for Type IV pili, such as those from P. aeruginosa (Wang et al., 2017), N. gonorrhoeae (Wang et al., 2017) and N. meningitidis (Kolappan et al., 2016) (Table 3). The symmetry of PpdD pili differs from that of PulG pseudopili assembled by the same T2SS, which showed ~10.2 Å axial rise and 83.2° of twist (Lopez-Castilla et al., 2017). The difference between pseudopilus and pilus helical parameters, as well as the similarity of these parameters between PpdD pili assembled by the two systems suggest that the PpdD subunit itself, and not the assembly machinery, is the main determinant of subunit packing and fibre structure.
Table 3.
Comparison of helical symmetry parameters among bacterial pili of the Tff superfamily.
| Pili | Rise (Å) | Twist (°) |
|---|---|---|
| Neisseria meningitidis type 4a pili | 10.3 | 100.8 |
| Neisseria gonorrhoeae type 4a pili | 10.1 | 100.8 |
| Pseudomonas aeruginosa type 4a pili | 10.5 | 87.3 |
| Escherichia coli type 4a pili | 11.2 | 96.0 |
| Klebsiella oxytoca T2SS pseudopili | 10.2 | 83.2 |
Pilin interactions with T4P assembly components
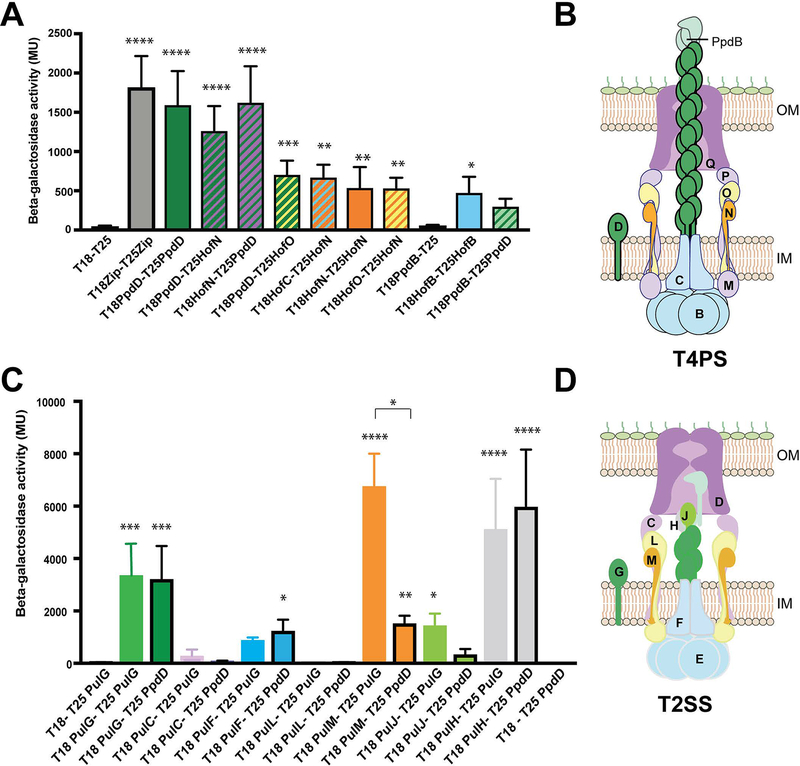
To examine how PpdD interacts with its cognate assembly apparatus, we employed the bacterial two-hybrid approach (BAC2H) (Karimova et al., 1998), used previously to study interactions of T4P assembly components in P. aeruginosa (Nguyen et al., 2015) and N. meningitidis (Georgiadou et al., 2012). The ppd and hof genes encoding full-length mature proteins were fused to genes encoding T18 and T25 fragments of the Bordetella pertussis CyaA toxin catalytic domain (Experimental procedures). We co-transformed the E. coli cya deletion strain DHT1 with selected plasmids and measured the reconstitution of adenylyl cyclase activity and induction of the chromosomal lacZ gene, dependent on interactions between the hybrid proteins. As shown in Fig. 7A, PpdD formed homo-dimers in this assay and interacted with the IM assembly components HofN and HofO. In addition, we detected homo-dimerization of HofB ATPase, whereas HofN and HofO formed heterodimers, as shown previously in T4P systems of N. meningitidis (Georgiadou et al., 2012) and P. aeruginosa (Leighton et al., 2015b). PpdD also interacted specifically with the minor pilin PpdB.
Figure 7.
