Summary:
Humans are repeatedly exposed to influenza virus via infections and vaccinations. Understanding how multiple exposures and pre-existing immunity impact antibody responses is essential for vaccine development. Given the recent prevalence of influenza H1N1 A/California/7/2009 (CA09), we examined the clonal composition and dynamics of CA09 hemagglutinin (HA)-reactive IgG repertoire over 5 years in a donor with multiple influenza exposures. The anti-CA09 HA polyclonal response in this donor comprised 24 persistent antibody clonotypes, accounting for 72.6 ± 10.0% of the anti-CA09 HA repertoire over 5 years. These persistent antibodies displayed higher somatic hypermutation relative to transient serum antibodies detected at one time point. Additionally, persistent antibodies predominantly demonstrated cross-reactivity and potent neutralization towards a phylogenetically distant H5N1 A/Vietnam/1203/2004 (VT04) strain, a feature correlated with HA stem recognition. This analysis reveals how ‘serological imprinting’ impacts responses to influenza and suggests that once elicited, cross-reactive antibodies targeting the HA stem can persist for years.
Graphical Abstract
eTOC blurb:
In recent years, there has been a great interest in understanding how repeated exposures to influenza can change the already-established host antibody repertoire. Lee et al. quantitatively demonstrate that over multiple years and repeated vaccinations the serum response to influenza is dominated by a small number of persistent antibodies.
Introduction
Influenza globally infects over a billion people and results in approximately half a million deaths annually (Lambert and Fauci, 2010). The influenza A virus, in particular, undergoes frequent changes that account for pandemics, epidemics and the majority of seasonal outbreaks worldwide (Hay, 2001), with the H1N1 A/California/7/2009 (CA09) strain being one of the latest examples (Wu and Wilson, 2017). The Influenza A viruses are classified into different subtypes based on the antigenic properties of their two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (Wright and Webster, 2001). HA is more immunogenic, and thus serves as the antigenic component of seasonal flu vaccines (Doherty et al., 2006).
The structure of HA consists of a head domain containing the receptor binding site (RBS) that binds to sialic acid moieties on the surface of target cells, and a stem domain which mediates membrane fusion at endosomal pH (Corti and Lanzavecchia, 2013; Skehel and Wiley, 2000). While the residues surrounding the RBS are subject to significant evolutionary drift and play an important role in the ability of influenza to evade pre-existing humoral immunity, the stem region is highly conserved, presumably due to both its limited accessibility and structural constraints imposed by the need for membrane fusion (Corti and Lanzavecchia, 2013). Extensive studies in humans have revealed that head-specific antibodies generally bind to a narrow range of viral subtypes (Lee et al., 2014; Schmidt, Aaron G. et al., 2015; Whittle et al., 2011), although exceptions of broad heterosubtypic recognition have been reported (Ekiert et al., 2012; McCarthy et al., 2018). In contrast, stem-binding antibodies are more likely to bind and neutralize a wide range of heterosubtypic strains (Corti et al., 2011; Joyce et al., 2016; Lee et al., 2016; Li et al., 2012; Pappas et al., 2014; Sui et al., 2009; Wei et al., 2010; Wrammert et al., 2011).
Due to the ubiquitous nature of influenza, humans are typically exposed to the virus at an early age (Bodewes et al., 2011), resulting in the production of antibodies to the encountered influenza strains (Kim et al., 2016; Schmidt, A.G. et al., 2015). Pre-existing immunity from these earlier infection(s) or from vaccination exerts a major, and possibly dominant, influence on the nature of the humoral response elicited upon subsequent challenge (Andrews et al., 2015; Angeletti et al., 2017; Lee et al., 2016), a phenomenon that has been described as ‘original antigenic sin’ (Francis, 1960), ‘back-boosting’ (Fonville et al., 2014), ‘antigenic seniority’ (Kucharski et al., 2015) or ‘HA immune imprinting’ (Gostic et al., 2016). The effect of pre-existing serological immunity can impede the elicitation of antibodies to new strains, although it may not be necessarily detrimental to protection (Lee et al., 2016; Linderman and Hensley, 2016).
Our current understanding of pre-existing humoral immunity is based primarily on bulk serological metrics that measure the aggregate properties of the circulating antibodies to historical influenza strains: ELISAs are used to detect the presence and abundance of influenza-specific antibodies while neutralization assays inform on the efficacy of the elicited response with regards to blocking viral binding and infection. Information on how the individual serum anti-influenza monoclonal antibodies (mAbs) that compose the polyclonal titer to influenza wax and wane over multiple years, during which an individual may be exposed to multiple vaccinations and/or infections, has been completely lacking. A molecular-level understanding of how pre-existing immunity influences the serological responses and further, how the serological antibody repertoire is reshaped by repeated vaccination over prolonged periods, is essential for developing more effective strategies for vaccination, including the design of ‘universal’ flu vaccines that can provide protection against a broad range of influenza strains (Impagliazzo et al., 2015; Krammer and Palese, 2015; Yassine et al., 2015).
Previously, Lanzavecchia and co-workers identified CA09 HA-binding mAbs through single cell analysis of peripheral plasma cells and memory B cells from one donor, who was born in 1952, over 6 years (Corti et al., 2010; Corti et al., 2011; Kallewaard et al., 2016; Pappas et al., 2014; Pinna et al., 2009). This collection of antibodies included several HA stem-binding mAbs with exceptional heterosubtypic neutralization breadth, such as FI6, which neutralizes all influenza A strains. However, it still remains unknown whether these mAbs are present in serum and if so, how much they contribute to the overall serological repertoire to CA09 HA, what other anti-HA antibodies are abundantly present in circulation, and finally how individual mAbs differ in their temporal dynamics in serum over time. Here we used Ig-Seq, a high-resolution, quantitative, proteomics-based serum antibody repertoire profiling technology (Lavinder et al., 2015; Lavinder et al., 2014; Lee et al., 2016; Williams et al., 2017; Wine et al., 2013) to longitudinally analyze the dynamics and persistence of circulating anti-CA09 HA antibodies from the same donor, over a period spanning 5 years and multiple exposures to CA09. We find that the donor’s serological repertoire to CA09 HA is largely unchanged over time and is dominated by a small number of persistent antibody clonotypes, which included several potent antibodies targeting highly conserved regions on the influenza virus.
Results
Longitudinal analysis of anti-CA09 HA serum IgG repertoire demonstrates high prevalence of persistent antibody clonotypes
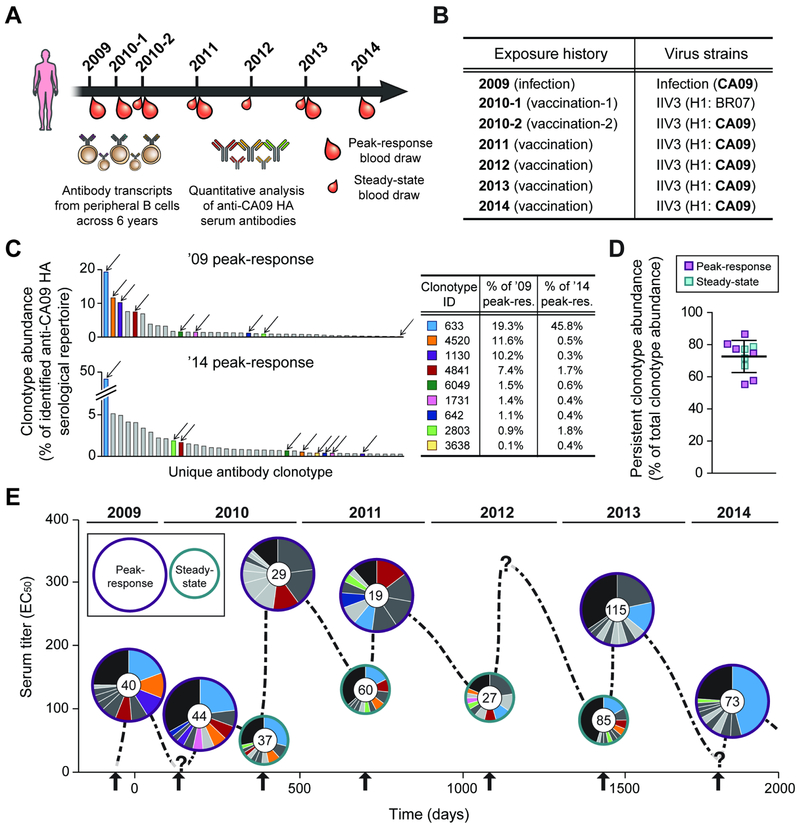
The experimental design, sample collection time points and the donor’s exposure history to CA09 HA are presented in Figure 1A and B, and in Table 1. Briefly, the earliest blood sample collected from the donor was ~21 days following infection with CA09 in 2009. Then early in 2010, the donor was vaccinated with trivalent inactivated influenza vaccine (IIV3) containing Brisbane/59/2007 as the H1 component, followed by another vaccination at the end of the year with IIV3 containing CA09; this second vaccination in 2010 (i.e. ’10-2 vaccination) marked the donor’s first re-exposure to CA09 HA since the infection. Every subsequent year between 2011 and 2014, the donor received IIV3 containing CA09.
