Abstract
Mechanical stimuli play a critical role in organ development, tissue homeostasis, and disease. Understanding how mechanical signals are processed in multicellular model systems is critical for connecting cellular processes to tissue- and organism-level responses. However, progress in the field that studies these phenomena, mechanobiology, has been limited by lack of appropriate experimental techniques for applying repeatable mechanical stimuli to intact organs and model organisms. Microfluidic platforms, a subgroup of microsystems that use liquid flow for manipulation of objects, are a promising tool for studying mechanobiology of small model organisms due to their size scale and ease of customization.
In this work, we describe design considerations involved in developing a microfluidic device for studying mechanobiology. Then, focusing on worms, fruit flies, and zebrafish, we review current microfluidic platforms for mechanobiology of multicellular model organisms and their tissues and highlight research opportunities in this developing field.
1. INTRODUCTION
This chapter discusses the current state and future opportunities in the field of microfluidics for mechanobiological studies of multicellular model organisms. We define the concept of mechanobiology and describe several model organisms and their advantages for mechanobiological studies. We discuss how microfluidic platforms could be used to manipulate and apply repeatable stimuli, examine design considerations for devices, and review prior work on microfluidics for mechanobiology of model organisms and their tissues. We hope this chapter will be helpful for engineers interested in developing tools for model organisms, and for biologists interested in learning how microfluidic technologies can benefit their research goals.
2. MECHANOBIOLOGY
Research in medicine and biology has largely focused on the biochemical nature of development, homeostasis, and disease, resulting in transformative insights (Broderick, Buchon, & Lemaitre, 2014; Ellis & Horvitz, 1986; Fire et al., 1998; Kok et al., 2015; Nüsslein-Volhard & Wieschaus, 1980; Walther et al., 2015). However, mechanical signaling also plays crucial roles in these processes (Janmey & Miller, 2011; Thompson, 1942; Vining & Mooney, 2017), including tissue patterning in development (Bardet et al., 2013; Hayashi & Carthew, 2004; Heisenberg & Bellaïche, 2013), balancing forces during homeostasis (Brown, Prajapati, McGrouther, Yannas, & Eastwood, 1998; Cahalan et al., 2015; Gomez, McLachlan, & Yap, 2011), and cancer metastasis (Guck et al., 2005; Spill, Reynolds, Kamm, & Zaman, 2016; Wei et al., 2015; Wirtz, Konstantopoulos, & Searson, 2011). Mechanobiology is the study of how cells, tissues, and organisms convert mechanical stimuli into information, process that information, and respond accordingly. The sense of touch is a clear example of the conversion of mechanical stimuli into information. For the purpose of this chapter, we focus on mechanobiology in the context of multicellular model organisms.
One of the enablers of the proliferation of mechanobiology investigations is technology development. For example, the invention of atomic force microscopy (Binnig, Quate, & Gerber, 1986; Marti et al., 1988; Sonnenfeld et al., 1986) created the possibility of mapping the topography as well as mechanical properties of nanoscale biological samples (Bustamante, Erie, & Keller, 1994; Hansma, Elings, Marti, & Bracker, 1988). The capabilities of atomic force microscopy continue to grow, and now include high-speed nanoscale imaging (Ando, Uchihashi, & Kodera, 2013) and single molecule force spectroscopy (Neuman & Nagy, 2008). Advances in microfluidic technology have similarly enabled new experiments, including investigations of mechanobiology.
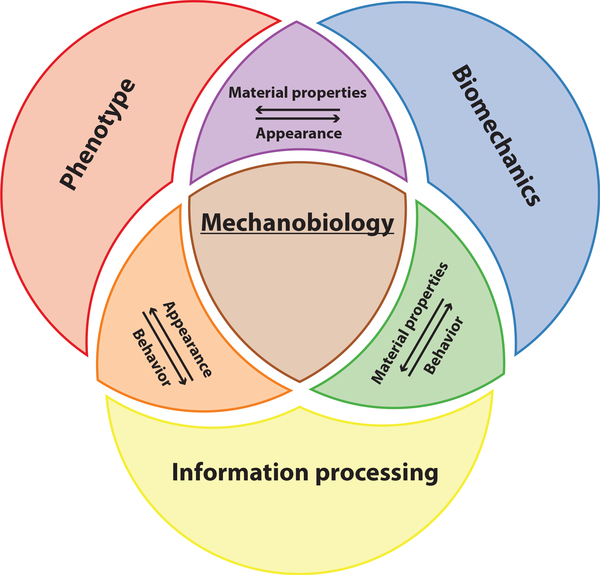
Importantly, the field of mechanobiology is not only focused on biomechanical properties of an organism but also involves understanding information processing and phenotypic change due to a mechanical stimulus. Our definition of mechanobiology thus sits at the intersection of phenotype, biomechanics, and information processing (Fig. 1). While biomechanical properties can be defined as phenotypes, we separate them out here because all mechanical stimuli are transmitted through physical materials. Thus, the biomechanical properties of an organism and its surroundings are inexorably linked to its response to mechanical stimuli. Box 1 describes the basics of how mechanical stimuli are measured.
FIG. 1.
Mechanobiology can be conceptually subdivided into biomechanics, phenotype, and information processing.
BOX 1. A NOTE ON MEASURING FORCE AND DISPLACEMENT.
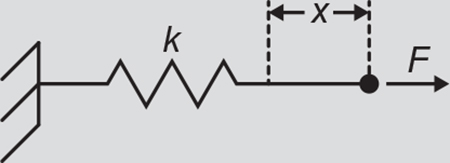
In mechanobiology, mechanical stimuli are often measured as displacement, x, and reported as a measurement of force, F. In the simplest case, force and displacement are related by a spring constant according to Hooke’s law: F = kx, where F is force in Newtons, x is the displacement of the spring in meters, and k is the spring constant in N/m (illustrated above). The spring constant is dependent on both geometry and material properties of the specimen, and mechanical models that take geometry into account can be used to derive the material properties (such as the elastic modulus, E with unitsof N/m2).
However, in many cases, displacement measurements between control and experimental conditions are sufficient for testing hypotheses and can be more accurate than force values. Each calibration measurement required to establish a mechanical model introduces uncertainty into the final force measurement. Moreover, Eastwood et al. (2015) found that the rate of displacement, rather than force, is more closely correlated to the response of touch receptor neurons in worms, and Petzold, Park, Mazzochette, Goodman, and Pruitt (2013) demonstrated that indentation of worms is a more proximal stimulus than force for touch receptor neurons. Together, these studies demonstrate cases where displacement is a more reliable indicator of stimulus strength than force. For more information, see reviews by Schoen, Pruitt, and Vogel (2013), Moeendarbary and Harris (2014), and Paluch et al. (2015).
The field of mechanobiology has become more prominent in recent decades, but it is not entirely new (Wolff, 2011). The study of touch goes back at least to the time of Aristotle (1990) and continues today (Fehrenbacher, 2015; Singh, Kishore, & Kaur, 2014). In modern times, scientists began to explore the effects of mechanical stimuli in contexts other than touch sensation. Physicians in the 19th century observed that bones can remodel their structure to support mechanical loads after a fracture (Wolff, 2010a, 2010b, 2011). More recently, Carter and Hayes (1976) continued this work and renewed the focus on mechanobiology of bone (reviewed by Carter, Beaupré, Giori, and Helms (1998) and Wall et al. (2017)). The field has since expanded to study other tissues and cell types. Pelham and Wang (1997) demonstrated that altering the stiffness of polyacrylamide substrates changes the locomotion and gene expression of cells. Lo, Wang, Dembo, and Wang (2000) followed this result with a study showing that cell migration can be influenced by stiffness gradients (reviewed by Discher, Janmey, and Wang (2005)). Engler, Sen, Sweeney, and Discher (2006) discovered that the stem cell lineages can be altered by the stiffness of the substrate on which they are grown, leading to further investigation of stem cell mechanobiology (reviewed by Lee, Knight, Campbell, and Bader (2011) and Ireland and Simmons (2015)). Paszek et al. (2005) demonstrated that the rigidity of tumors promotes the growth and malignancy of tissue. Thus, mechanobiology is relevant to cancer formation and propagation (reviewed by Huang and Ingber (2005), Jaalouk and Lammerding (2009), and Wirtz et al. (2011)). Aarabi et al. (2007) found that scar formation is initiated by mechanical forces applied to a wound, further underscoring the idea that mechanical signaling can affect clinical outcomes (reviewed by Gurtner, Werner, Barrandon, and Longaker (2008) and Ogawa and Huang (2016)). These milestones are not an exhaustive summary of mechanobiology, as the field encompasses a broad range molecular-, cellular-, tissue-, and organism-level investigation, which have been reviewed elsewhere (Ingber, 2003; Wang & Thampatty, 2006).
3. MULTICELLULAR MODEL ORGANISMS
Model organisms are widely-studied nonhuman systems in biological research, ranging from simple single cells (e.g., Escherichia coli) to animals like mice. Selection of a model organism is dictated by the complexity required to address the research question and the convenience of the experiment. For investigations of responses to mechanical stimuli in the context of differentiated and distinct organs, higher-order multicellular organisms provide an attractive platform. Caenorhabditis elegans (worms) (Frézal & Félix, 2015; Hulme, Shevkoplyas, Apfeld, Fontana, & Whitesides, 2007; Kaletta & Hengartner, 2006), Drosophila melanogaster (fruit flies) (Jennings, 2011; Pandey & Nichols, 2011; Ugur, Chen, & Bellen, 2016), and Danio rerio (zebrafish) (Dahm, Geisler, & Nusslein-Volhard, 2005; Dooley & Zon, 2000; van der Sar, Appelmelk, Vandenbroucke-Grauls, & Bitter, 2004) offer a high level of biological complexity with comparatively low laboratory maintenance. These organisms are cost-efficient, experimentally tractable, and compatible with microscopy-based techniques. Moreover, these multicellular organisms come with well-established toolboxes of genetic techniques, sequenced genomes, and a varying number of genes homologous to humans. Communities of model organism researchers provide resources, accessible online (FlyAtlas, n.d.; FlyBase, n.d.; WormBase, n.d.; WormBook, n.d.; ZFIN, n.d.) that support information exchange and facilitate research.
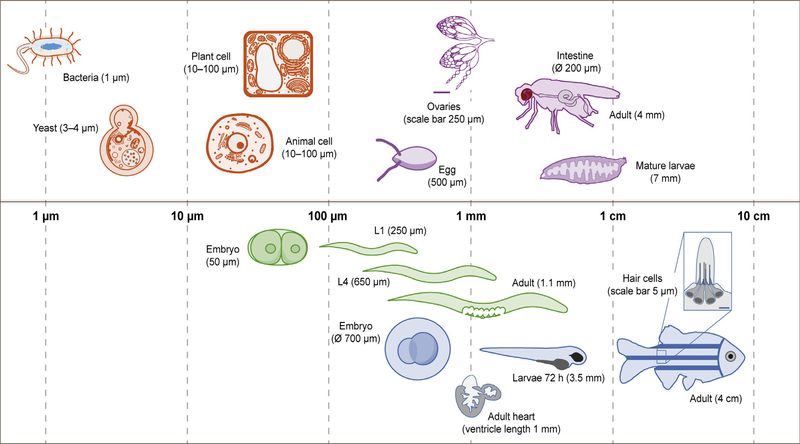
In this chapter, we focus on worms, fruit flies, and zebrafish and describe their individual features in the following sections. These multicellular model organisms can benefit from developments in microfluidic technologies, because they are the appropriate size and compatible with immersion in liquids for part or all of their life stages. Our experience with the design process for a microfluidic device for studying worms can thus be extended to these other model organisms. Fig. 2 compares the size of model organisms and single cells during different stages of their respective life cycles. We exclude mammalian systems due to their comparatively large size.
FIG. 2.
Approximate sizes of biological cells, tissues, and model organisms on a logarithmic scale. Biological cells (orange) are represented as a range because the size is dependent on cell type. Model organisms are organized by color, including nematodes (green), fruit flies (purple), and zebrafish (blue).
3.1. Caenorhabditis elegans
C. elegans is a species of small roundworms that develop from embryos to adults in less than 3 days when cultured at 20°C. It has four larval stages (L1–4) and lays eggs for 2–3 days after achieving adulthood (Corsi, Wightman, & Chalfie, 2015). Nearly all individuals in the species are hermaphrodites, making it easier to maintain homozygous genotypes, and worms can be recovered decades after freezing in liquid nitrogen, which can minimize the genetic variability of a population. Approximately 38% of worm genes are human orthologs, meaning they evolved from the same gene in a common ancestor (Hu et al., 2011).
Worms were the first animal to have a fully sequenced genome, and other genetic techniques, such as genetic mapping (Brenner, 1974), fluorescent protein expression (Chalfie, Tu, Euskirchen, Ward, & Prasher, 1994), and RNA interference (Fire et al., 1998) were pioneered in worms. One downside of using worms for mechanobiology research is their lack of a circulatory system (Corsi et al., 2015). Worms have a variety of mechanosensitive neurons and respond to mechanical stimuli by changing the direction of crawling (Goodman, 2006), making them useful for mechanobiology research.
3.2. Drosophila melanogaster
Similar to worms, fruit flies have a short generation time. The organism develops from embryo to adult in approximately 10 days when it is grown at 25°C. Fruit fly larvae go through three instar stages of increasing size before forming pupae, and eclose from pupae after metamorphosis is complete. Genetic crosses are facilitated because Drosophila are easily bred and a single female can lay 400 eggs (Greenspan, 2004). Approximately 40% of fruit fly genes are human orthologs (Hu et al., 2011).
Fruit flies are widely used for research, including studies in aging (Cannon et al., 2017; Fleming, Reveillaud, & Niedzwiecki, 1992; He & Jasper, 2014) and neurobiology (Feany & Bender, 2000; Nichols, 2006; Watanabe et al., 2017). In the context of mechanobiology, fruit flies are a promising model for investigations of organ architecture formation and function, as the animal is already a well-established model in developmental biology (Dai, Peterson, Kenney, Burrous, & Montell, 2017; Hiraoka et al., 1993; Pandey & Nichols, 2011). Research applications include morphogenesis and organogenesis using follicle cells (epithelium that encapsulates the developing egg), embryos, and imaginal discs (precursors to adult organs) (Dobens & Raftery, 2000; Haynie & Bryant, 1986; Lengyel & Iwaki, 2002). Other tissues of potential interest are the dorsal vessel and midgut. Recently, fruit flies became an invertebrate genetic model for cardiac disease (Cannon et al., 2017; Choma, Izatt, Wessells, Bodmer, & Izatt, 2006; Wolf et al., 2006) using the dorsal vessel that pumps hemolymph, although their open circulatory system does not entirely capture the complexity of vertebrate circulatory systems (Choma, Suter, Vakoc, Bouma, & Tearney, 2011). The adult midgut established the animal as a prime model for understanding principles of tissue-level processes that govern homeostasis in adult organs, following the discovery of intestinal stem cells (Jiang & Edgar, 2011; Liang, Balachandra, Ngo, & O’Brien, 2017; Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006).
3.3. Danio rerio
Zebrafish have a similar cultivation cost in the laboratory as flies (Lieschke & Currie, 2007), but they require 2–3 months longer for development, a timeline more comparable to the generation cycle of mice (Dahm et al., 2005). The development of zebrafish, indicated by hours postfertilization, is divided into four stages: (1) embryo (ends at 72 h postfertilization at 28.5°C), (2) larva (an individual that is no longer an embryo but has yet to become a juvenile), (3) juvenile (most adult characteristics developed, but absence of sexual maturity), and (4) adult (defined by the production of viable gametes) (Kimmel et al., 1995; Parichy, Elizondo, Mills, Gordon, & Engeszer, 2009). Like humans, zebrafish are vertebrates, and approximately 55%–70% of zebrafish genes are human orthologs (Howe et al., 2013; Hu et al., 2011). Zebrafish are an emerging model to study human diseases because of this genetic similarity to humans, the relative ease of genetic manipulation, and the optical clarity of embryos and larvae.