PpdD interactions with components of the T4PS and T2SSs. A, BAC2H analysis of interactions between PpdD and T4PS components, performed as described in Experimental procedures. Bar graphs represent mean β-galactosidase activities (in Miller units) measured from at least 6 independent co-transformants; error bars indicate standard deviation. Statistical analysis with ANOVA test in comparison with the negative control with significant probabilities indicated above bars: ****, p<0.0001, *** p<0.001, **, p<0.01; *, p<0.01. B, Cartoon model of the T4PS with different Hof -Ppd components indicated in single letter code (except PpdB, to avoid confusion with HofB), colour-coded corresponding to the bar graphs in (A). C, Bar graphs show mean β-galactosidase activities in a BAC2H assays, comparing interactions of T25-PulG (no contours) and T25-PpdD (black contours) with T18-fused T2SS components. Statistical analysis was performed with ANOVA test in comparison with the negative control. Significant probabilities (when applicable) are indicated above bars: ****, p<0.0001, *** p<0.001, **, p<0.01; *, p<0.01. Non-parametric Kruskal-Wallis and Dunn’s multiple comparison between pair groups identified a significantly lower degree of interaction between PulM-PpdD when compared to PulM-PulG. D, Cartoon model of the Pul T2SS with different Pul components indicated in single-letter code and colour-coded as in (C).
Assembly of PpdD T4 pilin by the heterologous T2SS suggests its productive interactions with one or several components of the PulG pseudopilus assembly system in the inner membrane. To identify these contacts, we compared interactions of PpdD with T2SS components to those of PulG, the cognate major pseudopilin of this system characterized previously (Nivaskumar et al., 2016). T18-PulG and T18-PpdD chimera were tested with several T25-Pul T2SS component hybrids (Experimental procedures) (Figs. 7C and D). As shown previously, the major pseudopilin PulG forms dimers in the membrane and interacts with PulM and PulF components of the assembly platform, as well as with the minor pseudopilins PulH and PulJ (Nivaskumar et al., 2016). PpdD showed interaction signals with PulF and PulH similar to those of PulG (Fig. 7D). Like PulG, PpdD did not interact with PulC or PulL IM AP components. In contrast to PulG, PpdD did not interact with the minor pseudopilin PulJ, which is part of the minor pseudopilin initiation complex in the T2SS (Korotkov & Hol, 2008, Cisneros et al., 2012a). PpdD interaction with PulM, the AP component involved in pseudopilin targeting to the assembly machinery (Nivaskumar et al., 2016, Santos-Moreno et al., 2017) was significantly weaker than that of PulG (Fig. 7D). Since the N-terminal domains of PulG and PpdD are highly conserved, this is likely due to differences between the periplasmic domains of these major Tff subunits.
Discussion
We report here the functional reconstitution of a 13-component trans-envelope complex involved in T4aP assembly in E. coli. Previously, similar reconstitutions have been achieved for pili of the T4b subclass encoded by single gene clusters (Sohel et al., 1996, Stone et al., 1996, Yuen et al., 2013). The reconstitution of the complete T4aP system from several operons, although somewhat more challenging, was greatly facilitated by the Gibson assembly approach, which allows assembly of multiple DNA fragments in a single step (Gibson et al., 2009). In the case of the ppd-hof operon, the successful cloning strategy consisted of minimizing the presence of noncoding regions, which may affect gene expression at post-transcriptional level. A similar approach may be used to reconstitute diverse nanomachines from model bacteria that are highly pathogenic or toxinogenic, non-cultivable or recalcitrant to genetic manipulation.
The reconstituted EHEC T4PS contains 13 components. Of note, the artificial ppd-hof operon does not encode an equivalent of a pilotin like PilF in P. aeruginosa (Koo et al., 2013) or PilW in Neisseria (Szeto et al., 2011) required for the correct targeting and assembly of the secretin in the OM. Although PilF homologue is absent in the E. coli genome, we cannot exclude that another E. coli OM lipoprotein or the Bam machinery itself support PilQ assembly. We show that, in addition to PpdD and a prepilin peptidase, HofC and four minor pilins together are essential for pilus assembly on the bacterial surface. The essential role of HofC homologue in P. aeruginosa PilC (Takhar et al., 2013) has been recently confirmed for N. meningitidis PilG (Goosens et al., 2017). In the latter study, a synthetic approach was employed to assemble meningococcal T4P in the periplasm of E. coli and only 8 proteins were essential. Besides the HofC equivalent PilG, these included the major pilin, prepilin peptidase, the HofB equivalent PilF and PilM, PilN, PilO and PilP assembly components. PilQ and PilW, which allow for pilus surface presentation, were not required for pilus assembly per se, but neither were the minor pilins (Goosens et al., 2017). This is in contrast to our results and to those showing that minor pilins in P. aeruginosa promote pilus assembly initiation (Nguyen et al., 2015). Although minor pseudopilins are not fully essential for pseudopilus assembly, they greatly improve its efficiency (Cisneros et al., 2012a). CryoEM tomography studies of Myxococcus T4PS suggests that pilins stabilize the AP complex (Chang et al., 2016). The fact that minor pilins are dispensable for pilus assembly in the reconstituted meningococcal T4PS could be explained therefore by the high-level expression of genes encoding PilE and assembly components controlled by the T7 promoter (Goosens et al., 2017). Nevertheless, minor pilins are essential for function in T4P and T2SS (Helaine et al., 2005, Giltner et al., 2010, Sauvonnet et al., 2000b, Cisneros et al., 2012b, Cehovin et al., 2013) and their binding to specific substrates has been shown in several systems (Cehovin et al., 2013, Douzi et al., 2011). Recent studies of Vibrio T4aP involved in competence strongly suggest that minor pilins mediate DNA binding via the pilus tip (Ellison et al., 2018).
