Abstract
Cu homeostasis depends on a tightly regulated network of proteins that transport or sequester Cu, preventing the accumulation of this toxic metal while sustaining Cu supply for cuproproteins. In Rhodobacter capsulatus, Cu-detoxification and Cu delivery for cytochrome c oxidase (cbb3-Cox) assembly depend on two distinct Cu-exporting P1B-type ATPases. The low-affinity CopA is suggested to export excess Cu and the high-affinity CcoI feeds Cu into a periplasmic Cu relay system required for cbb3-Cox biogenesis. In most organisms, CopA-like ATPases receive Cu for export from small Cu chaperones like CopZ. However, whether these chaperones are also involved in Cu export via CcoI-like ATPases is unknown. Here we identified a CopZ-like chaperone in R. capsulatus, determined its cellular concentration and its Cu binding activity. Our data demonstrate that CopZ has a strong propensity to form redox-sensitive dimers via two conserved cysteine residues. A ΔcopZ strain, like a ΔcopA strain, is Cu-sensitive and accumulates intracellular Cu. In the absence of CopZ, cbb3-Cox activity is reduced, suggesting that CopZ not only supplies Cu to P1B-type ATPases for detoxification but also for cuproprotein assembly via CcoI. This finding was further supported by the identification of a ~150 kDa CcoI-CopZ protein complex in native R. capsulatus membranes.
Keywords: copper homeostasis, copper chaperones, cbb3-type cytochrome c oxidase biogenesis, respiration, Rhodobacter capsulatus, P1b-type ATPases
Introduction
Copper (Cu) is an essential but potentially toxic micro-nutrient that serves as a cofactor for important enzymes, like cytochrome oxidases (Cox) or multicopper oxidases (laccases) (Rensing & McDevitt, 2013, Ekici et al., 2012a). Cu is a strong soft metal that can attack intracellular iron-sulfur centers of various proteins primarily under anoxic conditions (Macomber & Imlay, 2009). In oxic conditions, free copper ions are able to oxidize sulfhydryl groups and catalyse a Fenton-like reaction that can cause lipid peroxidation and protein damage (Chandrangsu et al., 2017, Arguello et al., 2013). Therefore, maintaining cellular copper homeostasis is essential for all organisms, and this process requires coordinated activity of Cu transporters, Cu chaperones and storage proteins (Ekici et al., 2012a, Trasnea et al., 2016b, Vita et al., 2016), which essentially reduce the amount of non-complexed Cu to a concentration below the detection limit (Arguello et al., 2013, Ekici et al., 2014, Rae et al., 1999).
The α-proteobacterium Rhodobacter capsulatus has developed into an attractive model organism for studying Cu homeostasis. In this species, multiple proteins were identified that target Cu to cuproproteins like the cbb3-type cytochrome c oxidase (cbb3-Cox), export excess Cu into the periplasm (Ekici et al., 2012a), or mediate Cu resistance (Rademacher et al., 2012, Rademacher & Masepohl, 2012). The cbb3-Cox contains a single Cu atom (CuB) within the catalytic centre of subunit CcoN, where oxygen is reduced to water (Gray et al., 1994). In contrast to the mitochondrial aa3-Cox and some ba3 Cox, cbb3-Cox lacks a second Cu centre (CuA) in subunit II, which serves as an electron acceptor from the soluble electron donor cytochrome c. Instead of a CuA-containing subunit, cbb3-Cox contains two membrane anchored c-type cytochromes, CcoP and CcoO, that receive and transfer electrons to the binuclear CuB-heme b3 catalytic centre in CcoN (Ekici et al., 2012a).
Cu trafficking to cbb3-Cox is initiated by Cu-uptake from the periplasm via the major facilitator superfamily (MFS) protein CcoA. In the absence of CcoA, cbb3-Cox activity is drastically reduced, but the activity can be restored by increasing the Cu concentration in the growth medium (Ekici et al., 2012b). Cytoplasmic Cu is exported back to the periplasm by the P1B-type ATPase CcoI (Koch et al., 2000, Ekici et al., 2012b). In the periplasm, Cu is transferred to cbb3-Cox via a periplasmic relay system involving the periplasmic Cu chaperones PccA (a PCuAC-homologue) and SenC (a Sco1-homologue) (Kulajta et al., 2006, Lohmeyer et al., 2012, Trasnea et al., 2016b, Trasnea et al., 2018). Similar to the phenotype of the ΔccoA strain, the very low cbb3-Cox activity of either senC- or pccA-deletion strains can be rescued by increasing the Cu concentrations in the growth medium (Trasnea et al., 2016b). In contrast, a ΔccoI strain shows no cbb3-Cox activity even at a surplus of Cu. The basis of this observation remains unclear, although it could indicate that Cu insertion into cbb3-Cox requires a cytoplasmic intermediate.
Many bacteria contain multiple Cu-exporting P1B-type ATPases with different functional specificities. CcoI-like P-type ATPases are characterized by a high Cu affinity but rather low efflux rates, consistent with their primary role in cuproprotein assembly (Raimunda et al., 2011). This is different for the CopA-like P-type ATPases, which have a rather low affinity but high efflux rates, in line with their primary role in Cu detoxification. The CopA-like ATPases do not bind free Cu for export but rather receive Cu from small cytoplasmic Cu-chaperones like CopZ (Gonzalez-Guerrero & Arguello, 2008), consistent with the fact that free Cu is virtually absent in living cells (Arguello et al., 2016, Rae et al., 1999). CopZ-like chaperones have been identified in all domains of life, and are characterized by a typical βαββαβ ferredoxin-like fold with a MxCxxC Cu binding site (Singleton & Le Brun, 2007). This binding site is very similar to the N-terminally located Cu-binding domains (N-MBDs) of P1B-type ATPases. Cu(I)-loaded CopZ has been shown to interact with this domain and to rapidly transfer Cu(I) to the N-MDB (Rosenzweig & Arguello, 2012). This Cu-exchange could serve as a Cu sensing mechanism (Arguello et al., 2016). While Cu binding to the N-MBD of the yeast P1B-type ATPase Ccc2p is required for Cu translocation (Morin et al., 2009), Cu translocation by A. fulgidus CopA occurs in vitro even when the N-MBDs are deleted (Gonzalez-Guerrero & Arguello, 2008). However, Cu transfer to A. fulgidus CopA still requires CopZ to deliver Cu to a second, membrane-integral Cu binding site (TM-MBS). This TM-MBS is formed by conserved residues within the transmembrane domains 6, 7 and 8, and is located close to a positively charged amino acid stretch that is suggested to serve as an additional contact site for CopZ and as putative Cu entrance side (Gourdon et al., 2011, Arguello et al., 2016). Mutating residues close to the Cu entrance side prevents Cu translocation in A. fulgidus (Padilla-Benavides et al., 2013), but not in S. pneumoniae CopA (Fu et al., 2013). Although the CopZ-CopA interaction seems to be required for sensing the intracellular Cu concentration via the N-MBD and for mediating Cu translocation via the TM-MBS (Arguello et al., 2016), it is still unclear how both of these events are connected. Furthermore, while the importance of the CopZ-CopA interaction for Cu detoxification has been clearly demonstrated, it is unknown whether CopZ also interacts with CcoI-type P1B-ATPases for insertion of Cu into cuproproteins like cbb3-Cox.
R. capsulatus contains both types of P1B-type ATPases, but a CopZ-like chaperone has not been identified so far. Here we describe the identification of R. capsulatus CopZ, its biochemical characterization, its impact on the Cu-detoxification pathway via CopA and on the cbb3-Cox assembly pathway via CcoI.
Results
Identification and characterization of R. capsulatus CopZ homologue
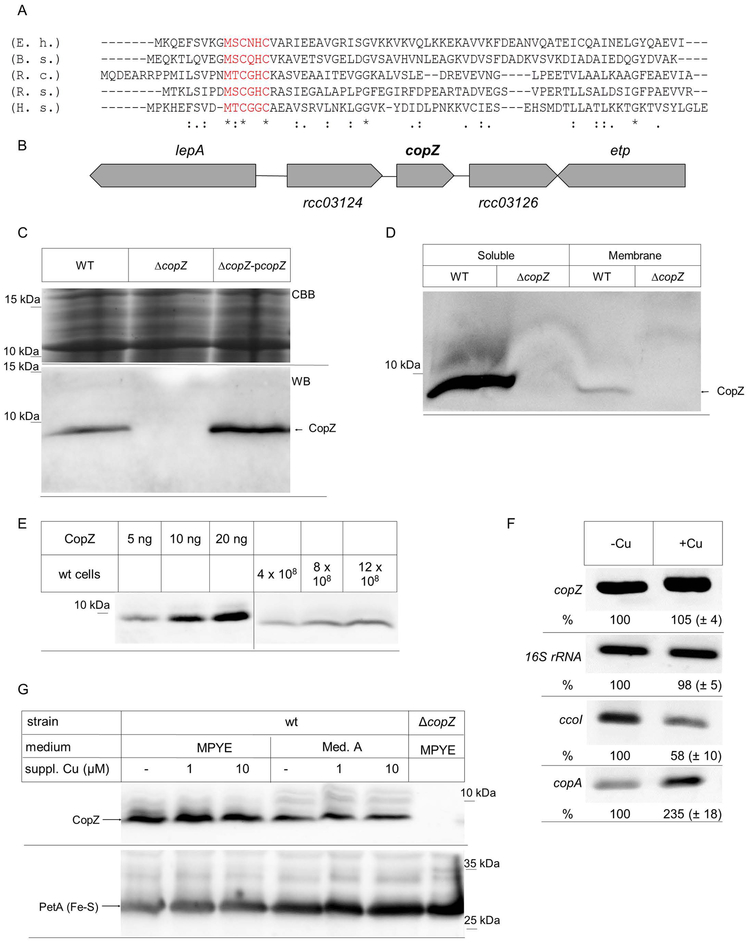
For identifying putative Cu chaperones in R. capsulatus, the available genome sequence was searched for open reading frames (ORFs) with significant amino acid sequence homology to known Cu chaperones. We identified a small ORF encoding a hypothetical protein of 62 amino acids, which exhibited approx. 30 % identity to CopZ of Bacillus subtilis, Enterococcus hirae or Rhodobacter sphaeroides (Fig. 1A). In particular, this ORF contained the conserved MxCxHC Cu-binding motif that is diagnostic for CopZ and its homologues, like yeast Atox1 or human Hah1 (Hatori et al., 2017).
Figure 1: Identification of a CopZ homologue in R. capsulatus.
(A) Amino acid alignment of the R. capsulatus CopZ (R.c.) with the respective CopZ-homologues of Enterococcus hirae (E.h.) Bacillus subtilis (B.s.), Rhodobacter sphaeroides (R.s.), and Homo sapiens (H.s.). The conserved Cu-binding motif is shown in red. (B) Genetic organization of copZ in R. capsulatus. lepA and etp presumably encode for a translation factor and a phosphotyrosine protein phosphatase, respectively. The open reading frames rcc03124 and rcc03126 encode for hypothetical proteins. (C) R. capsulatus cells were grown in MPYE medium, precipitated with trichloroacetic acid and the pellet was dissolved loading buffer. After SDS-PAGE, the gel was either stained directly with coomassie brilliant blue (CBB) as loading control, or was blotted and decorated with α-CopZ antibodies (WB). WT corresponds to MT1131, ΔcopZ to a MT1131 derivative carrying an insertion-deletion mutation within copZ and ΔcopZ-pcopZ to the ΔcopZ strain with a plasmid-encoded copZ. (D) MT1131 (WT) and ΔcopZ cell extracts were separated into a soluble fraction and a membrane fraction by ultracentrifugation. Subsequently, the material was separated by SDS-PAGE and decorated with α-CopZ antibodies. (E) The cellular concentration of CopZ in MT1131 grown on MPYE medium without further Cu supplementation was determined by quantitative western blotting, using defined amounts of purified CopZ as reference. Signal intensity was quantified by ImageJ and several independent experiments were performed and a representative western blot is shown. Note that the purified CopZ contained a His-tag and it therefore migrates slower on SDS-PAGE than the native CopZ. (F) RT-PCR analyses of mRNA levels in wild type cells grown on MPYE without and with Cu supplementation (10 μM Cu(II)). A representative gel of three independent experiments is shown. The 16S ribosomal RNA served as control and the ccoI and copA mRNA as reference. Quantification was performed with ImageJ and signal intensity of the mRNA level in cells without Cu supplementation was set to 100%. (G) The CopZ levels in whole cells grown either on enriched medium (MPYE) or minimal medium (MedA) were analysed by immunoblotting as described above. When indicated, CuSO4 was added to the growth medium. The levels of the Rieske Fe-S protein PetA served as loading control.