Zebrafish have mechanosensitive hair cells in the inner ear and the lateral line organ. These hair cells can regenerate, an ability that humans lack. In humans, damage or destruction of sensory hair cells in the inner ear can lead to debilitating hearing or balance deficits, especially in older adults (Kniss, Jiang, & Piotrowski, 2016). Larvae, adult zebrafish, and isolated hair cells can be used to study hearing loss in vertebrates. Zebrafish have a closed circulatory system with many similarities to mammals, making studies of cardiac development and function possible (Chan & Mably, 2011). In addition to hair cells, the central nervous system and the heart can regenerate, which makes zebrafish an excellent model system for vertebrate tissue regeneration (Gemberling, Bailey, Hyde, & Poss, 2013).
4. DESIGN CONSIDERATIONS FOR MICROFLUIDICS
The development of microfluidic devices began by taking advantage of fluid motion physics in the laminar flow regime (Whitesides, 2006). Devices capable of complex reactions that involve mixing and multiplexing were designed for chemical synthesis (Elvira, i Solvas, Wootton, & deMello, 2013; Hung et al., 2006; Lignos et al., 2016) and micro total analysis (Auroux, Koc, DeMello, Manz, & Day, 2004; Heiland et al., 2017; Patabadige et al., 2016).
Microfluidic devices for research involving biological cells soon followed (Chiu et al., 2017), because, in addition to fluid manipulation, microfluidics can manipulate objects at the scale of single cells (Fig. 2). Developments include microfluidic systems for cell culture (Halldorsson, Lucumi, Gómez-Sjöberg, & Fleming, 2015; Moreno et al., 2015; Rhee et al., 2005; Young & Beebe, 2010), sorting (Henry et al., 2016; Krüger et al., 2002; Siedlik, Varner, & Nelson, 2016; Ung et al., 2017; Warkiani, Wu, Tay, & Han, 2015), manipulation (Applegate, Squier, Vestad, Oakey, & Marr, 2004; Chen, Chen, Chen, Jang, & Wang, 2014; Ding et al., 2012; Yi, Li, Ji, & Yang, 2006; Yun, Kim, & Lee, 2013; Zhao, Cheng, Miller, & Mao, 2016), and bioanalysis (Chen, Li, et al., 2014; Mirasoli, Guardigli, Michelini, & Roda, 2014; Wheeler et al., 2003). The research field of microfluidics continues to expand as novel strategies for stimulation and analysis continuously emerge. Microfluidics thus enables investigations on the effects of mechanical stimuli such as shear (Kim et al., 2014; Shao et al., 2009), compression (Guo, Park, & Ma, 2012; Kim, Kang, Park, Yoon, & Park, 2007), and tension (Douville et al., 2011; Gossett et al., 2012) for a range of biological applications (Kurth, Eyer, & Franco-Obregón, 2012; Polacheck, Li, Uzel, & Kamm, 2013; Zheng, Nguyen, Wei, & Sun, 2013).
Due to the size scale of many model organisms, microfluidics extended its applications further to include multicellular systems, among them worms, fruit flies, and zebrafish. The technology has the capability to address some of the limitations of classic techniques, with the main advantages being higher experimental throughput and integration of manipulation, stimulation, and readout. Devices can be designed to sort, trap, immobilize, and otherwise manipulate organisms within a single device. In addition, microfluidics offer more control over magnitude and reproducibility of mechanical stimuli in comparison to classic assays (Nekimken, Mazzochette, Goodman, & Pruitt, 2017). Other instruments with even more precise control of stimuli also exist (Mazzochette et al., 2018; Park, Goodman, & Pruitt, 2007; Park, Petzold, Goodman, & Pruitt, 2011), but they are more costly, difficult to use, and lower throughput compared to microfluidics.
We designed and published a microfluidic device for mechanical stimulation of worms during high-resolution imaging (Nekimken, Fehlauer, et al., 2017), as well as a detailed protocol for fabricating and operating our device (Fehlauer et al., 2018). The device integrated a trapping channel adapted from a microfluidic platform for studying chemosensation in worms (Chronis, Zimmer, & Bargmann, 2007) with six individually addressable mechanical actuators for stimulating touch receptor neurons. The actuators consisted of thin, deformable walls on either side of the trap channel. When the chambers on the other side of the walls were pressurized, the thin wall inflated, indenting the worm. Experimental results are described in Section 5.1.1. The following sections focus on our design process for this microfluidic device, including actuator design, immobilization strategies, and fabrication constraints.
4.1. ACTUATORS
When designing the pneumatic actuators in our device, the key performance metric we chose was deflection distance of the actuation membrane, since this is often sufficient to quantify the magnitude of a mechanical stimulus (Box 1). Deflection distance is dependent on the pressure applied to channel, geometry of the actuators, and material properties of the device (Lee, Chan, Maung, Rezler, & Sundararajan, 2007; Nekimken, Fehlauer, et al., 2017). Material properties are discussed in Section 4.5. There are several practical challenges that limit these parameters.
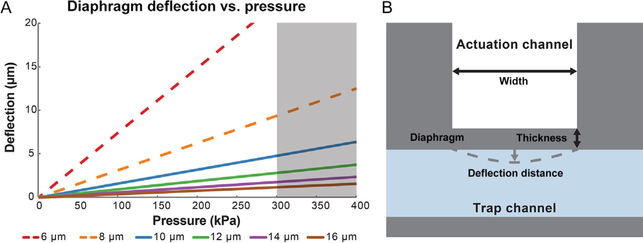
The maximum deflection of an actuation membrane (diaphragm) varies with its thickness to the power of negative 3 (t−3), therefore minimizing the thickness of the membrane resulted in better performance (Fig. 3). However, successfully creating high-aspect-ratio membranes was the most challenging part of the fabrication process, and feature sizes depend on mask resolution and exposure system available. Transparency masks offered sufficient resolution for prototyping trap designs (Section 4.2), but not for reliable fabrication of actuation membranes. Using a chrome mask and a collimated UV light source, we were able to reliably fabricate 10-μm-thick membranes with a 50 μm by 50 μm cross section. At times, we were successful in making 8-μm-thick membranes, we decided not to use this design because some membranes would fail to create a seal between the trap and the actuation channel, rendering the entire device defective. A more advanced lithography system or other processes such as deep reactive ion etching might successfully produce thinner actuation membranes, but we elected to keep the fabrication process simple.
FIG. 3.
The design space for actuator geometry was limited by fabrication constraints and the maximum pressure we could apply without tubing failure. (A) Using our exposure system, we were unable to reliably fabricate membranes (diaphragms) with a thickness of 6 μm or 8 μm (red and orange dashed lines). The connections to the pressure pump sometimes failed above 300 kPa (gray zone). We thus chose 10 μm as the thickness of the actuators because they met design constraints but still applied enough indentation to activate touch avoidance behavior. (B) Top-view schematic of one actuator with labeled geometric parameters, not to scale.
For our device, the pressure applied to the actuation channels was limited by the integrity of the tubing connecting the actuator and the pressure source. We punched holes in the PDMS device with a biopsy punch, then used polyethylene tubing with stainless steel pin connectors to interface with a pressure pump (Fehlauer et al., 2018). With this setup, the tubing reliably stayed in place when 300 kPa of pressure was applied, although we were able to increase this to 450 kPa in some cases (Nekimken, Fehlauer, et al., 2017). Actuation pressure could be increased further using a fixture and/or PDMS-compatible adhesive to hold the tubing in place.
The width and height of the actuator also impact the maximum deflection distance. The height of the actuator was set by the height of the trap (Section 4.2), because we wanted to limit device fabrication to a single layer to avoid alignment challenges (Section 4.5). We decided to design actuators of equal width and height, since membrane deflection is primarily affected by the smallest dimension, resulting in 50 by 50 μm actuators. For comparison, the size scale of this design fell between the diameter of a hair used in the classical hair touch assay (Nekimken, Mazzochette, et al., 2017) and glass beads (10 μm) glued to actuators for applying touch stimuli to worms (Park et al., 2007; Petzold et al., 2013, 2011).
4.2. IMMOBILIZATION
Immobilization of an object of interest is critical because microscopy-based readouts remain an important approach for evaluation of biological responses. Immobilization of worms in microfluidic devices can be accomplished by directing a worm into a snug-fitting channel that hold the worm in place (Hulme et al., 2007), deflection of the channel’s “ceiling” to compress a worm (Zeng, Rohde, & Yanik, 2008), or paralyzing a worm with carbon dioxide (Chokshi, Ben-Yakar, & Chronis, 2009), low temperature (Chung, Crane, & Lu, 2008), or anesthetic drugs (Morgan, Kayser, & Sedensky, 2007). Active immobilization strategies facilitate animal removal and accommodate size variations in a population, but require additional external components for implementation. We chose to use a passive approach that avoids the complications of additional moving parts as well as potential effects on the mechanics or information processing of the worm due to paralysis.
We tested two different strategies for passive immobilization of C. elegans during the development of our device (Fig. 4). Design A was a continuously tapering channel with evenly placed actuators along the full length of the channel. Design B was a form-fitting channel designed to accommodate the shape of a worm with a specific size. The height of the trap was optimized to a dimension approximately 5% less than the diameter of the worms (50 μm for young adults), successfully minimizing their movement without getting them permanently stuck.
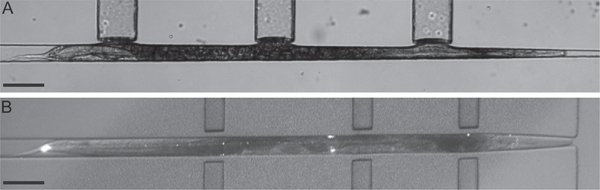
FIG. 4.
Passive trap designs for worm immobilization with actuators positioned along the length of the worm. (A) Continuous taper channel and (B) form-fitting channel. In design A the worm pushes the diaphragms toward the actuation channels. Scale bars 100 μm.
Design (A) adapted from Hulme, S. E., Shevkoplyas, S. S., Apfeld, J., Fontana, W., & Whitesides, G. M. (2007). A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab on a Chip, 7(11), 1515–1523. http://www.ncbi.nlm.nih.gov/pubmed/17960280. https://doi.org/10.1039/b707861g; Design (B) adapted from Chronis, N., Zimmer, M., & Bargmann, C. I. (2007). Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature Methods, 4(9), 727–731. http://www.nature.com/doifinder/10.1038/nmeth1075. https://doi.org/10.1038/nmeth1075.
The main advantage of design A was its ability to accommodate size variations of the animal, thus enabling studies of C. elegans at different life stages using the same microfluidic device. The disadvantages were clogging and low experimental throughput as a consequence of the tapering feature. The narrow opening at the end of the trap was easily clogged with debris. To remove worms from this device, the flow of buffer in the device had to be reversed to push a worm back to the inlet of the device. With design B, animals could be preloaded into a larger entrance chamber, significantly reducing preparation time per worm (approximately sixfold improvement). The smallest constriction of the trap was approximately half of the worm width, facilitating removal of animals and minimizing the likelihood of clogging. Following on-chip experiments, it was possible to recover organisms, thus enabling long-term studies of individuals (Fehlauer et al., 2018; Hulme et al., 2007; Nekimken, Fehlauer, et al., 2017).
4.3. FLOW RATE
The flow rate through the device can be important for (1) the sensitivity of the animal to shear stress and (2) the design of a fluidic network. Worms are not particularly sensitive to flow rates, so this was not a consideration for our device. However, other model organisms may be more responsive to shear flow. For example, as discussed in Section 5.3, mechanosensitivity of the lateral line of zebrafish larvae could be studied using this approach. In addition, the laminar flow profile and flow rate need to be considered in microfluidic platforms with an extended fluidic network, such as for drug or nutrient delivery.
4.4. MULTIPLEXING
Some devices for studying worms use multiplexing to increase experimental throughput by testing multiple worms in the same device (Hulme et al., 2007, 2010), but we found it difficult to remove worms from multiplexed devices. When one worm was successfully removed from such a device, the fluidic resistance of the vacant channel dropped. This redirected flowfrom channels that still hadworms in them, sothere was less pressure to push the remaining worms out.
4.5. FABRICATION
We fabricated microfluidic devices with the elastomer polydimethylsiloxane (PDMS) using well-established protocols (Jenkins, 2013; Xia & Whitesides, 1998). The process consists of two stages, (1) photolithography of the master mold and (2) replica molding and bonding of PDMS devices. Details can be found in Fehlauer et al. (2018) and Nekimken, Fehlauer, et al. (2017). Ease of fabrication was an important consideration in the design of the trapping channel and actuators. We decided to restrict our microfluidic platform to a single layer to eliminate the need for alignment and maintain the simplicity of fabricating devices from a single master mold, instead of a mold for each layer. This fabrication process also made bonding devices to coverslips fast, simple, and reliable, because only one bonding step was needed and no alignment was required.
Mechanical properties of PDMS can be modified by changing the ratio between the base polymer and the cross-linking agent (Wang, Volinsky, & Gallant, 2014). The work by Cho, Oakland, Lee, Schafer, and Lu (2018) took advantage of this material property to influence membrane deflection of the actuators. We chose to use the standard 10:1 ratio of base:curing agent, but we used devices within 1 month of fabrication to limit stiffening from continued cross-linking over time (Placet & Delobelle, 2015).
4.6. LAB AROUND THE CHIP
While microfluidic devices are sometimes touted as “lab on a chip” systems, this term neglects the external equipment that is required to operate the device, especially in a research environment. Operation of microfluidic devices with continuous flow requires external pressure pumps (e.g., syringe or solenoid) or integrated micropumps (Laser & Santiago, 2004). To apply mechanical stimuli in microfluidic devices to study mechanobiology in cells, tissues, and organisms, pneumatic or hydraulic equipment is required to generate the mechanical stimulus in addition to any pumps for flow control.
Our microfluidic device also required some external equipment for normal operation, and the detailed instructions can be found in Fehlauer et al. (2018). In this section, we describe why we chose to include the additional equipment in our experimental setup. We used a piezoelectric pressure control system (Elveflow OB-1 with 8 bar pressure unit) to control the pneumatic actuators because the system had a relatively fast response time and a simple user interface. As an alternative, we tested solenoid valves with a custom control system. However, they did not respond as fast as the piezoelectric technology, leading to attenuation of dynamic stimuli.
For fluid control, we used a gravity flow system supplemented with a peristaltic pump to provide a small amount of suction to the nose of the worm to hold it in place. The house vacuum with a simple regulator valve and liquid collector yielded similar results if a peristaltic pump was not available. The pump flowed buffer past the trap during the experiment, creating a small pressure drop in front of the worm from the continuously flowing fluid according to Bernoulli’s principle. The pressure drop had the added benefit of making it easy to dispose of worms, because the buffer carried the worm to the waste container when the worm was pushed through the exit of the trap. Alternatively, the worm could be collected for further experiments as it left the device.
Finally, a microscope was necessary for observing the results of an experiment. We measured the activation of mechanosensitive neurons using GCaMP (Chen et al., 2013), which changes fluorescence intensity when calcium concentration changes in the neuron. Mechanical stimulation of the neurons led to movement of the neuron away from the microscope’s focal plane. To correct for defocusing, we used an additional fluorophore in our intensity measurements. This, in turn, required two excitation wavelengths and a beam splitter for imaging the intensities of both of the fluorophores simultaneously (Fehlauer et al., 2018; Nekimken, Fehlauer, et al., 2017). Different experiments require different optical setups, and the choices made when designing an imaging system can have a major impact on the results of an experiment.