Fig. 1. Longitudinal analysis of CA09 HA-reactive serum antibodies from 2009 to 2014.
A. Serum samples were collected at the time points indicated and the serological repertoire at each time point was determined by Ig-Seq (see also Table 1). B. Exposure history of the donor to CA09. For each trivalent inactivated influenza vaccine (IIV3), H1 component is indicated. BR07, Brisbane/59/2007. C. Composition of the anti-CA09 HA serological repertoire at the first (’09 peak-response) and last (’14 peak-response) time points (see also Fig. S2 and Table S3). Each bar represents one of the 40 most abundant antibody clonotypes with height corresponding to the relative abundance of that clonotype. The 9 antibody clonotypes detected every year are indicated with arrows, and their relative abundances in the ’09 and ’14 peak-response sera are shown in the table. D. Relative abundance of the 24 persistent antibody clonotypes, detected in serum samples for ≥4 out of 5 years. Error-bars indicate s.d. (n = 10). E. The anti-CA09 HA antibody repertoire composition over 5 years (see also Table S2). y-axis shows the serum titer to CA09 HA. Within each pie chart, each of the top 10 most abundant antibody clonotypes is represented as a slice with an area proportional to the relative abundance of that clonotype. The 9 antibody clonotypes in C are shown using the same color scheme while the other 15 persistent antibody clonotypes are shown in darker grey. Anti-CA09 HA antibody clonotypes that are not among the top 10 most abundant antibodies are grouped together in the black slice. Inner circle in pie chart shows the number of antibody clonotypes identified in the serum sample. Arrows indicate exposure events. ‘?’, no serum sample was collected at that time point.
Table 1. Descriptions on the analyzed serum samples.
Titers were determined by ELISA, with 50% effective concentration (EC50) values representing titers. Averages are calculated as mean with error indicating s.d., repeated in triplicates.
| Sample name (collection date) |
Description | Note | Serum titer (EC50) |
|---|---|---|---|
| ’09 PR (Dec. 1, 2009) |
2009 peak-response (infection) |
~21 days post infection | 136 ± 4 |
| ’10-1 PR (May 6, 2010) |
2010-1 peak-response (vaccination-1) |
14 days post 2010-1 vaccination | 59 ± 1 |
| ’10-1 SS (Dec. 6, 2010) |
2010-1 steady-state (vaccination-1) |
229 days post 2010-1 vaccination (pre-vaccination for 2010-2 vaccination) |
36 ± 3 |
| ’10-2 PR (Dec. 20, 2010) |
2010-2 peak-response (vaccination-2) |
14 days post 2010-2 vaccination | 313 ± 9 |
| ’10-2 SS (Nov. 11, 2011) |
2010-2 steady-state (vaccination-2) |
340 days post 2010-2 vaccination (pre-vaccination for 2011 vaccination) |
117 ± 5 |
| ’11 PR (Nov. 28, 2011) |
2011 peak-response (vaccination) |
17 days post 2011 vaccination | 281 ± 4 |
| ’11 SS (Nov. 14, 2012) |
2011 steady-state (vaccination) |
369 days post 2011 vaccination (pre-vaccination for 2012 vaccination) |
102 ± 2 |
| ’12 SS (Dec. 3, 2013) |
2012 steady-state (vaccination) |
384 days post 2012 vaccination (pre-vaccination for 2013 vaccination) |
58 ± 1 |
| ’13 PR (Dec. 19, 2013) |
2013 peak-response (vaccination) |
16 days post 2013 vaccination | 252 ± 10 |
| ’14 PR (Dec. 10, 2014) |
2014 peak-response (vaccination) |
14 days post 2014 vaccination | 112 ± 5 |
CDR-H3 antibody clonotypes are identified using the Ig-Seq workflow. Serum IgG antibodies are purified by affinity chromatography using immobilized antigen, digested with trypsin, and analyzed through high-resolution liquid chromatography tandem mass spectrometry (LC-MS/MS). Peptide spectral matches are obtained using a custom database from the sequencing of the donor’s peripheral B cell heavy chain variable (VH) regions, and the identified CDR-H3 peptides establish clonotype presence, abundance and persistence across time points. The resulting dataset of longitudinal CA09 HA-reactive serum antibody repertoire spanning 5 years are tabulated in Table S2. Overall, we identified a total of 210 unique serum antibody clonotypes across the 10 samples.
Serum repertoire analysis demands careful interpretation due to the reliance on a donor-specific peripheral B cell sequence database for identification of an unknown number of non-germline IgG proteins (Boutz et al., 2014). To establish the degree to which the Ig-Seq workflow is able to comprehensively capture the serological repertoire from serum samples collected over multiple years, we developed a database-free method to assess whether the Ig-Seq results describe general dynamics of the anti-CA09-HA serum repertoire. This machine learning framework detects CDR-H3 peptide spectra through recognition of signature MS/MS peaks from the conserved CDR-H3 peptide C-terminus (Fig. S1A). These include both spectra identified through the database search and those that Ig-Seq did not identify. Grouping spectra that likely derive from the same precursor peptide then enables enumeration and quantitation of putative CDR-H3 peptides (Fig. S1B). Peptides which cannot be assigned a full sequence (because they cannot be matched to the database) do not contribute to our subsequent description of clonotype-level repertoire dynamics. On average, 71.6 ± 7.2% (mean ± s.d.; ranging from 58.9% to 80.5%) of 224 ± 108 (mean ± s.d.) putative CDR-H3 peptides were identified with high-confidence across all the samples; for the 20 most abundant in each sample, identification rates increased to 86.5 ± 4.7% (mean ± s.d.) (Table S1). This analysis reveals that Ig-Seq provides consistent and high coverage of the CDR-H3 peptide repertoire, particularly for the abundantly present peptides.
For each time point, the serological repertoire is plotted as a histogram in which each bar corresponds to a unique antibody clonotype, and the height of the bar indicates the percentage, or relative abundance, of that clonotype in the repertoire (Fig. 1C and Fig. S2A). Across all samples, the serum antibody repertoire to CA09 HA was oligoclonal (Lee et al., 2016), with ~50 antibody clonotypes (ranging from 19 to 115 antibody clonotypes) observed at each time point, and highly polarized with the top 10 clonotypes accounting for, on average, 73.1 ± 10.7% (mean ± s.d.) of all the detected serum CA09 HA-reactive circulating IgG (Fig. S2B). 9 anti-CA09 HA antibody clonotypes were observed in the serum every year (Fig. 1C); remarkably, one of the 9 antibody clonotypes, Clonotype-633, comprised on average 18.6 ± 12.3% (mean ± s.d.) of the identified serological repertoire to CA09 HA across all the samples (Table S3). There were 15 additional antibody clonotypes that were detected in the serum 4 out of 5 years. These included two antibody clonotypes that were not observed in the post-infection sample from 2009 (i.e. first sample analyzed) but emerged only after the first vaccination with CA09 (i.e. ’10-2 vaccination), as well as 13 antibody clonotypes that were not detected at an intermediate time point, possibly because they fell below the limit of detection by LC-MS/MS. We defined the set of 24 antibody clonotypes (9 seen every year and additional 15 seen on 4 of 5 years) as ‘persistent’. Strikingly, the persistent antibody clonotypes accounted for, on average, 72.6 ± 10.0% (mean ± s.d.) of the identified anti-CA09 HA serum antibodies (Fig. 1D). We further illustrate the overall molecular-level landscape of the anti-CA09 HA serum antibody repertoire (Fig. 1E), which quantitatively demonstrates how the longitudinal dominance of the persistent antibody clonotypes in the serum leads to a largely static repertoire to CA09 HA over multiple years and repeated immunizations.