CopZ-like chaperones are widely distributed in Gram-positive, but less frequent in Gram-negative bacteria (Cobine et al., 1999, Hearnshaw et al., 2009). In many Gram-positive bacteria CopZ is encoded in the copYAZ operon, where copY encodes a Cu-dependent transcription factor regulating the transcription of the Cu-exporting P1B-type ATPase CopA and of CopZ (Odermatt & Solioz, 1995, Singleton & Le Brun, 2007). In R. sphaeroides, a putative copZ homologue (Rsp_2891) is located immediately upstream of copA (Rsp_2890), but divergently transcribed (Peuser et al., 2011). However, R. capsulatus copZ is not genetically linked to other Cu-responsive genes. It is located downstream of RCAP_rcc03124, encoding a hypothetical protein of unknown function, and upstream of RCAP_rcc03126, encoding a hypothetical protein with homology to ornithine-arginine transport proteins. Further upstream and downstream are located the genes for the translation factor LepA and the phosphotyrosine protein phosphatase ETP (Fig. 1B).
For analyzing the role of CopZ in R. capsulatus, a chromosomal deletion strain was constructed (Materials and Methods). By immune detection using polyclonal antibodies against R. capsulatus CopZ an approx. 6 kDa protein was detected in wild type R. capsulatus, but not in the ΔcopZ strain. This 6 kDa band was also detected in the ΔcopZ strain expressing an ectopic copy of copZ carried by the low-copy plasmid pRK415 (Fig. 1C), in agreement with the predicted mass of 6.4 kDa for R. capsulatus CopZ.
CopZ does not contain any putative signal sequence or hydrophobic sequences long enough to account for a possible transmembrane domain. Thus, it is expected to be primarily a cytoplasmic protein, and this was confirmed by differential centrifugation of R. capsulatus cell extracts. CopZ was almost exclusively found in the cytoplasmic fraction, and only a very weak band was found associated with the intracytoplasmic membrane (ICM) fraction (Fig. 1D). For determining the cellular concentration of CopZ, different amounts of cells grown under semiaerobic conditions on enriched (MPYE) medium without supplementary Cu were precipitated using trichloroacetic acid (TCA), and their contents separated by SDS-PAGE, together with defined amounts of purified proteins for quantitative immunoblotting (Fig. 1E). The concentration of CopZ was determined to be 8.4 ± 0.3 ng/108 cells and based on these values and the predicted mass of CopZ (MW = 6.4 kDa) the cytoplasmic CopZ concentration was estimated to be ~ 6.9 μM, which is similar to that determined in E. hirae (1 μM) (Urvoas et al., 2004).
For analysing whether copZ expression is regulated by Cu, RT-PCR and immunoblotting experiments were performed. The total mRNA was isolated from wild type cells grown on enriched MPYE medium without Cu supplementation, or supplemented with 10 μM Cu. No significant differences in the copZ mRNA levels were detectable under both conditions, using the 16S rRNA levels as a control (Fig. 1F). In contrast, the copA mRNA levels were increased about two-fold upon Cu supplementation, while under these conditions, the ccoI mRNA levels were reduced, in agreement with previous reports (Ekici et al., 2014).
The CopZ levels were further analysed by immunoblotting in wild type cells that were grown on either enriched medium (MPYE) or minimal medium (Med. A) with and without Cu supplementation, using the Rieske-type Fe-S protein PetA as a loading control. The CopZ levels in Med A grown cells were slightly lower than in MPYE grown cells, but were not significantly influenced on either medium by addition of Cu (1 to 10 μM) (Fig. 1G). Thus, different from other species like R. sphaeroides (Peuser et al., 2011), the copZ expression in R. capsulatus does not seem to respond to Cu supplementation. The endogenous levels of CcoI or CopA proteins could not be probed by immunoblotting due to lack of specific antibodies.
Properties of R. capsulatus CopZ
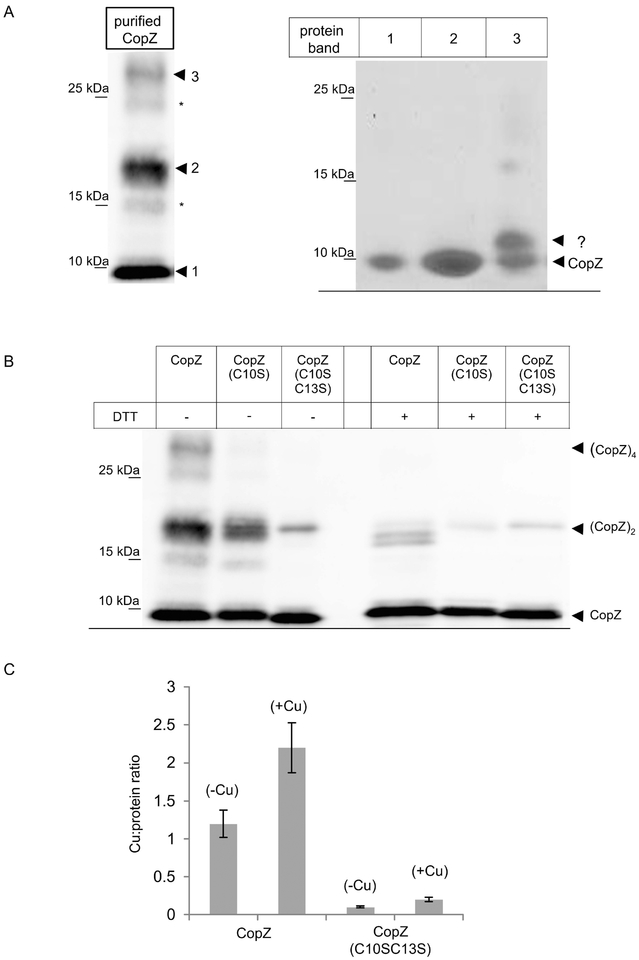
The CopZ homologue of B. subtilis has been shown to form dimers that contain up to 4 Cu atoms (Hearnshaw et al., 2009, Singleton et al., 2009, Kihlken et al., 2002, Kay et al., 2016), although the physiological relevance of the CopZ dimer is not clear (Boal & Rosenzweig, 2009, Banci et al., 2003). For analyzing the oligomeric state of R. capsulatus CopZ, the protein was expressed in E. coli and purified. When purified CopZ was separated on native gels under non-reducing conditions (-DTT), the CopZ antibodies recognized a prominent band slightly below the 10 kDa marker band (labelled 1), and a second band migrating slightly above the 15 kDa marker band (labelled 2), assumed to correspond to monomeric and dimeric CopZ, respectively (Fig. 2A, left panel). In addition, a weaker band above the 25 kDa marker band was detected (labelled 3), which could correspond to CopZ tetramers. Two additional weak bands at approx. 15 kDa and 25 kDa (indicated by *) were also detectable, and might reflect oxidized forms of CopZ oligomers with additional intramolecular disulfide-bridges (Fig. 2A, left panel). The putative CopZ oligomers were further analyzed, first by separating purified CopZ on native PAGE under non-reducing conditions as before, and then subjecting it to a second dimension SDS-PAGE under reducing conditions (Materials and Methods). Under these conditions, the initial 15 kDa band (band 2) now migrated also at approx. 6 kDa (like band 1), and no additional band was detected by Coomassie staining (Fig. 2A, right panel), confirming that the approx. 15 kDa band indeed reflected a CopZ dimer. Similarly, the band at above 25 kDa (band 3) also dissociated into the CopZ monomer, but Coomassie staining revealed the presence of an additional band, running just above the CopZ monomer (Fig. 2A, right panel,?). The identity of this band is unknown, but it could reflect either an incompletely reduced monomeric version, or an additional band representing an E. coli protein that interacts or co-purifies with CopZ. This finding was not pursued further.
Figure 2: CopZ is a Cu binding protein that forms redox-sensitive oligomers.
(A) Purified CopZ was separated on native, non-reducing PAGE and after western transfer was decorated with α-CopZ antibodies (left panel). The dominant CopZ species are indicated by numbers (1–3); two additional minor species are indicated by (*), but those were not further analyzed. The dominant species 1–3 were gel-extracted from the native gel and separated on a second dimension SDS-PAGE under reducing conditions (right panel). The second dimension was stained with coomassie brilliant blue. Indicated are CopZ and a second band (?) that was not characterized further. (B) Purified CopZ and its derivatives lacking either one (C10S) or both conserved cysteines of the Cu binding motif (C10S/C13S) were separated on native-PAGE either in the absence of the reducing agent DTT or in its presence. Indicated are the different oligomeric states of CopZ. (C) The Cu:protein ratio of purified CopZ was analyzed by atomic absorption spectroscopy. Wild type CopZ or the CopZ(C10S-C13S) mutant that lacked both conserved cysteine residues of the Cu binding motif, were directly analyzed (-Cu) or only after prior incubation with a 5-fold molar excess of Cu(I) and subsequent removal of unbound Cu(I) by gel filtration (+Cu). The molar Cu content was quantified by using a standard curve and the molar Cu:protein ratio was calculated. The values shown represent the mean of three independent experiments and the standard deviation is indicated by error bars.
The redox-sensitive oligomerization of CopZ was analyzed by using its purified mutant variants in which either a single Cys (i.e., C10) or both cysteines (i.e., C10/C13) of the putative Cu binding site were replaced by Ser. When separated on native PAGE under non-reducing conditions, wild type CopZ showed the three bands, which almost completely dissociated in the presence of DTT (Fig. 2B). For the CopZ(C10S) mutant, the putative CopZ tetramer was not detectable, but the putative CopZ dimer was only slightly reduced. Finally, for the CopZ(C10S/C13S) mutant, the CopZ dimer was significantly reduced and the tetramer was absent. In the presence of DTT, the tetramer was absent and the dimer significantly reduced in wild type and both CopZ mutant variants (Fig. 2B). Overall data show that R. capsulatus CopZ forms redox-sensitive oligomers primarily via intermolecular disulfide-bridge formation. The formation of the CopZ tetramer appears to be particularly sensitive to reduction, while the dimer seems to be more stable. Whether these different oligomeric states are physiologically relevant is currently unknown, but oligomerization of B. subtilis CopZ has been shown to influence the Cu:protein ratio (Hearnshaw et al., 2009).
This was analyzed for R. capsulatus CopZ by atomic absorption spectroscopy of purified wild type CopZ and the CopZ(C10S/C13S) mutant. When purified CopZ was analyzed without prior incubation with Cu(I), a Cu:protein ratio of approx. 1 was observed (Fig. 2C). However, when CopZ was incubated with 5-fold excess of Cu(I), and non-bound Cu was removed by gel filtration, a Cu:protein ratio of approx. 2 was observed. This is in line with the Cu4(CopZ)2 complex that was also observed in other species at increasing Cu concentrations (Kay et al., 2016, Hearnshaw et al., 2009). The CopZ(C10S/C13S) mutant served as control and showed only neglectable amounts of Cu binding, supporting that the Cys residues were required for this process.