5. MICROFLUIDICS FOR MECHANOBIOLOGY OF MODEL ORGANISMS
Many of the design considerations discussed above apply to designing new microfluidic devices for mechanobiology as well as to adapting existing devices for studying other model organisms. Prior work can inform future developments in this field, so reviewing existing devices is an important part of the process for designing new ones. To this end, this section focuses on existing microfluidic platforms for mechanobiological studies, as defined above (Fig. 1). Many microfluidic devices have been used to study other aspects of model organisms, but the potential for using these devices to study mechanobiology is not yet fully realized. For reviews on microfluidic devices for sorting and imaging of multicellular organisms, readers are referred to works by Ben-Yakar, Chronis, and Lu (2009), Crane, Chung, Stirman, and Lu (2010), Hwang and Lu (2013), Sivagnanam and Gijs (2013), and Porto, Rouse, San-Miguel, and Lu (2016).
Each of the following sections features an overview of microfluidic devices used to study a model organism and then describes specific devices used to study mechanobiology in that organism. Sections are organized in the order of the animal’s developmental stage: embryo, larva, adult, and explanted tissue. In contrast to worms, where the majority of research was performed on adults, microfluidic experiments on fruit flies and zebrafish focus on embryos and larvae. For fruit flies, this is a consequence of their physiological requirement for air, and, for zebrafish, it is due to their size. Each section concludes by highlighting some research opportunities tailored to each model organism.
5.1. Caenorhabditis elegans
5.1.1. Current state of the field
Microfluidic devices for studying C. elegans have focused on (1) enabling imaging of worms and (2) creating specialized environments to study behavior of worms. These are reviewed as a whole elsewhere (Bakhtina & Korvink, 2014; SanMiguel & Lu, 2013). Both classes of devices have been used to study mechanobiology of worms. Devices designed for epifluorescence imaging of a worm require some sort of immobilization, since the motile nature of worms makes high-resolution imaging of dim fluorophores difficult (Section 4).
Devices for studying C. elegans behavior typically consist of a structured environment that presents mechanical, optical, or chemical stimuli to worms while providing a means (typically imaging whole worms) for researchers to observe the worms’ behavior. Lockery et al. (2008) presented an early example of such a device where worms were placed in microfluidic chambers that contained micropillars of different size and density, then observed that the velocity of worms crawling through the device depends on pillar spacing. Subsequent researchers expanded on this device concept to measure the forces applied by worms during locomotion (Doll et al., 2009; Johari, Nock, Alkaisi, & Wang, 2013).
Initially, mechanobiological studies of C. elegans focused on the behavioral responses of worms to touch stimuli (Chalfie et al., 1985), but worms are now used to study mechanotransduction and mechanobiology of development on the cellular -and tissue-levels (Cram, 2014). The use of microfluidics for mechanobiology of worms is largely limited to studies of sensory neurons in adults rather than in developing larvae. To our knowledge, Cho et al. (2018) published the only device for testing touch sensitivity of larvae.
To study touch sensitivity of C. elegans larvae, Cho et al. (2018) presented a device adapted from previous devices for studying adult worms (Cho et al., 2017; Nekimken, Fehlauer, et al., 2017), with a fluorescent calcium sensor, GCaMP (Chen et al., 2013), as a readout for neuron activation. To mechanically stimulate a worm, the authors used actuators similar to those described in Section 3, but required a different polymer formulation, resulting in a lower modulus of elasticity to enable actuation suitable for small larvae. With this device, Cho et al. (2018) showed that the anterior ventral touch neuron can still be activated, despite the fact that the neuron is not incorporated into downstream neuronal circuits at the L2 stage. Similarly, a harsh touch neuron can be activated in L2 larvae even before its branched structure forms. Finally, the paper explores the mechanosensitivity of larvae during lethargus, a sleep-like state that occurs between larval stages in worms. In this case, anterior touch receptor neurons, as well as downstream interneuron, all failed to respond to a mechanical stimulus. Their findings indicate that lethargus involves inhibition of sensory neurons rather than attenuation of the signal by downstream neurons.
While no other microfluidic devices have been published for studying mechanobiology of worm embryos and larvae, other devices for sorting (Cornaglia et al., 2015; Sofela et al., 2018; Yang et al., 2017) and observing (Cornaglia et al., 2015; Hulme et al., 2010; Keil, Kutscher, Shaham, & Siggia, 2017) embryos and larvae have been presented. These devices have the potential to be used for studying mechanobiology if additional features are added to apply mechanical stimuli and observe the resulting information processing (Fig. 1). The devices described below accomplish this task for adult worms.
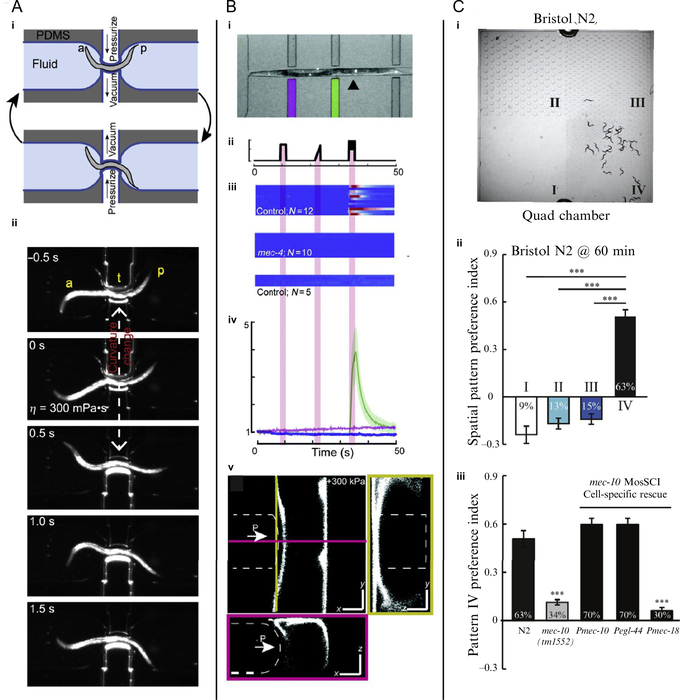
Wen et al. (2012) used microfluidics to study proprioception, a sensory system that uses mechanical cues to give organisms information about body position. In the study, the authors created microfluidic devices that control the curvature of a worm and then observed the worm’s resulting movements (Fig. 5A). In one of their devices, a worm traveled down a channel to a restriction flanked by two thin, deformable walls. These walls served as actuators when inflatable chambers on the opposite side of the wall from the worm were subjected to vacuum or positive pressure (Fig. 5Ai). By applying vacuum to one actuator and positive pressure to the other, the devices induced negative curvature on one side of the worm and positive curvature on the other. Using this device, Wen et al. (2012) observed that the induced curvature propagates toward the posterior of worms, independent of the curvature anterior to the actuators (Fig. 5Aii). They also used similar passive devices that constrain a worm to a straight posture or defined curvature, which prevented propagation of the worm’s normal sinusoidal motion past the constraint. Additional experiments combining microfluidic control of body curvature with optogenetic manipulation or optical measurement of calcium concentration in neurons showed that B-type cholinergic neurons are responsible for this proprioceptive coupling (Wen et al., 2012).
FIG. 5.
Microfluidic devices for C. elegans mechanobiology combine mechanical stimulation with behavioral and cellular readouts. (A) Device for controlling body curvature of worms (a: anterior, p: posterior). (i) Schematic of device and (ii) time series of a worm responding to change in body curvature. (B) Device for touch test of worms. (i) A worm immobilized in the device. (ii)–(iv) Stimulus profile, GCaMP fluorescence responses of individual worms, and average GCaMP response of touch receptor neurons. (v) Confocal images of a worm being indented in the device. (C) (i) Worms in quad-chamber for measuring texture preferences. (ii) Preference index for the spatial patterns. (iii) Disruption and rescue of preference for pillars in pattern IV.
Panel(A)reproducedandadaptedfromreferenceWen,Q.,Po,M.D.,Hulme,E.,Chen,S.,Liu,X.,Kwok,S.W.,… Samuel, A. D. (2012). Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron, 76(4), 750–761. http://linkinghub.elsevier.com/retrieve/pii/S0896627312008057. https://doi.org/10.1016/j.neuron.2012.08.039 with permission from Elsevier; Panel (B) reproduced and adapted from Nekimken, A. L., Fehlauer, H., Kim, A. A., Manosalvas-Kjono, S. N., Ladpli, P., Memon, F., … Krieg, M. (2017). Pneumatic stimulation of C. elegans mechanoreceptor neurons in a microfluidic trap. Lab on a Chip, 17(6), 1116–1127. http://xlink.rsc.org/?DOI=C6LC01165A. https://doi.org/10.1039/C6LC01165A with permission from the Royal Society of Chemistry, authors’ own work; Panel (C) reproduced and adapted from Han, B., Dong, Y., Zhang, L., Liu, Y., Rabinowitch, I., & Bai, J. (2017). Dopamine signaling tunes spatial pattern selectivity in C. elegans. eLife, 6(4), 431–436. http://journals.sagepub.com/doi/10.1177/2211068216669688; https://elifesciences.org/articles/22896. https://doi.org/https://doi.org/10.7554/eLife.22896 with permission from eLife under the CC BY license.
Nekimken, Fehlauer, et al. (2017) and Cho et al. (2017) used similar lateral pneumatic actuators for stimulation and a fluorescent calcium sensor, GCaMP (Chen et al., 2013), for readout of neuronal activity. With our device, which is described in Section 4, we found that the touch receptor neurons were also activated with a blue light stimulus, such as the excitation light for green fluorescent proteins. Worms lacking the protein LITE-1 were not activated by blue light and were used for subsequent experiments. In this device, step function and ramp stimuli failed to elicit significant activation of touch neurons, but a buzz stimulus with a sinusoid superimposed on a step led to observable activation of touch receptor neurons (Nekimken, Fehlauer, et al., 2017).
The device presented by Cho et al. (2017) used an active trapping strategy consisting of two pairs of laterally deformable actuators that hold a worm in place while a third pair between the two sets of trapping actuators applies mechanical stimuli to the worm. Using step function stimuli, they demonstrated that increases in stimulus magnitude or duration can increase the activation of touch receptor neurons. Additionally, they observed activation of a nociceptor in the worm (Chatzigeorgiouet al., 2010) when stimuli of higher magnitude were applied. Finally, Cho et al. (2017) looked at how drugs modulate touch sensitivity by performing a pilot screen with small molecules from a library of ligand candidates. Most of the compounds screened had deleterious effects on touch sensation to some degree, but some increased the touch sensitivity of worms.
McClanahan, Xu, and Fang-Yen (2017) also recently described a device for testing the touch sensitivity of crawling worms with a behavioral readout. Unlike the devices designed by Nekimken, Fehlauer, et al. (2017) and Cho et al. (2017), the actuators in this device stimulated worms by deforming the top of the channel rather than the side. Another distinctive feature of this device is that it contained several parallel worm channels, each with several actuators, enabling higher-throughput experiments. Image processing then determined the velocity of each worm’s centroid and an algorithm used a threshold to score changes in velocity as responses. Since worms were continuously crawling in the device, there was no active control over stimulus location, yielding a variety of stimulus locations for each trial. This enabled McClanahan et al. (2017) to explore the effect of stimulus location by grouping the stimuli into five bins based on where an individual stimulus was applied. They showed that egl-5 mutants, which lack functional posterior touch receptor neurons, tend to respond to posterior stimuli by moving toward the stimuli, rather than away from it like wild-type animals.
Park et al. (2008) used an agar device with micropillars to study how worms process information during locomotion. In contrast to other studies of worm locomotion in microfluidic chambers that focus on characterizing crawling and swimming in different environments (Lockery et al., 2008), this paper dissected mechanobiological pathways by showing the effect of knocking out genes that are essential for mechanosensation (Park et al., 2008). Worms crawling through the pillars lacking the ion channel subunits MEC-4 and MEC-10 moved less than half as fast as wild-type worms, on average. Their experiments thus quantified the dependence of worms’ locomotion on touch sensation using a behavioral readout.
Similarly, Han et al. (2017) studied the preference of worms for different mechanical environments by presenting four regions with different pillar spacing to worms and observing where the worms accumulated (Fig. 5C). After an hour, 63% of worms in the device were found in the quadrant with the smallest pillars and tightest spacing (175 μm pillar diameter, 116 μm pillar spacing). Subsequent experiments showed that the preference is regulated by a dopamine pathway activated by a mechanosensitive ion channel, TRP-4. Some alleles of TRP-4 suppress the preference for the smaller, denser pillars. Thus, by comparing the behavior of worms with different genotypes in their device, Han et al. (2017) were able to elucidate a mechanosensory pathway in worms responsible for discrimination of texture.
5.1.2. Research opportunities
The current literature on microfluidics for C. elegans mechanobiology is largely limited to behavioral and calcium-dependent fluorescence readouts. Electrophysiology, biomechanical characterization, and advanced fluorescence measurements, such as fluorescence resonance energy transfer (FRET), have received less attention in combination with microfluidics. A microfluidic device for electrophysiology of worms was presented, but it did not incorporate mechanical stimulation (Gonzales et al., 2017). Using these alternative readouts in existing devices for studying mechanobiology could lead to novel scientific findings.
Another opportunity for expanding the experimental capabilities in this field is the combination of multiple sensing modalities on the same device. Cho et al. (2017) demonstrated this possibility by showing that exposing a worm to small molecules in their device can increase or decrease its touch sensitivity, depending on the particular chemical compound (Cho et al., 2017). The effects of drugs on touch sensation are particularly interesting given the deleterious effects of chemotherapy on touch sensation (Fehrenbacher, 2015). Including additional modalities such as temperature control might yield further insights into how cells integrate multiple stimuli.
Moreover, mechanobiology is concerned with not just sensory biology, but also the effect of mechanical stimuli on development. While many studies explore the effect of mechanical cues within an organism on development Cram (2014), little is known about how external mechanical stimulation might perturb development. For example, it is not known how a worm would develop if an embryo was placed under increased hydrostatic pressure or compression. Microfluidics are an ideal technology to aid in answering these open questions, combining application of mechanical stimuli with other important considerations like delivery of nutrients.
5.2. Drosophila melanogaster
5.2.1. Current state of the field
Microfluidic devices for studying fruit flies include platforms for automation of embryonic assays (Chen et al., 2004; Chung et al., 2011; Levario, Zhan, Lim, Shvartsman, & Lu, 2013), defining temperature environments for morphogenetic studies (Dagani et al., 2007; Lucchetta, Lee, Fu, Patel, & Ismagilov, 2005; Lucchetta, Munson, & Ismagilov, 2006), and automating injections (Delubac et al., 2012). Recently, to study the mechanical properties of fruit fly follicle cells during oogenesis, Crest, Diz-Muñoz, Chen, Fletcher, and Bilder (2017) designed microscale flexible “egg cartons” to immobilize the cells while measuring the stiffness of basement membrane in live follicles using atomic force microscopy.
Drawing inspiration from devices originally designed to study C. elegans, such as Chokshi et al. (2009) and Wen et al. (2012), microfluidic platforms to study mechanobiology in Drosophila larvae are starting to emerge. Some of these devices immobilize fruit fly larvae for imaging purposes using mechanical constraints (Ghaemi, Rezai, Iyengar, & Selvaganapathy, 2015; Ghaemi, Rezai, Nejad, & Selvaganapathy, 2017), temperature (Chaudhury et al., 2017), or carbon dioxide (Ghannad-Rezaie, Wang, Mishra, Collins, & Chronis, 2012). Differences between worm and fruit fly larvae need to be considered for facile adaption of devices from one model organism to another. Fruit fly larvae are larger with a stiffer cuticle, and, moreover, the larvae are not as readily maintained in a fluid suspension and require some access to air (Chaudhury et al., 2017).