Biochemical and functional characterizations of the representative serum mAbs
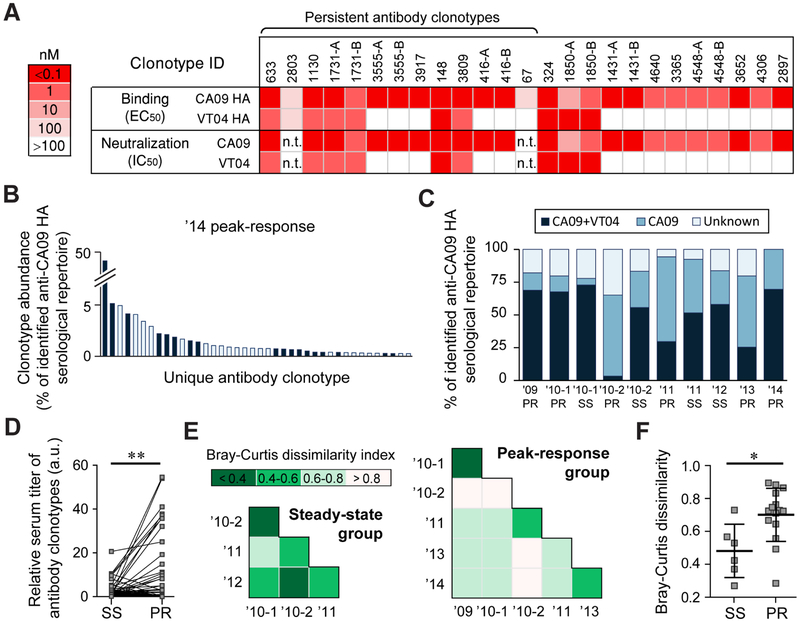
A panel of CA09 HA-reactive mAbs (representative mAbs from 19 clonotypes; for 6 clonotypes, two somatic variants were expressed and characterized for a total of 25 mAbs) detected in serum for which full-length heavy and light chain sequences were available from either single B cell or from high-throughput sequencing data were recombinantly expressed and studied in detail (Table S4). As expected, all 25 mAbs from the 19 clonotypes bound to CA09 HA with EC50 <100 nM, and 23 of 25 neutralized CA09 pseudovirus in microneutralization assays (Fig. 2A). 10 of 25 serum antibodies bound the highly divergent H5N1 A/Vietnam/1203/2004 (VT04). The HA head domain of VT04 has only 67.2% amino acid similarity to that of the CA09 HA; in contrast, the stem domains of these two HA proteins share 90.2% similarity (Fig. S3A). Out of the 10 serum antibodies that were cross-reactive for the CA09 and VT04 HAs, 9 also showed potent neutralization activity to VT04 pseudovirus with IC50 values in the range of 0.007-0.94 nM (0.0005-0.14 μg/ml). In light of earlier reports that all antibodies that neutralized both CA09 and VT04 viruses target HA stem (Corti et al., 2017), it is likely that the 9 serum antibodies above also bind to the stem region. For the mAbs without reactivity to VT04, we tested binding toward a panel of H1 hemagglutinins from strains that circulated from 1934 to 2007 (Fig. S3B). Only 1 of 11 mAbs showed detectable binding to other H1 HAs, suggesting the majority of the CA09-head-binding mAbs likely were elicited from exposure to CA09.
Fig. 2. Serum antibody binding specificities and repertoire similarities across time points.
A. Binding and neutralization activities of a panel of 25 recombinantly expressed serum antibodies. (representing 19 serum antibody clonotypes) (see also Table S4). ELISA was performed using recombinantly expressed HAs from H1N1 A/California/7/2009 (CA09) and H5N1 A/Vietnam/1203/2004 (VT04) influenza virus strains while the neutralization activities were measured using CA09 and VT04 pseudoviruses. In some case, two somatic variants from the same clonotypes were analyzed and denoted as A and B. n.t., not tested. B. The ’14 peak response serum antibody repertoire to CA09 HA with antibody clonotypes also detected in the affinity chromatography eluate with VT04 HA (see also Table S2) shown in dark blue. C. Fraction of the repertoire comprising of CA09+VT04 or CA09-selective antibody clonotypes. Antibodies for which the binding specificity was not determined are shown as ‘unknown’ (see also Fig. S3). D. Relative amount of the 24 persistent antibody clonotypes before and after vaccination. For three vaccinations (’10-1, ’11 and ’13), sera were collected both at before and after immunizations. Each data point pair corresponds to the amount (calculated as % abundance of individual antibody clonotype multiplied by serum titer at that time point) of each 24 persistent antibody clonotypes at steady-state (SS) and at peak-response (PR). Statistical significances were determined using the paired t test. a.u., arbitrary units. E. Pairwise Bray-Curtis dissimilarity analysis among repertoires from the steady-state serum samples (left) or peak-response serum samples (right). Lower indices indicate higher similarity between the samples. F. Pairwise Bray-Curtis dissimilarity indices among steady-state (SS) and peak-response (PR) serum samples; error-bars show s.d. (n = 6 and 15) (see also Fig. S4). Statistical significance by Mann-Whitney U test. For D and F, * p<0.05; ** p<0.01.
To further evaluate the degree to which anti-CA09 HA serum antibodies cross-react with VT04 HA (i.e. they are likely stem-binding), VT04 HA-reactive serum antibodies from the ’14 peak-response sample were isolated by affinity chromatography with immobilized VT04 HA and analyzed using Ig-Seq as above (Table S2). Antibody clonotypes detected in both the CA09 HA and VT04 HA affinity chromatography eluates (i.e. cross-reactive between CA09 HA and VT04 HA (CA09+VT04)), comprised 63.6% of the CA09 HA-reactive serological repertoire in the ’14 peak-response sample (Fig. 2B and Fig. S3C). We note that binding specificities of all the 25 recombinantly expressed mAbs were in perfect agreement with the expected specificities (i.e. CA09-selective or CA09+VT04) inferred by Ig-Seq. For the majority of the antibodies in the anti-CA09 HA serum repertoire, we were able to determine whether they selectively recognize CA09 HA or cross-react with VT04 HA (Fig. 2D).
Steady-state repertoires to CA09 HA are highly static across multiple years
In a recall response, transient antibody secreting cells (plasmablasts) peak in peripheral blood approximately 7-10 days following vaccination, leading to a sharp increase in circulating antibodies that peaks within 3-4 weeks (Bernasconi et al., 2002; Halliley et al., 2010; Wrammert et al., 2008). This plasmablast burst is mostly short-lived, and by day 90 the frequency of antigen-specific plasmablasts is extremely low (Ellebedy et al., 2016). B cells that have terminally differentiated into long-lived plasma cells (LL-PCs) migrate to specialized niches, primarily in the bone marrow, where they produce copious amounts of antibodies which constitute the steady-state serological memory (Bernasconi et al., 2002; Slifka et al., 1998). Our data illustrate that the infection with CA09 appears to have elicited CA09+VT04 antibodies which persisted after vaccination with IIV3 containing Brisbane/59/2007 (BR07) as the H1 component in 2010. Later in 2010, the donor was vaccinated again with IIIV3 containing CA09. 14 days after this second vaccination, the serum repertoire was dominated by antibodies that bound selectively to CA09 HA while at the same time the fraction of CA09+VT04 antibody clonotypes decreased by ~25-fold (from 73% to 3%) (Fig. 2C). However, when taking into consideration the large increase in the serum titer at the peak-response following vaccination (Table 1) arising from CA09 HA-selective antibodies, quantitative analysis reveals that the reduction in the absolute level of CA09+VT04 antibody clonotypes was relatively modest (from 26 a.u. to 10 a.u.) (Fig. S3C). This vaccination with CA09 appeared to have boosted the CA09+VT04 antibody clonotypes with relatively slower kinetics as evident by the increased amount of these antibodies at steady-state, 340 days post vaccination (i.e. ’10-2 steady-state; from 10 a.u. to 65 a.u.).
We determined the change in abundance of the persistent antibody clonotypes at the peak-response (i.e. at day 14-21) following vaccination. As expected, vaccination boosted the level of the persistent antibody clonotypes in serum (Fig. 2D). The magnitude of this boosting effect appeared to be stochastic for individual clonotypes and fluctuated from year to year in a manner that did not correlate with the level of that particular clonotype prior to vaccination; in contrast, the steady-state levels of persistent antibodies remained relatively constant over multiple years (Fig. S4).
Comparison of the composition of the serological repertoire at different time points using the Bray-Curtis dissimilarity index (Bray and Curtis, 1957) (Fig. 2E) revealed a higher degree of similarity (i.e. lower Bray-Curtis dissimilarity indices) for the antibodies in the steady-state serum repertoire compared to the peak-response group (mean Bray-Curtis dissimilarity index = 0.48 ± 0.16 s.d. for the pairwise comparisons of steady-state repertoires at different time points vs. 0.70 ± 0.16 s.d. for the peak-response repertoires) (Fig. 2F). For the peak-response group, a greater degree of serum repertoire similarity was observed only among the peak-responses in adjacent years (i.e., between ’09 & ’10-1; ’10-2 & ’11; and ’13 & ’14 samples). This was not the case for the ’10-1 and ’10-2 peak-response serum repertoires because the donor was vaccinated with IIV3 containing BR07 in ’10-1 and then with CA09 in ’10-2.