CopZ is important for maintaining Cu resistance
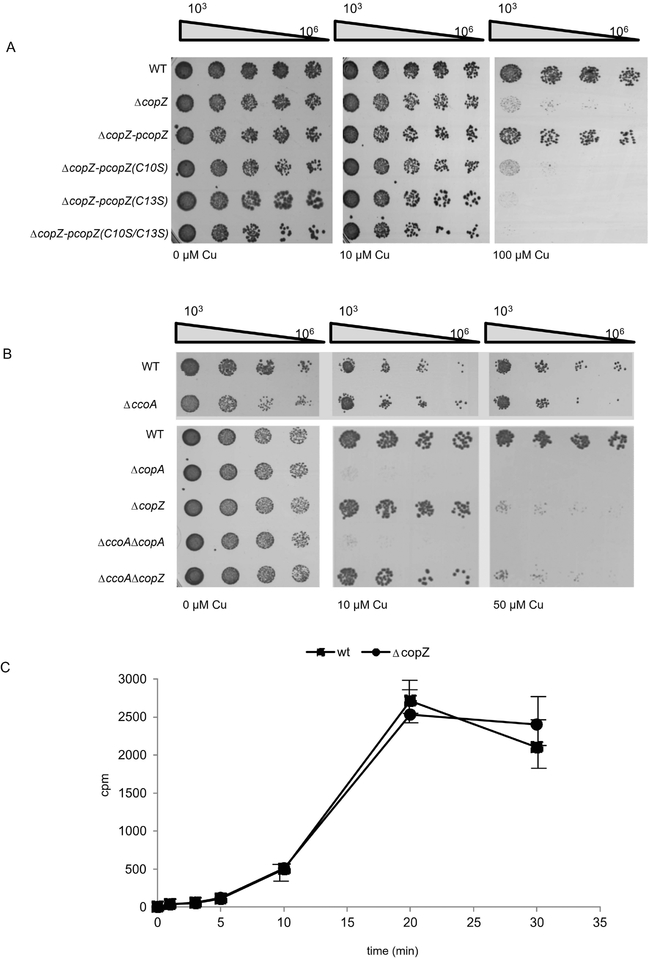
The contribution of CopZ to Cu resistance of cells was tested by analysing the Cu sensitivity of the ΔcopZ strain. Wild type R. capsulatus strains grow up to at least 100 μM Cu without significant growth reduction (Fig. 3A, right panel). However, the ΔcopZ strain failed to grow in the presence of 100 μM Cu, demonstrating that it is Cu sensitive. The enhanced Cu sensitivity of the ΔcopZ strain was rescued by expressing a plasmid-borne wild type allele of copZ (pcopZ) but not by expressing the single (C10S or C13S) or double (C10S/C13S) Cys mutants of CopZ (Fig. 3A). Thus, both cysteines of CopZ are required for conferring Cu resistance. However, considering that mutating Cys10 does not affect significantly CopZ dimerization, it appears that no direct correlation exists between CopZ dimerization and Cu binding.
Figure 3: CopZ is required for Cu tolerance in R. capsulatus.
(A) WT R. capsulatus and the ΔcopZ strain, complemented with plasmid-borne copies of different CopZ variants when indicated were grown on MPYE medium until they reached the exponential phase. They were subsequently diluted in fresh medium to an OD630 0.6 and diluted with sterile MPYE medium in a 96-well plate. 5 μl of the 103-106 diluted samples were spotted onto pre-warmed MPYE agar plates supplemented with different concentrations of CuSO4 and were analyzed after approx 24h incubation. (B) As in (A) but different R. capsulatus single and double mutants were analysed. Note, that the plate shown on the lower panel (wt and ΔccoA strain) was incubated only for 16 h for preventing the enrichment of revertants of the ΔccoA strain (Ekici et al., 2014). (C) Copper uptake was measured in whole cells by incubating cells with 64CuCl and measuring Cu content in whole cells by scintillation counting. The values shown represent the mean of three independent experiments and the standard deviation is indicated by error bars.
Intracytoplasmic Cu homeostasis in R. capsulatus is mainly controlled by the uptake-activity of the MFS protein CcoA, and the export activity of the P-type ATPase CopA (Ekici et al., 2014). The deletion of ccoA abolishes the production of R. capsulatus cbb3-Cox in the absence of supplementary Cu, but it does not significantly affect the Cu resistance properties of cells (Fig. 3B) (Ekici et al., 2012b), although the addition of Cu to the medium partially restores the growth defect and cbb3-Cox assembly. The situation is different for the ΔcopA strain, which is fully inhibited in the presence of 10 μM Cu, in line with its role in Cu extrusion. In contrast, the ΔcopZ strain grew without inhibition in the presence of 10 μM Cu, but was almost completely inhibited at 50 μM Cu (Fig. 3B, right panel). These data showed that a ΔcopZ strain is more Cu sensitive than the wild type, but less sensitive than the ΔcopA strain. This increase in Cu sensitivity was not due to increased Cu uptake in the absence of copZ, because 64Cu uptake kinetics in the wild type and ΔcopZ cells was comparable (Fig. 3C). Thus, although CopZ-like chaperones are known to deliver Cu to CopA for extrusion (Gonzalez-Guerrero & Arguello, 2008), at high Cu concentrations, Cu extrusion in R. capsulatus occurs at least partially even in the absence of CopZ, suggesting the presence of an alternative pathway.
The Cu sensitivity of a ΔccoA/ΔcopA double knock-out is similar to that of a ΔcopA single knock-out, although its slight growth defect and ability to produce a cbb3-Cox is rescued without supplementary Cu (Fig. 3B) (Ekici et al., 2014). Similarly, a ΔccoA/ΔcopZ double knock-out showed Cu sensitivity similar to that of a copZ single knock-out (Fig. 3B), while its ability to produce a cbb3-Cox is rescued only partially in the absence of Cu supplementation (see below). It therefore appears that the Cu sensitivity of R. capsulatus relies primarily on the export activity of CopA and its cognate Cu chaperone CopZ.
CopZ influences Cu content and distribution in R. capsulatus.
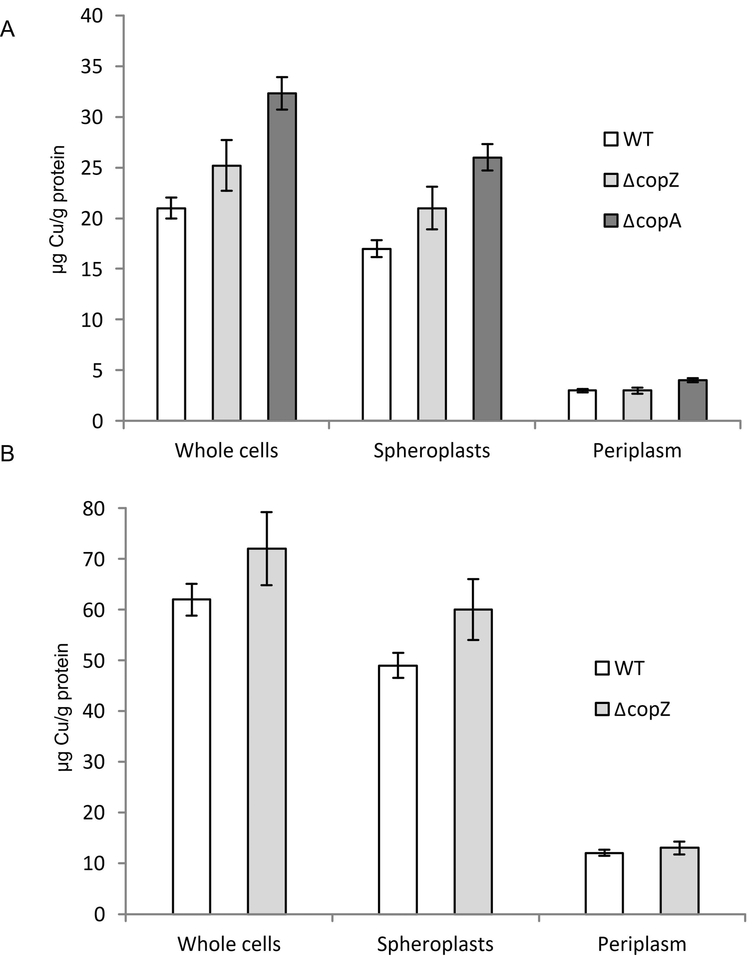
For analysing the influence of CopZ on Cu content and distribution between the cytoplasm and the periplasm, we determined the Cu content of whole cells, spheroplasts and periplasmic fractions by atomic absorption spectroscopy (AAS) (Trasnea et al., 2016a). In ΔcopZ cells grown in MPYE medium without any Cu supplementation, the total Cu content was slightly but reproducibly increased compared to the wild type cells, but it remained lower than that found in a ΔcopA strain (Fig. 4A). This increase was also seen in spheroplasts (reflecting the cytosol and the cytoplasmic membrane) of the ΔcopZ and ΔcopA strains, but not for the periplasmic fractions of the ΔcopZ strain. The ΔcopA strain also showed slightly increased Cu concentrations in the periplasmic fractions (Fig. 4A). This indicates that the absence of CopZ increases the cellular Cu concentration, primarily via a cytosolic Cu accumulation. Whether this surplus of cytosolic Cu is bound by other Cu-binding proteins or by low molecular weight compounds (e.g., glutathione) (Aliaga et al., 2016) is currently unknown. Importantly, the cellular Cu content in the absence of CopZ is lower than the Cu content in the absence of CopA, which likely explains the lower Cu sensitivity of the ΔcopZ strain.
Figure 4: CopZ influences Copper content and distribution in R. capsulatus.
(A) The Cu content R. capsulatus of whole cells, the spheroplast fraction (corresponding to the cytosol and the cytosolic membrane) and the periplasmic fraction was analysed by atomic absorption spectroscopy. Cells were grown on MPYE medium until they reached the exponential phase. Cells were then either directly analysed, or separated into the spheroplasts and periplasm fractions as described in Material and Methods. The Cu content was then calculated as μg Cu/g protein. Shown are the mean values of at least three independent experiments with the error bars indicating the standard deviation. (B) As in (A), but cells were grown in the presence of 10 μM Cu. Note, that the presence of 10 μM Cu prevents growth of the ΔcopA strain.
When cells were grown in the presence of additional 10 μM Cu, a similar effect was observed for the ΔcopZ strain (Fig. 4B), but the ΔcopA strain was not tested under these conditions as this Cu concentration is already toxic for it. As CopZ does not influence Cu uptake (Fig. 3C), the modest (approx. 20%) increase of the cytosolic Cu concentration is likely the result of a reduced Cu extrusion in the absence of CopZ.
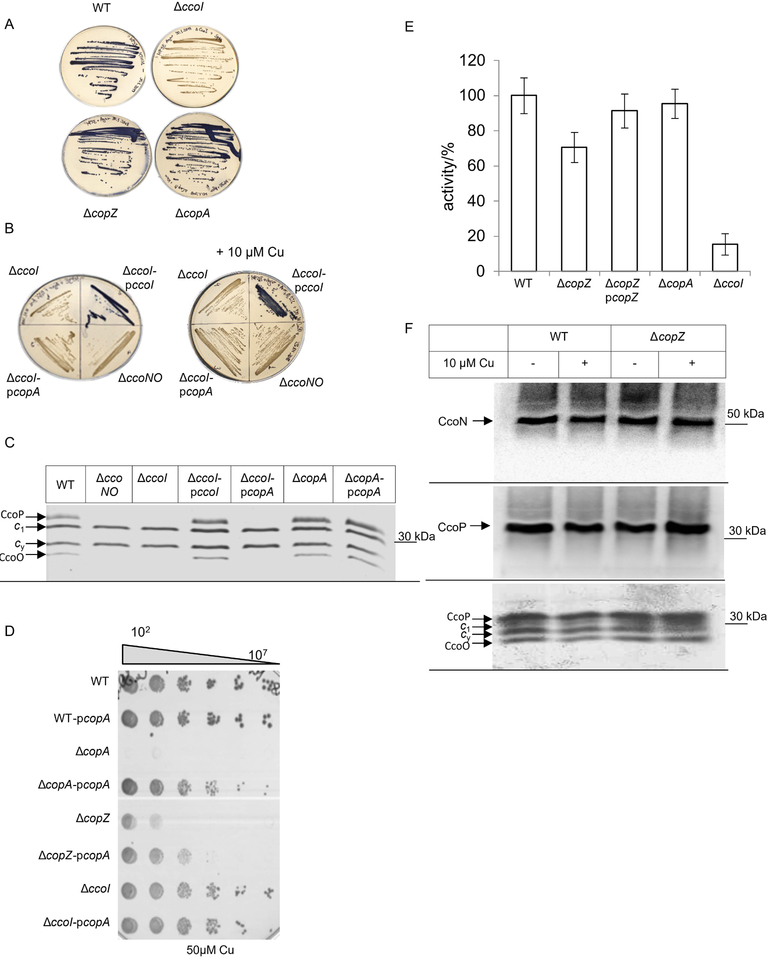
Comparative influence of CopZ, CopA and CcoI on cbb3-Cox activity
While CopZ is clearly required for maintaining Cu homeostasis in R. capsulatus and for preventing the accumulation of toxic Cu concentrations in the cytoplasm, it is unclear whether it is also involved in providing Cu for cuproprotein assembly. In R. capsulatus, the activity and stability of the cbb3-Cox is strictly linked to Cu availability, and in the absence of CcoI, which is a CopA homologue, only very low levels of cbb3-Cox activity is detectable (Koch et al., 2000, Kulajta et al., 2006). This feature can be monitored by the NADI-staining, which allows qualitative detection of cbb3-Cox activity in R. capsulatus colonies. In the presence of cbb3-Cox activity, the electron donor dimethyl-4-phenylenediamine (DMPD) is oxidized and forms together with α-naphthol the blue dye indophenol blue. In the presence of the NADI-solution, wild type cells (MT1131) turn blue within seconds, while the ΔccoI strain retains its original colour even after 30 min, indicative of the lack of cbb3-Cox activity (Fig. 5A). In contrast, colonies of the ΔcopZ and the ΔcopA strain also turn blue rapidly, indicating their ability to produce active cbb3-Cox (Fig. 5A). The lack of cbb3-Cox activity in the absence of CcoI is rescued only by a plasmid-borne copy of ccoI and not by copA (Fig. 5B), demonstrating that both P1B-type Cu exporting ATPases have non-overlapping functions. As CopA is expected to have a lower Cu affinity than CcoI (Raimunda et al., 2011), the NADI staining of appropriate strains was also performed on plates supplemented with 10 μM Cu. Under these conditions the plasmid-encoded copA still failed to complement the cbb3-Cox assembly defect in the ΔccoI strain (Fig. 5B).
