Abstract
Machado-Joseph disease, also known as Spinocerebellar ataxia type 3, is a fatal polyglutamine disease with no disease-modifying treatment. The selective serotonin reuptake inhibitor citalopram was shown in nematode and mouse models to be a compelling repurposing candidate for Machado-Joseph disease therapeutics. We sought to confirm the efficacy of citalopram to decrease ATXN3 aggregation in an unrelated mouse model of Machado-Joseph disease. Four-week-old YACMJD84.2 mice and non-transgenic littermates were given citalopram 8 mg/Kg in drinking water or water for 10 weeks. At the end of treatment, brains were collected for biochemical and pathological analyses. Brains of citalopram-treated YACMJD84.2 mice showed an approximate 50% decrease in the percentage of cells containing ATXN3-positive inclusions in the substantia nigra and three examined brainstem nuclei compared to controls. No differences in ATXN3 inclusion load were observed in deep cerebellar nuclei of mice. Citalopram effect on ATXN3 aggregate burden was corroborated by immunoblotting analysis. While lysates from brainstem and cervical spinal cord of citalopram-treated mice showed a decrease in all soluble forms of ATXN3 and a trend toward reduction of insoluble ATXN3, no differences in ATXN3 levels were found between cerebella of citalopram-treated and vehicle-treated mice. Citalopram treatment altered levels of select components of the cellular protein homeostatic machinery that may be expected to enhance the capacity to refold and/or degrade mutant ATXN3. The results here obtained in a second independent mouse model of Machado-Joseph disease further support citalopram as a potential drug to be repurposed for this fatal disorder.
Keywords: spinocerebellar ataxia, polyglutamine, neurodegeneration, therapy, proteinopathy
Introduction
Machado-Joseph disease (MJD) or Spinocerebellar ataxia type 3 (SCA3) is an adult onset polyglutamine (polyQ) disease and the most common autosomal dominant ataxia worldwide [1,2]. This disease is caused by an unstable expansion of a (CAG)n tract in the ATXN3 gene encoding a polyQ repeat near the C-terminus of the protein ataxin-3 (ATXN3) [3]. The length of ATXN3 CAG repeat ranges from 12 to 44 triplets in the general population, and from ~60 to 87 in affected individuals [4,5]. Gait ataxia and diplopia are commonly the first described symptoms [6,7], yet a clinical diagnosis of MJD is usually assigned to individuals with progressive cerebellar ataxia and pyramidal signs associated with a complex clinical presentation that can also include extrapyramidal signs and peripheral amyotrophy [7–9]. This heterogeneous symptomatology mirrors widespread neuropathology in multiple systems mainly involving the cerebellum, brainstem, substantia nigra, thalamus, basal ganglia, and spinal cord [7,10,11].
Development of disease-modifying drugs for MJD patients is an urgent, unmet need as treatments currently offered to patients are merely symptomatic and palliative [12]. Novel strategies and cellular targets for therapy have emerged as understanding of the mechanisms leading to neuronal dysfunction and degeneration in this disease become clearer over time [13]. Approaches thus far have focused on silencing mutant ATXN3 expression [14–19], reducing the abundance of the mutant protein ATXN3[20], decreasing ATXN3 cleavage [21,22], activating autophagy [23–26], and balancing energy buffering of cells [27].
Using unbiased drug screens, our two laboratories independently identified two existing drugs acting in serotonin signaling as candidates to be repurposed for Machado-Joseph disease: the selective serotonin reuptake inhibitor (SSRI) citalopram [28], and the atypical antipsychotic aripiprazole [20]. Here, joining efforts, we sought additional evidence to support increased serotonergic signaling by citalopram treatment as a promising therapeutic route for MJD. We previously showed that citalopram reduced aggregation of mutant ATXN3, restored motor function, and attenuated neuronal loss in CMVMJD135 (Q135) transgenic mice, which express a full-length isoform of ATXN3 containing an expanded polyQ tract with 135 glutamines [28].
In the current study we test the ability of citalopram to decrease ATXN3 aggregation in a second mouse model of disease that more closely represents the human target: the YACMJD84.2 (Q84) transgenic mice, which harbor the full-length human ATXN3 disease gene, including all the regulatory regions, with an expansion size within the human disease range [29].
Materials and Methods
Experimental design
The study was conceived to test the efficacy of citalopram in decreasing human mutant ATXN3 aggregation at the pathological (primary outcome) and molecular (secondary outcome) levels in brains of YACMJD84.2 (Q84) mice (C57Bl/6 background) [29]. Assessment of efficacy of citalopram on motor performance in Q84 mice was not a goal of this study. We performed a 10-week in vivo trial using hemizygous Q84 mice. Mice were assembled randomly in two groups: one received citalopram dissolved in drinking water, and the other received drinking water (vehicle). We powered our study to assess our primary and secondary outcomes. Based on previously documented ATXN3 levels assessed by immunoblotting [20], sample size was calculated assuming a power of 0.99 and P <0.05 (two-tailed) to achieve 50% ATXN3 reduction (n=10). All experiments were performed at the University of Michigan and slides with immunostained brain slices were sent to the University of Minho for assessment of intranuclear ATXN3 aggregation by a rater blinded to treatment.
Q84 mouse genotyping
Genotyping was performed using DNA isolated from tail biopsy at the time of weaning, as previously described [29]. The ATXN3 CAG repeat size in Q84 mice (Supplementary Tables 1 and 2) was determined at Laragen Inc. by gene fragment analysis using primers hATAXN3forFam (5’ACAGCAGCAAAAGCAGCAA) and hATAXN3rev (5’CCAAGTGCTCCTGAACTGGT).
Treatment of hemizygous Q84 mice with citalopram
All animal procedures were approved by the University of Michigan Committee on the Use and Care of Animals (Protocol PRO00006371). Q84 transgenic mice have been maintained in individual ventilated cages under the same animal husbandry since 2007. Q84 mouse colony was backcrossed with C57Bl/6 mice (Jackson’s Laboratories) in 2014. Mice were housed in cages with corncob bedding and enviropaks (Lab Supply) for enrichment with a maximum number of five animals (range 3–5) and maintained in a standard 12-hours light/dark cycle with food and water ad libitum. Cages with hemizygous Q84 mice and accompanying wild type littermates (wt) were randomly selected to receive either bottles containing citalopram dissolved in drinking water or vehicle alone (drinking water). Citalopram hydrobromide (8 mg/kg) (CAS 59729–32-7, Lündbeck, Denmark) was orally administered to a cohort of 10 Q84 (six males and four females) and eight wt mice (three males and five females), and regular drinking water was given to 10 Q84 (six males and four females) and six wt mice (four males and two females) (Supplementary Table 1). After 10 weeks of treatment, mice were anesthetized with ketamine/xylazine, perfused transcardially with cold PBS, and brains were collected. The right hemisphere was post-fixed for 3 days at 4°C in 4% PFA, immersed in 30% sucrose/ PBS, and sectioned on a sledge microtome (SM200R Leica Biosystems). Free-floating 40 μm sagittal sections were collected and stored in cryoprotectant solution at −20°C. The left hemisphere was divided into cervical spinal cord, brainstem, cerebellum and forebrain areas and saved at −80°C.