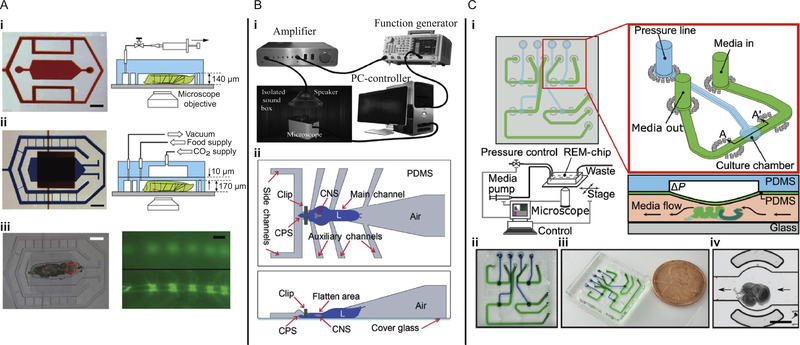
Ghannad-Rezaie et al. (2012) demonstrated two different designs of microfluidic devices for short- and long-term studies of cellular responses to neural injuries in larvae. The authors developed a novel approach for immobilization of larvae that reduces stress in the larval body by employing mechanical forces and/or carbon dioxide gas. To deliver carbon dioxide, they implemented a second layer on top of the microfluidic chamber (Fig. 6A). Both devices for short- and long-term investigations sealed reversibly to a coverslip. For long-term studies, the device enabled time lapse imaging of axonal sprouting following injury to dendrites with laser axotomy. Recently, the two-layer architecture was adapted to develop a device that combines liquid coolant with mechanical compression for immobilization of larvae for high-resolution imaging in vivo (Chaudhury et al., 2017).
FIG. 6.
Microfluidic devices for mechanobiology of Drosophila melanogaster combine mechanical stimulation with imaging. (A) Photographs and schematics of microfluidic devices for (i) short- and (ii) long-term immobilization of larvae for studying cellular responses to neural injuries (scale bars 1 mm). (iii) Bright-field (left, scale bar 1 mm) and fluorescent images (right, scale bar 20 μm) of the larva body with a fluorescently (GFP) labeled ventral cord. (B) (i) Experimental setup for examining the auditory response of fruit fly larvae. (ii) Schematic top and side views of the FlexiChip used to immobilize larvae for quick manual animal loading. (C) Regulated Environment for Microorgans (REM) chip, (i) schematic design of the chip with the inset showing detail of a single chamber unit, and the operation diagram for wing disc compression, (ii) and (iii) photographs of the device with individual fluidic (green) and pressure (blue) channels, and (iv) wing disc loaded into a culture chamber of the device (scale bar 400 μm).
Panel (A) reproduced and adapted from reference Ghannad-Rezaie, M., Wang, X., Mishra, B., Collins, C., & Chronis, N. (2012). Microfluidic chips for in vivo imaging of cellular responses to neural injury in Drosophila larvae. PLoS ONE, 7(1), e29869. https://doi.org/10.1371/journal.pone.0029869 with permission from PloS ONE under the CC BY licensel; Panel (B) reproduced and adapted from reference Ghaemi, R., Rezai, P., Iyengar, B. G., & Selvaganapathy, P. R. (2015). Microfluidic devices for imaging neurological response of Drosophila melanogaster larva to auditory stimulus. Lab on a Chip, 15(4), 1116–1122. http://xlink.rsc.org/?DOI=C4LC01245C. https://doi.org/10.1039/C4LC01245C with permission from the Royal Society of Chemistry under the CC BY license; Panel (C) reproduced and adapted from reference Narciso, C. E., Contento, N. M., Storey, T. J., Hoelzle, D. J., & Zartman, J. J. (2017). Release of applied mechanical loading stimulates intercellular calcium waves in Drosophila wing discs. Biophysical Journal, 113(2), 491–501. http://www.sciencedirect.com/science/article/pii/S0006349517306264. https://doi.org/10.1016/j.bpj.2017.05.051 with permission from Elsevier.
To study the auditory response of larvae, Ghaemi et al. (2015) demonstrated two designs, one for automated loading and one with a manual loading option. The manual FlexiChip is shown in Fig. 6B, along with their experimental setup for auditory stimulation. Their devices reduced movement of the central nervous system to permit monitoring of the response to acoustic signals using fluorescence imaging. The authors proposed further study of neuronal responses to other external cues with this platform.
Zhang, Dong, and Liu (2017) reported a system setup for robotic micromanipulation of fruit fly larvae, consisting primarily of a fluorescence microscope, an immobilization device, micromanipulators, and a glass pipette mounted on a microscale force sensor. The manipulation procedure was automated to recognize the pipette tip and the centroid of the first larva’s head in the immobilization device as the target location for applying mechanical stimulation. The platform was developed to facilitate identification of new mechanotransduction mechanisms in larvae.
Narciso, Contento, Storey, Hoelzle, and Zartman (2017) demonstrated one of the first microfluidic devices for probing mechanosensitive responses in fruit fly organs. The microfluidic device was used to create a “Regulated Environment for Microorgans,” termed REM-chip, and it was designed to support studies on explanted organs by mimicking the in vivo environment. The detailed schematic of the REM-chip is depicted in Fig. 6C. Briefly, the organ tissue was gently loaded into a fluidic channel where growth media could be delivered. The second layer of the chip applies compressive stress at intersections across deformable diaphragms. Interestingly, the authors found that mechanical unloading can initiate intercellular calcium waves in fruit fly wing discs, similar to those observed in vivo during organ development.
He, Si, Huang, Samuel, and Perrimon (2018) investigated mechanoregulation of stem cell differentiation in the adult midgut. To examine cellular responses, the authors presented an ex vivo approach. By compressing explanted midguts in a microfluidic device, He et al. (2018) studied calcium signaling in cells expressing an ion channel sensitive to mechanical stimuli, PIEZO. The design of the chip was previously applied to study C. elegans by Wen et al. (2012), described in Section 5.1.1 and shown in Fig. 5A.
5.2.2. Research opportunities
Microfluidic devices to study mechanobiology in fruit flies have followed in the footsteps of those for C. elegans. However, because adult fruit flies breathe air and fly, devices for whole-organism studies are restricted to embryos and larvae. An interesting opportunity is the development of devices that not only combine microfluidics with mechanobiology but also adapt to the model organism as it grows in the different stages of its life cycle, facilitating development studies of multicellular organisms.
Microfluidic technologies for ex vivo mechanobiological studies are an important future direction, since explanted tissues and organs can provide valuable knowledge on biological processes, as already illustrated in works by Narciso et al. (2017) and He et al. (2018). This particular area would benefit from standardized protocols for maintaining healthy explants.
Furthermore, as technological developments in the field of microfluidics continue to advance, they enable integration of new readouts and stimuli, such as exposure to chemical gradients or electrical fields. Since, in principle, it is possible to recover larvae for subsequent analyses following stimulation, this opens up new doors for studies on aging, genomics, and proteomics.
5.3. Danio rerio
5.3.1. Current state of the field
The small freshwater vertebrate Danio rerio (zebrafish) is an emerging model to study mechanobiology in microfluidic devices. So far, microfluidics for studying zebrafish focus on methods for high-throughput drug screening. One pair of adult fish can produce many eggs, allowing a high number of replicates for drug screening with automated microfluidics (Gibert, Trengove, & Ward, 2013; Parng, Seng, Semino, & McGrath, 2002). The fish embryo toxicity assay is a promising alternative approach to classical ecotoxicity testing with adult fish (Lammer, Carr, et al., 2009; Lammer, Kamp, et al., 2009; Strähle et al., 2012). The transparency of embryos and larvae makes the effects of drug application on the developing animal easy to monitor. Heart rate can be tracked in larvae 22–24 h postfertilization and the cardiovasculatory system is fully developed 36 h postfertilization (Kimmel et al., 1995; Yanik, Rohde, & Pardo-Martin, 2011). Mechanosensitive hair cells in the lateral line are fully developed and visible 48 h postfertilization (Kimmel et al., 1995), making zebrafish an ideal model to study mechanobiology.
Most microfluidic devices for zebrafish have focused on trapping embryos or larvae for long-term imaging (reviewed by Yang et al. (2016)). The biggest challenges in the design of a zebrafish microfluidic-based chip compared to other model organisms are (1) the zebrafish is a freshwater organism and needs sufficient perfusion for fresh water exchange to deliver oxygen and maintain consistent pH without damaging the animal, and (2) embryos develop at an optimal temperature of 28–29°C (Westerfield, 2000), so a heating system may be necessary for the health of the fish.
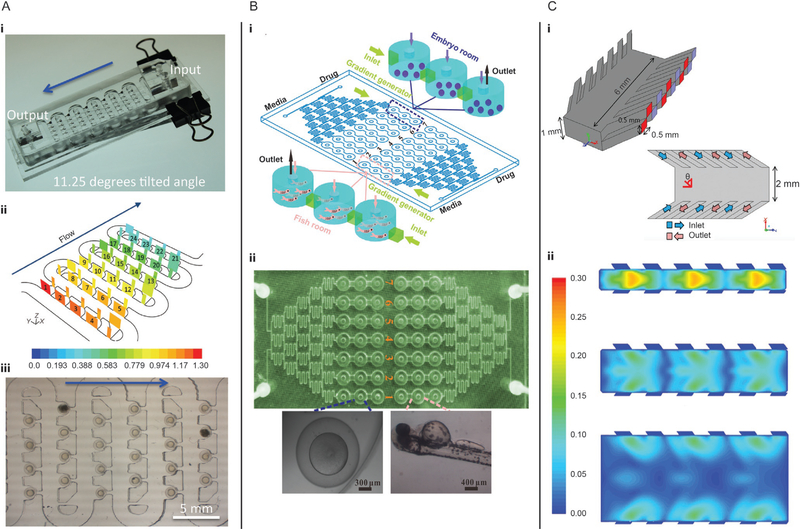
Akagi et al. (2012) developed a microfluidic device to study a large number of embryos simultaneously (Fig. 7A). The device consisted of several connected rows with trapping channels containing hydrodynamic deflectors and a small suction channel. Embryos were loaded one-by-one via hydrodynamic forces. The first embryo was trapped in the first empty trap channel, blocking that channel so subsequent embryos rolled freely toward the next available trap channel. Mounting the device on an elevated stage improved the trapping success from 80% when the hydrodynamic forces alone trapped the embryo, to nearly 100% when a combination of both hydrodynamic and gravitational forces held the embryos in place. In this device, the embryos can be kept for long-term imaging until they hatch and leave the trap. The device was positioned on a stage heater to maintain optimal culturing temperature. With the same device, Akagi et al. (2012) performed an angiogenesis assay to measure the development of new blood vessels and tested the effect of a selective blocker for angiogenesis, the inhibitor AV951. The transparency of the larvae allowed for a clear visualization of the presence or absence of newly forming blood vessels.
FIG. 7.
Microfluidic devices for studying zebrafish have the potential to be used for mechanobiology, but have mostly been used for other applications. (A) Tilted device for combined gravitational and hydrodynamic trapping. (i) Photograph and (ii) simulation of hydrodynamic forces for trapping embryos. (iii) Embryos trapped in the device for long-term imaging. (B) Microfluidic device for drug-delivery to embryos and larvae. (i) Schematic of the device with media inlet, drug inlet, gradient generator and seven series of fish tanks. (ii) Photograph of the device. (C) Microfluidic device to apply shear stress to zebrafish larvae. (i) Isometric and top-view schematics of optimized device design (ν = 60 degrees). (ii) Simulation of fluid velocity magnitude in m/s at channel widths of 0.5, 1.0, and 2.0 mm with an inlet and outlet angles ν set to 45 degrees.
Panel (A) reproduced and adapted from reference Akagi, J., Khoshmanesh, K., Evans, B., Hall, C. J., Crosier, K. E., Cooper, J. M., … Wlodkowic, D. (2012). Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS ONE, 7(5), 12–15. https://doi.org/10.1371/journal.pone.0036630 with permission from PloS ONE under the CC BY license; Panel (B) reproduced and adapted from Li, Y., Yang, F., Chen, Z., Shi, L., Zhang, B., Pan, J., … Yang, H. (2014). Zebrafish on a chip: A novel platform for real-time monitoring of drug-induced developmental toxicity. PLoS ONE, 9(4), e94792. https://doi.org/10.1371/journal.pone.0094792 with permission from PloS ONE under the CC BY license; Panel (C) reproduced and adapted from reference Kwon, H.-J., Xu, Y., Solovitz, S. A., Xue, W., Dimitrov, A. G., Coffin, A. B., & Xu, J. (2014). Design of a microfluidic device with a non-traditional flow profile for on-chip damage to zebrafish sensory cells. Journal of Micromechanics and Microengineering, 24(1), 017001. http://stacks.iop.org/0960-1317/24/i=1/a=017001?key=crossref. 898bd18771a566d89de4176cbca5c https://doi.org/10.1088/0960-1317/24/1/017001 with permission from IOP publishing.
To study different drug concentrations in both embryo and larvae, Li et al. (2014) developed a device consisting of two identical units each divided into two parts: (1) three replicates of seven open culture rooms for embryos or larvae and (2) a concentration gradient generator for dose-dependent drug application (Fig. 7B). Yang et al. (2011) and Choudhury et al. (2012) previously described similar designs with concentration gradient generators. They all had a perfusion system to replace manual exchange of media and prevent pH changes. Li et al. (2014) increased the number of replicates per well. Each well was connected to a plug, allowing easy removal of dead embryos or larvae. Indicators of health were recorded by imaging the device, including the lack of a heartbeat, blood circulation, and motility, which together characterize a lethal or teratogenic effect. The goal of Li et al. (2014) was to investigate the toxicity on embryos and larvae of an antiasthmatic drug which increases zebrafish heart rate with increased drug concentration. In parallel, they also investigated the effect of perfusion velocity on hatching success of embryos, estimating a flow rate of 5 μL/min for optimal hatching success. Although their main goal focused on the effects of drugs on healthy embryos, this device is also suitable to study the effect of shear stress on hatching and organ development.
Kwon et al. (2014) presented a microfluidic device to apply shear stress to the lateral line of larvae (Fig. 7C), which detects mechanical stimuli induced by movements of the surrounding fluid. The hair cells in the lateral line system are functionally analogous to hair cells in the inner ear of mammals, except they have the capacity to regenerate. Better understanding of the molecular and cellular basis of hearing loss and regeneration of hair cells in zebrafish may aid in designing therapies to induce regeneration in mammals (Kniss et al., 2016). The design of Kwon et al. (2014) focused on manipulating boundary layers using sidewall inlets and outlets. They used computational fluid dynamics simulations with and without a fish-shaped obstruction to optimize the flow profile in their device. Their simulations showed that a 2.0 mm wide channel with the inlet/outlet pairs set to 45 degrees was optimal without a fish, and a 1.0 mm wide channel with a 60 degrees inlet/outlet angle created the lowest pressure (0.3 Pa) on the fish head while maintaining a reasonably strong shear stress (1.9 Pa) on the lateral line hair cells. To study hair cell damage, shear stress at the boundary must be sufficient to induce damage with a reduced flow rate in the center of the device to prevent damage to the rest of the fish.
5.3.2. Research opportunities
Microfluidic devices have potential for studying mechanobiology of zebrafish, especially larvae. So far, the effects of shear stress on zebrafish have only been studied to identify optimal flow rates to avoid damaging larvae (Li et al., 2014). Thus, future studies could address the effect of shear stress on cell damage, regeneration, and organ development more thoroughly, as suggested by Kwon et al. (2014). Organ development has also been studied under static mechanical input using microfabricated platforms (Chen & Chen, 2013). Chen and Chen (2013) studied the effects of silicon nanowires on heart rate and organ development. Using silicon substrates with and without hydrophobic or hydrophilic nanowires, they identified an increase in heart rate on substrates with rougher textures. These textures and other mechanical stimuli could be integrated into a microfluidic device to study their impact on embryo development. Adapting devices originally designed for C. elegans and Drosophila will also advance experimental capabilities for zebrafish. Although progress in studying zebrafish mechanobiology with microfluidics is so far limited, microfluidic platforms have the potential to advance the study of zebrafish mechanobiology.