Reconstitution of E. coli T4aPS allowed us to compare the symmetry of fibres produced by the cognate assembly machinery with those assembled by the T2SS. Regardless of the system that promoted their polymerization, PpdD pili showed dominant azimuthal rotation of 96° and axial rise of 11.2 Å. Minor pseudopilins priming assembly of PpdD fibres in the T2SS, or minor pilins priming assembly in the cognate T4PS had no influence on this symmetry. Conversely, assembly of the cognate substrate PulG of the K. oxytoca T2SS resulted in different fibres with the twist of 83.2° and rise of 10.2 Å (Lopez-Castilla et al., 2017). Together, these results show that major (pseudo)pilin subunits, through their specific packing and interfaces, determine the overall symmetry parameters.
Although K. oxytoca T2SS promotes assembly of PpdD, it does not assemble other major pilins including P. aeruginosa PilA or the gonococcal PilE (Kohler et al., 2004), suggesting selective substrate recognition in Tff nanomachines. Although non-exhaustive, our BAC2H results provide insights into the basis of this heterologous assembly. In T2SS, PpdD interacts strongly with PulH, which shares 19% identity with its cognate minor pilin PpdB (Fig. S3A), perhaps explaining why the Klebsiella T2SS and E. coli T4aPS minor (pseudo)pilins are partially interchangeable (Cisneros et al., 2012b). Strong sequence conservation of PulG and PpdD N-terminal segments (Fig. S3B) is consistent with their similar interactions with PulF and shared ability to interact with PulM. Indeed, important determinants of PulM interaction were mapped to the N-terminal region of PulG (Nivaskumar et al., 2016, Santos-Moreno et al., 2017). However, PulM interaction with PpdD was significantly weaker than that with PulG, suggesting that globular domains of these proteins are also involved. In support of this idea, periplasmic domain of Thermus thermophilus PilA binds to PilM-PilN complex in vitro (Karuppiah et al., 2013). Two AP components of the EHEC system, HofN and HofO, are the main interacting partners of PpdD and share the same topology with PulM. Like PulM, with which it shares 25% identity (Fig. S3C), HofN is highly positively charged (pKi >10), whereas PulG and PpdD are acidic (pKi of 4.4 and 4.7, respectively). Further biochemical and structural analyses will be required to understand the molecular basis of pilin selectivity, likely crucial for assembly.
Upon induction of the ppd-hof expression on plasmid pMS39 about 50% of PpdD was assembled into pili, which is comparable to the efficiency observed for other T4PSs expressed from the host chromosomes (Giltner et al., 2010). Bypassing the need for Sxy and excluding the yggR gene from the artificial operon allowed us to reconstitute pilus assembly, dissociated from retraction. Since competence pili appear to be less abundant compared to T4P involved in motility and adherence (Ellison et al., 2018), this strategy facilitated pilus purification for ultrastructural studies. However, to assess the function of EHEC T4aP in DNA uptake or twitching motility, future studies should include the antagonistic YggR ATPase to reconstitute a fully functional and dynamic system. Studies in V. cholerae demonstrated the long-proposed role of pilus retraction in DNA uptake (Seitz & Blokesch, 2013), but also showed that low levels of retraction and transformation can be observed in pilT mutants (Ellison et al., 2018).
In V. cholerae, T4PS is induced by chitin oligosaccharides (Meibom et al., 2005), which activate transcription of a small RNA that stimulates sxy translation (Yamamoto et al., 2011). Although there is no evidence for a similar translational regulation of sxy in E. coli (Jaskolska & Gerdes, 2015), specific starvation or other signals appear to induce sxy expression and its regulon in EHEC strains leading to pilus assembly (Xicohtencatl-Cortes et al., 2007). The hofMNOPQ operon is required for growth on extracellular DNA as carbon source, which might also act as an inducer (Palchevskiy & Finkel, 2006). In addition, Sxy- and HofQ-dependent DNA uptake has been observed in E. coli K-12 (Sinha & Redfield, 2012). T4PS might be involved in DNA repair and horizontal gene transfer in mixed bacterial populations and biofilms, where extracellular DNA is abundant (Ibáñez de Aldecoa et al., 2017). Recently, by using time-lapse fluorescence imaging, DNA uptake mediated by the Sxy-controlled T4aP has been observed in Vibrio and mutational analysis implicated minor pilins in DNA binding to the pilus tip (Ellison et al., 2018). Whether T4aP from E. coli and related species also mediate DNA binding, uptake and exchange is a fascinating question that deserves further study (Veening & Blokesch, 2017). Although the genes encoding T4aP are present and conserved in all enterobacteria, their biological role and conditions that induce their expression are poorly understood. Reconstitution of a complex multi-component pilus assembly system from an enterobacterial class III pathogen in a non-pathogenic E. coli strain should facilitate future studies of its biological function and assembly mechanism.