Immunostaining
Sagittal brain sections were subjected to antigen retrieval and incubated using the Vector MOM immunodetection kit (Vector Laboratories, Burlingame, CA, USA) whenever a mouse primary antibody was used. Sections were incubated with mouse anti-ataxin-3 (1H9) (1:1000; MAB5360 Millipore, Billerica, MA, USA). Immunolabeling was detected by incubation with a biotinylated anti-mouse antibody followed by ABC coupled to horseradish peroxidase (Vector Laboratories, Burlingame, CA, USA) and DAB substrate (Vector Laboratories, Burlingame, CA, USA). Slides with mouse brain slices immunostained for ATXN3 were coded and sent to the University of Minho where they were imaged and assessed for ATXN3 aggregation using a bright-field Olympus BX51 stereological microscope (Olympus). Cells containing dark-brown intranuclear ATXN3-positive inclusions in the pontine, facial, vestibular and deep cerebellar nuclei (DCN), as well as in the substantia nigra compacta of vehicle- or citalopram-treated Q84 animals (n = 6–8 mice for each condition, 1–4 slices per mouse) were counted in the full depth of the slices and normalized for total area using the Visiopharm integrator system software.
Western blotting
Protein extracts from different mouse brain areas were obtained by lysis in RIPA buffer containing protease inhibitors (Complete, Roche Diagnostics), followed by sonication and centrifugation. Supernatants (soluble protein fractions) from cervical spinal cord, brainstem, and cerebellum were collected and total protein concentration was determined using the BCA method (Pierce). Total protein lysates from soluble fractions (100 μg from brainstem, and cerebellum) were resolved on 10% SDS-PAGE gels, and corresponding PVDF membranes were incubated overnight at 4 °C with primary antibodies: mouse anti-HSP70 (1:500; SPA810, Enzo Life Sciences), rabbit anti-HSP90α (1:1000; ab2928, Abcam), mouse anti-HSP90β (1:1000; ADI-SPA842, Enzo Life Sciences), rabbit anti-HSP40 (1:1000; #4868, Cell Signaling), mouse anti-p62/SQSTM1 (1:1000; H00008878-M01, Novus Biologicals), rabbit anti-Beclin1 (1:1000; ab207612, Abcam), rabbit anti-LC3 (1:500; PM036, MBL International Corporation), mouse anti-GAPDH (1:5000; MAB374, Millipore), and rabbit anti-MJD [30] (1:30000). Bound primary antibodies were visualized by incubation with a peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (1:10000; Jackson Immuno Research Laboratories) followed by treatment with ECL-plus reagent (Western Lighting, PerkinElmer) and exposure to autoradiography films. Band intensity was quantified by densitometry using ImageJ.
Filter Trap Assay
Pellets corresponding to insoluble protein fractions were washed twice with RIPA buffer, resuspended in 300 μL of Laemmli buffer 2X, sonicated during 30 sec, and boiled at 100°C for 15 min. After 20 min of centrifugation at 13000 rpm, the supernatant was collected and saved at −80°C. Insoluble protein lysates were quantified and loaded (50 μg for cerebellum and brainstem, and 30 μg spinal cord) in a filter trap assay apparatus containing a 0.20 μm acetate cellulose membrane (Sterlitech) that was then incubated overnight with anti-MJD (1:10000) at 4°C.
Gene expression analysis
Total RNA from cervical spinal cords of vehicle or citalopram-treated Q84 mice was extracted as previously [20]. Total cDNA was generated from 1 μg of total RNA per sample using iScript™ cDNA synthesis kit (Bio-RAD). Levels of human ATXN3, mouse Atxn3 and Gapdh transcripts were assessed by quantitative real-time polymerase chain reaction as previously [20].
Statistical Analysis
Levels of proteins and counts of cells containing ATXN3 intranuclear inclusions were compared using Student’s t-test (comparison of two groups) whenever distributions were normal and homogeneous. In the other cases, comparisons were assessed using the non-parametric Mann-Whitney U test. A P <0.05 was considered statistically significant in all analyses. Data were analyzed using IBM SPSS Statistics 22 software.
Results
Citalopram reduces ATXN3 intranuclear inclusions in brainstem nuclei and substantia nigra but not in deep cerebellar nuclei (DCN) of Q84 transgenic mice
This study builds on the discovery that citalopram decreases ATXN3 intranuclear inclusions in diverse brainstem nuclei of Q135 transgenic mice [28]. Here, we combined efforts to assess the efficacy of citalopram in ameliorating ATXN3 aggregation in brains of a second mouse model of MJD, the hemizygous Q84 mice [29]. While the mouse trial and all the molecular and immunostaining experiments were carried out at the University of Michigan, imaging and counts of cells containing ATXN3 intranuclear inclusions were executed blindly to treatment at the University of Minho. Cages containing four-week-old Q84 mice and wt littermates were randomly assigned to receive either a bottle containing citalopram dissolved in drinking water (estimated concentration of 8 mg/Kg) or a bottle of regular drinking water (vehicle) during 10 weeks (Fig. 1A, Supplementary Table 1). On the last day of treatment, mouse brains were collected for protein and pathological analyses (Fig.1A). While all animals were included in the experimental molecular and pathological assessments, mice number 1, 2, and 17 were excluded from data analyses as their main CAG repeat sizes were found to be approximately 10 repeats lower that the other mice (Supplementary Table 1).
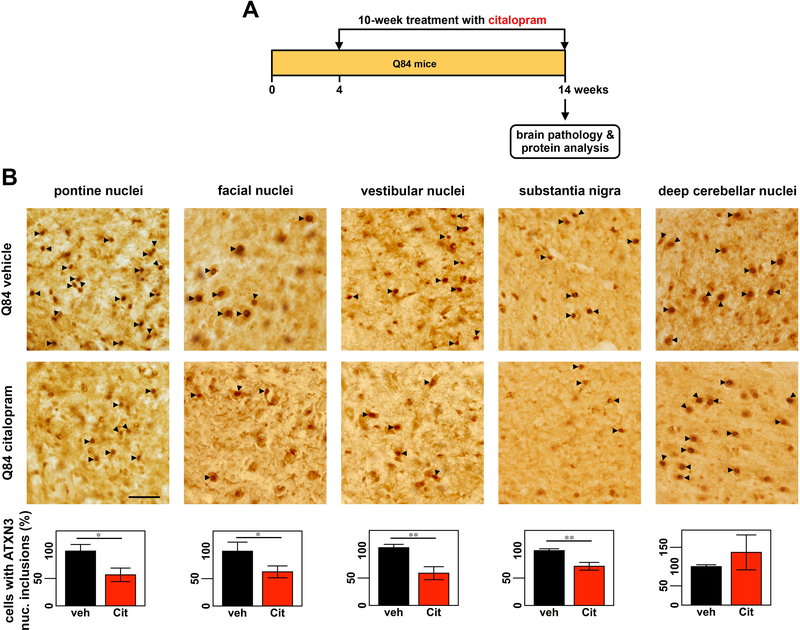
Figure 1: Citalopram decreases ATXN3-positive intranuclear inclusions in the midbrain and hindbrain of Q84 mice.