Comparisons between the antibody clonotypes with different temporal dynamics in serum
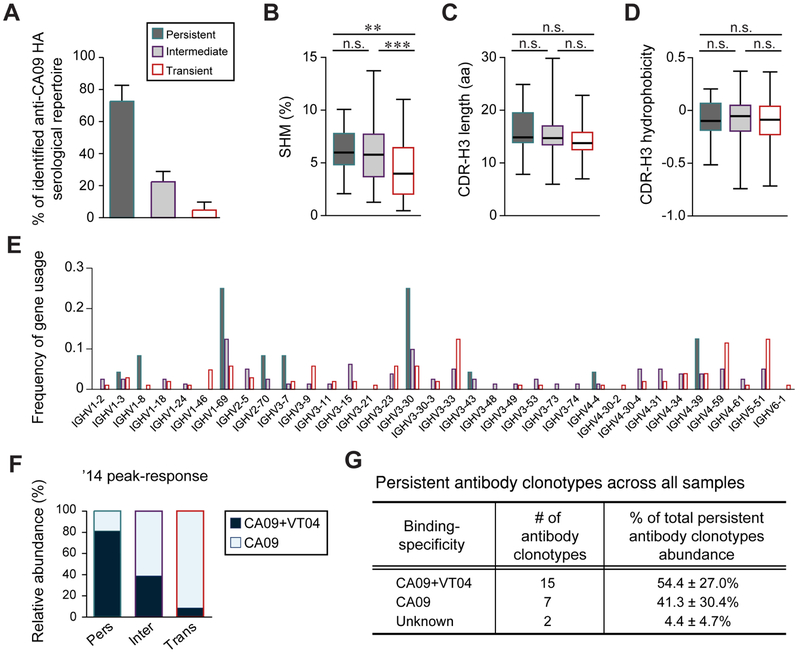
As defined above, persistent antibody clonotypes were those detected at least 4 of 5 years in serum (Fig. 1D). We additionally defined antibody clonotypes that were detected in serum only at one time point as ‘transient’ and the remaining antibody clonotypes that were observed in at least two time points as ‘intermediate’. We note that the intermediate group contained Clonotype-324, which includes FI6, an antibody with remarkable neutralization potency currently in clinical evaluation as an influenza therapeutic. Clonotype-324 was first detected in serum following the ’10-1 vaccination and substantially increased in abundance following the ’11 vaccination before decaying away to below the limit of detection (5 ng/ml) in ’13 (Table S3). Overall, 81 antibody clonotypes were assigned to the intermediate group, accounting for 22.7 ± 6.4% (mean ± s.d.) across all time points, while 105 antibody clonotypes belonged to the transient group accounting on average for a total of only 4.7 ± 5.0% (mean ± s.d.) of the serological repertoire (Fig. 3A).
Fig. 3. Characterization of persistent, intermediate and transient antibody clonotypes (see also Table S2).
A. Average fraction of persistent, intermediate and transient antibody clonotypes as fraction of the serological repertoire. B-D. IGHV somatic hypermutation mutation (SHM) rates (B), CDR-H3 length (C), and CDR-H3 hydrophobicity (Eisenberg index) (D) are shown where boxes extend from the 25th to 75th percentiles with the whiskers indicating min to max. Statistical significance was determined using the Mann-Whitney U test (** p<0.01; *** p<0.001). E. Frequency of gene usage. F. Relative abundance of CA09+VT04 and CA09-selective antibody clonotypes in the ’14 peak-response sample for each temporal dynamic group. G. Relative abundance of CA09+VT04 and CA09-selective persistent antibody clonotypes. Unknown, binding specificity not determined.
Somatic hypermutation mutation (SHM) rates of the immunoglobulin heavy chain variable (IGHV) gene in the persistent, 6.3 ± 2.1% (mean ± s.d.; p-value: 0.0014), and intermediate, 5.7 ± 2.6% (mean ± s.d.; p-value: 0.0008), groups were significantly higher than those in the transient group, 4.4 ± 2.7% (mean ± s.d.), suggesting that antibodies detected in circulation for over one year were derived from B cells that had experienced a greater degree of affinity maturation (Fig. 3B). There were no significant differences in CDR-H3 length or hydrophobicity between the groups (Fig. 3C and D), but IGHV1-69 and IGHV3-30 were more frequently used among persistent antibodies; previous studies have shown that stem-binding heterosubtypic antibodies frequently use IGHV1-69 or IGHV3-30, and in fact IGHV1-69 with polymorphic phenylalanine at position 54 are predisposed to stem-binding (Dreyfus et al., 2013; Pappas et al., 2014; Sui et al., 2009). On the other hand, IGHV3-33, IGHV4-59 and IGHV5-51 were more prevalent in the transiently antibody group (Fig. 3E).
The majority of persistent antibody clonotypes were CA09+VT04 while transient antibody clonotypes were predominantly specific to CA09 HA (Fig. 3F). Averaged across all the time points, 54.4 ± 27.0% (mean ± s.d.) of persistent antibodies were CA09+VT04 and, as discussed above are highly likely to bind stem while 41.3 ± 30.4% (mean ± s.d.) bound selectively to CA09 HA (Fig. 3G).
Discussion
Several studies have reported that infection with influenza virus induces long-lived protective serological immunity more effectively than vaccination with the seasonal influenza vaccine (He et al., 2015; Moody et al., 2011). Infection with CA09 strain, in particular, has been shown to be more likely to induce stem-binding antibodies cross-reacting to divergent strains (Nachbagauer et al., 2017; Wrammert et al., 2011). This is likely due to the highly divergent sequence of the head region of CA09 HA relative to recent historical seasonal H1N1 strains (Garten et al., 2009), which might have led to the preferential elicitation and/or boosting of antibodies targeting the conserved epitopes in the stem domain. Here we analyzed in molecular detail the composition of the serological antibody repertoire to H1 in a donor that was infected with CA09 and then vaccinated, first with a different H1 and subsequently (i.e. from the ’10-2 vaccination onwards) with CA09 for 5 consecutive years.
Numerous bulk serology studies have highlighted the significance of the phenomenon of ‘serological imprinting’ whereby an early influenza exposure event greatly impacts and curtails ELISA and HAI titers to related viruses (Fonville et al., 2014; Francis, 1960; Gostic et al., 2016; Kucharski et al., 2015). Through the combination of longitudinal sampling and molecular serology using Ig-Seq, we now show that serological imprinting is associated with the presence of a small set (in this case only 24) antibody clonotypes that persist in circulation for years. The impact of persistent antibodies on the serum response is underscored by the finding that the top most abundant persistent antibody clonotype (Clonotype-633) alone contributed on average about 19% of the serum response across 5 years and at least in one instance (’14 peak-response) accounted for nearly 50% of the identified anti-CA09 HA repertoire (Fig. 1C and Table S3).
We find that the peak response 2-3 weeks following boost vaccination with CA09 (i.e. after the ’10-2 immunization) is accompanied by the appearance of a wave of transient CA09 HA-selective antibodies that in turn expand the clonotypic diversity of the repertoire (Fig. S3C and Fig. 2C). While this burst of CA09 HA-selective antibodies was observed after multiple immunizations over several years, in each instance transient antibodies decayed substantially over the ensuing 6-9 months. Even during the peak-response when a sizable portion of the antibodies in serum are produced by recently differentiated, circulating antigen-specific plasmablasts (in addition to the serological memory antibodies that continue to be produced by LL-PCs), >50% of the serum titer was contributed by persistent antibodies (Fig. 1C). Once transient antibodies from the plasmablast burst had been cleared from circulation, the ensuing steady-state serological repertoire was not significantly impacted by each preceding boost vaccination and remained relatively unchanged from year to year (Fig. 2F).
The long-term protection against influenza depends heavily on the potency and breadth of the persistent antibodies. We established that Infection with CA09 infection triggers the emergence of high levels of CA09+VT04 antibodies which accounted for the majority of the serum titer to CA09 HA within weeks after infection. Once established, the CA09+VT04 antibodies were boosted to various degrees following vaccination with either a different strain (in which case the absolute titer of CA09+VT04 increased with slower kinetics) or with CA09. The biochemical and functional characterization of the repertoire showed that a substantial fraction of the persistent antibody clonotypes bound and neutralized both CA09 and VT04 strains (Fig. 2A), suggesting that these clonotypes recognize the conserved stem region of HA. It is worth noting that in the 10-2 peak-response repertoire, the majority of the elicited antibodies were CA09-specific, a finding that is in excellent agreement with a previous study (Andrews et al., 2015) which reported that the first exposure to CA09 results in a stem-focused peak plasmablast repertoire whereas the second exposure results in a HA head-focused response. Our data reveal that, once elicited cross-reactive stem-binding antibodies with significant breadth and potency can persist in serum for many years. This observation supports the feasibility of designing next-generation, ‘universal’ influenza vaccines capable of eliciting HA stem-reacting, broad, heterosubtypic immunity that lasts for many years and circumvents the need for seasonal vaccination (Angeletti and Yewdell, 2017; Impagliazzo et al., 2015; Krammer and Palese, 2015; Yassine et al., 2015).