Experimental procedures
Bacterial strains and culture conditions
The E. coli strains used in this study are listed in Table 1. Bacteria were grown at 30ºC in LB or minimal M9 medium containing 0.5% glycerol as the carbon source (Miller, 1972). Media were supplemented with 0.5 mM IPTG or 0.2 % D-maltose. Antibiotics were added at following concentrations: ampicillin (Ap), 100 μg.mL−1, chloramphenicol (Cm), 25 μg.mL−1, kanamycin (Km), 25 μg.mL−1 and zeocin (Ze), 8 μg.mL-1. An indicator of β-galactosidase activity 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) was added to LB media at 40 μg.mL-1.
Table 1.
Bacterial strains used in this study
| Name | Genotype | Source/reference |
|---|---|---|
| PAP7460 | Δ(lac-argF)U169 araD139 relA1 rpsL150 ΔmalE444 malG501 [F’ (lacIQ ΔlacZM15 pro+ Tn10)] | Laboratory collection |
| PAP105 | Δ(lac-pro) F’(laclQ ΔlacZM15 pro+ Tn10). | Laboratory collection |
| BL21 | F− ompT hsdSB (rB−mB−) gal dcm (DE3) | Bill Studier |
| BW25113 F’ lacIQ | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (Datsenko & Wanner, 2000) |
| DH5α F’lacIQ | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | Laboratory collection |
| DHT1 | Δcya | (Dautin et al., 2000) |
Plasmid constructions
The list of plasmids used in this study is given in Table 2. To build the synthetic operon the three gene clusters were PCR amplified from the chromosomal DNA of strain EDL933, together with ~100-bp flanking non-coding sequences using primers (1 to 6) listed in Supplemental Table S1. A fragment of pBR322 vector containing ApR and ori region was PCR-amplified with primers 7 and 8 (Table S1). The primers were designed to generate fragments with 20-bp overlap, following the Gibson assembly manual (New England Biolabs). Following DpnI digestion of the PCR fragments generated with Phusion enzyme, the 4-fragment Gibson assembly reaction was performed as suggested by the manufacturer. The reaction mix was transformed into ultracompetent bacteria of strain DH5α. The resulting plasmid pMS10 was purified and fully sequenced (GATC).
Plasmid pMS37 was obtained by successively replacing each gene cluster with a corresponding fragment lacking the noncoding DNA, leaving only the Shine-Dalgarno regions of the first gene in each operon. These minimal operon fragments were PCR-amplified from the EDL933 DNA using primers 9 – 14 (Table S1) and digested with DpnI. Fragments were individually replaced by directed cloning, by ligating the new PCR fragments with the electro-eluted vector sequences digested with the same enzymes: AflII-NcoI for fragment ppdD-hofBC, NcoI-AscI for fragment hofMNOPQ, and AscI-NotI restriction sites for fragment ppdAB-ygdB-ppdC to yield plasmid pMS37. Plasmid pMS37 was digested with NotI and XhoI to introduce the gspO gene, PCR amplified from the MG1655 DNA using primers 15 and 16 to give pMS38. Addition of the lacZ promoter (PCR amplified from pSU18 vector using primers 17, 18) into pMS38 using EcoRI-ClaI sites produced the plasmid pMS39. Introduction of the pulC promoter (PCR-amplified from plasmid pCHAP8185 using primers 19, 20) into pMS38 using the same restriction sites produced the plasmid pMS40. Plasmid pMS43 was obtained by cloning the 1623-bp BsiWI-NotI fragment from plasmid pMS39 into pMS10 digested with the same enzymes.
To generate plasmid pMS41, the hofB-hofC fragment, PCR-amplified with primers hofB AflII 5 and hofC N NcoI 3, was cloned into pMS39 digested with AflII and NcoI. Plasmid pMS47 was generated by cloning the ppdD-hofB fragment with primers ppdD Afl 5 and HofB Nco 3 into the pMS39 AflII-NcoI A fragment of 9507 bp. To generate plasmid pMS45 plasmid pMS39 was digested with AscI and NotI and the resulting 10028-bp fragment was treated with T4 polymerase before being religated.
For the bacterial two-hybrid constructs, ppd and hof genes were fused in frame to the C-terminal end of cyaA fragments in plasmids pUT18c and pKT25 by cloning PCR-amplified ppd/hof genes flanked by Kpn and EcoRI sites. For pilin genes, only the sequences encoding mature (prepilin peptidase processed) protein regions were included. The list of primers (Eurofins) used for these constructs is given in Table S1. All clones were verified by sequencing (GATC and Eurofins).