(A) Four-week-old Q84 mice were orally treated with citalopram ~8 mg/kg or drinking water (vehicle) for 10 weeks, when they were euthanized and brains were collected for pathological assessment and protein analysis. (B) Brain slices were immunostained with anti-ATXN3 antibody (1H9) and cells containing ATXN3-positive intranuclear (nuc.) inclusions (black arrowheads) were counted in pontine nuclei, facial nuclei, vestibular nuclei, substantia nigra, and deep cerebellar nuclei (DCN) (n = 6–8 mice per condition, 1–4 slices per mouse). Graphs representing the quantification of cell count for each region show a robust decrease in number of cells containing ATXN3 intranuclear inclusions in all regions of citalopram-treated Q84 mice compared to vehicle-treated mice, except for the DCN. Scale bar= 50 μm. Bars represent the average percentage of cells with ATXN3 inclusions relative to vehicle-treated Q84 mice (± SEM), *P < 0.05 and **P < 0.01, normalized for total area using Visiopharm software.
We first assessed the efficacy of citalopram in reducing ATXN3 aggregation by counting cells that contained ATXN3-positive intranuclear inclusions in brain regions known to be affected in MJD patients [10,31]. Brain sections from citalopram-treated Q84 mice showed a significant decrease in cells containing ATXN3 intranuclear inclusions in pontine, vestibular and facial nuclei, and substantia nigra compacta to, respectively, 56%, 60%, 62%, and 71% of levels found in controls (Fig. 1B). In contrast, no differences in ATXN3 intranuclear inclusion load were observed in DCN of mice from the two treatment groups (Fig.1B). Thus, the efficacy of citalopram in Q84 mice under these experimental conditions appears to be region-selective, reducing ATXN3 aggregate pathology in various examined nuclei of the midbrain and hindbrain but not in the cerebellar nuclei.
Levels of all ATXN3 protein forms are decreased in the brainstem and spinal cord, but not cerebellum, of Q84 mice treated with citalopram
The pathological observation of a region-specific effect of citalopram to reduce ATXN3 aggregation in Q84 mouse brains was corroborated by protein analysis. Compared to controls protein lysates from brainstem and cervical spinal cord of citalopram-treated Q84 mice showed a decrease of all soluble forms of human and mouse ATXN3 (Fig. 2A,B) and, respectively, a trend toward reduction and significantly reduced insoluble ATXN3 (Fig. 2D,E). Efficacy of citalopram to reduce ATXN3 seems to be downstream of gene transcription level as abundance of human ATXN3 and mouse Atxn3 transcripts was similar in cervical spinal cords of vehicle and citalopram-treated Q84 mice (Supplementary Fig. 1). Levels of soluble and insoluble ATXN3 were unchanged in cerebellar lysates of citalopram-treated mice (Fig. 2C,F). Treatment of wt littermate mice with citalopram did not affect levels of endogenous mouse Atxn3 in any of the above-mentioned regions (Supplementary Fig. 2).
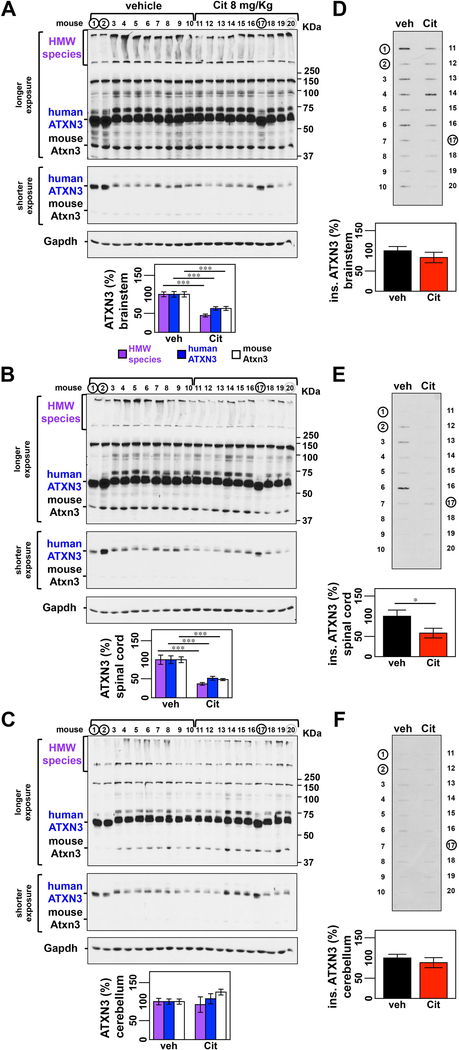
Figure 2: ATXN3 levels are reduced in the brainstem and spinal cord but not the cerebellum of Q84 mice treated with citalopram.
(A, B) Anti-ATXN3 immunoblotting of soluble protein extracts shows decreased HMW ATXN3, mutant human ATXN3, and endogenous mouse ATXN3 abundance in the brainstem of citalopram-treated mice (numbers 11–20) compared to vehicle-treated mice (numbers 1–10) (males 1–6 and 11–16; females 7–10 and 17–20). (B) Soluble protein extracts from spinal cord also show decrease levels of all forms of ATXN3. (C) Soluble proteins from cerebella of the same mice do not reveal any difference between treatment groups. Graphs show quantification of protein bands by densitometry. Bars represent the average percentage of protein species relative to vehicle-treated mice, corrected for Gapdh (± SEM). (D, E) Filter trap assay shows a trend toward reduction of insoluble ATXN3 in brainstem (P = 0.232) and in the spinal cord (P = 0.095) of citalopram-treated mice. (F) No differences in insoluble ATXN3 were found for cerebellar lysates (P = 0.421). Bars in the graph represent the average level of insoluble ATXN3 relative to vehicle-treated mice (± SEM). Statistical significance is indicated as *P < 0.05, **P < 0.01 and ***P < 0.001. Black circles indicate mice, whose results were excluded from statistical analysis because they harbored significantly lower CAG repeat numbers compared with the average (CAG)n of the mouse group. Grey circle represents a mouse whose band density was excluded from statistical analysis for being positioned in the last lane of the gel and consequently of unreliable value due to technical reasons.