Figure 5: CopZ is required for full activity of cbb3-Cox.
(A) The cbb3-Cox activity in R. capsulatus wild type and mutant cells was determined by the NADI reaction on MPYE plates. In the presence of cbb3 Cox activity, the blue dye indophenol blue is formed within 15 sec. (B) As in (A); when indicated cells were grown on MPYE plates supplemented with 10 μM Cu. ΔccoNO corresponds to a cbb3-Cox deficient R. capsulatus strain. (C) The membrane-bound c-type cytochrome profile of membranes isolated from the indicated strains was determined by heme-staining on SDS-PAGE. CcoP and CcoO correspond to the membrane-bound c-type cytochrome subunits of cbb3-Cox, while c1 is the subunit of cyt bc1 complex and cy a membrane-bound electron donor. (D) Cu sensitivity assay of different R. capsulatus strains, containing a plasmid-encoded copA copy when indicated. The assay was performed as described in the legend to Fig. 3. (E) cbb3-Cox activity was determined in isolated intracytoplasmic membranes (ICMs) by measuring the ascorbate-TMPD (N,N,N,N-tetramethyl-p-phenylendiamine) oxidase activity. The activiy of wild type ICMs was set to 100 % and the mean values of at least three independent experiments are shown. The error bar reflects the standard deviation. (F) The cbb3-Cox subunit profile of wild type and ΔcopZ ICMs. When indicated, cells were grown in the presence of 10 μM Cu. The CcoN and CcoP subunits were detected by western blotting using polyclonal antibodies against CcoN or CcoP, respectively (upper two panels). In addition, the membrane-bound c-type cytochrome profile was determined by heme-staining on SDS-PAGE (lower panel) as in (C).
TMBZ-mediated heme-staining allows for the detection of the two c-type cytochrome subunits CcoP and CcoO of cbb3-Cox and the c-type cytochrome profile was monitored in membranes of the ΔccoI and ΔcopA strains. Wild type membranes and membranes of a ΔccoNO strain served as control. In wild type membranes, CcoP, CcoO, as well as cyt c1 of the cytochrome bc1 complex and the membrane-bound electron carrier cyt cy were detectable (Fig. 5C). Membranes of the ΔccoI and ΔccoNO strains lacked both CcoP and CcoO, while the levels of cyt c1 and cyt cy were unchanged. The c-type cytochrome profile of the ΔcopA strain was like wild type. Plasmid encoded ccoI but not plasmid encoded copA rescued the phenotype of the ΔccoI strain, further demonstrating that CopA is not required for cbb3-Cox assembly and that CopA cannot substitute for the CcoI function.
As a control for monitoring whether the plasmid encoded copA is active, the Cu sensitivity of the ΔcopA and the ΔcopA pcopA strains were tested in the presence of 50 μM Cu. This showed that plasmid-encoded copA rescues the Cu sensitivity of the ΔcopA strain and demonstrate that plasmid encoded copA is functional. (Fig. 5D). Therefore, although CopA exports Cu into the periplasm, it cannot supply Cu for cbb3-Cox assembly. It is noteworthy that the expression of a plasmid-borne copy of copA also increased the Cu tolerance of the ΔcopZ strain but not that of the ΔccoI strain (Fig. 5D), further indicating that Cu-export via CopA is still functional in the absence of CopZ and that absence of CcoI does not significantly contribute to the extrusion of excess Cu.
NADI staining is a fast but not very sensitive method for determining cbb3-Cox activity in R. capsulatus, and therefore this activity was measured quantitatively using a fibre optic oxygen meter. In membranes of the ΔcopZ strain, an approx. 30% decrease of cbb3-Cox activity was observed, and this was rescued by expressing a plasmid-borne copy of copZ (Fig. 5E). As controls, the ΔcopA strain showed no significant decrease in cbb3-Cox activity, while the ΔccoI strain had extremely low activity, in line with previous reports (Koch et al., 1998a, Koch et al., 2000).
The basis of the reduced cbb3-Cox activity in the ΔcopZ strain was further analysed by monitoring the steady-state stability of the catalytic subunit CcoN and the di-heme subunit CcoP in membranes. By immunoblot analyses no significant difference was seen in the amounts of CcoN and CcoP between wild type and ΔcopZ in membranes from cells grown in the absence or presence of additional 10 μM Cu (Fig. 5F, top and middle). TMBZ-mediated heme-staining confirmed the unchanged presence of CcoP and CcoO, the c-type cytochrome subunits of cbb3-Cox, of cyt c1 and of the membrane-bound electron carrier cyt cy in both wild type and ΔcopZ membranes (Fig. 5F, bottom). Altogether the data indicated that, in contrast to the ΔccoI strain, the absence of CopA does not impair the activity or stability of cbb3-Cox (Fig 5C), but the absence of CopZ reduces cbb3-Cox activity by approx. 30% (Fig. 5E)
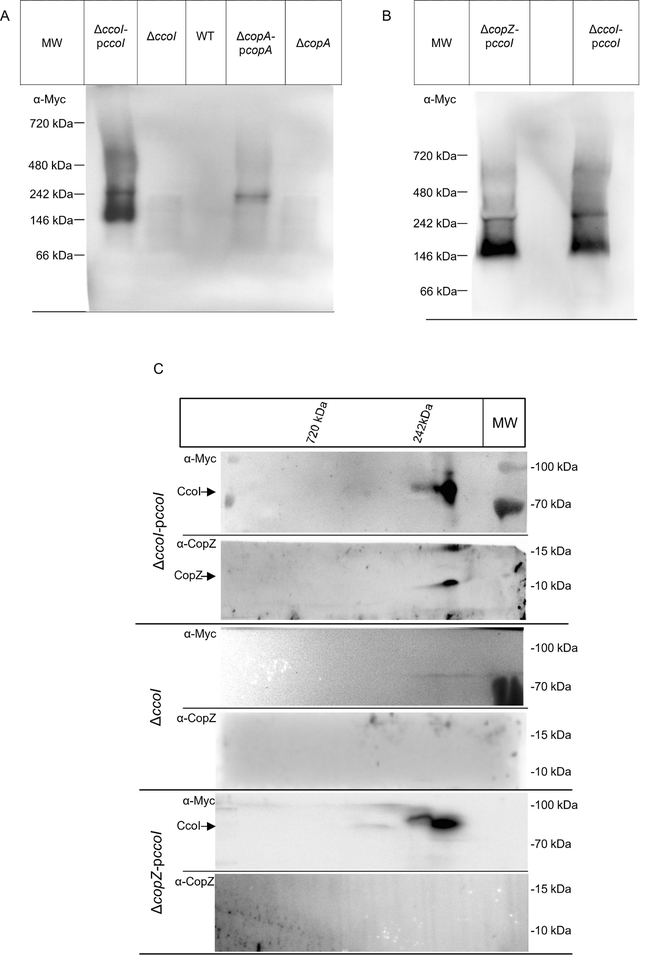
CcoI forms a protein complex that contains CopZ.
The above presented data strongly support a role of CopZ during CopA-dependent Cu detoxification, and also link CopZ to Cu trafficking for cbb3-Cox assembly, likely via CcoI. This issue was further analysed by Blue-Native-PAGE (BN-PAGE), which allows the detection of protein complexes in native membranes. Membranes of cells expressing either ccoI or copA, each with a C-terminal Myc-His-tag were DDM-solubilized and separated on BN-PAGE without prior purification (Materials and Methods). After immune detection with α-Myc antibodies, an approx. 150 kDa complex was detected in cells expressing ccoI-Myc that was not present in the wild type or ΔccoI cells (Fig. 6A). In addition, two weaker complexes, one at approx. 250 kDa and the other at approx. 600 kDa, were detected. In CopA-containing membranes, only one complex at approx. 250 kDa was detected that migrated slightly faster than the ~250 kDa CcoI complex. We noted that the expression level of copA-Myc from the pBAD vector in R. capsulatus was weaker than that of ccoI-Myc, and it is possible that additional CopA complexes would be visible at higher copA expression levels. Nevertheless, these data indicated that CcoI and CopA form distinct membrane assemblies in the R. capsulatus membrane.
Fig. 6: CopZ forms a complex with the P1B-type ATPase CcoI.
(A) ICMs of the indicated strains were solubilized, and separated without further purification by BN-PAGE. After western transfer, potential CcoI and CopA complexes were visualized with antibodies against their C-terminal Myc-tags. (B) As in (A), but probing for CcoI complexes in membranes of the ΔcopZ strain. (C) ICMs of the ΔccoI strain complemented with pccoI (upper two panels), the ΔccoI strain (middle two panels) and of ΔcopZ pccoI (lower two panels) were solubilized and separated on BN-PAGE as in (A). The BN-PAGE gel lane was cut out, subsequently equilibrated in equilibration buffer containing SDS and urea and subjected to a 2nd dimension SDS-PAGE. After western transfer, the membrane was horizontally cut and the upper part was decorated with α-Myc antibodies and the lower part with α-CopZ antibodies. Representative gels of at least three biological replicates are shown.
For identifying whether CopZ is part of the CcoI complexes, ccoI was also expressed in the ΔcopZ strain. All three complexes were visible even in the absence of CopZ, suggesting that CopZ is not essential for the stability of the CcoI complexes (Fig. 6B). However, we noted that the 150 kDa complex migrated slightly faster in the absence of CopZ (Fig. 6B), which might indicate that CopZ is part of this complex. For analysing this further, the gel lanes of the BN-PAGE were excised, denatured in SDS/urea-containing equilibration buffer and then subjected to a second dimension SDS-PAGE, followed by immune detection with α-Myc and α-CopZ antibodies. CcoI was detected by α-Myc antibodies at about 80 kDa, in line with its predicted mass of 79 kDa (Fig. 6C). The CcoI band on SDS-PAGE corresponded to the approx. 150 kDa complex seen in BN-PAGE, but was trailing to the left, possibly reflecting the approx. 250 kDa complex. No CcoI signal was observed from the approx. 600 kDa complex, most likely due to the low abundance of this complex. Importantly, α-CopZ antibodies recognized two bands at approx. 10 and 15 kDa, migrating within the same 2D lane as the 150 kDa CcoI complex (Fig. 6C, top). These two bands are in line with the CopZ monomer and dimer species that have been seen before (c.f. Fig. 2). The presence of the CopZ dimer under these conditions is likely the result of incomplete reduction of CopZ during the equilibration of the BN-PAGE gel slice. As a control, membranes from ΔccoI cells were also separated on similar BN-PAGE/SDS-PAGE, and in this case, no band was recognized by either α-Myc or α-CopZ antibodies (Fig. 6C, middle). For membranes of the ΔcopZ pccoI strain, α-Myc antibodies detected CcoI on the second dimension as a component of the 150 kDa complex, whereas α-CopZ antibodies failed to recognize any specific signal (Fig. 6C, bottom). Thus, overall data indicated that the 150 kDa complex likely contains both CcoI and CopZ, although CopZ does not seem to be required for complex stability. The presence of any additional protein(s) in this complex, and the composition of the weaker approx. 250 kDa and 600 kDa CcoI complexes were not further studied here, and will be pursued in future experiments.