Persistent antibodies were much more likely to be derived from IGHV1-69 and IGHV3-30, consistent with earlier observations that broad breadth stem-binding antibodies are predominantly derived from these two IGHV germline genes. Antibody clonotypes that were detectable in serum for >1 year (Fig. 3A) had a higher degree of SHM relative to the transient antibodies (Fig. 3B) implying that these antibodies were derived from cells that had undergone more extensive affinity maturation; on the other hand, CA09-seletive antibodies were likely elicited more recently, and then reinforced after the first vaccine with CA09, since the epitopes in the head were substantially different from previously circulated strains.
Two critical questions for improving vaccine design that remain to be addressed include: (i) how does the static repertoire develop early in life (i.e., through the first several exposures to influenza) and (ii) how can we significantly reshape already-imprinted serological repertoires? The last question is particularly important for the universal vaccine to be effective; identifying various avenues for overcoming potential restrictions imposed by the imprinted antibody repertoire could be key for the design of an effective vaccine that can benefit the broader population regardless of their exposure history and pre-existing immunological memory.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, George Georgiou (gg@che.utexas.edu).
Experimental Model and Subject Details
Human Subject
Peripheral blood mononuclear cells (PBMCs) and sera were obtained from a donor (FI) postinfection, pre- and post-vaccination time points selected for this study. Donor was in good health during the time of sample collection before and after each vaccination. The donor is a female born in 1952. The study protocols were approved by Cantonal Ethical Committee of Cantone Ticino. Informed consent was obtained from the participant.
Method Details
PBMC Isolation
Blood was taken before and after infection or vaccination over a time period of 5 years. 60 ml peripheral blood were taken from individuals and heparinized. PBMCs were extracted from heparinized blood by using a Ficoll-Gradient and frozen in 10% (v/v) DMSO/90% (v/v) FBS. B cells were isolated from cryopreserved PBMCs.
High-throughput Sequencing of VH
VH-only transcripts were amplified from PBMCs from 7 days post-vaccination in 2014. 500 ng of total RNA was used for reverse transcription according to the manufacturer’s instructions using Superscript III Enzyme (Life Technologies) and oligo-dT primer. VH repertoires were PCR-amplified using FastStart Taq DNA polymerase (Sigma-Aldrich) with gene specific primers as previously described (Ippolito et al., 2012). The sequencing library was sequenced using the Illumina MiSeq platform.
VH: VL-paired Sequencing
Paired heavy and light chain sequencing of single B-cells from the donor’s days 14 and 21 post-vaccination in 2014 were carried out as previously described (DeKosky et al., 2015; McDaniel et al., 2016). Briefly, B-cells were isolated as single cells inside emulsion droplets using a custom flow-focusing apparatus. Droplets contained lysis buffer and poly(dT) conjugated magnetic beads to capture heavy and light chain mRNA. Magnetic beads were collected and emulsified to serve as template for emulsion overlap extension RT-PCR, in which VH and VL amplicons were physically linked. The VH:VL amplicons were extracted from the emulsions, amplified with a nested PCR, and sequenced using the Illumina MiSeq platform. VH and VL regions were amplified separately for full-length VH and VL analysis for the recombinant expression of monoclonal antibodies using the Illumina MiSeq platform as previously described (Lavinder et al., 2014; McDaniel et al., 2016).
Preparation of Total IgG from Sera
Each serum sample analyzed in this study was passed through a 1 ml Protein G Plus agarose (Pierce) affinity column in gravity mode. Serum flow-through was collected and passed through the column three times. The column was washed with 15 cv of PBS prior to elution with 5 cv of 100 mM glycine-HCl, pH 2.7. The eluate, containing total IgG from serum, was immediately neutralized with 2 ml of 1 M Tris-HCl, pH 8.0. Purified IgG was digested into F(ab’)2 with 10 μg of IdeS protease (Genovis) per 1 mg of IgG in PBS for 4 hr on inverter at 37°C.
Antigen-enrichment and MS Sample Preparation
CA09 (A/California/07/2009) rHA provided by Dr. Stephen Harrison (Harvard Medical School) or VT04 (A/Vietnam/1203/2004) rHA (NR-10510) obtained through BEI Resources, NIAID, NIH was immobilized on N-hydroxysuccinimide (NHS)–activated agarose resins (Pierce) by overnight rotation at 4°C. The coupled agarose resins were washed with PBS, and unreacted NHS groups were blocked with 1 M ethanolamine, pH 8.3 for 30 min at RT. The beads were further washed with PBS and packed into a 0.5 ml chromatography column (Clontech). The column was equilibrated with 10 cv of PBS.
For each sample, F(ab’)2 was applied to the individual antigen affinity columns in gravity mode. Flow-through was collected and reapplied to the column three times, and the column was washed with 10 cv of PBS and 10 cv of diluted PBS (1:2 in ddH20). Antigen-enriched F(ab’)2 was eluted with 2% (v/v) formic acid in 0.5 ml fractions. 20 μl from each elution fraction, neutralized with NaOH/Tris, and flow-through were assayed by indirect ELISA with the rHA. Depletion of ELISA signal in each flow-through sample was checked using 1:5000 diluted goat anti-human IgG (Fab specific) HRP-conjugated secondary antibodies (Sigma-Aldrich; A0293-1ML). Elution fractions showing an ELISA signal were pooled and concentrated under vacuum to a volume of ~3 μl before neutralizing with NaOH/Tris. The neutralized elution samples were concentrated to 50 μl under vacuum.
For each enrichment, elution and flow-through samples were denatured in 50% (v/v) 2,2,2-trifluoroethanol (TFE), 50 mM ammonium bicarbonate, and 10 mM dithiothreitol (DTT) at 60°C for 1 hr, then alkylated by incubation with 32 mM iodoacetamide (Sigma) for 1 hr at RT in the dark. Alkylation was quenched by the addition of 20 mM DTT. Samples were diluted 10-fold with 50 mM ammonium bicarbonate and digested with trypsin (1:30 trypsin/protein) for 16 hr at 37°C. Formic acid was added to 1% (v/v) to quench the digestion, and the sample volume was reduced to ~100 μl under vacuum. Peptides were then bound to a C18 Hypersep SpinTip (Thermo Scientific), washed three times with 0.1% formic acid, and eluted with 60% acetonitrile, 0.1% formic acid. C18 eluate was dried under vacuum centrifugation and resuspended in 50 μl in 5% acetonitrile, 0.1% formic acid.
LC-MS/MS Analysis
Samples were analyzed by liquid chromatography-tandem mass spectrometry on a Dionex Ultimate 3000 RSLCnano uHPLC system (Thermo Scientific) coupled to an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). Peptides were first loaded onto an Acclaim PepMap RSLC NanoTrap column (Dionex; Thermo Scientific) prior to separation on a 75 μm×15 cm Acclaim PepMap RSLC C18 column (Dionex; Thermo Scientific) using a 5–40% (v/v) acetonitrile gradient over 160 or 240 min at 300 nl/min. Eluting peptides were injected directly into the mass spectrometer using a nano-electrospray source. The LTQ Orbitrap Velos Pro was operated in data-dependent mode with parent ion scans (MS1) collected at 60,000 resolutions. Monoisotopic precursor selection and charge state screening were enabled. Ions with charge ≥+2 were selected for collision-induced dissociation fragmentation spectrum acquisition (MS2) in the ion trap, with a maximum of 20 MS2 scans per MS1. Dynamic exclusion was active with a 45-s exclusion time for ions selected more than twice in a 30-s window. Each sample was run three times to generate technical replicate datasets.
MS/MS Data Analysis
Protein sequence databases were constructed using the VH sequences obtained from the donor. Specifically, HA-specific antibody sequences obtained across 6 years (Corti et al., 2010; Corti et al., 2011; Kallewaard et al., 2016; Pappas et al., 2014; Pinna et al., 2009) were combined with high-throughput sequencing of VH transcripts from 2014 days 7, 14 and 21 post-vaccination B cells with ≥2 reads. These sets of VH sequences were concatenated to a database of background proteins comprising non-donor derived VL sequences (HD1 (Lavinder et al., 2014)), a consensus human protein database (Ensembl 73, longest sequence/gene), and a list of common protein contaminants (MaxQuant). Spectra were searched against the database using SEQUEST (Proteome Discoverer 1.4; Thermo Scientific). Searches considered fully-tryptic peptides only, allowing up to two missed cleavages. A precursor mass tolerance of 5 ppm and fragment mass tolerance of 0.5 Da were used. Modifications of carbamidomethyl cysteine (static) and oxidized methionine (dynamic) were selected. High-confidence peptide-spectrum matches (PSMs) were filtered at a false discovery rate of <1% as calculated by Percolator (q-value <0.01, Proteome Discoverer 1.4; Thermo Scientific).