Central nervous system (CNS) areas of Q84 mice show differential expression patterns of major components of macroautophagy and chaperone machineries
As observed in a C. elegans model of MJD [28], citalopram-mediated reduction of mutant ATXN3 aggregation/abundance in brains of transgenic mice of this disease is probably mediated by components of serotonergic signaling. Serotonin is known to modulate the protein quality control (PQC) machinery by which cells clear misfolded proteins to maintain homeostasis [32,33]. PQC machineries that help cells clearing misfolded proteins are often dysregulated in proteinopathies contributing to the cellular dysfunction observed in these diseases. While macroautophagy, the proteasome and the chaperone machinery have been shown to be involved in the clearance of mutant ATXN3 [34,23–25,35–41], these cellular machineries have also been reported to be dysregulated in MJD patients and animal models of this disease including the Q84 mice [34,42,20,24,25,43,44]. However, a comprehensive evaluation of temporal/spatial dysregulation of PQC machineries in MJD animal models, including the Q84 mice, is still lacking.
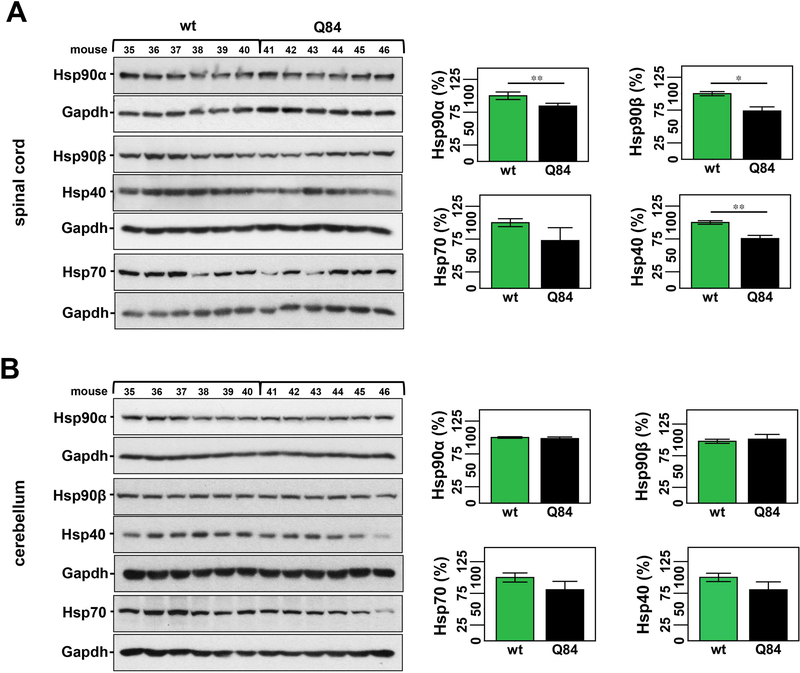
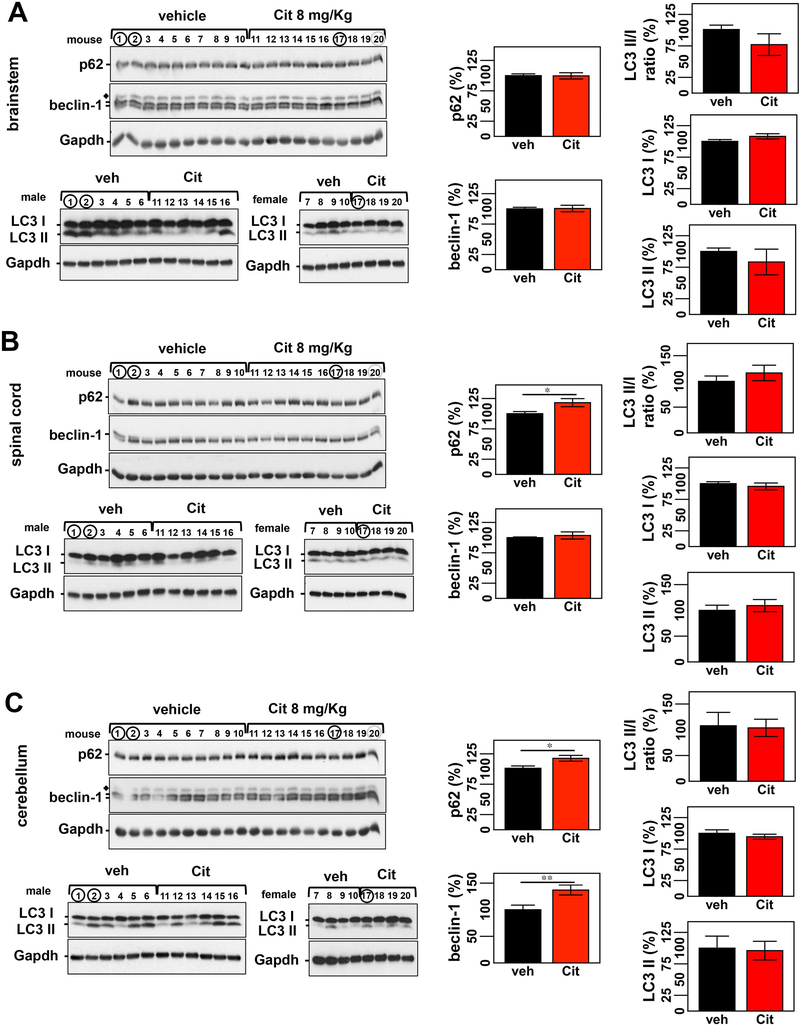
Accordingly, we measured levels of major components of macroautophagy (LC3, p62, and beclin-1) and the chaperone machinery (Hsp40, Hsp70, Hsp90α, and Hsp90β) in the brainstem, cervical spinal cord and cerebellum of Q84 and control mice of 12 weeks of age, which is close to the age of mice included in the citalopram trial at end-stage (14-weeks old). We previously assessed levels of key molecular chaperones in the brainstem of 12-week-old Q84 mice and wt littermates (Supplementary Table 2) and observed increased Hsp90β and decreased Hsp40 levels in Q84 brainstem [20]. Here, we evaluated the expression levels of molecular chaperones in two other areas typically affected in MJD patients - cerebellum and cervical spinal cord - in the same cohort of Q84 and control mice used in our previous study (Supplementary Table 2) [20]. Decreased levels of Hsp40 were found in all studied areas of Q84 mice: 25% reduction in the spinal cord and a trend towards a reduction in the cerebellum of Q84 mice compared to wt littermates (Fig. 3A, B). No other examined chaperone showed different levels in the cerebellum of Q84 mice compared to littermate non-transgenic mice (Fig. 3B). In spinal cord, however, Hsp90α and Hsp90β were decreased in Q84 mice relative to controls (Fig. 3A). Hsp70 levels were unchanged in all three studied regions of Q84 mice (Fig. 3A,B).
Figure 3: 12 week-old Q84 mice show decreased levels of key molecular chaperones in the spinal cord but not cerebellum compared to wt littermates.