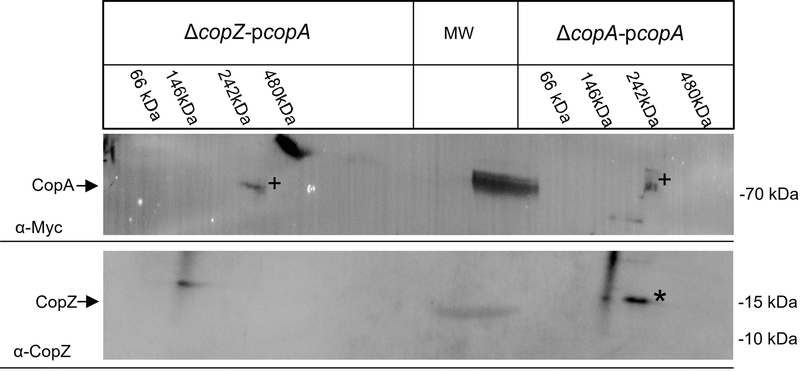
CopA containing membranes were also subjected to the second dimension for analysing whether CopZ is part of the 250 kDa complex seen in Fig 6A. After separation of the corresponding BN-PAGE lane on SDS-PAGE, α-Myc antibodies recognized CopA at approx. 80 kDa both in the presence and absence of CopZ (Fig 7, top, +). Thus similar to the CcoI-complexes, the stability of the CopA complex was not significantly influenced by the absence of CopZ. However, in comparison to CcoI, the CopA signal was weaker due to the lower CopA expression in R. capsulatus (Fig. 6A) and required longer exposure. This probably explains why also the 70 kDa marker band and a non-characterized band at approx. 100 kDa in the ΔcopZ-pcopA sample were recognized. α-CopZ antibodies recognized a band at approx. 15 kDa, which was not detected in the ΔcopZ strain expressing copA (Fig. 7, bottom, *). α-CopZ antibodies recognized additionally a smeary signal both in the presence and absence of CopZ, which likely reflects unspecific recognition due to the long exposure. In summary, these data demonstrate that the Cu-chaperone CopZ forms complexes with CcoI and also with CopA, even though their interactions are expected to be highly transient and for the putative CopA-CopZ complex at the detection limit. Still, considering all data, we conclude that CopZ is not only engaged in Cu-detoxification via CopA, but is also involved in Cu delivery to cbb3-Cox via CcoI in R. capsulatus.
Fig. 7: CopZ forms also a complex with the P1B-type ATPase CopA.
ICMs of ΔcopZ pcopA and ΔcopA pcopA were solubilized, and first separated by BN-PAGE. The BN-PAGE lanes were then cut and subjected to a 2nd dimension SDS-PAGE, followed by immune detection, as described for Fig. 6. CopA and CopZ are indicated by (+) and (*), respectively.
Discussion
Copper homeostasis depends on an intricate network of Cu binding proteins that serve as sensors, chaperones, transporters and dedicated cuproprotein assembly factors (Arguello et al., 2013, Singleton & Le Brun, 2007, Cobine et al., 2006, Stewart et al., 2018). In combination with Cu storage proteins, initially found only in eukaryotes but now also identified in bacteria (Dennison et al., 2018, Parmar et al., 2018), these proteins basically reduce the concentration of free Cu below the detection limit and thus prevent potential toxicity. CopZ-like Cu-chaperones have been identified as major determinants for Cu detoxification because they can bind Cu with femtomolar affinity (Xiao et al., 2011) and can transfer it to P1B-type ATPases for extrusion. This has been shown for Atox1, which delivers Cu to ATP7B in the trans-Golgi network of human cells (Lutsenko, 2016) or for CopZ, which delivers Cu to CopA in the bacterial membrane (Gonzalez-Guerrero & Arguello, 2008, Odermatt & Solioz, 1995). In contrast to the CopA-like ATPases, the CcoI/FixI-like ATPases transport Cu primarily for cbb3-Cox assembly (Preisig et al., 1996, Koch et al., 1998a, Kulajta et al., 2006) and whether they also receive Cu from CopZ or any other Cu chaperone is unknown.
In the current study, a CopZ-like chaperone was identified in the R. capsulatus genome. It shows high homology to other members of the CopZ chaperone family and binds Cu, consistent with the presence of the invariant MxCxxC Cu binding motif. The deletion of CopZ increased the Cu sensitivity of R. capsulatus cells, in line with a role in Cu detoxification via the CopA mediated Cu extrusion pathway. Cu detoxification was not observed when the cysteine residues of the CopZ Cu binding motif were replaced by serine, verifying the importance of the MxCxxC motif in Cu binding. We note that the Cu sensitivity of the ΔcopZ strain was less pronounced than the Cu sensitivity of the ΔcopA strain, suggesting that at higher Cu concentrations, CopA might bind Cu directly without the need for a Cu chaperone. A chaperone-independent Cu transport via CopA is supported by data showing Cu transport in inverted membrane vesicles in the absence of CopZ (Gonzalez-Guerrero et al., 2010). Alternatively, CopA in R. capsulatus might receive Cu from another CopZ-like chaperone of lower affinity (Fig. 8). Two very similar CopZ homologs are present in Pseudomonas (Quintana et al., 2017), but we did not identify a second copZ-like gene in R. capsulatus. Thus, if other chaperones are able to deliver Cu to CopA in R. capsulatus, then they remain to be identified. The deletion of copZ in R. capsulatus resulted in increased cellular Cu accumulation, primarily in the cytoplasm, which also supports a defect in Cu extrusion, because absence of CopZ had no detectable influence on CcoA-mediated Cu uptake, similar to the observations in mammalian cells, where the down-regulation of Atox1 has no effect on the rate of Cu entry (Maryon et al., 2013).
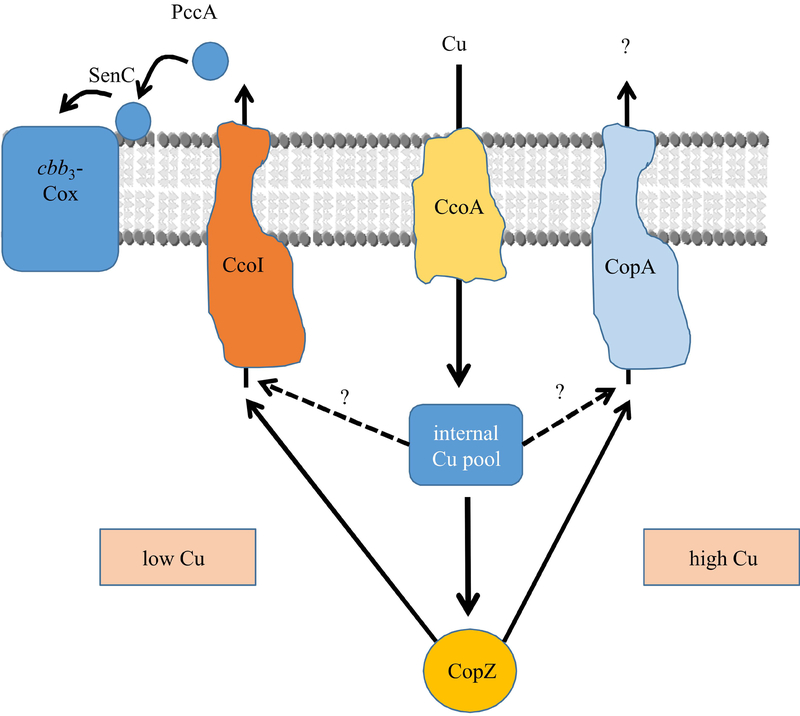
Fig. 8: The dual role of CopZ in R. capsulatus Cu homeostasis.
Cu uptake in R. capsulatus is primarily mediated by the major facilitator superfamily protein CcoA. Cytosolic Cu is present in a poorly defined internal Cu pool, and is rapidly transferred to CopZ. At low Cu concentrations, Cu supply to cbb3-Cox is maintained by both the internal Cu pool (dashed line) and by CopZ-bound Cu (solid line), both feeding Cu into the CcoI-dependent Cu export. Subsequently, Cu is bound by two periplasmic Cu chaperones, PccA and SenC, and transferred to cbb3-Cox. Thus, in the absence of CopZ, Cu supply to cbb3-Cox is reduced, but still possible via the internal Cu pool. Under these conditions, CopA does not significantly drain the internal Cu pool due to its lower Cu affinity.
At high Cu concentrations, Cu supply for cbb3-Cox assembly is also maintained by the internal Cu pool and by CopZ. But CopZ will predominantly feed Cu into the CopA-dependent export (solid line), and due to the combined activity of CopZ and CopA, the cellular Cu concentration will rapidly decrease and reach again low Cu concentrations. However, in the absence of CopZ, CopA can still receive some amounts of Cu from the increased internal Cu pool (dashed line), hence Cu detoxification is at least partially sustained even in the absence of CopZ. This working model rationalizes why the absence of CopZ confers less pronounced Cu sensitivity than the absence of CopA and why it decreases but not abolishes the cbb3-Cox activity.
Data on the role of CopZ-like chaperones in cuproprotein assembly are scarce. The co-expression of CopZ increased the activity of a heterologously expressed CotA laccase (Gunne et al., 2013), indicating a potential role of CopZ in Cu insertion into CotA. On the other hand, a deletion of copZ in B. subtilis did not influence aa3-type cytochrome oxidase (aa3-Cox) activity (Radford et al., 2003), in agreement with the occurrence of dedicated Cu chaperones, like CtaG or Sco1, for aa3-Cox assembly (Bengtsson et al., 2004, Hill & Andrews, 2012). However, in Rhodobacter species, the cbb3-Cox and aa3-Cox assembly pathways do not engage identical Cu uptake and trafficking conduits (Khalfaoui-Hassani et al., 2016, Khalfaoui-Hassani et al., 2018). The cbb3-Cox activity in the ΔcopZ strain is reduced to about 70% of the wild type activity, whereas the deletion of copA does not reduce cbb3-Cox activity, and the expression of a plasmid-encoded copy of copA in the ΔccoI strain does not rescue its cbb3-Cox assembly defect. On the other hand, different from the deletion of copA, the deletion of ccoI does not influence the Cu resistance profile of R. capsulatus, showing that CcoI and CopA have non-overlapping functions. The situation is different for CopZ, which appears to be involved in both Cu detoxification and cbb3-Cox assembly as its absence affects both Cu sensitivity and cbb3-Cox activity of R. capsulatus. An interesting possibility is that at low Cu concentrations the CopZ-dependent Cu delivery to CcoI might be particularly important because the high-affinity of CopZ for Cu and the affinity of CcoI for Cu-loaded CopZ would ensure that sufficient amounts of Cu are fed into the Cu delivery system for cbb3-Cox assembly. Only at higher Cu concentrations, CopZ would be dispensable for Cu trafficking to CcoI, but particularly important for Cu delivery to CopA for extrusion of excess Cu (Fig. 8). This would also explain why in the absence of CopZ, cbb3-Cox activity is reduced but not abolished, as cytoplasmic Cu concentrations are increased. The Cu KD values for CopZ homologues have been determined to be in the range of 10−15M (Banci et al., 2010). Cu-transporting ATPases have a similar KD (approx. 10−17 M) (Banci et al., 2010, Kay et al., 2017). These values would support a possible Cu transfer from CopZ to CopA/CcoI.