Pilus assembly assay
Pilus assembly assays were performed as described previously (Campos et al., 2010) (Luna Rico et al., 2018). Briefly, E. coli strains containing indicated plasmids were grown at 30°C for 48–72 hr on LB or M9 agar supplemented as required with inducers and antibiotics. Bacteria were scraped off the plates and resuspended in LB and normalized to 1 OD600 ml-1. Suspensions were vortexed for 1 min to detach surface pili and centrifuged at ~12000 x g for 5 minutes at 4°C. The pelleted bacteria were resuspended in SDS sample buffer at 10 OD600 mL-1. The supernatants were spun at ~12000 x g for another 10 minutes and 0.7 ml was transferred to a clean tube and submitted to precipitation with 10 % trichloroacetic acid. After incubation for 30 minutes on ice and centrifugation at ~12000 x g for 30 minutes at 4°C, the pellets were washed twice with cold acetone, air-dried and resuspended in SDS sample buffer. Equivalent amounts of cell and sheared fractions were analysed by denaturing polyacrylamide gel electrophoresis (PAGE) on 10% slab gels using the Tris-Tricin separation system (Schagger & von Jagow, 1987). Proteins were transferred onto nitrocellulose membranes (ECL, Amersham) using the Pierce Power Blotter system (Thermo) and revealed with polyclonal anti-PppD antisera diluted 1:2000, unless otherwise indicated (Sauvonnet et al., 2000a). Secondary goat anti-rabbit antibodies coupled to HRP (GE) were diluted to 1:20 000. Blots were revealed by ECL2 (Thermo) and fluorescence signal was recorded on Typhoon FLA9000 phospho-imager (GE). Images were processed with Adobe Photoshop.
Immunofluorescence microscopy
Sampled for IF analysis of pili were prepared as described previously (Cisneros et al., 2012b, Luna Rico et al., 2018). Bacteria were grown for 48 hours at 30°C on LB agar supplemented with antibiotics and inducer (0.4% maltose or 0.5 mM IPTG, as indicated) were carefully resuspended in PBS at 1 OD600.ml−1 and immobilized on coverslips coated with poly-L-lysine. After 30 minutes of fixation using 3.7% formaldehyde at room temperature, reactions were quenched with 1M Tris-HCl pH 8.0 and samples were blocked with 1% bovine serum albumin (BSA) in PBS. Pili were detected using an anti-PpdD antibody (1:1000) and a secondary anti-rabbit IgG coupled to Alexa Fluor 488 (1:200); bacteria were stained with 4’,6-diamidino-2-phenylindole (DAPI). Samples were observed with an Axio Imager A2 microscope (Zeiss) and images were acquired with the AxioCam MRm digital camera connected to the microscope. Images were analysed with Zen 2012 software (Zeiss) and processed with Adobe Photoshop.
Preparation of pili
Ten 90-mm LB plates containing Ap, Cm and IPTG were inoculated with BW25113 F’lacIQ strain containing plasmid pCHAP8565 and either pCHAP8184 (encoding T2SS) or pMS41 (encoding T4PS). Bacteria were cultured for 72 hours at 30°C and harvested in LB medium. Bacterial suspensions were subjected to vortex agitation for 5 min and then passed 10 times through a 21-gauge needle to detach surface pili. Bacteria were removed by centrifugation for 10 min at 10000 g. Supernatants were further centrifuged in Eppendorf tubes for 10 min at 16000xg to pellet the remaining bacteria. Pili from the supernatant fractions were recovered by ultracentrifugation for 1 hour at 100000xg at 4°C. Pili were taken up in 0.1 mL of 50 mM HEPES pH 7.2, 50 mM NaCl and kept on ice before EM analysis.
Electron microscopy
For the negative stain EM imaging, 3 μl of the PpdD pili sample were applied to previously plasma-cleaned carbon-coated copper grids. After incubating for 30 seconds, the excess sample were removed and stained with 2% uranyl acetate (w / v) before imaging with FEI Tecnai F20 electron microscope at 120 kV. Motioncor2 (Li et al., 2013) was used for motion correcting all the images, following by using CTFFIND3 (Mindell and Grigorieff, 2003) for estimation of the defocus and astigmatism. Filament boxes were selected and extracted by using e2helixboxer within EMAN2 (Tang et al., 2007) on images multiplied by the CTF. The Spider software package (Frank et al., 1996) was used for all further image processing.