5.4. OTHER MODELS
While the three model organisms discussed above are some of the most commonly used, there are other model organisms that are suited for addressing other mechanobiological questions. Below, we highlight two microfluidic devices, one designed to study root growth in the plant Camellia japonica and another for monitoring muscular activity in Hydra vulgaris, and comment on how these devices could be used for further mechanobiological studies.
Agudelo et al. (2013) designed a microfluidic device to study the pollen tube growth of the plant C. japonica. Their device distributes pollen grains to microchannels of varying geometries. The pollen grains then extended growth tubes into the channels with different growth characteristics due to the geometry of the channel, flow rate through the channel, and chemical composition of fluid in the channel. The authors found that high flow rates can damage the pollen grain, and that kinks in the growth channels can help the pollen grain anchor itself in place. Either of these observations could be the basis of a mechanobiology study on root development or mechanosensing at the tip of pollen tubes, but the authors focused on the effect of air-water interfaces and chemical cues in this study.
H. vulgaris is a freshwater polyp and model organism for tissue regeneration. Hydra are small (0.5–15 mm) and transparent. Their neurons continually replenished and are more dynamic than in other model organisms (Badhiwala, Gonzales, Vercosa, Avants, & Robinson, 2018). Badhiwala et al. (2018) presented three polymer-based microfluidic devices to study the fragile Hydra: (1) an hour-glass chamber for electrophysiology that constrained the body column from large movements, (2) a wheel-and-spoke perfusion chamber that constrained locomotion, and (3) a behavioral micro-arena with micropillars that limited movements and locomotion in the vertical plane but allowed all movements in the horizontal plane. They observed that the Hydra reacted to shear stress induced by a high flow rate with body contractions or tentacle swaying. Here, they tried to avoid mechanical stimulation by using a reduced flow rate and instead studied the response to different chemical stimuli. However, this design seems suitable to investigate shear stress responses and future designs could integrate microactuators for applying mechanical compression in addition to shear stimuli.
6. CONCLUSION
Microfluidic technologies are opening doors to mechanobiological studies of multicellular model organisms and their tissues. Worms, fruit flies, and zebrafish are excellent candidates for mechanobiological study in microfluidics because they share fundamental characteristics with more complex organisms, are amenable to genetic techniques, can be cultivated in the lab with relative simplicity, and have the proper size to fit in microfluidics. Further integration of mechanical actuation into existing devices will be key to supporting many more applications. Devices and features developed for one model organism can be easily adapted to another, expanding experimental capabilities without starting from scratch. Thus, microfluidic technologies will continue to contribute to advancing our understanding of mechanobiology in development, homeostasis, and disease.
ACKNOWLEDGMENTS
We thank Dave Wallace for assistance with graphics and Sandra N. Manosalvas-Kjono, Purim Ladpli, and Farah Memon for helpful discussions. This work was supported in part by the National Institutes of Health under grants R01EB006745, R01GM116000, R01NS047715, R21HL13099301, and F31NS100318, National Science Foundation under grants EFRI MIKS 1136790 and CMMI 166243, Stanford Bio-X IIP, a gift from the G. Harold & Leila Y. Mathers Foundation, and fellowships from the Swedish Research Council (VR) under grant 2017-06156 and the German Research Foundation (DFG, 313913559).
REFERENCES
- Aarabi S, Bhatt KA, Shi Y, Paterno J, Chang EI, Loh SA,…Gurtner GC (2007). Mechanical load initiates hypertrophic scar formation through decreased cellular apoptosis. The FASEB Journal, 21(12), 3250–3261. 10.1096/fj.07-8218com. [DOI] [PubMed] [Google Scholar]
- Agudelo CG, Nezhad AS, Ghanbari M, Naghavi M, Packirisamy M, & Geitmann A (2013). TipChip: A modular, MEMS-based platform for experimentation and phenotyping of tip-growing cells. Plant Journal, 73(6), 1057–1068. 10.1111/tpj.12093. [DOI] [PubMed] [Google Scholar]
- Akagi J, Khoshmanesh K, Evans B, Hall CJ, Crosier KE, Cooper JM,… Wlodkowic D (2012). Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS ONE, 7(5), 12–15. 10.1371/journal.pone.0036630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Uchihashi T, & Kodera N (2013). High-Speed AFM and applications to biomolecular systems. Annual Review of Biophysics, 42(1), 393–414. https://doi.org/10.1146/annurev-biophys-083012-130324 . 10.1146/annurev-biophys-083012-130324http://www.annualreviews.org/doi/10.1146/annurev-biophys-083012-130324. http://www.annualreviews.org/doi/10.1146/annurev-biophys-083012-130324 . [DOI] [PubMed] [Google Scholar]
- Applegate RW, Squier J, Vestad T, Oakey J, & Marr DWM (2004). Optical trapping, manipulation, and sorting of cells and colloids in microfluidic systems with diode laser bars. Optics Express, 12(19), 4390 https://doi.org/10.1364/OPEX.12.004390 . 10.1364/OPEX.12.004390https://www.osapublishing.org/abstract.cfm?URI=oe-12-19-4390. https://www.osapublishing.org/abstract.cfm?URI=oe-12-19-4390 . [DOI] [PubMed] [Google Scholar]
- Aristotle (1990). On the soul. Great Books of the Western World, 7, 631–672. [Google Scholar]
- Auroux P-A, Koc Y, DeMello A, Manz A, & Day PJR (2004). Miniaturised nucleic acid analysis. Lab on a Chip, 4(6), 534 https://doi.org/10.1039/b408850f . 10.1039/b408850fhttp://xlink.rsc.org/?DOI=b408850f. http://xlink.rsc.org/?DOI=b408850f . [DOI] [PubMed] [Google Scholar]
- Badhiwala KN, Gonzales DL, Vercosa DG, Avants BW, & Robinson JT (2018). Microfluidics for electrophysiology, imaging, and behavioral analysis of hydra. bioRxiv, 1–40. 10.1101/257691. [DOI] [PubMed] [Google Scholar]
- Bakhtina NA, & Korvink JG (2014). Microfluidic laboratories for C. elegans enhance fundamental studies in biology. RSC Advances, 4(9), 4691–4709. https://doi.org/10.1039/C3RA43758B . 10.1039/C3RA43758Bhttp://xlink.rsc.org/?DOI=C3RA43758B. http://xlink.rsc.org/?DOI=C3RA43758B . [DOI] [Google Scholar]
- Bardet P-L, Guirao B, Paoletti C, Serman F, Leopold V, Bosveld F,…Bellaïche Y (2013). PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Developmental Cell, 25(5), 534–546. https://doi.org/10.1016/j.devcel.2013.04.020 . https://www.sciencedirect.com/science/article/pii/S1534580713002499 . 10.1016/j.devcel.2013.04.020https://www.sciencedirect.com/science/article/pii/S1534580713002499http://linkinghub.elsevier.com/retrieve/pii/S1534580713002499. https://www.sciencedirect.com/science/article/pii/S1534580713002499 . http://linkinghub.elsevier.com/retrieve/pii/S1534580713002499 . [DOI] [PubMed] [Google Scholar]
- Ben-Yakar A, Chronis N, & Lu H (2009). Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Current Opinion in Neurobiology, 19(5), 561–567. https://doi.org/10.1016/j.conb.2009.10.010 . https://www.sciencedirect.com/science/article/pii/S0959438809001470 . 10.1016/j.conb.2009.10.010https://www.sciencedirect.com/science/article/pii/S0959438809001470http://linkinghub.elsevier.com/retrieve/pii/S0959438809001470. https://www.sciencedirect.com/science/article/pii/S0959438809001470 . http://linkinghub.elsevier.com/retrieve/pii/S0959438809001470 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, & Gerber Ch (1986). Atomic force microscope. Physical Review Letters, 56(9), 930–933. 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics, 77(1), 71–94. 10.1002/cbic.200300625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, & Lemaitre B (2014). Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio, 5(3). 10.1128/mBio.01117-14. e01117–e01114. http://www.ncbi.nlm.nih.gov/pubmed/24865556. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4045073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RA, Prajapati R, McGrouther DA, Yannas IV, & Eastwood M (1998). Tensional homeostasis in dermal fibroblasts: Mechanical responses to mechanical loading in three-dimensional substrates. Journal of Cellular Physiology, 175(3), 323–332. https://doi.org/10.1002/(SICI)1097-4652(199806)175:3¡323::AID-JCP10¿3.0.CO;2-6 . http://doi.wiley.com/10.1002/%28SICI%291097-4652%28199806%29175%3A3%3C323%3A%3AAID-JCP10%3E3.0.CO%3B2-6. http://doi.wiley.com/10.1002/%28SICI%291097-4652%28199806%29175%3A3%3C323%3A%3AAID-JCP10%3E3.0.CO%3B2-6 . [DOI] [PubMed] [Google Scholar]
- Bustamante C, Erie DA, & Keller D (1994). Biochemical and structural applications of scanning force microscopy. Current Opinion in Structural Biology, 4(5), 750–760. 10.1016/S0959-440X(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Cahalan SM, Lukacs V, Ranade SS, Chien S, Bandell M, & Patapoutian A (2015). Piezo1 links mechanical forces to red blood cell volume. eLife, 4, e07370 https://doi.org/10.7554/eLife.07370 . http://www.ncbi.nlm.nih.gov/pubmed/26001274 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4456639 . 10.7554/eLife.07370http://www.ncbi.nlm.nih.gov/pubmed/26001274http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4456639https://elifesciences.org/articles/07370. http://www.ncbi.nlm.nih.gov/pubmed/26001274 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4456639 . https://elifesciences.org/articles/07370 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon L, Zambon AC, Cammarato A, Zhang Z, Vogler G, Munoz M,…Bodmer R (2017). Expression patterns of cardiac aging in Drosophila. Aging Cell, 16(1), 82–92. https://doi.org/10.1111/acel.12559 . 10.1111/acel.12559http://doi.wiley.com/10.1111/acel.12559. http://doi.wiley.com/10.1111/acel.12559 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, & Hayes W (1976). Bone compressive strength: The influence of density and strain rate. Science, 194(4270), 1174–1176. https://doi.org/10.1126/science.996549 . 10.1126/science.996549http://www.sciencemag.org/cgi/doi/10.1126/science.996549. http://www.sciencemag.org/cgi/doi/10.1126/science.996549 . [DOI] [PubMed] [Google Scholar]
- Carter DR, Beaupré GS, Giori NJ, & Helms JA (1998). Mechanobiology of skeletal regeneration. Clinical Orthopaedics and Related Research, 355(Suppl.), 41–55. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=9917625&retmode=ref&cmd=prlinks%5Cn. http://www.ncbi.nlm.nih.gov/pubmed/9917625%5Cn. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, & Brenner S (1985). The neural circuit for touch sensitivity in Caenorhabditis elegans. The Journal of Neuroscience, 5(4), 956–964.https://doi.org/3981252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward W, & Prasher D (1994). Green fluorescent protein as a marker for gene expression. Science, 263(5148), 802–805. https://doi.org/10.1126/science.8303295 . 10.1126/science.8303295http://www.sciencemag.org/cgi/doi/10.1126/science.8303295%7B&%7Did=8303295%7B&%7Dretmode=ref%7B&%7Dcmd=prlinks. http://www.sciencemag.org/cgi/doi/10.1126/science.8303295%7B&%7Did=8303295%7B&%7Dretmode=ref%7B&%7Dcmd=prlinks . [DOI] [PubMed] [Google Scholar]
- Chan J, & Mably JD (2011). Dissection of cardiovascular development and disease pathways in zebrafish. (Vol. 100), Elsevier Inc; 10.1016/B978-0-12-384878-9.00004-2. [DOI] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Yoo S, Watson JD, Lee W-H, Spencer WC, Kindt KS,… Schafer WR (2010). Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nature Neuroscience, 13(7), 861–868. https://doi.org/10.1038/nn.2581 . 10.1038/nn.2581http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2975101&tool=pmcentrez&rendertype=abstract. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2975101&tool=pmcentrez&rendertype=abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AR, Insolera R, Hwang R-D, Fridell Y-W, Collins C, & Chronis N (2017). On chip cryo-anesthesia of Drosophila larvae for high resolution in vivo imaging applications. Lab on a Chip, 17(13), 2303–2322. https://doi.org/10.1039/C7LC00345E . 10.1039/C7LC00345Ehttp://xlink.rsc.org/?DOI=C7LC00345E. http://xlink.rsc.org/?DOI=C7LC00345E . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Zappe S, Sahin O, Zhang XJ, Fish M, Scott M, & Solgaard O (2004). Design and operation of a microfluidic sorter for Drosophila embryos. Sensors and Actuators B: Chemical, 102(1), 59–66. https://doi.org/10.1016/J.SNB.2003.10.015 . 10.1016/J.SNB.2003.10.015http://www.sciencedirect.com/science/article/pii/S0925400503007718. http://www.sciencedirect.com/science/article/pii/S0925400503007718 . [DOI] [Google Scholar]
- Chen C-Y, & Chen C-Y (2013). Influences of textured substrates on the heart rate of developing zebrafish embryos. Nanotechnology, 24(26), 265101 https://doi.org/10.1088/0957-4484/24/26/265101 . 10.1088/0957-4484/24/26/265101http://stacks.iop.org/0957-4484/24/i=26/a=265101?key=crossref.a3006efcf759d371d32ac0381121850a. http://stacks.iop.org/0957-4484/24/i=26/a=265101?key=crossref.a3006efcf759d371d32ac0381121850a . [DOI] [PubMed] [Google Scholar]