Western blot analysis of total soluble protein lysates from spinal cords of Q84 mice show decreased levels of Hsp90α, Hsp90β, and Hsp40 relative to controls (A). No significant changes were observed for any assessed chaperones in cerebellar extracts of Q84 mice (B). (A & B) Left panels show immunoblots of indicated proteins in soluble fractions of spinal cord or cerebellum from Q84 and control littermates (Supplementary Table 2). Right panels show graphs representing the quantification of protein bands by densitometry. Bars represent the average percentage of protein species relative to wt mice, corrected for Gapdh (± SEM). Statistical significance is indicated as *P < 0.05 and **P < 0.01.
Protein lysates from the same group of mice (Supplementary Table 2) were also evaluated for abundance of key players in macroautophagy. Whereas no changes of p62, beclin-1 and different isoforms of LC3 were observed in the spinal cord (Supplementary Fig. 3B), p62 and beclin-1 levels varied significantly in the brainstem and cerebellum of Q84 mice compared to controls: i) increased p62 and beclin-1 levels, 13% and 15%, respectively, in the brainstem (Supplementary Fig. 3A); and ii) reduced p62 (20%) in cerebellum (Supplementary Fig. 3C).
In summary, the three affected areas in 12-week old Q84 mice - brainstem, spinal cord and cerebellum - display different patterns of dysregulation of the molecular chaperone machinery and macroautophagy.
Citalopram-treated Q84 mice show region-specific changes in the abundance of key chaperones and macroautophagy proteins
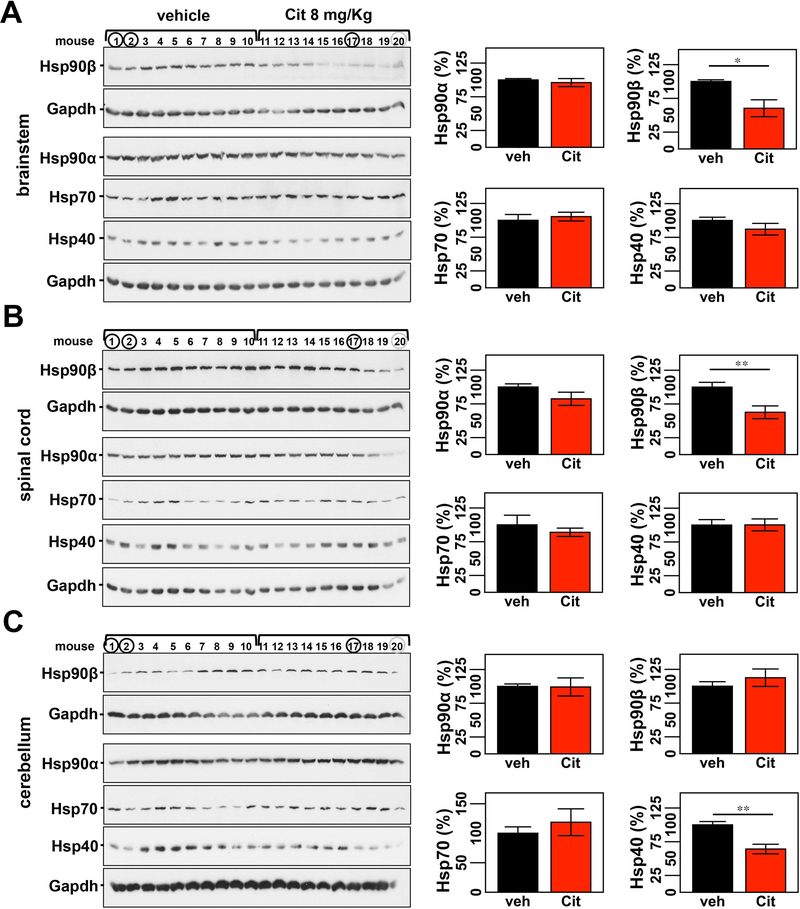
The above results showing altered components of the chaperone machinery and macroautophagy in brains of Q84 mouse led us to investigate whether citalopram treatment affected the abundance of these proteins that could mediate the folding and/or clearance of ATXN3. Citalopram treatment modulated levels of key chaperones and macroautophagy proteins in Q84 mouse brains (Figs. 4 and 5). Among the assessed chaperones: 1) levels of Hsp90β were significantly decreased to 60% and 63%, respectively, of control levels in brainstem and spinal cord (Fig. 4. A,B); and 2) Hsp40 was decreased to 64% of control levels in cerebella of citalopram-treated mice (Fig. 4C). On another hand, Q84 mice treated with citalopram revealed increases of 18% of p62 in both cerebellum and spinal cord, and 37% of beclin-1 in the cerebellum relative to vehicle-treated mice (Fig. 5B,C).
Figure 4: Citalopram reduces levels of Hsp90β in the brainstem and spinal cord and abundance of Hsp40 in the cerebellum of Q84 mice.
Citalopram-treated mice show significantly decreased levels of Hsp90β in brainstem (A) and spinal cord (B) extracts, and reduced amount of Hsp40 in cerebellar lysates (C). (A, B, and C) Left panels show immunoblots of indicated proteins in soluble fractions of brainstem, spinal cord and cerebellum. Right panels display quantification of band intensity, with values normalized to Gapdh. Bars represent the average percentage of protein relative to vehicle-treated mice (± SEM). Statistical significance is indicated as **P < 0.01. Mice 1, 2, and 20 were excluded from statistical analysis.
Figure 5: Q84 mice treated with citalopram reveal slight alterations of macroautophagy components in spinal cord and cerebellum.
Assessment of macroautophagy markers shows increased levels of p62 in the spinal cord (B), and of p62 and beclin-1 in the cerebellum (C) of citalopram-treated mice compared to controls. Citalopram treatment did not affect levels of LC3 in any of the three regions (A-C). (A, B, and C) Left panels show immunoblots of indicated proteins in soluble fractions of brainstem, spinal cord and cerebellum. Right panels display quantification of band intensity, with values normalized to Gapdh. Bars represent the average percentage of protein relative to vehicle-treated mice (± SEM). Statistical significance is indicated as *P < 0.05 and **P < 0.01. Mice 1, 2, and 20 were excluded from statistical analysis. Diamond (◆) represents p62 bands.
In summary, treatment of Q84 mice with citalopram for 10 weeks modulates levels of certain components of protein homeostasis involved in the clearance/folding of misfolded proteins.
Discussion
While no preventive therapy for MJD is yet available, our two laboratories have independently identified available drugs acting in the serotonin-signaling pathway as potential candidates to repurpose for this fatal disease [20,28]. In particular, the SSRI citalopram was shown to reduce the load of ATXN3 inclusions in brains of CMVMJD135 mice and ameliorate the motor phenotype presented by these mice [28].