Although such a dual role of CopZ-like chaperones in Cu detoxification and cuproprotein assembly has so far not been described for bacteria, the eukaryotic CopZ homologues generally execute such a dual role. The CopZ-homologue Atox1 delivers Cu to ATP7B for the assembly of cuproproteins in the trans-Golgi network, like ceruloplasmin or dopamine-β-hydroxylase. At elevated Cu concentrations and after vesicular traffic of ATP7B to the basolateral membrane, Atox1 then delivers Cu for extrusion (Lutsenko, 2016). A dual role in Cu export and Cu delivery to cuproporteins has also been described for the bacterial periplasmic chaperone CueP (Fenlon & Slauch, 2017, Osman et al., 2013). The proposal of a dual role of CopZ in R. capsulatus is also supported by the observation that CopZ can be detected in complex with CcoI and CopA in the R. capsulatus membrane. The transient association of CopZ with CopA and CcoI could also explain why a small amount of CopZ was found in the membrane fraction. In ΔccoI membranes, the amount of membrane-associated CopZ was not significantly changed, which is in line with the hypothesis that CopZ can transiently associate with either protein. In membranes of the ΔccoI strain expressing pCcoI, we noticed increased CopZ membrane binding, further supporting that CcoI and CopZ interact. However, we cannot completely exclude that this membrane association is the result of CopZ aggregation. The size of the observed complex of ~ 150 kDa suggests that besides CcoI (80 kDa) and CopZ (6 kDa), additional proteins might be a part of this complex. The composition of this and the other larger CcoI complexes, as well as that of the CopA complex, are currently under investigation.
CcoI and CopA in R. capsulatus are highly homologous, and mainly differ in their respective N-and C-termini. Assuming that CcoI starts at the first in-frame methionine residue, it contains an additional glutaredoxin-like CPxC motif upstream of the typical CxxC metal binding domain (MBD) motif found in both CcoI- and CopA-like P1B-type Cu-ATPases. The role of this additional CPxC motif is currently unknown. However, mutating the second cysteine residue of the canonical MBD motif resulted in reduced cbb3-Cox assembly (Koch et al., 2000), indicating that it is important for CcoI activity. One additional difference between CcoI and CopA in R.capsulatus is that CcoI contains only seven predicted transmembrane domains (TMs), instead of the eight TMs found in CopA-like ATPases (Gourdon et al., 2011). Despite these differences, a Gly-Gly-kink (residues 188 & 189 in CcoI) on the cytosolic face of TM2, which exposes a positively charged platform where the Cu-loaded CopZ interacts with CopA (Padilla-Benavides et al., 2013) is present in R. capsulatus CcoI. Immediately downstream of this kink, two positively charged residues (K191 & R194), which could serve as an interaction surface, are also present. In addition, the Cu entrance site composed of a methionine and two negatively charged residues (M197, D242 and E373 in CcoI), as well as a conserved tyrosine (Y224 in CcoI) as Cu exit site, are conserved in R. capsulatus CcoI. Thus, the critical determinants that are required for CopZ interaction seem to be conserved in CcoI. However, the interaction surface between P1B-type Cu-ATPases and CopZ-like chaperones appears to be rather small (Banci et al., 2009), which further explains why their interaction is likely only transient.
It is striking that despite the similarity between CcoI and CopA, both proteins cannot substitute for each other. The lower affinity of CopA for free Cu (Gonzalez-Guerrero et al., 2010) is unlikely to explain this functional difference if both CopA and CcoI were to receive Cu from CopZ. Likewise, the faster turnover rate of CopA would not prevent the usage of the exported Cu for cbb3-Cox assembly. It is possible that CcoI has some additional chaperone activity that is required for cbb3-Cox assembly. An alternative explanation is that Cu is transferred from both CopA and CcoI to dedicated periplasmic Cu chaperones, as has been shown for E. coli CopA that transfers Cu directly to CusF (Padilla-Benavides et al., 2014). In the case of R. capsulatus two periplasmic chaperones, PccA and SenC, link Cu export via CcoI to cbb3-Cox assembly (Lohmeyer et al., 2012, Trasnea et al., 2018, Trasnea et al., 2016b), although which one is the direct acceptor remains to be determined. The constraints of CopA and CcoI to interact with only dedicated Cu chaperones in the periplasm could be important for maintaining homeostasis between cuproprotein assembly and controlling Cu toxicity.
Experimental Procedures
Bacterial strains and growth conditions
The E. coli strains harbouring plasmids were grown at 37˚ C in LB medium supplemented with 50 μg/mL ampicillin or 30 μg/mL tetracycline. R. capsulatus wild type (MT1131) and mutant strains were grown in enriched medium (MPYE) at 35˚ C under semi-aerobic conditions (Koch et al., 1998b), appropriate antibiotics were added to the growth media when needed. Metal-free MPYE medium or metal-free H2O was obtained by mixing 100 ml of each with 5 g of Chelex-100 resin and stirring for 1 hour at room temperature. The Chelex-100 resin was then removed by filtration.
Cu sensitivity assays on plates
R. capsulatus cells were grown on MPYE media under semi-aerobic conditions (100 mL of growth medium in a 250 mL flask) at 35° C, 110 rpm until they reached the logarithmic phase (OD685 of 0.7 – 0.8). The pre-cultures were diluted to OD685 0.2 and incubated further until they reached again the logarithmic phase. Cell cultures were adjusted to OD685 0.6 and subsequently diluted with sterile MPYE medium in multiple sequential steps in a 96-well plate ranging, resulting in 100 to 106 final dilutions. 5 μL of selected dilutions were spotted onto pre-warmed MPYE agar plates supplemented with different concentrations of CuSO4, as needed. The liquid drops were allowed to dry for approximately 20 min; afterwards the plates were incubated at 35° C for approx. 24h, with the exception of the ΔccoA strain, which was incubated only for approx. 16h to reduce the risk of selecting revertants.
Molecular genetic techniques
Standard molecular genetic techniques were performed as described previously (Sambrook & Russel, 2001, Koch et al., 1998b). To construct a chromosomal knockout allele of copZ, the gene transfer agent (GTA) was used as described earlier (Koch et al., 1998b). Genomic DNA of R. capsulatus was extracted from 10 ml cultures of wild type (MT1131) strain, which were grown in MPYE medium by using the DNAeasy Blood & Tissue kit (Qiagen Inc.). The isolated DNA was analysed qualitatively and quantitatively by agarose gel electrophoresis, and by Nanodrop spectrophotometer, respectively.
To construct a deletion-insertion allele of copZ (RCAP_rcc03125), a 1.2 Kbp DNA fragment, covering the putative copZ gene and approx. 500 nucleotides each upstream and downstream of it, was PCR amplified using wild type R. capsulatus (MT1131) chromosomal DNA as a template, and the primers CopZ_SacI_For (5’ACC GAC CGA GCT CAC AAT CTC A3´) and CopZ_KpnI_Rev (5’TAA GCT TCG GTA CCC TGG GCA A3´) containing 5′-SacI and 3′-KpnI restriction sites. This 1.2 Kbp fragment was digested with SacI and KpnI and cloned into the same sites of pBlueScript II KS (+) and pRK415 to yield the plasmids pBlueScript-copZ and pRK415-copZ, respectively. A BglII restriction site was inserted into the copZ allele in pBlueScript-copZ using the primer: CopZ_Insert_BglII_For (5’GCC GCA GAT CTT GTT GGG A 3´) and CopZ_Insert_ BglII_Rev (5’CAT TGC AAG GCC AGC GTG 3´), yielding the plasmid pBlueScript-copZ-BglII. The kanamycin resistance cassette of pMA117 (which contains a 5’ BamHI site in front of the Kanamycin cartridge) was amplified using the primers Kan_pMA117_For (5’CGA GGC CCT TTC GTC TTC-3´ and Kan_BamHI_pMA117_Rev (5’TTG AAG GAT CCC AAG GGC 3´) adding a 3’ BamHI restriction site. The PCR product was digested with BamHI and ligated into the BglII-digested pBlueScript-copZ-BglII, resulting in pBlueScript-Δ(copZ::Kan). The SacI-KpnI fragment of pBlueScript-Δ(copZ::Kan) was cloned into the respective cloning sites of pRK415 to yield pRK415-ΔcopZ. This plasmid was used to generate the ΔcopZ knockout by GTA crosses with the wild-type (MT1131) to yield the ΔcopZ strain.
Plasmid pBlueScript-copZ was used as a template with the primer CopZ_pET19b_NdeI_ For (5’GCC CCA TAT GAT CCT GTC CG3´) and CopZ_pET19b-XhoI_Rev (5’ATC TGG CCT CGA GGA TGA CCT CGG CC3´) to create pET19b-His-copZ carrying a His-tagged version of copZ. This latter plasmid was also used as a template for constructing CopZ variants in which the Cys residues at positions 10 and 13 were replaced by Ser using the CopZ_C10S_For (5’GCA ATG GCC GGA GGT CAT GTT3´) with CopZ_C10S_Rev (5’AAG GCC AGC GTG GAA GCC G3´) and CopZ_C10S-C13S_For (5’CTT GGA ATG GCC GGA GGT CAT3´) with CopZ_C10S-C13S_Rev (5’GCC AGC GTG GAA GCC G3´) primers.
For constructing an inducible expression system for CcoI, the ccoI gene of plasmid pCW3 (Koch et al., 2000) was amplified via PCR using the Primer 1a (5’GGC TAA CAG GAG GAA TTA ACA TGA CCG CTG CCC CCC TTG CGT C3`) and Primer 1b (5’AGT TTT TGT TCG GGC CCA AGC TTC AGC CGC ATC GAG TTC AGC G3`). The PCR fragment was ligated into the NcoI- and HindIII-digested pBAD A Myc-His (Invitrogen) vector using the Gibson assembly protocol (Gibson et al., 2009). The obtained plasmid pBAD-ccoI-Myc-His containing a C-terminal Myc-His tagged ccoI was transformed into E. coli DH5a cells and verified by DNA sequencing. The entire pBAD-ccoI-Myc-His was then inserted into the plasmid pRK415 by first amplifying pBAD-ccoI-Myc-His via PCR using the Primer 2a (5’CTC TAG AGT CGA CCT GCA GGC ATG CAG CAG CAG ATC AAT TCG CGC GCG AAG3’) and Primer 2b (5’GAA ACA GCT ATG ACC ATG ATT ACG CCA CTC GCG CGT TTC GGT GAT GAC GGT G3’). The PCR fragment was ligated into a PstI-digested pRK415 using the Gibson assembly protocol, yielding the plasmid pRK415-pBAD-ccoI-Myc-His, which was then transformed into the E. coli strain DH5a or HB101, and subsequently transferred into appropriate R. capsulatus strains by triparental conjugation (Koch et al., 1998b). Essentially the same strategy was used for constructing pRK415-pBAD-copA-Myc-His expressing CopA, using the primers CopA-fw (5’CGG GGT ACC CCG ATG GCC AGC ACC AGT TTC3’) and copA-Rev (5’CGG AAT TCC GTC CGG CGG CGG CGC GGC GCAG3’). The PCR product was cloned into KpnI and EcoRI digested pBAD A Myc-His resulting in plasmid pBAD-copA-His. The entire plasmid was amplified via PCR using the primers (5’GGA TCC TCT AGA GTC GAC CTG CAC GAG GAA GCG GAA GAG CGC CTG CAT GCG-3’) and (5’CCA TGA TTA CGC CAA GCT TGC ATG CCT GCA CTC ACT GAC TCG CTG CGC TCG GTC GTTC3’) and inserted into pRK415 as described above, yielding pRK415-pBAD-copA-Myc-His. After transformation into E. coli HB101, the plasmid was transferred into appropriate R. capsulatus strains by triparental conjugation. Protein expression from both plasmids was induced by the addition of 0.5 % arabinose, and expression was monitored by immunoblots using antibodies against the Myc and His tags.
RNA isolation and RT-PCR
Semi-aerobic cultures of R. capsulatus in MPYE medium were grown to mid log phase (OD625 0.8–1.0). Approximately 1× 109 cells were centrifuged, incubated in TE buffer (10mM Tris, 0,5mM EDTA, pH 7) containing 10 mg/mL lysozyme for 15 minutes at 37˚C and passed 5 times through a needle with 0.8 μm diameter into RNase free tubes. Isolation of total RNA was achieved by using the GE Healthcare Mini Spin kit following the suggested protocol. 2μg of total RNA was treated with DNaseI for 30 min at 25˚C in the presence of RNase inhibitor RNasin. 50 ng of treated RNA was used in RT-PCR reactions with the One Step RT-PCR kit (Quiagen, Hilden, Germany). The following primers were used CopZ-RT-For (5’ ATGCAAGACGAAGCGCGGAGG 3’) and CopZ-RT-Rev (5’ TCAGGCGATGACCTCGGCCTC 3’); CcoI-RT-For (5’ TCCTGCTAGTCGGCCGC-TACC 3’) and CcoI-RT-Rev (5’ TGCCCGAAGTCACCTCGCC 3’); CopA-RT-For (5’ CGATGT-CTATTTCGAGGCGGC 3’) and CopA-RT-Rev (5’ CCCGGGGCCAGGTCTACC 3’). Primers used for 16S rRNA have been described (Ekici et al., 2014). Samples were separated on a 1.2% agarose gel and visualized by SYBR green staining.