Cryo-EM imaging and helical symmetry comparison
PpdD pili sample (3 μl) assembled from T2SS and T4PS were applied to plasma-cleaned lacey carbon grids. The grids were plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI). The images were recorded on a Titan Krios microscope at 300kV, using a Falcon III direct electron detector at magnification of 75,000X with a total dose of ~45 electrons/Å2. Motioncor2 was used for motion correcting all the images, following by using CTFFIND3 for estimation of the defocus and astigmatism. Filament boxes were selected and extracted by using e2helixboxer within EMAN2 on images multiplied by the CTF. The Spider software package was used for all further image processing. Segments with a very large out-of-plane tilt as well as with symmetry far from the average were discarded.
Bacterial two-hybrid (BAC2H) analysis
Competent cells of strain DHT1 (Dautin et al., 2000) were co-transformed with pUT18C and pKT25 plasmid derivatives and bacteria were grown for 48 h at 30°C on LB plates containing Ap and Km. Six colonies were picked at random and inoculated into 5 ml cultures in LB containing Km and Ap, grown overnight and inoculated the next day into fresh medium containing 1 mM IPTG. Bacteria were cultured to mid-log phase and β-galactosidase activity was measured as described (Miller, 1972). At least 2 independent experiments were performed with several randomly picked transformants. Bar graphs represent mean values and error bars indicate standard deviation. The non-parametric Kruskal-Wallis followed by Dunn’s multiple comparison tests were used in statistical analysis using the Graphpad Prism 7 software.
Supplementary Material
Acknowledgements
This work was funded by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), the French Agence Nationale de la Recherche grant ANR-14-CE09–0004 and NIH grant GM122510 (to E.H.E). We thank Daniel Ladant and members of the BIM, NMR of Biomolecules and Structural Bioinformatics Units in Institut Pasteur for support and helpful discussions. We are grateful to Nadia Izadi-Pruneyre for advice and critical reading of the manuscript. A.L.R. is a fellow of the Pasteur - Paris University (PPU) International PhD program.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL & Burrows LL, (2009) PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol 394: 128–142. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL & Mori H, (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome B, Jubete Y, Martinez E & de la Cruz F, (1991) Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102: 75–78. [DOI] [PubMed] [Google Scholar]
- Berry JL & Pelicic V, (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39: 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JL, Xu Y, Ward PN, Lea SM, Matthews SJ & Pelicic V, (2016) A comparative structure/function analysis of two type IV pili DNA receptors defines a novel mode of DNA binding. Structure. 24: 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH & Falkow S, (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2: 95–113. [PubMed] [Google Scholar]
- Cameron ADS & Redfield RJ, (2006) Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucl. Acids Res 34: 6001–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA & Francetic O, (2010) Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci USA 107: 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ & Pelicic V, (2013) Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA 110: 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L & Jensen GJ, (2016) Architecture of the type IVa pilus machine. Science 351: aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros DA, Bond PJ, Pugsley AP, Campos M & Francetic O, (2012a) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J 31: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros DA, Pehau-Arnaudet G & Francetic O, (2012b) Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol Microbiol 86: 805–818. [DOI] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT & Tainer JA, (2003) Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell 11: 1139–1150. [DOI] [PubMed] [Google Scholar]
- Datsenko KA & Wanner BL, (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Karimova G, Ullmann A & Ladant D, (2000) Sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J Bacteriol 182: 7060–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Amorim GC, Cisneros DA, Delepierre M, Francetic O & Izadi-Pruneyre N, (2014) 1H, 15N and 13C resonance assignments of PpdD, a type IV pilin from enterohemorrhagic Escherichia coli. Biomol NMR Assign. 8: 43–46. [DOI] [PubMed] [Google Scholar]
- Douzi B, Ball G, Cambillau C, Tegoni M & Voulhoux R, (2011) Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J Biol Chem 286: 40792–40801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Dalia TN, Ceballos AV, Wang JC, Biais N, Brun YV & Dalia AB, (2018) Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat Microbiol 3773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB & Brun YV, (2017) Obstruction of pilus retraction stimulates bacterial surface sensing. Science 368: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O, Belin D, Badaut C & Puglsey AP, (2000) Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. The EMBO J 19: 6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O, Lory S & Puglsey AP, (1998) A second prepilin peptidase gene in Escherichia coli K-12. Mol Microbiol 27: 763–775. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, and Leith A (1996). SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct Biol 116: 190–199. [DOI] [PubMed] [Google Scholar]
- Fullner KJ & Mekalanos JJ, (1999) Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun 67: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou M, Castagnini M, Karimova G, Ladant D & Pelicic V, (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol 84: 857–873. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd & Smith HO, (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Giltner CL, Habash M & Burrows LL, (2010) Pseudomonas aeruginosa minor pilins are Incorporated into Type IV Pili. J Mol Biol 398: 444–461. [DOI] [PubMed] [Google Scholar]