Iso/Leu sequence variants were collapsed into single peptide groups. For each scan, PSMs were ranked first by posterior error probability (PEP), then q-value, and finally XCorr. Only unambiguous top-ranked PSMs were kept; scans with multiple top-ranked PSMs (equivalent PEP, q-value, and XCorr) were designated ambiguous identifications and removed. The average mass deviation (AMD) for each peptide was calculated as described (Boutz et al., 2014) using data from elution only. Peptides with AMD >1.5 ppm were removed. Additionally, only peptides identified in ≥2 replicate injections for at least one elution sample were kept as high-confidence identifications.
Peptide abundance was calculated from the extracted-ion chromatogram (XIC) peak area, as described (Lavinder et al., 2014), using peak area values generated by the Precursor Ions Area Detector node in Proteome Discoverer. For each peptide, a total XIC area was calculated as the sum of all unique XIC areas of associated precursor ions, and the average XIC area across replicate injections was calculated for each sample. For each antigen dataset, the eluate and flowthrough abundances were compared and peptides with ≥10-fold higher signal in the elution sample were considered to be antigen-specific.
Peptide-to-clonotype Indexing and Mapping
VH sequences were grouped into clonotypes based on single-linkage hierarchical clustering as described (Lavinder et al., 2014). Cluster membership required ≥90% identity across the CDR-H3 amino sequence as measured by edit distance. High-confidence peptides identified by MS/MS analysis were mapped to clonotype clusters, and peptides uniquely mapping to a single clonotype were considered ‘informative’. Abundance of each antibody clonotype was calculated by summing the XIC areas of the informative peptides mapping to ≥4 amino acids of the CDR-H3 region (i.e. CDR-H3 peptides). For each antibody clonotype, the most abundantly detected CDR-H3 peptide (in case of multiple somatic variants being detected) was used as the representative CDR-H3 sequence, and the CDR-H3 length and hydrophobicity (Eisenberg index) were calculated based on the representative CDR-H3 sequence. Somatic hypermutation (SHM) rates for individual clonotypes were calculated by averaging the SHM rates of all the VH sequences within each clonotype that contain the detected CDR-H3 sequences.
Database-free Enumeration of CDR-H3 Peptides
General limitations of protein mass spectrometry preclude identification of all proteins in sufficiently complex samples, and it is no different for serum antibody repertoire analysis. For a protein or clonotype to be confidently identified, it must yield peptides upon digestion that are (i) unique to that protein, (ii) capable of producing mass spectra and (iii) unambiguously identifiable from their associated spectra. To elaborate on the second point, proteolytic generation of extremely short or long peptides and production of peptides with unusual physicochemical properties can prevent their observation. Furthermore, short peptides are more likely to match multiple protein sequences, especially for repertoire proteomics where antibodies have so much sequence conservation. We use trypsin for IgG digestion because tryptic residues are highly conserved immediately preceding the CDR-H3 and following the FR4. This means most antibodies produce CDR-H3-containing peptides theoretically amenable to proteomic characterization. Finally, repertoire characterization suffers from CDR-H3 peptide spectra that are misidentified or that cannot be identified. Unidentified spectra can, however, be classified independently of database searching, though full peptide sequence recovery is typically impossible.
Here, we provide an estimate of the total number of canonical CDR-H3 spectra in the influenza dataset, that is, the spectra of IgG peptides spanning some or all of the CDR-H3 and J gene and terminating in the conserved ‘ASTK’ motif. These CDR-H3+J peptide spectra display a fragmentation ion peak signature which allows their recognition even without identification through database searching. We implemented a machine learning framework for this task, using a random forest (RF) decision tree ensemble for spectrum classification (Fig. S1A), and trained it against a library of spectra collected from 65 mass spectrometry injections spanning this and five additional non-influenza studies. The positive training set included all 32,140 spectra with high-confidence CDR-H3+J PSMs, identified using the same search process as described above. A matching set of 64,280 negative training examples were pulled from the same MS experiments after CDR-H3+J spectra were removed. Half were selected at random. The remaining spectra were selected using a random sampling process, weighted so that the precursor mass distribution of this second set matched that of the CDR-H3+J spectrum pool. Each spectrum in the positive and negative sets was converted into an attribute vector1 prior to RF training. Most attributes in each vector describe normalized peak intensities from the corresponding MS/MS spectrum. Peaks were placed in integer mass bins, and the bins were indexed in the forward direction starting from 0 Da and in reverse starting from the observed precursor mass. Both sets were included as attributes, as were peaks indexed to account for charge 2+ fragment ions. The remaining attributes for each RF training example (spectrum) comprise the precursor mass and charge, and the full set of output variables from msmsEval, a software application that scores spectral quality for each spectrum and assigns a probability that it is identifiable through database searching (Wong et al., 2007).
Through an iterative attribute selection process, the computer automatically identified a small subset of attributes to use for the final spectrum classifier. RF were first trained using the entire set of spectral attributes for each spectrum, and the importance of each attribute was returned as a side effect of the training. Those most useful for spectrum classification were used in a second round of RF training, and attributes were iteratively eliminated through five successive rounds until the 70 most important remained. At the same time, each round of training allowed detection and removal of potential CDR-H3+J spectra from the set of negative-class training examples. Such spectra could be real CDR-H3+J spectra that went unidentified through the database search process, and their removal is important for creating a stronger classifier. A final RF of 1000 decision trees was constructed using the reduced attribute vectors and the reduced set of training examples. Application of this RF to the 1.64×106 MS/MS in the influenza dataset identified which spectra are likely derived from CDR-H3+J peptides. We then mapped these spectra to precursor features for CDR-H3 peptide enumeration and quantitation (Fig. S1B), extending to unidentified putative CDR-H3 peptides the same quantitation methods that Proteome Discoverer v1.4 (PD) applies to identified peptides.
The Precursor Ions Area Detector node in PD performs feature-based peptide quantitation on spectra identified through the main PD search. Features are defined from signal in the MS1 spectra, irrespective of any associated MS/MS or PSMs, and each describes the peak intensity profile, or XIC, of an ion and its isotopic variants over the time it elutes from the LC. The integrated intensity, or feature area, can be linked to any corresponding spectra, and ideally, the total set of features includes one for each precursor peptide and charge state. Through this, PD quantifies the identified peptides and assigns the ‘Area’ value visible in the exported search results.
PD also saves the complete set of features in the sqlite-formatted *.msf file. We wrote a script, available on request, to directly query MSF files and extract features linked to the putative CDR-H3+J spectra. Because each feature is reasonably expected to derive from a single precursor, the related spectra are merged. Each group is treated as evidence of a distinct CDR-H3 precursor and retains the area of the corresponding feature. Finally, CDR-H3 precursors with the same RT and mass, but potentially different precursor charge, are merged across replicate injections. The resulting peptide-level CDR-H3+J enumeration for each sample is summarized in Table S1.
Recombinant Abs Expression and Purification
Selection of antibody sequences for recombinant expression was based on the combination of VH:VL-paired databases and proteomics data. First, we identified antibody clonotypes identified in the proteomics analysis and searched for the same clonotype in the VH:VL-paired database. Full-length heavy and light chain sequences were then determined from the paired sequencing database.
These genes were purchased as gBlocks (Integrated DNA Technologies) and cloned into the pcDNA3.4 vector (Invitrogen). Heavy and light chain plasmids for each monoclonal antibody were transfected into Expi293 cells (Invitrogen) at a 1:3 ratio. After incubating for 5 days at 37°C with 8% CO2, the supernatant containing secreted antibodies was collected by centrifugation at 500×g for 15 min at RT. Supernatant was passed over a column with 0.5 ml Protein A agarose resin (Thermo Scientific) three times to ensure efficient capture. After washing the column with 20 cv of PBS, antibodies were eluted with 3 ml 100 mM glycine-HCl, pH 2.7 and immediately neutralized with 1 ml 1 M Tris-HCl, pH 8.0. Antibodies were buffer-exchanged into PBS utilizing Amicon Ultra-30 centrifugal spin columns (Millipore).