Preparation of intracytoplasmic membranes (ICM) and Blue Native Page analyses
Inverted cytoplasmic membranes from wild-type R. capsulatus (MT1131) and its mutants were prepared as described previously (Koch et al., 1998a, Pawlik et al., 2010). For Blue Native (BN) PAGE analyses (Wittig et al., 2006), ICMs (~ 200 μg of total protein) were incubated at 25°C for 15 minutes with continuous shaking (550 rpm Eppendorf Thermomixer) in the lysis buffer (50 mM NaCl, 50 mM imidazole/HCl, pH 7.0, 5 mM 6-aminohexanoic acid) containing 1% n-dodecyl-β-maltoside (Thermo Scientific Inc.). Solubilized proteins were isolated by a 15 min centrifugation step at 70,000 rpm in a TLA 100.3 rotor. The supernatant was mixed with 6 μL of loading dye (5% Coomassie blue G250 in 0.5 M 6-aminohexanoic acid) and 17 % glycerol, and loaded onto 4–16 % gradient BN-PAGE gels (Life Technologies).
For the second dimension SDS-PAGE, the entire lane of a first dimension BN-PAGE was excised and incubated for 30 min at room temperature in equilibration buffer (50 mM Tris-HCl pH 6.8, 2% (w/v) SDS, 6 M urea, 30% (v/v) glycerol). The lane was then mounted on top of a 5 – 15% gradient SDS-PAGE or a 16% Tris-Tricine gel (Wittig et al., 2006). After western blotting, the membrane was cut into two pieces and the upper part was decorated with α-Myc antibodies and the lower part with α-CopZ antibodies.
Non-denaturing gels
Samples were resuspended in native loading buffer (50 mM Tris-HCl, pH 6.8, 10 % glycerol, 0.01 % bromophenol blue), denatured at 25°C for 5 min and separated by native-PAGE (375 mM Tris pH 8.8, 50 % of Rotiphorese® NF-Acrylamide/Bis-solution 30 %, TEMED, APS). For two-dimensional PAGE, purified CopZ was separated by native-PAGE as a first dimension. The gel was stained with colloidal Coomassie, and the CopZ bands were excised and incubated in the denaturation buffer (50 mM Tris-HCl pH 6.8, 2 % SDS (w/v), 6 M Urea, 30 % glycerol, 1 M DTT) for 30 min at 37°C and continuous shaking (180 rpm). The sample was separated by reducing SDS-PAGE as the second dimension.
Quantification of CopZ in vivo
Quantification of CopZ was basically performed as described previously for SenC and PccA (Trasnea et al., 2018). In brief, R. capsulatus cells were lysed by addition of trichloroacetic acid (TCA) and precipitated proteins were separated by SDS-PAGE and compared to similarly treated known amounts of purified CopZ. Chemiluminescence based immune detection was performed using anti-CopZ polyclonal antibodies raised in rabbits against purified CopZ. The signals corresponding to the purified proteins and the whole cells were quantified using Image J. Quantification is based on at least three independent biological replicates with two technical replicates each. The concentration of CopZ in whole cells was determined to be 8.4 ± 0.3 ng/108 cells. Assuming that the cytosol plus the inner membrane of one R. capsulatus cell has a volume of 1.9 × 10−16 L (Gaju et al., 1989, Sundararaj et al., 2004), the molar concentration of CopZ (MW = 6.4 kDa) in the cytoplasm was calculated to be ~ 6.9 μM.
Immune-detection methods and heme staining
For immune-detection of proteins on SDS- or BN-PAGE, proteins were electroblotted on onto a PVDF Immobilon-P membrane (GE Healthcare) (BN-PAGE), or onto a nitrocellulose membrane (GE Healthcare) (SDS-PAGE). Polyclonal antibodies against CcoN and CcoP (Koch et al., 1998a) were used with either peroxidase-conjugated goat anti-rabbit (Caltag Laboratories, Burlingame, CA) or alkaline phosphatase-conjugated goat anti-rabbit antibodies (Sigma, St. Louis, USA) antibodies as secondary antibodies with ECL (GE Healthcare, Munich, Germany). Antibodies against the His- or Myc tags (clone 9E10) were purchased from Millipore (Temecula, USA). Antibodies against CopZ were raised in rabbits (Gramsch Laboratories, Schwabhausen, Germany) using the protein purified from E. coli. Heme staining revealing the peroxidase activity of membrane-bound c-type cytochromes was performed as described previously (Koch et al., 1998b)
CopZ purification and quantification of the Cu content
For purifying CopZ, a N-terminally His-tagged variant (HisCopZ) and its derivatives (HisCopZ-C10S and HisCopZ-C10S-C13S) were expressed from pET19b derived plasmids in E. coli BL21(DE3). E. coli cells were grown aerobically in LB medium with apropriate antibiotics, and induced at an OD600 of 0.6 by adding freshly prepared IPTG (1 mM). After two hours of incubation at 30° C with 170 rpm shaking, cells were harvested and washed once with 15 mL of resuspension buffer (20 mM Tris-HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2) per g of cell mass. Following centrifugation, cells were resuspended in 1 mL of washing buffer (resuspension buffer with 10 % glycerol) per g cell mass. Complete protease inhibitor mixture (Roche) and phenylmethylsulfonyl fluoride (PMSF, final concentration 0.5 mM) were added, and cells were disrupted by using a French pressure cell. Cell debris were removed by centrifugation at 28.790 x g for 30 min using the Sorvall SS-34 rotor (Evolution RC6 centrifuge, Thermo Scientific Inc.). The supernatant was then ultracentrifuged for one hour at 192,840 x g using a Type 50.2 Ti rotor (Beckmann-Coulter, Krefeld, Germany). The supernatant thus obtained was loaded onto a polypropylene column containing TALON® Metal Affinity Resin (Clonetech), equilibrated with washing buffer and incubated for two hours. After five washing steps with five times column volume of washing buffer each, elution was initiated using the elution buffer (washing buffer with increasing imidazole concentration, 100–300 mM) and fractions were collected. Eluted proteins were buffer-exchanged to washing buffer using PD10 columns (GE Healthcare). Protein purity was checked by SDS-PAGE.
For determining the Cu(I) content of CopZ, 1 μM purified CopZ was incubated with a 5-fold molar excess of CuSO4 in washing buffer containing 10 mM ascorbate for 10 min at room temperature with gentle mixing (Gonzalez-Guerrero & Arguello, 2008). Unbound Cu was removed by passing the sample through a Sephadex-G25 column (PD10, GE Healthcare) and the Cu content in CopZ was determined using an atomic absorption spectrophotometer Perkin-Elmer 4110 ZL Zeeman and TraceCert Copper AAS standards (Sigma-Aldrich) as reference (Trasnea et al., 2016a).
64Cu uptake assays
64Cu uptake assays were basically performed as described earlier (Ekici et al., 2014). In brief, R. capsulatus cells were grown on MPYE medium overnight and harvested at OD630 ~ 0.8, washed once with fresh MPYE medium, and resuspended in 1 mL MPYE medium. The total number of cells was determined spectrophotometrically (1 OD630 unit = 7.5 × 108 cells/mL) and all cultures were normalized to the same number of cells per milliliter of MPYE medium. The assay mixture contained 100–200 μL of MPYE culture and approx. 107 cpm 64CuCl2 completed to a final volume of 500 μL with citrate buffer (50 mM sodium citrate, pH 6.5; 5% glucose), and the reaction was started by addition of CuCl2. At different time points, 50 μL samples were transferred to a pre-cooled Eppendorf tube containing 50 μL of 1 mM non-radioactive CuCl2 and 50 μL of 50 mM EDTA (pH 6.5). Samples were centrifuged for 5 min at 10,000 rpm, and the cell pellet was washed twice with 100 μL ice-cold 50 mM EDTA solution before resuspension in scintillation liquid and determination of the radioactivity by a scintillation counter. 64Cu was obtained as CuCl2 from the National Centre for Nuclear Research, Otwock, Poland and was delivered by Hartmann Analytic GmbH, (Braunschweig, Germany). It was dissolved in washing buffer to desired concentrations.
Oxygen uptake assays
Ascorbate-TMPD (N,N,N,N-tetramethyl-p-phenylendiamine) oxidase activity was measured with a fiber optic oxygen meter (Fibox 3; PreSens GmbH, Regensburg, Germany) at 28° C in a closed reaction chamber. R. capsulatus membranes were disolved in ICM buffer (50 mM TeaAc pH 7.5, 1 mM EDTA, 1 mM DTT and 0.5 mM PMSF) to a final concentration of about 0.1 μg/μL. The reaction was initiated by the addition of 10 μL of 1 M sodium ascorbate (10 mM final concentration) and 5 μL of 24 mM TMPD (N,N,N,N–tetramethyl-p-phenylendiamine, final concentration 0.2 mM). Oxygen consumption was recorded at 28° C using the OxyView 3.5.1 software (PreSens GmbH) and terminated after several minutes of recording by the addition of 2 μL NaCN (0.1 mM final concentration). cbb3-Cox activity was determined by subtraction of the respiration rate remaining after addition of NaCN (200 mM) from that initiated by ascorbate-TMPD.
Measurement of cellular copper accumulation
R. capsulatus cells were grown in MPYE media under aerobic conditions (100 mL of growth medium in 250 mL flask) at 35° C, 110 rpm to an OD685 of 1. Afterwards, cells were harvested by centrifugation at 4° C, 5,000 rpm for 20 min, and cell pellets washed with 50 mL of metal-depleted MPYE and re-centrifuged. The pellet was then resuspended in 1 mL of metal-depleted H2O, and a small aliquot (about 10 to 20 μL) was taken for protein concentration determination by the Lowry assay. Exactly 1 mL of cell suspension was transferred to a new 15 mL tube to which 0.75 mL of 53 % nitric acid was added and incubated at 80 °C for 1 hour followed by overnight incubation at 60 °C to yield a clear liquid. The reaction was terminated by the addition of 300 μL H2O2. Cu-free water was added up to a final volume of 10 mL, and samples were measured by using an atomic absorption spectrophotometer Perkin-Elmer 4110 ZL Zeeman. In case any insoluble impurities were observed within the sample, they were filtered through a 0.45 μm filter.
R. capsulatus spheroplasts were prepared as described previously (Trasnea et al., 2016a). Formation of spheroplasts was monitored microscopically, using an Olympus BX51 microscope at a 100 x magnification with a numerical aperture of 1.4. The spheroplasts were pelleted by centrifugation at 5,000 rpm for 20 min, and copper contents of the pellet and the supernatant were determined after acid digestions, as described above for the intact cells.
Acknowledgements
We thank Huiying Li and Hanna Wetzel for their help with constructing the CcoI and CopA expression plasmids. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG GRK2202 & IRTG1478) to HGK, from the Motivate College of the University Freiburg Medical School to MU and DM, from the Deutsche Akademische Austauschdienst (DAAD) to PIT, and was partly supported by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of Department of Energy [DOE DE-FG02–91ER20052], and partly by the National Institute of Health [NIH GM 38237] grants to FD.