- Gold VA, Salzer R, Averhoff B & Kuhlbrandt W, (2015) Structure of a type IV pilus machinery in the open and closed state. Elife 4. doi: 10.7554/eLife.07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosens VJ, Busch A, Georgiadou M, Castagnini M, Forest KT, Waksman G & Pelicic V, (2017) Reconstitution of a minimal machinery capable of assembling periplasmic type IV pili. Proc Natl Acad Sci USA 114: E4978–E4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S, Carbonelle E, Prouvensier L, Beretti J-L, Nassif X & Pelicic V, (2005) PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol 55: 65–77. [DOI] [PubMed] [Google Scholar]
- Helaine S, Dyer DH, Nassif X, Pelicic V & Forest KT, (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci USA 104: 15888–15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospenthal M, Costa TRD & Waksman G, (2017) A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol 15: 365–379. [DOI] [PubMed] [Google Scholar]
- Ibáñez de Aldecoa AL, Zafra O & González-Pastor JE, (2017) Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Frontiers Microbiol 8: 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolska M & Gerdes K, (2015) CRP-dependent positive autoregulation and proteolytic degradation regulate competence activator Sxy of Escherichia coli. Mol Microbiol 95: 833–845. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A & Ladant D, (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppiah V, Collins RF, Thistlethwaite A, Gao Y & Derrick JP, (2013) Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis. Proc Natl Acad Sci USA 110: E4638–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Schafer K, Muller S, Vignon G, Diederichs K, Philippsen A, Ringler P, Pugsley AP, Engel A & Welte W, (2004) Structure and assembly of the pseudopilin PulG. Mol Microbiol 54: 647–664. [DOI] [PubMed] [Google Scholar]
- Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH & Craig L, (2016) Structure of the Neisseria meningitidis Type IV pilus. Nat Commun 7: 13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo J, Lamers RP, Rubinstein JL, Burrows LL & Howell LP, (2016) Structure of the Pseudomonas aeruginosa type IVa pilus secretin at 7.4 A. Structure 24: 1778–1787. [DOI] [PubMed] [Google Scholar]
- Koo J, Tang T, Harvey H, Tammam S, Sampaleanu LM, Burrows LL & Howell LP, (2013) Functional mapping of PilF and PilQ in the Pseudomonas aeruginosa type IV pilus system. Biochemistry 52: 2914−2923. [DOI] [PubMed] [Google Scholar]
- Korotkov KV & Hol WG, (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol 15: 462–468. [DOI] [PubMed] [Google Scholar]
- Ledesma MA, Ochoa SA, Cruz A, Rocha-Ramírez LM, Mas-Oliva J, Eslava CA, Girón JA & Xicohtencatl-Cortes J, (2010) The Hemorrhagic Coli Pilus (HCP) of Escherichia coli O157:H7 is an inducer of proinflammatory cytokine secretion in intestinal epithelial cells. PLoS One 5: e12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton TL, Buensuceso RN, Howell PL & Burrows LL, (2015a) Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17: 4148–4163. [DOI] [PubMed] [Google Scholar]
- Leighton TL, Dayalani N, Sampaleanu LM, Howell PL & Burrows LL, (2015b) Novel role for PilNO in type IV pilus retraction revealed by alignment subcomplex mutations. J. Bacteriol. 197: 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, U. M., Gur T, McWilliam H, Squizzato S, Park YM, Buso N & Lopez R, (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucl Acids Res 43(W1): W580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, and Cheng Y (2013). Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10: 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Castilla A, Thomassin J-L, Bardiaux B, Zheng W, Nivaskumar M, Yu X, Nilges M, Egelman EH, Izadi-Pruneyre N & Francetic O, (2017) Structure of the calcium-dependent type 2 secretion pseudopilus. Nat Microbiol 2: 1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna Rico A, Thomassin J-L & Francetic O, (2018) Analysis of bacterial pilus assembly by shearing and immunofluorescence microscopy. Methods Mol Biol 1764: 291–305. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV & Albers SV, (2016) Diversity and Evolution of Type IV pili Systems in Archaea. Frontiers Microbiol 7: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M, Tammam S, Khan A, Burrows LL & Howell PL, (2017) The molecular mechanism of the type IVa pilus motors. Nat Commun 8: 15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K, Blokesch M, Dolganov N, Wu C & Schoolnik G, (2005) Chitin induces natural competence in Vibrio cholerae. Science 310: 1824–1827. [DOI] [PubMed] [Google Scholar]
- Melville S & Craig L, (2013) Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev 77: 323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH, (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. [Google Scholar]
- Mindell JA, and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347. [DOI] [PubMed] [Google Scholar]
- Misic AM, Satyshur KA & Forest KT, (2010) P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J Mol Biol 400: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R, Ageorges V, Rojas-Lopez M, Schmidt H, Weiss A, Bertin Y, Forano E, Jubelin G, Henderson IR, Livrelli V, Gobert AP, Rosini R, Soriani M & Desvaux M, (2016) A secretome view of colonisation factors in Shiga toxin-encoding Escherichia coli (STEC): from enterohaemorrhagic E. coli (EHEC) to related enteropathotypes. FEMS Microbiol Rev 363 pii: fnw179. [DOI] [PubMed] [Google Scholar]