- Chen N-C, Chen C-H, Chen M-K, Jang L-S, & Wang M-H (2014). Singlecell trapping and impedance measurement utilizing dielectrophoresis in a parallelplate microfluidic device. Sensors and Actuators B: Chemical, 190, 570–577. https://doi.org/10.1016/J.SNB.2013.08.104 . 10.1016/J.SNB.2013.08.104https://www.sciencedirect.com/science/article/pii/S0925400513010423. https://www.sciencedirect.com/science/article/pii/S0925400513010423 . [DOI] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A,…Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499(7458), 295–300. https://doi.org/10.1038/nature12354 . 10.1038/nature12354http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3777791&tool=pmcentrez&rendertype=abstract. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3777791&tool=pmcentrez&rendertype=abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li P, Huang P-H, Xie Y, Mai JD, Wang L,…Huang TJ (2014). Rare cell isolation and analysis in microfluidics. Lab on a Chip, 14(4), 626 https://doi.org/10.1039/c3lc90136j . 10.1039/c3lc90136jhttp://xlink.rsc.org/?DOI=c3lc90136j. http://xlink.rsc.org/?DOI=c3lc90136j . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu DT, deMello AJ, Di Carlo D, Doyle PS, Hansen C, Maceiczyk RM, & Wootton RCR (2017). Small but perfectly formed? Successes, challenges, and opportunities for microfluidics in the chemical and biological sciences. Chem, 2(2), 201–223. https://doi.org/10.1016/J.CHEMPR.2017.01.009 . 10.1016/J.CHEMPR.2017.01.009https://www.sciencedirect.com/science/article/pii/S2451929417300335. https://www.sciencedirect.com/science/article/pii/S2451929417300335 . [DOI] [Google Scholar]
- Cho Y, Oakland DN, Lee SA, Schafer WR, & Lu H (2018). On-chip functional neuroimaging with mechanical stimulation in Caenorhabditis elegans larvae for studying development and neural circuits. Lab on a Chip, 18, 29–31. https://doi.org/10.1039/C7LC01201B . http://pubs.rsc.org/en/Content/ArticleLanding/2018/LC/C7LC01201B . 10.1039/C7LC01201Bhttp://pubs.rsc.org/en/Content/ArticleLanding/2018/LC/C7LC01201Bhttp://xlink.rsc.org/?DOI=C7LC01201B. http://pubs.rsc.org/en/Content/ArticleLanding/2018/LC/C7LC01201B . http://xlink.rsc.org/?DOI=C7LC01201B . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Porto D, Hwang H, Grundy L, Schafer WR, & Lu H (2017). Automated and controlled mechanical stimulation and functional imaging in vivo in C. elegans. Lab on a Chip, 17, 2609–2618. https://doi.org/10.1039/C7LC00465F . 10.1039/C7LC00465Fhttp://pubs.rsc.org/en/Content/ArticleLanding/2017/LC/C7LC00465F. http://pubs.rsc.org/en/Content/ArticleLanding/2017/LC/C7LC00465F . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokshi TV, Ben-Yakar A, & Chronis N (2009). CO2 and compressive immobilization of C. elegans on-chip. Lab on a Chip, 9(1), 151–157. 10.1039/b807345g. [DOI] [PubMed] [Google Scholar]
- Choma MA, Izatt SD, Wessells RJ, Bodmer R, & Izatt JA (2006). Images in cardiovascular medicine: In vivo imaging of the adult Drosophila melanogaster heart with real-time optical coherence tomography. Circulation, 114(2), 35–36. https://doi.org/10.1161/CIRCULATIONAHA.105.593541 . 10.1161/CIRCULATIONAHA.105.593541http://www.ncbi.nlm.nih.gov/pubmed/16831991. http://www.ncbi.nlm.nih.gov/pubmed/16831991 . [DOI] [PubMed] [Google Scholar]
- Choma MA, Suter MJ, Vakoc BJ, Bouma BE, & Tearney GJ (2011). Physiological homology between Drosophila melanogaster and vertebrate cardiovascular systems. Disease Models & Mechanisms, 4(3), 411–420. https://doi.org/10.1242/dmm.005231 . 10.1242/dmm.005231http://dmm.biologists.org/cgi/doi/10.1242/dmm.005231. http://dmm.biologists.org/cgi/doi/10.1242/dmm.005231 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D, van Noort D, Iliescu C, Zheng B, Poon K-L, Korzh S,…Yu H (2012). Fish and Chips: A microfluidic perfusion platform for monitoring zebrafish development. Lab on a Chip, 12(5), 892–900. https://doi.org/10.1039/C1LC20351G . 10.1039/C1LC20351Ghttp://xlink.rsc.org/?DOI=C1LC20351G. http://xlink.rsc.org/?DOI=C1LC20351G . [DOI] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, & Bargmann CI (2007). Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature Methods, 4(9), 727–731. https://doi.org/10.1038/nmeth1075 . 10.1038/nmeth1075http://www.nature.com/doifinder/10.1038/nmeth1075. http://www.nature.com/doifinder/10.1038/nmeth1075 . [DOI] [PubMed] [Google Scholar]
- Chung K, Crane MM, &Lu H (2008). Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nature Methods, 5(7), 637–643. 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, & Lu H (2011). A microfluidic array for large-scale ordering and orientation of embryos. Nature Methods, 8(2), 171–176. https://doi.org/10.1038/nmeth.1548 . http://www.nature.com/doifinder/10.1038/nmeth.1548 . 10.1038/nmeth.1548http://www.nature.com/doifinder/10.1038/nmeth.1548http://www.nature.com/articles/nmeth.1548. http://www.nature.com/doifinder/10.1038/nmeth.1548 . http://www.nature.com/articles/nmeth.1548 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaglia M, Mouchiroud L, Marette A, Narasimhan S, Lehnert T, Jovaisaite V,… Gijs MAM (2015). An automated microfluidic platform for C. elegans embryo arraying, phenotyping, and long-term live imaging. Scientific Reports, 5(1), 10192 https://doi.org/10.1038/srep10192 . 10.1038/srep10192http://www.nature.com/articles/srep10192. http://www.nature.com/articles/srep10192 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Wightman B, & Chalfie M (2015). A transparent window into biology: A primer on Caenorhabditis elegans. Genetics, 200(2), 387–407. https://doi.org/10.1534/genetics.115.176099 . http://www.genetics.org/cgi/doi/10.1534/genetics.115.176099 . 10.1534/genetics.115.176099http://www.genetics.org/cgi/doi/10.1534/genetics.115.176099http://www.genetics.org/lookup/doi/10.1534/genetics.115.176099. http://www.genetics.org/cgi/doi/10.1534/genetics.115.176099 . http://www.genetics.org/lookup/doi/10.1534/genetics.115.176099 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram EJ (2014). Mechanotransduction in C. elegans morphogenesis and tissue function In Engler AJ & Kumar S (Eds.), Progress in molecular biology and translational science: Vol. 126, (pp. 281–316). Elsevier Inc; [DOI] [PubMed] [Google Scholar]
- Crane MM, Chung K, Stirman J, & Lu H (2010). Microfluidics-enabled phenotyping, imaging, and screening of multicellular organisms. Lab on a Chip, 10(12), 1509 https://doi.org/10.1039/b927258e . 10.1039/b927258ehttp://xlink.rsc.org/?DOI=b927258e. http://xlink.rsc.org/?DOI=b927258e . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crest J, Diz-Muñoz A, Chen D-Y, Fletcher DA, & Bilder D (2017). Organ sculpting by patterned extracellular matrix stiffness. eLife, 6, e24958 https://doi.org/10.7554/eLife.24958 . 10.7554/eLife.24958https://elifesciences.org/articles/24958. https://elifesciences.org/articles/24958 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagani GT, Monzo K, Fakhoury JR, Chen C-C, Sisson JC, & Zhang X (2007). Microfluidic self-assembly of live Drosophila embryos for versatile high-throughput analysis of embryonic morphogenesis. Biomedical Microdevices, 9(5), 681–694. 10.1007/s10544-007-9077-z. [DOI] [PubMed] [Google Scholar]
- Dahm R, Geisler R, & Nusslein-Volhard C (2005). Zebrafish (Danio rerio) genome and genetics In Encyclopedia of molecular cell biology and molecular medicine: Vol. 15, (pp. 593–626). John Wiley & Sons. [Google Scholar]
- Dai W, Peterson A, Kenney T, Burrous H, & Montell DJ (2017). Quantitative microscopy of the Drosophila ovary shows multiple niche signals specify progenitor cell fate. Nature Communications, 8(1), 1244 https://doi.org/10.1038/s41467-017-01322-9 . 10.1038/s41467-017-01322-9http://www.nature.com/articles/s41467-017-01322-9. http://www.nature.com/articles/s41467-017-01322-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delubac D, Highley CB, Witzberger-Krajcovic M, Ayoob JC, Furbee EC, Minden JS, & Zappe S (2012). Microfluidic system with integrated microinjector for automated Drosophila embryo injection. Lab on a Chip, 12(22), 4911 https://doi.org/10.1039/c2lc40104e . 10.1039/c2lc40104ehttp://xlink.rsc.org/?DOI=c2lc40104e. http://xlink.rsc.org/?DOI=c2lc40104e . [DOI] [PubMed] [Google Scholar]
- Ding X, Lin S-CS, Kiraly B, Yue H, Li S, Chiang I-K,…Huang TJ(2012).On-chipmanipulation of single microparticles, cells, and organisms using surface acoustic waves. Proceedings of the National Academy of Sciences of the United States of America, 109(28), 11105–11109. https://doi.org/10.1073/pnas.1209288109 . http://www.ncbi.nlm.nih.gov/pubmed/22733731 . 10.1073/pnas.1209288109http://www.ncbi.nlm.nih.gov/pubmed/22733731http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3396524. http://www.ncbi.nlm.nih.gov/pubmed/22733731 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3396524 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey P, & Wang Y-L (2005). Tissue cells feel and respond to the stiffness of their substrate. Science (New York, NY), 310(5751), 1139–1143. https://doi.org/10.1126/science.1116995 . http://www.sciencemag.org/cgi/doi/10.1126/science.1116995 . 10.1126/science.1116995http://www.sciencemag.org/cgi/doi/10.1126/science.1116995http://www.ncbi.nlm.nih.gov/pubmed/16293750. http://www.sciencemag.org/cgi/doi/10.1126/science.1116995 . http://www.ncbi.nlm.nih.gov/pubmed/16293750 . [DOI] [PubMed] [Google Scholar]
- Dobens LL, & Raftery LA (2000). Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Developmental Dynamics, 218(1), 80–93. https://doi.org/10.1002/(SICI)1097-0177(200005)218:1¡80::AID-DVDY7¿3.0.CO;2-8 . http://doi.wiley.com/10.1002/%28SICI%291097-0177%28200005%29218%3A1%3C80%3A%3AAID-DVDY7%3E3.0.CO%3B2-8. http://doi.wiley.com/10.1002/%28SICI%291097-0177%28200005%29218%3A1%3C80%3A%3AAID-DVDY7%3E3.0.CO%3B2-8 . [DOI] [PubMed] [Google Scholar]
- Doll JC, Harjee N, Klejwa N, Kwon R, Coulthard SM, Petzold B,…Pruitt BL (2009). SU-8 force sensing pillar arrays for biological measurements. Lab on a Chip, 9(10), 1449–1454. https://doi.org/10.1039/b818622g . 10.1039/b818622ghttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2818990&tool=pmcentrez&rendertype=abstract. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2818990&tool=pmcentrez&rendertype=abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley K,&Zon LI(2000).Zebrafish:Amodelsystemforthestudyofhumandisease. Current Opinion in Genetics & Development, 10(3), 252–256. https://doi.org/10.1016/S0959-437X(00)00074-5 . 10.1016/S0959-437X(00)00074-5https://www.sciencedirect.com/science/article/pii/S0959437X00000745. https://www.sciencedirect.com/science/article/pii/S0959437X00000745 . [DOI] [PubMed] [Google Scholar]
- Douville NJ, Zamankhan P, Tung Y-C, Li R, Vaughan BL, Tai C-F,… Takayama S (2011). Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab on a Chip, 11(4), 609–619. https://doi.org/10.1039/C0LC00251H . 10.1039/C0LC00251Hhttp://xlink.rsc.org/?DOI=C0LC00251H. http://xlink.rsc.org/?DOI=C0LC00251H . [DOI] [PubMed] [Google Scholar]
- Eastwood AL, Sanzeni A, Petzold BC, Park S-J, Vergassola M, Pruitt BL, & Goodman MB (2015). Tissue mechanics govern the rapidly adapting and symmetrical response to touch. Proceedings of the National Academy of Sciences, 15(50), E6955–E6963. 10.1073/pnas.1514138112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, & Horvitz HR (1986). Genetic control of programmed cell death in the nematode C. elegans. Cell, 44(6), 817–829. https://doi.org/10.1016/0092-8674(86)90004-8 . https://www.sciencedirect.com/science/article/pii/0092867486900048 . 10.1016/0092-8674(86)90004-8https://www.sciencedirect.com/science/article/pii/0092867486900048http://linkinghub.elsevier.com/retrieve/pii/0092867486900048. https://www.sciencedirect.com/science/article/pii/0092867486900048 . http://linkinghub.elsevier.com/retrieve/pii/0092867486900048 . [DOI] [PubMed] [Google Scholar]
- Elvira KS, i Solvas XC, Wootton RCR, & deMello AJ (2013). The past, present and potential for microfluidic reactor technology in chemical synthesis. Nature Chemistry, 5(11), 905–915. https://doi.org/10.1038/nchem.1753 . 10.1038/nchem.1753http://www.nature.com/articles/nchem.1753. http://www.nature.com/articles/nchem.1753 . [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, & Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell, 126(4), 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Feany MB, & Bender WW (2000). A Drosophila model of Parkinson’s disease. Nature, 404(6776), 394–398. https://doi.org/10.1038/35006074 . 10.1038/35006074http://www.nature.com/articles/35006074. http://www.nature.com/articles/35006074 . [DOI] [PubMed] [Google Scholar]
- Fehlauer H, Nekimken AL, Kim AA, Pruitt BL, Goodman MB, & Krieg M (2018). Using a microfluidics device for mechanical stimulation and high resolution imaging of C. elegans. Journal of Visualized Experiments, (132). 10.3791/56530, e56530–e56530, https://www.jove.com/video/56530/using-microfluidics-device-for-mechanical-stimulation-high-resolution. [DOI] [PMC free article] [PubMed]
- Fehrenbacher JC (2015). Chemotherapy-induced peripheral neuropathy Progress in molecular biology and translational science: Vol. 131, (pp. 471–508). Elsevier Inc; [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, & Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391(6669), 806–811. https://doi.org/10.1038/35888 . 10.1038/35888http://www.nature.com/articles/35888. http://www.nature.com/articles/35888 . [DOI] [PubMed] [Google Scholar]
- Fleming JE, Reveillaud I, & Niedzwiecki A (1992). Role of oxidative stress in Drosophila aging. Mutation Research/DNAging, 275(3–6), 267–279. https://doi.org/10.1016/0921-8734(92)90031-J . 10.1016/0921-8734(92)90031-Jhttps://www.sciencedirect.com/science/article/pii/092187349290031J. https://www.sciencedirect.com/science/article/pii/092187349290031J . [DOI] [PubMed] [Google Scholar]
- FlyAtlas (n.d.). http://flyatlas.org/.
- FlyBase (n.d.). http://flybase.org/.