Virus Neutralization Assays
Virus microneutralization assays of the monoclonal antibodies were carried out as previously described (Pappas et al., 2014). H1N1 virus microneutralization assays were performed by preincubating a serial dilution of antibodies with 2000 TCID50/ml of A/H1N1/California/07/2009 virus for 1 hr at 37°C and then used to infect cultured Madin-Darby canine kidney (MDCK) cells, in MEM infection medium (MEM+Glutamax+Eagle Salts, Kanamycin 1%, TPCK-treated Trypsin 2μg/ml) (Gibco). H5N1 pseudovirus microneutralization assays were performed by preincubating a serial dilution of antibodies with H5N1 A/Vietnam/1194/04 pseudovirus produced as previously described (Temperton et al., 2007) for 1 hr at 37°C and then infected Human embryonic kidney 293 (293T) cells in DMEM infection medium (DMEM+Glutamax+Eagle Salts, PenStrep 1%, Hyclone fetal bovine serum albimun 10%) (Gibco) cultured in white Opaque 96-well Microplates Culture Plate (PerkinElmer). For H1N1 virus microneutralization, readout was performed after 3 days of infection-medium culture by assaying the released neuraminidase activity conversion of fluorescent substrate solution (2’-(4-Methylumbelliferyl)-alpha-D-N-aceylneuramainic acid sodium salt hydrate (Sigma)) in reaction buffer. Substrate was added to transferred supernatants in black Opaque 96-well plates (Perkin Elmer), and after 1 hr of incubation at 37°C determined at EX 355nm/EM 460nm with an Envision Fluorometer (PerkinElmer). For H5N1 pseudovirus microneutralization assays, readout was performed after 3 days of infection-medium culture by assaying the luciferase activity measuring catalyzed relative light units (RLU). Cells were lysed with Steadylite solution (Steadylite plus luminescent reagent) and after 1 hr of incubation at 37°C, the cell lysates were determined on a luminometer microplate reader (Veritas, Turner Biosystems). The neutralization titer (50% inhibitory concentration (IC50)) is expressed as the antibody concentration that reduced the fluorescence signal by 50% compared with cell virus only control wells. The IC50 values were calculated by interpolation of neutralization curves fitted with a four-parameter nonlinear regression with a variable slope. All antibodies were tested in quadruplicates.
ELISAs
50% effective concentration (EC50) values based on ELISA were used to determine the binding apparent affinities of the recombinant monoclonal antibodies. CA09 (A/California/07/2009), PR34 (A/Puerto Rico/8/1934) and MA90 (A/Massachusetts/1/1990) rHAs were provided by Dr. Stephen Harrison (Harvard Medical School), VT04 (A/Vietnam/1203/2004) rHA (NR-10510) was obtained through BEI Resources, and AL51 (A/Albany/12/1951; 40464-V08B), USSR77 (A/USSR/92/1977; 40134-V08B), NC99 (A/New Caledonia/20/99; 11683-V08H), SI06 (A/Solomon Islands/3/2006; 11708-V08H), and BR07 (A/Brisbane/59/2007; 11052-V08H) rHAs were purchased from Sino Biological. First, costar 96-well ELISA plates (Corning) were coated overnight at 4°C with 4 μg/ml recombinant HAs and washed and blocked with 2% milk in PBS for 2 hr at RT. After blocking, serially diluted recombinant antibodies or serum samples bound to the plates for 1 hr, followed by 1:5000 diluted goat anti-human IgG Fc HRP-conjugated secondary antibodies (Jackson ImmunoResearch; 109-035-008) for 1 hr. For detection, 50 μl TMB-ultra substrate (Thermo Scientific) was added before quenching with 50 μl 1 M H2SO4. Absorbance was measured at 450 nm using a Tecan M200 plate reader. Data were analyzed and fitted for EC50 using a 4-parameter logistic nonlinear regression model in the GraphPad Prism software. All ELISA assays were performed in triplicate.
Quantification and Statistical Analysis
Increases in the relative amounts of each persistent antibody clonotype in the matched pre- and post-vaccination samples were evaluated using the paired t test (n=67). Mann-Whitney U test was used to analyze differences in the pairwise Bray-Curtis dissimilarity indices among steady-state and peak-response samples (n=6 and n=15 for steady-states and peak-responses, respectively). Differences in SHM rates, CDR-H3 length, and CDR-H3 hydrophobicity were evaluated using the Mann-Whitney U test (n=24, n=81, and n=105 for persistent, intermediate, and transient antibody clonotype groups, respectively). All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA). All the statistical tests performed are described in the figure legends, and correlations were considered significant at a p-value of <0.05 (* p<0.05; ** p<0.01; *** p<0.001).
Data and Software Availability
The proteomics data reported in this paper are tabulated in the Table S2, and the raw proteomic data and high-throughput sequences have been deposited in MassIVE (massive.ucsd.edu) under accession ID MSV000083120, and in ProteomeXchange (http://www.proteomexchange.org) under accession ID PXD011636. Nucleotide sequences of individual monoclonal antibodies characterized in this paper are deposited at GenBank (accession numbers MK353354-MK353401).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG Fc, HRP | Jackson ImmunoResearch | Cat#109-035-008 |
| Goat anti-human IgG Fab, HRP | Sigma-Aldrich | Cat#A0293-1ML |
| Bacterial and Virus Strains | ||
| H1N1 A/California/7/2009 virus | Pappas et al. 2014 | N/A |
| H5N1 A/Vietnam/1194/2004 pseudovirus | Temperton et al. 2007 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IdeS protease | Genovis | Cat#A0-FR1-050 |
| 2,2,2-trifluoroethanol | Acros Organics | Cat#139751000 |
| Iodoacetamide | Sigma-Aldrich | Cat#I1149-5G |
| Trypsin | Thermo Fisher Scientific | Cat#PR-V5280 |
| C18 Hypersep SpinTip | Thermo Fisher Scientific | Cat#60109-412 |
| A/Vietnam/1203/2004 recombinant hemagglutinin | BEI Resources | NR-10510 |
| A/Albany/12/1951 recombinant hemagglutinin | Sino Biological | Cat#40464-V08B |
| A/USSR/92/1977 recombinant hemagglutinin | Sino Biological | Cat#40134-V08B |
| A/New Caledonia/20/99 recombinant hemagglutinin | Sino Biological | Cat#11683-V08H |
| A/Solomon Islands/3/2006 recombinant hemagglutinin | Sino Biological | Cat#11708-V08H |
| A/Brisbane/59/2007 recombinant hemagglutinin | Sino Biological | Cat#11052-V08H |
| SuperScript III polymerase | Thermo Fisher Scientific | Cat#12574018 |
| FastStart polymerase | Sigma-Aldrich | Cat#4738292001 |
| Poly(dT) conjugated magnetic beads | NEB | Cat#S1419S |
| Deposited Data | ||
| Raw and analyzed sequencing and proteomics data | This paper | MassIVE (MSV000083120), ProteomeXchange (PXD011636), and Table S2 |
| Monoclonal antibody nucleotide sequences | This paper | MK353354-MK353401 |
| Software and Algorithms | ||
| Proteome Discoverer 1.4 | Thermo Fisher Scientific | N/A |
Highlights:
Longitudinal profiling of anti-H1N1 serum antibodies (Ab) reveals persisting Ab
The persistent Ab on average account for >70% of the serum responses over 5 years
Most persistent Abs bind and neutralize a highly divergent H5N1 viral strain
Cross-neutralizing anti-influenza Abs can persist in the circulation
Acknowledgments
We are grateful to Dr. Sebastian Schatzle and Dr. Yariv Wine for discussion and advice, and Dr. Stephen Harrison and Dr. Aaron Schmidt (Harvard Medical School) for insightful discussions and providing rHA.