- Ng D, Harn T, Altindal T, Kolappan S, Marles JM, Lala R, Spielman I, Gao Y, Hauke CA, Kovacikova G, Verjee Z, Taylor RK, Biais N & Craig L, (2016) The Vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog 12 e1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y, Sugiman-Marangos S, Harvey H, Bell SD, Charlton CL, Junop MS & Burrows LL, (2015) Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem 290: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M, Bouvier G, Campos M, Nadeau N, Yu X, Egelman EH, Nilges M & Francetic O, (2014) Distinct docking and stabilization steps of the pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers. Structure 22: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M, Santos-Moreno J, Malosse C, Nadeau N, Chamot-Rooke J, Tran Van Nhieu G & Francetic O, (2016) Pseudopilin residue E5 is essential for recruitment by the type 2 secretion system assembly platform. Mol Microbiol 101: 924–941. [DOI] [PubMed] [Google Scholar]
- Palchevskiy V & Finkel SE, (2006) Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188: 3902–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED & Tainer JA, (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378: 32–38. [DOI] [PubMed] [Google Scholar]
- Pelicic V, (2008) Type IV pili: e pluribus unum? Mol Microbiol 68: 827–837. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett GI, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta E, Potamousis K, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA & Blattner FR, (2001) Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409: 529–533 [DOI] [PubMed] [Google Scholar]
- Santos-Moreno J, East A, Bond PJ, Tran Van Nhieu G & Francetic O, (2017) Polar N-terminal residues conserved in type 2 secretion pseudopilins determine subunit targeting and membrane extraction during fibre assembly. J Mol Biol 429: 1746–1765. [DOI] [PubMed] [Google Scholar]
- Sauvonnet N, Gounon P & Puglsey AP, (2000a) PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J Bacteriol 182: 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N, Vignon G, Pugsley AP & Gounon P, (2000b) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J 19: 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H & von Jagow G, (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Henkel B & Karch H, (1997) A gene cluster closely related to type II secretion pathway operons of Gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett 148: 265–272. [DOI] [PubMed] [Google Scholar]
- Seitz P & Blokesch M, (2013) DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci USA 110: 17987–17992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers L, Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, & Higgins DG. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Cameron ADS & R. R.J., (2009) Sxy induces a CRP-S regulon in Escherichia coli. J Bacteriol 191: 5180–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S & Redfield RJ, (2012) Natural DNA uptake by Escherichia coli. PLoS One 7: e35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohel I, Puente JL, Ramer SW, Bieber D, Wu C-Y & Schoolnik GK, (1996) Enteropathogenic Escherichia coli: Identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178: 2613–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding CN, Schreiber HL, Zheng W, Dodson KW, Hazen JE, Conover MS, Wang F, Svenmarker P, Luna-Rico A, Francetic O, Andersson M, Hultgren S, and Egelman EH (2018) Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions. eLife. pii e31662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone KD, Zhang H-Z, Carlson LK & Donnenberg MS, (1996) A cluster of fourteen genes from enteropathogenic Escherchia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20: 325–337. [DOI] [PubMed] [Google Scholar]
- Strom MS & Lory S, (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem 266: 1656–1664. [PubMed] [Google Scholar]
- Strom MS, Nunn DN & Lory S, (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA 90: 2404–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Dessen A & Pelicic V, (2011) Structure/function analysis of Neisseria meningitidis PilW, a conserved protein playing multiple roles in type IV pilus biology. Infect. Immun 79: 3028−3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhar HK, Kemp K, Kim M, Howell PL & Burrows LL, (2013) The platfrm protein is essential for type IV pilus biogenesis. J Biol Chem 288: 740–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammam S, Sampaleanu LM, Koo J, Manoharan K, Daubaras M, Burrows LL & Howell PL, (2013) PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J Bacteriol 195: 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct Biol 157: 38–46. [DOI] [PubMed] [Google Scholar]
- Tomich M, Planet PJ & Figurski DH, (2007) The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 5: 363–375. [DOI] [PubMed] [Google Scholar]
- Veening JW & Blokesch M, (2017) Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat Rev Microbiol 15: 621–629. [DOI] [PubMed] [Google Scholar]
- Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH & Craig L, (2017) Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV Pili at sub-nanometer resolution. Structure. 25: 1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, Park H-S, Hayes SF, van Putten JPM & Koomey M, (1998) Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95: 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Ledesma MA, Jordan D, Francetic O, Kaper JB, Puente JL & Girón JA, (2007) Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J Clin. Investig. 117: 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Saldana Z, Ledesma MA, Puente JL & Giron JA, (2009) The type 4 pili of Enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol 191: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M & Watanabe H, (2011) Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol 193: 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen ASW, Kolappan S, Ng D & Craig L, (2013) Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic Escherichia coli. Mol Microbiol 90: 898–918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.