In this collaborative study, we confirmed that citalopram administered pre-symptomatically is effective in reducing the aggregation and abundance of ATXN3 in brains of a second mouse model of MJD, the YACMJD84.2 (Q84), which expresses the full human disease gene [29]. As in the earlier trial performed in CMVMJD135 mice [28], we carried out a chronic pre-symptomatic treatment of Q84 mice with citalopram, though only for 10 weeks rather than 30 weeks. As in the CMVMJD135 mice [28], citalopram given to Q84 mice before detectable intranuclear ATXN3 aggregation led to a significant decrease in the accumulation of ATXN3-positive intranuclear inclusions in several brainstem nuclei and spinal cord. Citalopram treatment showed, however, different effects on the levels of soluble mutant ATXN3 in the two mouse models. While this SSRI was not effective in reducing levels of soluble mutant ATXN3 in brainstems of CMVMJD135 mice [28], it led to a reduction in all forms of soluble mutant human ATXN3 and mouse Atxn3 in the brainstem and spinal cord of Q84 mice. This difference in citalopram treatment effect between the two mouse models may be explained by differences in the biochemical nature of mutant ATXN3 proteins in brain extracts of Q84 and CMVMJD135 mice. Whereas CMVMJD135 mouse brains express a single human mutant ATXN3 isoform with a polyQ hyperexpansion (~135 glutamines) that is barely detectable in soluble extracts and mostly forms insoluble intranuclear inclusions, Q84 brains express several isoforms of easily detectable soluble and insoluble mutant ATXN3 with a polyQ expansion in the known disease range (70–80 glutamines) [29,35]. The efficacy of citalopram to modulate the folding/degradation of soluble forms of mutant ATXN3 may be greater for the shorter, less aggregation-prone polyQ lengths found in Q84 mice. Indeed citalopram decreased levels of endogenous mouse Atxn3 in brainstem and spinal cord of Q84 mice suggesting that citalopram modulates pathways that clear both human and mouse ATXN3 in these two brain regions. Further evidence that supports the existence of common clearance mechanisms of human and mouse ATXN3 that are impaired in Q84 brains comes from the fact that mouse Atxn3 in Q84 brainstems is increased to about 270% of wt levels (Supplementary Fig. 4) [20]. With the potential effect of citalopram at the transcription level excluded, this SSRI may then improve the function of such protein control mechanisms and promote a better folding of mutant ATXN3 and clearance of both human and mouse ATXN3 in specific brain regions of Q84 mice.
Like the brainstem and spinal cord, the cerebellum shows a high degree of degeneration in MJD patients, particularly in the DCN [6,31]. For successful translation to patients afflicted with MJD, citalopram ideally will ameliorate signs of this disease in the cerebellum as well as the brainstem and spinal cord. In CMVMJD135 mice treated for 30 weeks, citalopram had a positive effect in the cerebellum by restoring cerebellar calbindin staining [28] although it is unknown if it reduced cerebellar ATXN3 aggregation. Further evidence of efficacy of citalopram in the cerebellum of CMVMJD135 mice comes from the fact that mice treated post-symptomatically for 17 weeks not only showed restoration of calbindin levels but also a decrease in ATXN3 intranuclear aggregates in the DCN (Esteves et al., submitted for publication to Molecular Neurobiology). In the present study, however, citalopram treatment for a shorter time period (10 weeks) did not lead to a reduction in the cerebellar load of ATXN3 intranuclear inclusions or ATXN3 soluble or insoluble forms in Q84 mice. A follow-up trial in which Q84 mice are subjected to prolonged treatment with this SSRI beyond 10 weeks would answer whether our failure to see an effect in the cerebellum is due to the limited duration of treatment.
The divergent impact of citalopram treatment on various brain regions may depend on the degree of regional neuropathology and disease severity at a given time point. We observed that the brainstem, cervical spinal cord and cerebellum of 12-week old Q84 mice differ in the state of dysregulation of major components of macroautophagy and chaperone machinery. For example, while reduced amount of Hsp40 was a common signature to all three studied brain regions, changes in the abundance of cytoplasmic forms of Hsp90 in Q84 mice were confined to the brainstem and spinal cord. Whereas Hsp90β was increased in the brainstem [20], both Hsp90α and Hsp90β were decreased in the cervical spinal cord of Q84 mice. Others and we have previously shown that decreased abundance of Hsp90, or its inhibition by 17-DMAG, correlates directly with reduction of mutant ATXN3 levels and improvement of pathological and motor phenotypes in animal models of MJD [20,45,35,46]. Indeed, in this pre-clinical trial of citalopram in Q84 mice we confirmed the existence of a direct correlation of HMW species of ATXN3 to Hsp90β abundance in brainstem and spinal cord (Supplementary Fig. 5A, B). These results suggest that modulation of serotonergic signaling by citalopram regulates specific components of protein homeostasis that could help the folding and/or clearance of toxic ATXN3. In particular this study provides strengthening evidence that Hsp90, mainly the constitutive form Hsp90β, could represent a therapeutic target for MJD.
The efficacy of citalopram in reducing mutant ATXN3 aggregation in brains of CMVMJD135 [28] and Q84 mice (this study) when administered pre-symptomatically appears to be greater than was observed in CMVMJD135 mice treated after the onset of motor symptoms (Esteves et al., submitted for publication to Molecular Neurobiology). Together, these studies suggest that treatment with citalopram or perhaps other SSRIs in MJD might be most effective if initiated at early stages of disease.
This study 1) confirms the efficacy of an approved drug, citalopram, in decreasing ATXN3 aggregation in an independent mouse model of MJD, 2) supports the view that various brain regions, and perhaps specific types of CNS cells, respond differently to mutant ATXN3-mediated toxicity, and 3) underscores the importance of evaluating the efficacy of citalopram, or other future therapeutic agents, in all brain regions affected in the disease.
While the current study does not establish a role for the treatment of Machado-Joseph disease patients with citalopram, our confirmation of this drug’s efficacy in reducing ATXN3 pathology in a second rodent model of disease, coupled with its known safety record in humans, offers strong support for future efforts to perform human clinical trials to establish a role for citalopram, and possibly other SSRIs, in the treatment of this severely impairing disease.
Supplementary Material
Acknowledgements
The authors thank Lündbeck for providing citalopram hydrobromide for the mouse trial. This work was funded by Becky Babcox Research Fund/ pilot research award G015617, University of Michigan to M.C.C. and NINDS/NIH R01NS038712 to H.L.P. The work performed at the University of Minho was funded by the European Regional Development Funds (FEDER), through the Competitiveness Factors Operational Programme (COMPETE), and by National funds, through the Foundation for Science and Technology (FCT), under the scope of the project POCI-01-0145-FEDER-007038. This article was developed under the scope of the project NORTE-01-0145-FEDER-000013, supported by the Northern Portugal Regional Operational Program (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the FEDER. This work was also supported by FCT and COMPETE through the projects [PTDC/SAU-GMG/112617/2009] (to P.M.) and [EXPL/BIM-MEC/0239/2012] (to A.T.C.); by FCT through the project [POCI-01-0145-FEDER-016818 (PTDC/NEU-NMC/3648/2014)] (to P.M.); by National Ataxia Foundation (to P.M. and to A.T.C.); and by Ataxia UK (to P.M.). S.D.S. and A.T.C. were supported by fellowships from FCT, SFRH/BD/78388/2011 and SFRH/BPD/102317/2014, respectively. FCT fellowships are co-financed by POPH, QREN, Governo da República Portuguesa and EU/FSE.