- Frézal L, & Félix M-A (2015). C. elegans outside the Petri dish. eLife, 4, e05849 https://doi.org/10.7554/eLife.05849 . 10.7554/eLife.05849https://elifesciences.org/articles/05849. https://elifesciences.org/articles/05849 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Bailey TJ, Hyde DR, & Poss KD (2013). The zebrafish as a model for complex tissue regeneration. Trends in Genetics, 29(11), 611–620. 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemi R, Rezai P, Iyengar BG, & Selvaganapathy PR (2015). Microfluidic devices for imaging neurological response of Drosophila melanogaster larva to auditory stimulus. Lab on a Chip, 15(4), 1116–1122. https://doi.org/10.1039/C4LC01245C . 10.1039/C4LC01245Chttp://xlink.rsc.org/?DOI=C4LC01245C. http://xlink.rsc.org/?DOI=C4LC01245C . [DOI] [PubMed] [Google Scholar]
- Ghaemi R, Rezai P, Nejad FR, & Selvaganapathy PR (2017). Characterization of microfluidic clamps for immobilizing and imaging of Drosophila melanogaster larva’s central nervous system. Biomicrofluidics, 11(3), 034113 10.1063/1.4984767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannad-Rezaie M, Wang X, Mishra B, Collins C, & Chronis N (2012). Microfluidic chips for in vivo imaging of cellular responses to neural injury in Drosophila larvae. PLoS ONE, 7(1), e29869 10.1371/journal.pone.0029869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert Y, Trengove MC, & Ward AC (2013). Zebrafish as a genetic model in pre-clinical drug testing and screening. Current Medicinal Chemistry, 20(19), 2458–2466. https://doi.org/10.2174/0929867311320190005 . 10.2174/0929867311320190005http://www.ncbi.nlm.nih.gov/pubmed/23521675. http://www.ncbi.nlm.nih.gov/pubmed/23521675 . [DOI] [PubMed] [Google Scholar]
- Gomez GA, McLachlan RW, & Yap AS (2011). Productive tension: Force-sensing and homeostasis of cell-cell junctions. Trends in Cell Biology, 21(9), 499–505. https://doi.org/10.1016/j.tcb.2011.05.006 . 10.1016/j.tcb.2011.05.006http://www.ncbi.nlm.nih.gov/pubmed/21763139. http://www.ncbi.nlm.nih.gov/pubmed/21763139 . [DOI] [PubMed] [Google Scholar]
- Gonzales DL, Badhiwala KN, Vercosa DG, Avants BW, Liu Z, Zhong W, & Robinson JT (2017). Scalable electrophysiology in intact small animals with nanoscale suspended electrode arrays. Nature Nanotechnology, 12(7), 684–691. 10.1038/nnano.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M (2006). Mechanosensation. WormBook, 1–14. https://doi.org/10.1895/wormbook.1.62.1 . 10.1895/wormbook.1.62.1http://www.wormbook.org/chapters/www_mechanosensation/mechanosensation.html. http://www.wormbook.org/chapters/www_mechanosensation/mechanosensation.html . [DOI] [PMC free article] [PubMed]
- Gossett DR, Tse HTK, Lee SA, Ying Y, Lindgren AG, Yang OO,…Di Carlo D (2012). Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proceedings of the National Academy of Sciences, 109(20), 7630–7635. https://doi.org/10.1073/pnas.1200107109 . http://www.ncbi.nlm.nih.gov/pubmed/22547795 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3356639 . 10.1073/pnas.1200107109http://www.ncbi.nlm.nih.gov/pubmed/22547795http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3356639http://www.pnas.org/cgi/doi/10.1073/pnas.1200107109. http://www.ncbi.nlm.nih.gov/pubmed/22547795 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3356639 . http://www.pnas.org/cgi/doi/10.1073/pnas.1200107109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ (2004). Fly pushing: The theory and practice of Drosophila genetics. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M,…Bilby C (2005). Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophysical Journal, 88(5), 3689–3698. https://doi.org/10.1529/biophysj.104.045476 . https://www.sciencedirect.com/science/article/pii/S0006349505734172 . 10.1529/biophysj.104.045476https://www.sciencedirect.com/science/article/pii/S0006349505734172http://linkinghub.elsevier.com/retrieve/pii/S0006349505734172. https://www.sciencedirect.com/science/article/pii/S0006349505734172 . http://linkinghub.elsevier.com/retrieve/pii/S0006349505734172 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Park S, & Ma H (2012). Microfluidic micropipette aspiration for measuring the deformability of single cells. Lab on a Chip, 12(15), 2687 https://doi.org/10.1039/c2lc40205j . 10.1039/c2lc40205jhttp://xlink.rsc.org/?DOI=c2lc40205j. http://xlink.rsc.org/?DOI=c2lc40205j . [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, & Longaker MT (2008). Wound repair and regeneration. Nature, 453(7193), 314–321. 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Halldorsson S, Lucumi E, Gómez-Sjöberg R, & Fleming RMT (2015). Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosensors and Bioelectronics, 63, 218–231. https://doi.org/10.1016/J.BIOS.2014.07.029 . 10.1016/J.BIOS.2014.07.029https://www.sciencedirect.com/science/article/pii/S0956566314005302. https://www.sciencedirect.com/science/article/pii/S0956566314005302 . [DOI] [PubMed] [Google Scholar]
- Han B, Dong Y, Zhang L, Liu Y, Rabinowitch I, & Bai J (2017). Dopamine signaling tunes spatial pattern selectivity in C. elegans. eLife, 6(4), 431–436. https://doi.org/10.7554/eLife.22896 . http://journals.sagepub.com/doi/10.1177/2211068216669688 . 10.7554/eLife.22896http://journals.sagepub.com/doi/10.1177/2211068216669688https://elifesciences.org/articles/22896. http://journals.sagepub.com/doi/10.1177/2211068216669688 . https://elifesciences.org/articles/22896 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma P, Elings V, Marti O, & Bracker C (1988). Scanning tunneling microscopy and atomic force microscopy: Application to biology and technology. Science, 242(4876), 209–216. 10.1126/science.3051380. [DOI] [PubMed] [Google Scholar]
- Hayashi T, & Carthew RW (2004). Surface mechanics mediate pattern formation in the developing retina. Nature, 431(7009), 647–652. https://doi.org/10.1038/nature02952 . 10.1038/nature02952http://www.nature.com/doifinder/10.1038/nature02952. http://www.nature.com/doifinder/10.1038/nature02952 . [DOI] [PubMed] [Google Scholar]
- Haynie JL, & Bryant PJ (1986). Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. Journal of Experimental Zoology, 237(3), 293–308. 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- He L, Si G, Huang J, Samuel ADT, & Perrimon N (2018). Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature, 555, 103–106. https://doi.org/10.1038/nature25744 . 10.1038/nature25744http://www.nature.com/doifinder/10.1038/nature25744. http://www.nature.com/doifinder/10.1038/nature25744 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, & Jasper H (2014). Studying aging in Drosophila. Methods (San Diego, Calif.), 68(1), 129–133. https://doi.org/10.1016/j.ymeth.2014.04.008 . http://www.ncbi.nlm.nih.gov/pubmed/24751824 . 10.1016/j.ymeth.2014.04.008http://www.ncbi.nlm.nih.gov/pubmed/24751824http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4066732. http://www.ncbi.nlm.nih.gov/pubmed/24751824 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4066732 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiland JJ, Warias R, Lotter C, Mauritz L, Fuchs PJW, Ohla S,…Belder D (2017). On-chip integration of organic synthesis and HPLC/MS analysis for monitoring stereoselective transformations at the micro-scale. Lab on a Chip, 17(1), 76–81. https://doi.org/10.1039/C6LC01217E . 10.1039/C6LC01217Ehttp://xlink.rsc.org/?DOI=C6LC01217E. http://xlink.rsc.org/?DOI=C6LC01217E . [DOI] [PubMed] [Google Scholar]
- Heisenberg C-P, & Bellaïche Y (2013). Forces in tissue morphogenesis and patterning. Cell, 153(5), 948–962. https://doi.org/10.1016/J.CELL.2013.05.008 . 10.1016/J.CELL.2013.05.008https://www.sciencedirect.com/science/article/pii/S0092867413005734. https://www.sciencedirect.com/science/article/pii/S0092867413005734 . [DOI] [PubMed] [Google Scholar]
- Henry E, Holm SH, Zhang Z, Beech JP, Tegenfeldt JO, Fedosov DA, & Gompper G (2016). Sorting cells by their dynamical properties. Scientific Reports, 6(1), 34375 https://doi.org/10.1038/srep34375 . 10.1038/srep34375http://www.nature.com/articles/srep34375. http://www.nature.com/articles/srep34375 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, & Sedat JW (1993). The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. The Journal of Cell Biology, 120(3), 591–600. https://doi.org/10.1083/JCB.120.3.591 . http://www.ncbi.nlm.nih.gov/pubmed/8425892 . 10.1083/JCB.120.3.591http://www.ncbi.nlm.nih.gov/pubmed/8425892http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2119536. http://www.ncbi.nlm.nih.gov/pubmed/8425892 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2119536 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature, 496(7446), 498–503. https://doi.org/10.1038/nature12111 . 10.1038/nature12111http://www.nature.com/nature/journal/vaop/ncurrent/full/nature12111.html?utm_source=feedly. http://www.nature.com/nature/journal/vaop/ncurrent/full/nature12111.html?utm_source=feedly . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, & Mohr SE (2011). An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics, 12, 357 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, & Ingber DE (2005). Cell tension, matrix mechanics, and cancer development. Cancer Cell, 8(3), 175–176. 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, & Whitesides GM (2007). A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab on a Chip, 7(11), 1515–1523. https://doi.org/10.1039/b707861g . 10.1039/b707861ghttp://www.ncbi.nlm.nih.gov/pubmed/17960280. http://www.ncbi.nlm.nih.gov/pubmed/17960280 . [DOI] [PubMed] [Google Scholar]
- Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, & Whitesides GM (2010). Lifespan-on-a-chip: Microfluidic chambers for performing lifelong observation of C. elegans. Lab on a Chip, 10(5), 589–597. https://doi.org/10.1039/B919265D . 10.1039/B919265Dhttp://xlink.rsc.org/?DOI=B919265D. http://xlink.rsc.org/?DOI=B919265D . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L-H, Choi KM, Tseng W-Y, Tan Y-C, Shea KJ, & Lee AP (2006). Alternating droplet generation and controlled dynamic droplet fusion in microfluidic device for CdS nanoparticle synthesis. Lab on a Chip, 6(2), 174 https://doi.org/10.1039/b513908b . http://www.ncbi.nlm.nih.gov/pubmed/16450024 . 10.1039/b513908bhttp://www.ncbi.nlm.nih.gov/pubmed/16450024http://xlink.rsc.org/?DOI=b513908b. http://www.ncbi.nlm.nih.gov/pubmed/16450024 . http://xlink.rsc.org/?DOI=b513908b . [DOI] [PubMed] [Google Scholar]
- Hwang H, & Lu H (2013). Microfluidic tools for developmental studies of small model organisms -nematodes, fruit flies, and zebrafish. Biotechnology Journal, 8(2), 192–205. 10.1002/biot.201200129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D (2003). Mechanobiology and diseases of mechanotransduction. Annals of Medicine, 35(8), 564–577. https://doi.org/10.1080/07853890310016333 . 10.1080/07853890310016333http://www.tandfonline.com/doi/full/10.1080/07853890310016333. http://www.tandfonline.com/doi/full/10.1080/07853890310016333 . [DOI] [PubMed] [Google Scholar]
- Ireland RG, & Simmons CA (2015). Human pluripotent stem cell mechanobiology: Manipulating the biophysical microenvironment for regenerative medicine and tissue engineering applications. STEM CELLS, 33(11), 3187–3196. 10.1002/stem.2105. [DOI] [PubMed] [Google Scholar]
- Jaalouk DE, & Lammerding J (2009). Mechanotransduction gone awry. Nature Reviews Molecular Cell Biology, 10(1), 63–73. http://www.nature.com/doifinder/10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, & Miller RT (2011). Mechanisms of mechanical signaling in development and disease. Journal of Cell Science, 124(Pt. 1), 9–18. https://doi.org/10.1242/jcs.071001 . http://www.ncbi.nlm.nih.gov/pubmed/21172819 . 10.1242/jcs.071001http://www.ncbi.nlm.nih.gov/pubmed/21172819http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3001405. http://www.ncbi.nlm.nih.gov/pubmed/21172819 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3001405 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G (2013). Rapid prototyping of PDMS devices using SU-8 lithography In Jenkins G & Mansfield C (Eds.), Microfluidic diagnostics. methods in molecular biology (methods and protocols) (pp. 153–168). Totowa, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Jennings BH (2011). Drosophila A versatile model in biology & medicine. Materials Today, 14(5), 190–195. https://doi.org/10.1016/S1369-7021(11)70113-4 . 10.1016/S1369-7021(11)70113-4https://www.sciencedirect.com/science/article/pii/S1369702111701134. https://www.sciencedirect.com/science/article/pii/S1369702111701134 . [DOI] [Google Scholar]
- Jiang H, & Edgar BA (2011). Intestinal stem cells in the adult Drosophila midgut. Experimental Cell Research, 317(19), 2780–2788. https://doi.org/10.1016/J.YEXCR.2011.07.020 . 10.1016/J.YEXCR.2011.07.020https://www.sciencedirect.com/science/article/pii/S0014482711003065. https://www.sciencedirect.com/science/article/pii/S0014482711003065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johari S, Nock V, Alkaisi MM, & Wang W (2013). On-chip analysis of C. elegans muscular forces and locomotion patterns in microstructured environments. Lab on a Chip, 13(9), 1699–1707. https://doi.org/10.1039/C3LC41403E . http://pubs.rsc.org/en/content/articlelanding/2013/lc/c3lc41403e%5Cn . http://pubs.rsc.org/en/Content/ArticleLanding/2013/LC/c3lc41403e%5Cn . http://pubs.rsc.org/en/content/articlepdf/2013/lc/c3lc41403e . 10.1039/C3LC41403Ehttp://pubs.rsc.org/en/content/articlelanding/2013/lc/c3lc41403e%5Cnhttp://pubs.rsc.org/en/Content/ArticleLanding/2013/LC/c3lc41403e%5Cnhttp://pubs.rsc.org/en/content/articlepdf/2013/lc/c3lc41403ehttp://xlink.rsc.org/?DOI=c3lc41403e. http://pubs.rsc.org/en/content/articlelanding/2013/lc/c3lc41403e%5Cn . http://pubs.rsc.org/en/Content/ArticleLanding/2013/LC/c3lc41403e%5Cn . http://pubs.rsc.org/en/content/articlepdf/2013/lc/c3lc41403e . http://xlink.rsc.org/?DOI=c3lc41403e . [DOI] [PubMed] [Google Scholar]
- Kaletta T, & Hengartner MO (2006). Finding function in novel targets: C. elegans as a model organism. Nature Reviews Drug Discovery, 5(5), 387–399. https://doi.org/10.1038/nrd2031 . 10.1038/nrd2031http://www.nature.com/articles/nrd2031. http://www.nature.com/articles/nrd2031 . [DOI] [PubMed] [Google Scholar]
- Keil W, Kutscher LM, Shaham S, & Siggia ED (2017). Long-term high-resolution imaging of developing C. elegans larvae with microfluidics. Developmental Cell, 40(2), 202–214. https://doi.org/10.1016/j.devcel.2016.11.022 . 10.1016/j.devcel.2016.11.022http://linkinghub.elsevier.com/retrieve/pii/S1534580716308358. http://linkinghub.elsevier.com/retrieve/pii/S1534580716308358 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Choi YJ, Hwang J-H, Kim AR, Cho HJ, Hwang ES,…Hong J-H (2014). Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS ONE, 9(3), e92427 10.1371/journal.pone.0092427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Kang JH, Park S-J, Yoon E-S, & Park J-K (2007). Microfluidic biomechanical device for compressive cell stimulation and lysis. Sensors and Actuators B: Chemical, 128(1), 108–116. https://doi.org/10.1016/J.SNB.2007.05.050 . 10.1016/J.SNB.2007.05.050https://www.sciencedirect.com/science/article/pii/S0925400507003899. https://www.sciencedirect.com/science/article/pii/S0925400507003899 . [DOI] [Google Scholar]
- Kimmel CB, Kimmel CB, Ballard WW, Ballard WW, Kimmel SR, Kimmel SR, …Schilling TF (1995). Stages of embryonic development of the zebrafish. Developmental Dynamics, 203(3), 253–310. https://doi.org/10.1002/aja.1002030302 . 10.1002/aja.1002030302http://www.ncbi.nlm.nih.gov/pubmed/8589427. http://www.ncbi.nlm.nih.gov/pubmed/8589427 . [DOI] [PubMed] [Google Scholar]