Funding: This work was supported by NIH grant P01 AI089618 (G.G.), DTRA contract HDTRA1-12-C-0105 (G.G.), Clayton Foundation (G.G.), and European Research Council (Grand no. 670955) BROADimmune (A.L.).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Here, the terms ‘attribute’ and ‘attribute vector’ are synonymous with ‘feature’ and ‘feature vector’, the nomenclature commonly used in machine learning literature. A feature (attribute) is an explanatory variable of the input (MS/MS spectra), one of potentially many which describe each observation (spectrum). We use ‘attribute’ here to disambiguate it from our subsequent usage of ‘feature’ in the context of mass spectrometry.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews SF, Kaur K, Pauli NT, Huang M, Huang Y, and Wilson PC (2015). High Preexisting Serological Antibody Levels Correlate with Diversification of the Influenza Vaccine Response. Journal of Virology. 89(6), 3308–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat Immunol. 18(4), 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, and Yewdell JW (2017). Is It Possible to Develop a “Universal” Influenza Virus Vaccine?: Outflanking Antibody Immunodominance on the Road to Universal Influenza Vaccination. Cold Spring Harbor Perspectives in Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, and Lanzavecchia A (2002). Maintenance of Serological Memory by Polyclonal Activation of Human Memory B Cells. Science. 298(5601), 2199. [DOI] [PubMed] [Google Scholar]
- Bodewes R, de Mutsert G, van der Klis FRM, Ventresca M, Wilks S, Smith DJ, Koopmans M, Fouchier RAM, Osterhaus ADME, and Rimmelzwaan GF (2011). Prevalence of Antibodies against Seasonal Influenza A and B Viruses in Children in Netherlands. Clinical and Vaccine Immunology. 18(3), 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz DR, Horton AP, Wine Y, Lavinder JJ, Georgiou G, and Marcotte EM (2014). Proteomic Identification of Monoclonal Antibodies from Serum. Analytical Chemistry. 86(10), 4758–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, and Curtis JT (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecological Monographs. 27(4), 325–349. [Google Scholar]
- Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, and Lanzavecchia A (2017). Tackling influenza with broadly neutralizing antibodies. Current Opinion in Virology. 24, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, and Lanzavecchia A (2013). Broadly Neutralizing Antiviral Antibodies. Annual Review of Immunology. 31(1), 705–742. [DOI] [PubMed] [Google Scholar]
- Corti D, Suguitan AL Jr., Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, et al. (2010). Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. The Journal of Clinical Investigation. 120(5), 1663–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. (2011). A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science. 333(6044), 850–856. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, and Georgiou G (2015). In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 21(1), 86–91. [DOI] [PubMed] [Google Scholar]
- Doherty PC, Turner SJ, Webby RG, and Thomas PG (2006). Influenza and the challenge for immunology. Nat Immunol. 7(5), 449–455. [DOI] [PubMed] [Google Scholar]
- Dreyfus C, Ekiert DC, and Wilson IA (2013). Structure of a Classical Broadly Neutralizing Stem Antibody in Complex with a Pandemic H2 Influenza Virus Hemagglutinin. Journal of Virology. 87(12), 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O/'Neil RE, Faynboym AM, et al. (2012). Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 489(7417), 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy AH, Jackson KJL, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, et al. (2016). Defining antigen-specific plasmablast and memory B cell subsets in blood following viral infection and vaccination of humans. Nature immunology. 17(10), 1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, H. LNM, T. PQ, et al. (2014). Antibody landscapes after influenza virus infection or vaccination. Science. 346(6212), 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TJ (1960). On the Doctrine of Original Antigenic Sin. Proc Am Philos Soc. 104(6), 572–578. [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, et al. (2009). Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science. 325(5937), 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic KM, Ambrose M, Worobey M, and Lloyd-Smith JO (2016). Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 354(6313), 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliley JL, Kyu S, Kobie JJ, Walsh EE, Falsey AR, Randall TD, Treanor J, Feng C, Sanz I, and Lee FE-H (2010). Peak Frequencies of Circulating Human Influenza-specific Antibody Secreting Cells Correlate with Serum Antibody Response after Immunization. Vaccine. 28(20), 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Gregory V, Douglas AR, and Lin YP (2001). The evolution of human influenza viruses. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 356(1416), 1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X-S, Holmes TH, Sanyal M, Albrecht RA, García-Sastre A, Dekker CL, Davis MM, and Greenberg HB (2015). Distinct Patterns of B-Cell Activation and Priming by Natural Influenza Virus Infection Versus Inactivated Influenza Vaccination. The Journal of Infectious Diseases. 211(7), 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. (2015). A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 349(6254), 1301–1306. [DOI] [PubMed] [Google Scholar]
- Ippolito GC, Hoi KH, Reddy ST, Carroll SM, Ge X, Rogosch T, Zemlin M, Shultz LD, Ellington AD, VanDenBerg CL, et al. (2012). Antibody Repertoires in Humanized NOD-scid-IL2Rγnull Mice and Human B Cells Reveals Human-Like Diversification and Tolerance Checkpoints in the Mouse. PLoS ONE. 7(4), e35497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Wheatley AK, Thomas PV, Chuang G-Y, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. (2016). Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell. 166(3), 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallewaard Nicole L., Corti D, Collins Patrick J., Neu U, McAuliffe Josephine M., Benjamin E, Wachter-Rosati L, Palmer-Hill Frances J., Yuan Andy Q., Walker Philip A., et al. (2016). Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell. 166(3), 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Liepkalns J, Reber AJ, Lu X, Music N, Jacob J, and Sambhara S (2016). Prior infection with influenza virus but not vaccination leaves a long-term immunological imprint that intensifies the protective efficacy of antigenically drifted vaccine strains. Vaccine. 34(4), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, and Palese P (2015). Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 14(3), 167–182. [DOI] [PubMed] [Google Scholar]
- Kucharski AJ, Lessler J, Read JM, Zhu H, Jiang CQ, Guan Y, Cummings DAT, and Riley S (2015). Estimating the Life Course of Influenza A(H3N2) Antibody Responses from Cross-Sectional Data. PLOS Biology. 13(3), e1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert LC, and Fauci AS (2010). Influenza Vaccines for the Future. New England Journal of Medicine. 363(21), 2036–2044. [DOI] [PubMed] [Google Scholar]
- Lavinder JJ, Horton AP, Georgiou G, and Ippolito GC (2015). Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Current Opinion in Chemical Biology. 24, 112–120. [DOI] [PubMed] [Google Scholar]
- Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, et al. (2014). Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proceedings of the National Academy of Sciences. 111(6), 2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee C-H, Lavinder JJ, et al. (2016). Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. 22(12), 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, and Wilson IA (2014). Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-M, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng N-Y, Lee J-H, Huang M, Qu X, Edupuganti S, et al. (2012). Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proceedings of the National Academy of Sciences. 109(23), 9047–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman SL, and Hensley SE (2016). Antibodies with ‘Original Antigenic Sin’ Properties Are Valuable Components of Secondary Immune Responses to Influenza Viruses. PLOS Pathogens. 12(8), e1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018). Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity. 48(1), 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel JR, DeKosky BJ, Tanno H, Ellington AD, and Georgiou G (2016). Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat. Protocols. 11(3), 429–442. [DOI] [PubMed] [Google Scholar]
- Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, et al. (2011). H3N2 Influenza Infection Elicits More Cross-Reactive and Less Clonally Expanded Anti-Hemagglutinin Antibodies Than Influenza Vaccination. PLoS ONE. 6(10), e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachbagauer R, Choi A, Hirsh A, Margine I, Iida S, Barrera A, Ferres M, Albrecht RA, García-Sastre A, Bouvier NM, et al. (2017). Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nature immunology. 18, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. (2014). Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 516(7531), 418–422. [DOI] [PubMed] [Google Scholar]
- Pinna D, Corti D, Jarrossay D, Sallusto F, and Lanzavecchia A (2009). Clonal dissection of the human memory B-cell repertoire following infection and vaccination. European Journal of Immunology. 39(5), 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao H-X, Moody MA, Haynes BF, and Harrison SC (2015). Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell reports. 13(12), 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt Aaron G., Therkelsen Matthew D., Stewart S, Kepler Thomas B., Liao H-X, Moody MA, Haynes Barton F., and Harrison Stephen C. (2015). Viral Receptor-Binding Site Antibodies with Diverse Germline Origins. Cell. 161(5), 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, and Wiley DC (2000). Receptor Binding and Membrane Fusion in Virus Entry: The Influenza Hemagglutinin. Annual Review of Biochemistry. 69(1), 531–569. [DOI] [PubMed] [Google Scholar]
- Slifka MK, Antia R, Whitmire JK, and Ahmed R (1998). Humoral Immunity Due to Long-Lived Plasma Cells. Immunity. 8(3), 363–372. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L.-m., Santelli E, Stec B, Cadwell G, Ali M, et al. (2009). Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 16(3), 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperton NJ, Hoschler K, Major D, Nicolson C, Manvell R, Hien VM, Ha D, de Jong M, Zambon M, Takeuchi Y, et al. (2007). A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza and other respiratory viruses, 1(3), 105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-J, Boyington JC, McTamney PM, Kong W-P, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang Z-Y, et al. (2010). Induction of Broadly Neutralizing H1N1 Influenza Antibodies by Vaccination. Science. 329(5995), 1060–1064. [DOI] [PubMed] [Google Scholar]
- Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences. 108(34), 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LD, Ofek G, Schätzle S, McDaniel JR, Lu X, Nicely NI, Wu L, Lougheed CS, Bradley T, Louder MK, et al. (2017). Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Science Immunology. 2(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, et al. (2013). Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proceedings of the National Academy of Sciences. 110(8), 2993–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JWH, Sullivan MJ, Cartwright HM, and Cagney G (2007). msmsEval: tandem mass spectral quality assignment for high-throughput proteomics. BMC Bioinformatics. 8(1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of Experimental Medicine. 208(1), 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, et al. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 453(7195), 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, and Webster RG (2001). Fields Virology (Lippincott, Williams & Wilkins). [Google Scholar]
- Wu NC, and Wilson IA (2017). A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. Journal of Molecular Biology. 429(17), 2694–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei C-J, Kanekiyo M, Kong W-P, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 21(9), 1065–1070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data reported in this paper are tabulated in the Table S2, and the raw proteomic data and high-throughput sequences have been deposited in MassIVE (massive.ucsd.edu) under accession ID MSV000083120, and in ProteomeXchange (http://www.proteomexchange.org) under accession ID PXD011636. Nucleotide sequences of individual monoclonal antibodies characterized in this paper are deposited at GenBank (accession numbers MK353354-MK353401).