References
- 1.Costa Mdo C, Paulson HL (2012) Toward understanding Machado-Joseph disease. Prog Neurobiol 97 (2):239–257. doi:S0301-0082(11)00212-7 [pii] 10.1016/j.pneurobio.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schols L, Bauer P, Schmidt T, Schulte T, Riess O (2004) Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol 3 (5):291–304 [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, et al. (1994) CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet 8 (3):221–228. doi: 10.1038/ng1194-221 [DOI] [PubMed] [Google Scholar]
- 4.Lima M, Costa MC, Montiel R, Ferro A, Santos C, Silva C, Bettencourt C, Sousa A, Sequeiros J, Coutinho P, Maciel P (2005) Population genetics of wild-type CAG repeats in the Machado-Joseph disease gene in Portugal. Hum Hered 60 (3):156–163 [DOI] [PubMed] [Google Scholar]
- 5.Maciel P, Costa MC, Ferro A, Rousseau M, Santos CS, Gaspar C, Barros J, Rouleau GA, Coutinho P, Sequeiros J (2001) Improvement in the molecular diagnosis of Machado-Joseph disease. Arch Neurol 58 (11):1821–1827. doi:noc10138 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Coutinho P, Andrade C (1978) Autosomal dominant system degeneration in Portuguese families of the Azores Islands. A new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology 28 (7):703–709 [DOI] [PubMed] [Google Scholar]
- 7.Coutinho P, Sequeiros J (1981) [Clinical, genetic and pathological aspects of Machado-Joseph disease]. J Genet Hum 29 (3):203–209 [PubMed] [Google Scholar]
- 8.Paulson HL (2007) Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin Neurol 27 (2):133–142 [DOI] [PubMed] [Google Scholar]
- 9.Sequeiros J, Coutinho P (1993) Epidemiology and clinical aspects of Machado-Joseph disease. Adv Neurol 61:139–153 [PubMed] [Google Scholar]
- 10.Rub U, Schols L, Paulson H, Auburger G, Kermer P, Jen JC, Seidel K, Korf HW, Deller T (2013) Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog Neurobiol 104:38–66. doi: 10.1016/j.pneurobio.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 11.Scherzed W, Brunt ER, Heinsen H, de Vos RA, Seidel K, Burk K, Schols L, Auburger G, Del Turco D, Deller T, Korf HW, den Dunnen WF, Rub U (2012) Pathoanatomy of cerebellar degeneration in spinocerebellar ataxia type 2 (SCA2) and type 3 (SCA3). Cerebellum 11 (3):749–760. doi: 10.1007/s12311-011-0340-8 [DOI] [PubMed] [Google Scholar]
- 12.Bettencourt C, Lima M (2011) Machado-Joseph Disease: from first descriptions to new perspectives. Orphanet J Rare Dis 6:35. doi:1750-1172-6-35 [pii] 10.1186/1750-1172-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Liu H, Fischhaber PL, Tang TS (2015) Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance. Prog Neurobiol 132:34–58. doi: 10.1016/j.pneurobio.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 14.Alves S, Nascimento-Ferreira I, Auregan G, Hassig R, Dufour N, Brouillet E, Pedroso de Lima MC, Hantraye P, Pereira de Almeida L, Deglon N (2008) Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One 3 (10):e3341. doi: 10.1371/journal.pone.0003341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves S, Nascimento-Ferreira I, Dufour N, Hassig R, Auregan G, Nobrega C, Brouillet E, Hantraye P, Pedroso de Lima MC, Deglon N, de Almeida LP (2010) Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum Mol Genet 19 (12):2380–2394. doi:ddq111 [pii] 10.1093/hmg/ddq111 [DOI] [PubMed] [Google Scholar]
- 16.do Carmo Costa M, Luna-Cancalon K, Fischer S, Ashraf NS, Ouyang M, Dharia RM, Martin-Fishman L, Yang Y, Shakkottai VG, Davidson BL, Rodriguez-Lebron E, Paulson HL (2013) Toward RNAi Therapy for the Polyglutamine Disease Machado-Joseph Disease. Mol Ther 21 (10):1898–1908. doi:mt2013144 [pii] 10.1038/mt.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobrega C, Nascimento-Ferreira I, Onofre I, Albuquerque D, Hirai H, Deglon N, de Almeida LP (2013) Silencing mutant ataxin-3 rescues motor deficits and neuropathology in machado-joseph disease transgenic mice. PLoS One 8 (1):e52396. doi: 10.1371/journal.pone.0052396PONE-D-12-17854 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Lebron E, Costa MD, Luna-Cancalon K, Peron TM, Fischer S, Boudreau RL, Davidson BL, Paulson HL (2013) Silencing Mutant ATXN3 Expression Resolves Molecular Phenotypes in SCA3 Transgenic Mice. Mol Ther 21 (10):1909–1918. doi:mt2013152 [pii] 10.1038/mt.2013.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore LR, Rajpal G, Dillingham IT, Qutob M, Blumenstein KG, Gattis D, Hung G, Kordasiewicz HB, Paulson HL, McLoughlin HS (2017) Evaluation of Antisense Oligonucleotides Targeting ATXN3 in SCA3 Mouse Models. Mol Ther Nucleic Acids 7:200–210. doi: 10.1016/j.omtn.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa MD, Ashraf NS, Fischer S, Yang Y, Schapka E, Joshi G, McQuade TJ, Dharia RM, Dulchavsky M, Ouyang M, Cook D, Sun D, Larsen MJ, Gestwicki JE, Todi SV, Ivanova MI, Paulson HL (2016) Unbiased screen identifies aripiprazole as a modulator of abundance of the polyglutamine disease protein, ataxin-3. Brain 139 (November):2891–2908. doi: 10.1093/brain/aww228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoes AT, Goncalves N, Koeppen A, Deglon N, Kugler S, Duarte CB, Pereira de Almeida L (2012) Calpastatin-mediated inhibition of calpains in the mouse brain prevents mutant ataxin 3 proteolysis, nuclear localization and aggregation, relieving Machado-Joseph disease. Brain 135 (Pt 8):2428–2439. doi:aws177 [pii] 10.1093/brain/aws177 [DOI] [PubMed] [Google Scholar]
- 22.Simoes AT, Goncalves N, Nobre RJ, Duarte CB, Pereira de Almeida L (2014) Calpain inhibition reduces ataxin-3 cleavage alleviating neuropathology and motor impairments in mouse models of Machado-Joseph disease. Hum Mol Genet 23 (18):4932–4944. doi: 10.1093/hmg/ddu209 [DOI] [PubMed] [Google Scholar]
- 23.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC (2010) Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain 133 (Pt 1):93–104. doi:awp292 [pii] 10.1093/brain/awp292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nascimento-Ferreira I, Nobrega C, Vasconcelos-Ferreira A, Onofre I, Albuquerque D, Aveleira C, Hirai H, Deglon N, Pereira de Almeida L (2013) Beclin 1 mitigates motor and neuropathological deficits in genetic mouse models of Machado-Joseph disease. Brain 136 (Pt 7):2173–2188. doi: 10.1093/brain/awt144 [DOI] [PubMed] [Google Scholar]