References
- Aliaga ME, Lopez-Alarcon C, Bridi R, and Speisky H (2016) Redox-implications associated with the formation of complexes between copper ions and reduced or oxidized glutathione. Journal of inorganic biochemistry 154: 78–88. [DOI] [PubMed] [Google Scholar]
- Arguello JM, Patel SJ, and Quintana J (2016) Bacterial Cu+-ATPases: models for molecular structure-function studies. Metallomics : integrated biometal science 8: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JM, Raimunda D, and Padilla-Benavides T (2013) Mechanisms of copper homeostasis in bacteria. Frontiers in cellular and infection microbiology 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Calderone V, Della-Malva N, Felli IC, Neri S, Pavelkova A, and Rosato A (2009) Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. The Biochemical journal 422: 37–42. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, and Palumaa P (2010) Affinity gradients drive copper to cellular destinations. Nature 465: 645–648. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, and Del Conte R (2003) Solution structure of apo CopZ from Bacillus subtilis: further analysis of the changes associated with the presence of copper. Biochemistry 42: 13422–13428. [DOI] [PubMed] [Google Scholar]
- Bengtsson J, von Wachenfeldt C, Winstedt L, Nygaard P, and Hederstedt L (2004) CtaG is required for formation of active cytochrome c oxidase in Bacillus subtilis. Microbiology (Reading, England) 150: 415–425. [DOI] [PubMed] [Google Scholar]
- Boal AK, and Rosenzweig AC (2009) Structural biology of copper trafficking. Chemical reviews 109: 4760–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Rensing C, and Helmann JD (2017) Metal homeostasis and resistance in bacteria. Nature reviews. Microbiology 15: 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P, Wickramasinghe WA, Harrison MD, Weber T, Solioz M, and Dameron CT (1999) The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS letters 445: 27–30. [DOI] [PubMed] [Google Scholar]
- Cobine PA, Pierrel F, and Winge DR (2006) Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochimica et biophysica acta 1763: 759–772. [DOI] [PubMed] [Google Scholar]
- Dennison C, David S, and Lee J (2018) Bacterial copper storage proteins. The Journal of biological chemistry 293: 4616–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Pawlik G, Lohmeyer E, Koch H-G, and Daldal F (2012a) Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochimica et biophysica acta 1817: 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Turkarslan S, Pawlik G, Dancis A, Baliga NS, Koch HG, and Daldal F (2014) Intracytoplasmic Copper homeostasis controls Cytochrome c oxidase production. mBio 5: e01055–01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Yang H, Koch HG, and Daldal F (2012b) Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenlon LA, and Slauch JM (2017) Cytoplasmic Copper Detoxification in Salmonella Can Contribute to SodC Metalation but Is Dispensable during Systemic Infection. Journal of bacteriology 199: e00437–00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Tsui HC, Bruce KE, Sham LT, Higgins KA, Lisher JP, Kazmierczak KM, Maroney MJ, Dann CE 3rd, Winkler ME, and Giedroc DP (2013) A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nature chemical biology 9: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaju N, Guerrero R, and Pedros-Alio C (1989) Measurement of cell volume of phototrophic bacteria in pure cultures and natural samples: phase contrast, epifluorescence and particle sizing. FEMS Microbiol. Ecology 62: 295–302. [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, and Arguello JM (2008) Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proceedings of the National Academy of Sciences of the United States of America 105: 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, Raimunda D, Cheng X, and Arguello JM (2010) Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Molecular microbiology 78: 1246–1258. [DOI] [PubMed] [Google Scholar]
- Gourdon P, Liu XY, Skjorringe T, Morth JP, Moller LB, Pedersen BP, and Nissen P (2011) Crystal structure of a copper-transporting PIB-type ATPase. Nature 475: 59–64. [DOI] [PubMed] [Google Scholar]
- Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, and Daldal F (1994) Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry 33: 3120–3127. [DOI] [PubMed] [Google Scholar]
- Gunne M, Al-Sultani D, and Urlacher VB (2013) Enhancement of copper content and specific activity of CotA laccase from Bacillus licheniformis by coexpression with CopZ copper chaperone in E. coli. Journal of biotechnology 168: 252–255. [DOI] [PubMed] [Google Scholar]
- Hatori Y, Inouye S, and Akagi R (2017) Thiol-based copper handling by the copper chaperone Atox1. IUBMB life 69: 246–254. [DOI] [PubMed] [Google Scholar]
- Hearnshaw S, West C, Singleton C, Zhou L, Kihlken MA, Strange RW, Le Brun NE, and Hemmings AM (2009) A tetranuclear Cu(I) cluster in the metallochaperone protein CopZ. Biochemistry 48: 9324–9326. [DOI] [PubMed] [Google Scholar]
- Hill BC, and Andrews D (2012) Differential affinity of BsSCO for Cu(II) and Cu(I) suggests a redox role in copper transfer to the Cu(A) center of cytochrome c oxidase. Biochimica et biophysica acta 1817: 948–954. [DOI] [PubMed] [Google Scholar]
- Kay KL, Hamilton CJ, and Le Brun NE (2016) Mass spectrometry of B. subtilis CopZ: Cu(i)-binding and interactions with bacillithiol. Metallomics : integrated biometal science 8: 709–719. [DOI] [PubMed] [Google Scholar]
- Kay KL, Zhou L, Tenori L, Bradley JM, Singleton C, Kihlken MA, Ciofi-Baffoni S, and Le Brun NE (2017) Kinetic analysis of copper transfer from a chaperone to its target protein mediated by complex formation. Chem. Commun 53: 1397–1400. [DOI] [PubMed] [Google Scholar]
- Khalfaoui-Hassani B, Verissimo AF, Shroff NP, Ekici S, Trasnea PI, Utz M, Koch HG, and Daldal F, (2016) Biogenesis of Cytochrome c Complexes: From heme insertion of Redox Cofactors to Assembly of different subunits. Springer, Dordrecht. [Google Scholar]
- Khalfaoui-Hassani B, Wu H, Blaby-Haas CE, Zhang Y, Sandri F, Verissimo AF, Koch HG, and Daldal F (2018) Widespread Distribution and Functional Specificity of the Copper Importer CcoA: Distinct Cu Uptake Routes for Bacterial Cytochrome c Oxidases. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihlken MA, Leech AP, and Le Brun NE (2002) Copper-mediated dimerization of CopZ, a predicted copper chaperone from Bacillus subtilis. The Biochemical journal 368: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Hwang O, and Daldal F (1998a) Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. Journal of bacteriology 180: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Myllykallio H, and Daldal F (1998b) Using Genetics to explore Cytochrome function and Structure in Rhodobacter. Methods in enzymology 297: 81–94. [Google Scholar]
- Koch HG, Winterstein C, Saribas AS, Alben JO, and Daldal F (2000) Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. Journal of molecular biology 297: 49–65. [DOI] [PubMed] [Google Scholar]
- Kulajta C, Thumfart JO, Haid S, Daldal F, and Koch HG (2006) Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. Journal of molecular biology 355: 989–1004. [DOI] [PubMed] [Google Scholar]
- Lohmeyer E, Schroder S, Pawlik G, Trasnea PI, Peters A, Daldal F, and Koch HG (2012) The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochimica et biophysica acta 1817: 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S (2016) Copper trafficking to the secretory pathway. Metallomics : integrated biometal science 8: 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, and Imlay JA (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proceedings of the National Academy of Sciences of the United States of America 106: 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon EB, Molloy SA, and Kaplan JH (2013) Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am J Physiol Cell Physiol 304: C768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin I, Gudin S, Mintz E, and Cuillel M (2009) Dissecting the role of the N-terminal metal-binding domains in activating the yeast copper ATPase in vivo. The FEBS journal 276: 4483–4495. [DOI] [PubMed] [Google Scholar]
- Odermatt A, and Solioz M (1995) Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. The Journal of biological chemistry 270: 4349–4354. [DOI] [PubMed] [Google Scholar]
- Osman D, Patterson CJ, Bailey K, Fisher K, Robinson NJ, Rigby SE, and Cavet JS (2013) The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Molecular microbiology 87: 466–477. [DOI] [PubMed] [Google Scholar]
- Padilla-Benavides T, George Thompson AM, McEvoy MM, and Arguello JM (2014) Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. The Journal of biological chemistry 289: 20492–20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Benavides T, McCann CJ, and Arguello JM (2013) The mechanism of Cu+ transport ATPases: interaction with CU+ chaperones and the role of transient metal-binding sites. The Journal of biological chemistry 288: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar JH, Quintana J, Ramirez D, Laubenbacher R, Arguello JM, and Mendes P (2018) An important role for periplasmic storage in Pseudomonas aeruginosa copper homeostasis revealed by a combined experimental and computational modeling study Molecular microbiology in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik G, Kulajta C, Sachelaru I, Schröder S, Waidner B, Hellwig P, Daldal F, and Koch H-G (2010) The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. Journal of bacteriology 192: 6378–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuser V, Glaeser J, and Klug G (2011) The RSP_2889 gene product of Rhodobacter sphaeroides is a CueR homologue controlling copper-responsive genes. Microbiology (Reading, England) 157: 3306–3313. [DOI] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, and Hennecke H (1996) The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Archives of microbiology 165: 297–305. [DOI] [PubMed] [Google Scholar]
- Quintana J, Novoa-Aponte L, and Arguello JM (2017) Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. The Journal of biological chemistry 292: 15691–15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher C, Hoffmann MC, Lackmann JW, Moser R, Pfander Y, Leimkuhler S, Narberhaus F, and Masepohl B (2012) Tellurite resistance gene trgB confers copper tolerance to Rhodobacter capsulatus. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine 25: 995–1008. [DOI] [PubMed] [Google Scholar]
- Rademacher C, and Masepohl B (2012) Copper-responsive gene regulation in bacteria. Microbiology (Reading, England) 158: 2451–2464. [DOI] [PubMed] [Google Scholar]
- Radford DS, Kihlken MA, Borrelly GP, Harwood CR, Le Brun NE, and Cavet JS (2003) CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx-type ATPase CopA. FEMS microbiology letters 220: 105–112. [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, and O’Halloran TV (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science (New York, N.Y.) 284: 805–808. [DOI] [PubMed] [Google Scholar]
- Raimunda D, Gonzalez-Guerrero M, Leeber BW 3rd, and Arguello JM (2011) The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine 24: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, and McDevitt SF (2013) The copper metallome in prokaryotic cells. Met Ions Life Sci 12: 417–450. [DOI] [PubMed] [Google Scholar]
- Rosenzweig AC, and Arguello JM (2012) Toward a molecular understanding of metal transport by P(1B)-type ATPases. Current topics in membranes 69: 113–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, and Russel DW, (2001) Molecular Cloning. In.: Cold Spring Harbor Laboratory, pp. [Google Scholar]
- Singleton C, Hearnshaw S, Zhou L, Le Brun NE, and Hemmings AM (2009) Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. The Biochemical journal 424: 347–356. [DOI] [PubMed] [Google Scholar]
- Singleton C, and Le Brun NE (2007) Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine 20: 275–289. [DOI] [PubMed] [Google Scholar]
- Stewart LJ, Thaqi D, Kobe B, McEwan AG, Waldron KJ, and Djoko KY (2018) Handling of nutrient copper in the bacterial envelope. Metallomics : integrated biometal science. [DOI] [PubMed] [Google Scholar]
- Sundararaj S, Guo A, Habibi-Nazhad B, Rouani M, Stothard P, Ellison M, and Wishart DS (2004) The CyberCell database (CCDB): a comprehensive, self-updated, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic acids research 32: 293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasnea PI, Andrei A, Marckmann D, Utz M, Khalfaoui-Hassani B, Selamoglu N, Daldal F, and Koch HG (2018) A Copper Relay System Involving Two Periplasmic Chaperones Drives cbb3-Type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. ACS chemical biology 13: 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasnea PI, Marckmann D, Utz M, and Koch HG (2016a) Measurement of cellular copper in Rhodobacter capsulatus by atomic absorption spectroscopy. bio-protocol 6: 1948–1957. [Google Scholar]
- Trasnea PI, Utz M, Khalfaoui-Hassani B, Lagies S, Daldal F, and Koch HG (2016b) Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3 -type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Molecular microbiology 100: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urvoas A, Moutiez M, Estienne C, Couprie J, Mintz E, and Le Clainche L (2004) Metal-binding stoichiometry and selectivity of the copper chaperone CopZ from Enterococcus hirae. European journal of biochemistry / FEBS 271: 993–1003. [DOI] [PubMed] [Google Scholar]
- Vita N, Landolfi G, Basle A, Platsaki S, Lee J, Waldron KJ, and Dennison C (2016) Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Scientific reports 6: 39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, and Schagger H (2006) Blue native PAGE. Nature protocols 1: 418–428. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Brose J, Schimo S, Ackland SM, La Fontaine S, and Wedd AG (2011) Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. The Journal of biological chemistry 286: 11047–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]