- 25.Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, Auregan G, Onofre I, Alves S, Dufour N, Colomer Gould VF, Koeppen A, Deglon N, Pereira de Almeida L (2011) Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain 134 (Pt 5):1400–1415. doi:awr047 [pii] 10.1093/brain/awr047 [DOI] [PubMed] [Google Scholar]
- 26.Ou Z, Luo M, Niu X, Chen Y, Xie Y, He W, Song B, Xian Y, Fan D, OuYang S, Sun X (2016) Autophagy Promoted the Degradation of Mutant ATXN3 in Neurally Differentiated Spinocerebellar Ataxia-3 Human Induced Pluripotent Stem Cells. Biomed Res Int 2016:6701793. doi: 10.1155/2016/6701793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duarte-Silva S, Neves-Carvalho A, Soares-Cunha C, Silva MJ, Teixeira-Castro A, Vieira R, Silva-Fernandes A, Maciel P (2018) Neuroprotective Effects of Creatine in the CMVMJD135 Mouse Model of Spinocerebellar Ataxia Type 3. Movement Disorders in press. doi: 10.1002/mds.27292 [DOI] [PubMed] [Google Scholar]
- 28.Teixeira-Castro A, Jalles A, Esteves S, Kang S, da Silva Santos L, Silva-Fernandes A, Neto MF, Brielmann RM, Bessa C, Duarte-Silva S, Miranda A, Oliveira S, Neves-Carvalho A, Bessa J, Summavielle T, Silverman RB, Oliveira P, Morimoto RI, Maciel P (2015) Serotonergic signalling suppresses ataxin 3 aggregation and neurotoxicity in animal models of Machado-Joseph disease. Brain 138 (Pt 11):3221–3237. doi: 10.1093/brain/awv262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cemal CK, Carroll CJ, Lawrence L, Lowrie MB, Ruddle P, Al-Mahdawi S, King RH, Pook MA, Huxley C, Chamberlain S (2002) YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet 11 (9):1075–1094 [DOI] [PubMed] [Google Scholar]
- 30.Paulson HL, Das SS, Crino PB, Perez MK, Patel SC, Gotsdiner D, Fischbeck KH, Pittman RN (1997) Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann Neurol 41 (4):453–462. doi: 10.1002/ana.410410408 [DOI] [PubMed] [Google Scholar]
- 31.Seidel K, Siswanto S, Brunt ER, den Dunnen W, Korf HW, Rub U (2012) Brain pathology of spinocerebellar ataxias. Acta Neuropathol 124 (1):1–21. doi: 10.1007/s00401-012-1000-x [DOI] [PubMed] [Google Scholar]
- 32.Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, Liu Y, Dillin A (2016) Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell 166 (6):1553–1563 e1510. doi: 10.1016/j.cell.2016.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatum MC, Ooi FK, Chikka MR, Chauve L, Martinez-Velazquez LA, Steinbusch HW, Morimoto RI, Prahlad V (2015) Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr Biol 25 (2):163–174. doi: 10.1016/j.cub.2014.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, Squitieri F, Hardenberg MC, Imarisio S, Menzies FM, Rubinsztein DC (2017) Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545 (7652):108–111. doi: 10.1038/nature22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva-Fernandes A, Duarte-Silva S, Neves-Carvalho A, Amorim M, Soares-Cunha C, Oliveira P, Thirstrup K, Teixeira-Castro A, Maciel P (2014) Chronic treatment with 17-DMAG improves balance and coordination in a new mouse model of Machado-Joseph disease. Neurotherapeutics 11 (2):433–449. doi: 10.1007/s13311-013-0255-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira PF, Cerca F, Santos SD, Saraiva MJ (2006) Endoplasmic reticulum stress associated with extracellular aggregates. Evidence from transthyretin deposition in familial amyloid polyneuropathy. J Biol Chem 281 (31):21998–22003 [DOI] [PubMed] [Google Scholar]
- 37.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM (1999) Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet 23 (4):425–428. doi: 10.1038/70532 [DOI] [PubMed] [Google Scholar]
- 38.Blount JR, Tsou WL, Ristic G, Burr AA, Ouyang M, Galante H, Scaglione KM, Todi SV (2014) Ubiquitin-binding site 2 of ataxin-3 prevents its proteasomal degradation by interacting with Rad23. Nat Commun 5:4638. doi:ncomms5638 [pii] 10.1038/ncomms5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N (2005) Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem 280 (12):11635–11640. doi:M412042200 [pii] 10.1074/jbc.M412042200 [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI (2004) Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J 23 (3):659–669. doi: 10.1038/sj.emboj.76000817600081 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Jia N, Fei E, Wang Z, Liu C, Zhang T, Fan J, Wu M, Chen L, Nukina N, Zhou J, Wang G (2007) p45, an ATPase subunit of the 19S proteasome, targets the polyglutamine disease protein ataxin-3 to the proteasome. J Neurochem 101 (6):1651–1661. doi:JNC4460 [pii] 10.1111/j.1471-4159.2007.04460.x [DOI] [PubMed] [Google Scholar]
- 42.Chou AH, Yeh TH, Ouyang P, Chen YL, Chen SY, Wang HL (2008) Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol Dis 31 (1):89–101. doi:S0969-9961(08)00067-3 [pii] 10.1016/j.nbd.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 43.Sittler A, Muriel MP, Marinello M, Brice A, den Dunnen W, Alves S (2017) Deregulation of autophagy in postmortem brains of Machado-Joseph disease patients. Neuropathology. doi: 10.1111/neup.12433 [DOI] [PubMed] [Google Scholar]
- 44.Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL (1999) Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet 8 (4):673–682. doi:ddc073 [pii] [DOI] [PubMed] [Google Scholar]
- 45.He WT, Zheng XM, Zhang YH, Gao YG, Song AX, van der Goot FG, Hu HY (2016) Cytoplasmic Ubiquitin-Specific Protease 19 (USP19) Modulates Aggregation of Polyglutamine-Expanded Ataxin-3 and Huntingtin through the HSP90 Chaperone. PLoS One 11 (1):e0147515. doi: 10.1371/journal.pone.0147515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira-Castro A, Ailion M, Jalles A, Brignull HR, Vilaca JL, Dias N, Rodrigues P, Oliveira JF, Neves-Carvalho A, Morimoto RI, Maciel P (2011) Neuron-specific proteotoxicity of mutant ataxin-3 in C. elegans: rescue by the DAF-16 and HSF-1 pathways. Hum Mol Genet 20 (15):2996–3009. doi:ddr203 [pii] 10.1093/hmg/ddr203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.