- Kniss JS, Jiang L, & Piotrowski T (2016). Insights into sensory hair cell regeneration from the zebrafish lateral line. Current Opinion in Genetics and Development, 40(10), 32–40. 10.1016/j.gde.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Kok FO,Shin M,Ni C-W,Gupta A,Grosse AS,vanImpel A,…Lawson ND(2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental Cell, 32(1), 97–108. https://doi.org/10.1016/j.devcel.2014.11.018 . https://www.sciencedirect.com/science/article/pii/S1534580714007357 . 10.1016/j.devcel.2014.11.018https://www.sciencedirect.com/science/article/pii/S1534580714007357http://linkinghub.elsevier.com/retrieve/pii/S1534580714007357. https://www.sciencedirect.com/science/article/pii/S1534580714007357 . http://linkinghub.elsevier.com/retrieve/pii/S1534580714007357 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Singh K, O’Neill A, Jackson C, Morrison A, & O’Brein P (2002). Development of a microfluidic devicefor fluorescence activated cell sorting. Journal of Micromechanics and Microengineering, 12(4), 486–494. https://doi.org/10.1088/0960-1317/12/4/324 . 10.1088/0960-1317/12/4/324http://stacks.iop.org/0960-1317/12/i=4/a=324?key=crossref.8427c40b462b65fc7188ca531b4dddff. http://stacks.iop.org/0960-1317/12/i=4/a=324?key=crossref.8427c40b462b65fc7188ca531b4dddff . [DOI] [Google Scholar]
- Kurth F, Eyer K, & Franco-Obregón A (2012). A new mechanobiological era: Microfluidic pathways to apply and sense forces at the cellular level. Current Opinion in Chemical Biology, 16(3–4), 400–408. https://doi.org/10.1016/J.CBPA.2012.03.014 . 10.1016/J.CBPA.2012.03.014https://www.sciencedirect.com/science/article/pii/S136759311200049X. https://www.sciencedirect.com/science/article/pii/S136759311200049X . [DOI] [PubMed] [Google Scholar]
- Kwon H-J, Xu Y, Solovitz SA, Xue W, Dimitrov AG, Coffin AB, & Xu J (2014). Design of a microfluidic device with a non-traditional flow profile for on-chip damage to zebrafish sensory cells. Journal of Micromechanics and Microengineering, 24(1), 017001 https://doi.org/10.1088/0960-1317/24/1/017001 . 10.1088/0960-1317/24/1/017001http://stacks.iop.org/0960-1317/24/i=1/a=017001?key=crossref.898bd18771a566d89de4176cbca5ce71. http://stacks.iop.org/0960-1317/24/i=1/a=017001?key=crossref.898bd18771a566d89de4176cbca5ce71 . [DOI] [Google Scholar]
- Lammer E,Carr GJ,Wendler K,Rawlings JM,Belanger SE,&Braunbeck Th(2009). Is the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) a potential alternative for the fish acute toxicity test? Comparative Biochemistry and Physiology - C Toxicology and Pharmacology, 149(2), 196–209. 10.1016/j.cbpc.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Lammer E, Kamp HG, Hisgen V, Koch M, Reinhard D, Salinas ER,… Braunbeck T. h. (2009). Development of a flow-through system for the fish embryo toxicity test (FET) with the zebrafish (Danio rerio). Toxicology In Vitro, 23(7), 1436–1442. 10.1016/j.tiv.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Laser DJ, &Santiago JG(2004).A review of micropumps. Journal of Micromechanics and Microengineering, 14(6), R35–R64. https://doi.org/10.1088/0960-1317/14/6/R01 . 10.1088/0960-1317/14/6/R01http://stacks.iop.org/0960-1317/14/i=6/a=R01?key=crossref.137857bfcf5ea513064ec576b484af8f. http://stacks.iop.org/0960-1317/14/i=6/a=R01?key=crossref.137857bfcf5ea513064ec576b484af8f . [DOI] [Google Scholar]
- Lee DA, Knight MM, Campbell JJ, & Bader DL (2011). Stem cell mechanobiology. Journal of Cellular Biochemistry, 112(1), 1–9. 10.1002/jcb.22758. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Chan JC-Y, Maung KJ, Rezler E, & Sundararajan N (2007). Characterization of laterallydeformable elastomer membranes for microfluidics. Journal of Micromechanics and Microengineering, 17, 843–851. 10.1088/0960-1317/17/5/001. [DOI] [Google Scholar]
- Lengyel JA, & Iwaki D (2002). It takes guts: The Drosophila hindgut as a model system for organogenesis. Developmental Biology, 243(1), 1–19. https://doi.org/10.1006/DBIO.2002.0577 . 10.1006/DBIO.2002.0577https://www.sciencedirect.com/science/article/pii/S0012160602905774. https://www.sciencedirect.com/science/article/pii/S0012160602905774 . [DOI] [PubMed] [Google Scholar]
- Levario TJ, Zhan M, Lim B, Shvartsman SY, & Lu H (2013). Microfluidic trap array for massively parallel imaging of Drosophila embryos. Nature Protocols, 8(4), 721–736. https://doi.org/10.1038/nprot.2013.034 . 10.1038/nprot.2013.034http://www.nature.com/doifinder/10.1038/nprot.2013.034. http://www.nature.com/doifinder/10.1038/nprot.2013.034 . [DOI] [PubMed] [Google Scholar]
- Li Y, Yang F, Chen Z, Shi L, Zhang B, Pan J,…Yang H (2014). Zebrafish on a chip: A novel platform for real-time monitoring of drug-induced developmental toxicity. PLoS ONE, 9(4), e94792 10.1371/journal.pone.0094792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Balachandra S, Ngo S, & O’Brien LE (2017). Feedback regulation of steady-state epithelial turnover and organ size. Nature, 548(7669), 588–591. https://doi.org/10.1038/nature23678 . 10.1038/nature23678http://www.nature.com/doifinder/10.1038/nature23678. http://www.nature.com/doifinder/10.1038/nature23678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, & Currie PD (2007). Animal models of human disease: Zebrafish swim into view. Nature Reviews Genetics, 8(5), 353–367. 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lignos I, Stavrakis S, Nedelcu G, Protesescu L, deMello AJ, & Kovalenko MV (2016). Synthesis of cesium lead halide perovskite nanocrystals in a droplet-based microfluidic platform: Fast parametric space mapping. Nano Letters, 16(3), 1869–1877. https://doi.org/10.1021/acs.nanolett.5b04981 . 10.1021/acs.nanolett.5b04981http://pubs.acs.org/doi/10.1021/acs.nanolett.5b04981. http://pubs.acs.org/doi/10.1021/acs.nanolett.5b04981 . [DOI] [PubMed] [Google Scholar]
- Lo C-M, Wang H-B, Dembo M, & Wang Y. l. (2000). Cell movement is guided by the rigidity of the substrate. Biophysical Journal, 79(1), 144–152. https://doi.org/10.1016/S0006-3495(00)76279-5 . 10.1016/S0006-3495(00)76279-5http://linkinghub.elsevier.com/retrieve/pii/S0006349500762795. http://linkinghub.elsevier.com/retrieve/pii/S0006349500762795 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockery SR, Lawton KJ, Doll JC, Faumont S, Coulthard SM, Thiele TR,… Pruitt BL (2008). Artificial dirt: Microfluidic substrates for nematode neurobiology and behavior. Journal of Neurophysiology, 99(6), 3136–3143. 10.1152/jn.91327.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, & Ismagilov RF (2005). Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature, 434(7037), 1134–1138. https://doi.org/10.1038/nature03509 . 10.1038/nature03509http://www.nature.com/articles/nature03509. http://www.nature.com/articles/nature03509 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Munson MS, & Ismagilov RF (2006). Characterization of the local temperature in space and time around a developing Drosophila embryo in a microfluidic device. Lab on a Chip, 6(2), 185 https://doi.org/10.1039/b516119c . 10.1039/b516119chttp://xlink.rsc.org/?DOI=b516119c. http://xlink.rsc.org/?DOI=b516119c . [DOI] [PubMed] [Google Scholar]
- Marti O, Ribi HO, Drake B, Albrecht TR, Quate CF, & Hansma PK (1988). Atomic force microscopy of an organic monolayer. Science (New York, NY), 239(4835), 50–52. https://doi.org/10.1126/SCIENCE.3336773 . 10.1126/SCIENCE.3336773http://www.ncbi.nlm.nih.gov/pubmed/3336773. http://www.ncbi.nlm.nih.gov/pubmed/3336773 . [DOI] [PubMed] [Google Scholar]
- Mazzochette EA, Nekimken AL, Loizeau F, Whitworth J, Huynh B, Goodman MB, & Pruitt BLL (2018). The tactile receptive field of freely moving Caenorhabditis elegans nematodes. Integrative Biology. 10.1039/C8IB00045J (accepted for publication). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan PD, Xu JH, & Fang-Yen C (2017). Comparing Caenorhabditis elegans gentle and harsh touch response behavior using a multiplexed hydraulic microfluidic device. Integrative Biology, 9(10), 800–809. https://doi.org/10.1039/C7IB00120G . 10.1039/C7IB00120Ghttp://xlink.rsc.org/?DOI=C7IB00120G. http://xlink.rsc.org/?DOI=C7IB00120G . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, & Perrimon N (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature, 439(7075), 475–479. https://doi.org/10.1038/nature04371 . 10.1038/nature04371http://www.nature.com/articles/nature04371. http://www.nature.com/articles/nature04371 . [DOI] [PubMed] [Google Scholar]
- Mirasoli M, Guardigli M, Michelini E, & Roda A (2014). Recent advancements in chemical luminescence-based lab-on-chip and microfluidic platforms for bioanalysis. Journal of Pharmaceutical and Biomedical Analysis, 87, 36–52. https://doi.org/10.1016/J.JPBA.2013.07.008 . 10.1016/J.JPBA.2013.07.008https://www.sciencedirect.com/science/article/pii/S073170851300304X. https://www.sciencedirect.com/science/article/pii/S073170851300304X . [DOI] [PubMed] [Google Scholar]
- Moeendarbary E, & Harris AR (2014). Cell mechanics: Principles, practices, and prospects. Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 6(5), 371–388. https://doi.org/10.1002/wsbm.1275 . 10.1002/wsbm.1275http://doi.wiley.com/10.1002/wsbm.1275. http://doi.wiley.com/10.1002/wsbm.1275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno EL, Hachi S, Hemmer K, Trietsch SJ, Baumuratov AS, Hankemeier T,… Fleming RMT (2015). Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab on a Chip, 15(11), 2419–2428. https://doi.org/10.1039/C5LC00180C . 10.1039/C5LC00180Chttp://xlink.rsc.org/?DOI=C5LC00180C. http://xlink.rsc.org/?DOI=C5LC00180C . [DOI] [PubMed] [Google Scholar]
- Morgan PG, Kayser E-B, & Sedensky MM (2007). C. elegans and volatile anesthetics. WormBook, 1–13. https://doi.org/10.1895/wormbook.1.140.1 . 10.1895/wormbook.1.140.1http://www.wormbook.org/chapters/www_anesthetics/anesthetics.html. http://www.wormbook.org/chapters/www_anesthetics/anesthetics.html . [DOI] [PMC free article] [PubMed]
- Narciso CE, Contento NM, Storey TJ, Hoelzle DJ, & Zartman JJ (2017). Release of applied mechanical loading stimulates intercellular calcium waves in Drosophila wing discs. Biophysical Journal, 113(2), 491–501. https://doi.org/10.1016/j.bpj.2017.05.051 . 10.1016/j.bpj.2017.05.051http://www.sciencedirect.com/science/article/pii/S0006349517306264. http://www.sciencedirect.com/science/article/pii/S0006349517306264 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekimken AL, Fehlauer H, Kim AA, Manosalvas-Kjono SN, Ladpli P, Memon F, …Krieg M (2017). Pneumatic stimulation of C. elegans mechanoreceptor neurons in a microfluidic trap. Lab on a Chip, 17(6), 1116–1127. https://doi.org/10.1039/C6LC01165A . 10.1039/C6LC01165Ahttp://xlink.rsc.org/?DOI=C6LC01165A. http://xlink.rsc.org/?DOI=C6LC01165A . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekimken AL, Mazzochette EA, Goodman MB, & Pruitt BL (2017). Forces applied during classical touch assays for Caenorhabditis elegans. PLOS ONE, 12(5), e0178080 10.1371/journal.pone.0178080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, & Nagy A (2008). Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nature Methods, 5(6), 491–505. 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CD (2006). Drosophila melanogaster neurobiology, neuropharmacology, and how the fly can inform central nervous system drug discovery. Pharmacology & Therapeutics, 112(3), 677–700. https://doi.org/10.1016/J.PHARMTHERA.2006.05.012 . 10.1016/J.PHARMTHERA.2006.05.012https://www.sciencedirect.com/science/article/pii/S0163725806001124. https://www.sciencedirect.com/science/article/pii/S0163725806001124 . [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, & Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature, 287(5785), 795–801. https://doi.org/10.1038/287795a0 . 10.1038/287795a0http://www.nature.com/doifinder/10.1038/287795a0. http://www.nature.com/doifinder/10.1038/287795a0 . [DOI] [PubMed] [Google Scholar]
- Ogawa R, & Huang C (2016). Mechanobiology and mechanotherapy for cutaneous scarring In Rawlinson SCF (Ed.), Mechanobiology (pp. 255–265). Hoboken, NJ: John Wiley & Sons, Inc; 10.1002/9781118966174.ch16. [DOI] [Google Scholar]
- Ohlstein B, & Spradling A (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature, 439(7075), 470–474. https://doi.org/10.1038/nature04333 . 10.1038/nature04333http://www.nature.com/articles/nature04333. http://www.nature.com/articles/nature04333 . [DOI] [PubMed] [Google Scholar]
- Paluch EK, Nelson CM, Biais N, Fabry B, Moeller J, Pruitt BL,…Federle W (2015). Mechanotransduction: Use the force(s). BMC Biology, 13, 47 https://doi.org/10.1186/s12915-015-0150-4 . 10.1186/s12915-015-0150-4http://www.biomedcentral.com/1741-7007/13/47. http://www.biomedcentral.com/1741-7007/13/47 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, & Nichols CD (2011). Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacological Reviews, 63(2), 411–436. https://doi.org/10.1124/pr.110.003293 . http://www.ncbi.nlm.nih.gov/pubmed/21415126 . 10.1124/pr.110.003293http://www.ncbi.nlm.nih.gov/pubmed/21415126http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3082451. http://www.ncbi.nlm.nih.gov/pubmed/21415126 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3082451 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, & Engeszer RE (2009). Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Developmental Dynamics, 238(12), 2975–3015. 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Hwang H, Nam S-W, Martinez F, Austin RH, & Ryu WS (2008). Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment. PLoS ONE, 3(6), e2550 10.1371/journal.pone.0002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Goodman MB, & Pruitt BL (2007). Analysis of nematode mechanics by piezo-resistive displacement clamp. Proceedings of the National Academy of Sciences, 104(44), 17376–17381. 10.1073/pnas.0702138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Petzold BC, Goodman MB, & Pruitt BL (2011). Piezoresistive cantilever force-clamp system. The Review of Scientific Instruments, 82(4), 43703 https://doi.org/10.1063/1.3574362 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3091308&tool=pmcentrez&rendertype=abstract . 10.1063/1.3574362http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3091308&tool=pmcentrez&rendertype=abstracthttp://www.ncbi.nlm.nih.gov/pubmed/25006998. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3091308&tool=pmcentrez&rendertype=abstract . http://www.ncbi.nlm.nih.gov/pubmed/25006998 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parng C, Seng WL, Semino C, & McGrath P (2002). Zebrafish: A preclinical model for drug screening. ASSAY and Drug Development Technologies, 1(1), 41–48. https://doi.org/10.1089/154065802761001293 . 10.1089/154065802761001293http://www.liebertonline.com/doi/abs/10.1089/154065802761001293. http://www.liebertonline.com/doi/abs/10.1089/154065802761001293 . [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A,… Weaver VM (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell, 8(3), 241–254. https://doi.org/10.1016/j.ccr.2005.08.010 . 10.1016/j.ccr.2005.08.010http://linkinghub.elsevier.com/retrieve/pii/S1535610805002680. http://linkinghub.elsevier.com/retrieve/pii/S1535610805002680 . [DOI] [PubMed] [Google Scholar]
- Patabadige DEW, Jia S, Sibbitts J, Sadeghi J, Sellens K, & Culbertson CT (2016). Micro total analysis systems: Fundamental advances and applications. Analytical Chemistry, 88(1), 320–338. https://doi.org/10.1021/acs.analchem.5b04310 . 10.1021/acs.analchem.5b04310http://pubs.acs.org/doi/10.1021/acs.analchem.5b04310. http://pubs.acs.org/doi/10.1021/acs.analchem.5b04310 . [DOI] [PubMed] [Google Scholar]
- Pelham RJ, & Wang Y-L (1997). Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences, 94(25), 13661–13665. 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold BC, Park S-J, Mazzochette EA, Goodman MB, & Pruitt BL (2013). MEMS-based force-clamp analysis of the role of body stiffness in C. elegans touch sensation. Integrative Biology, 5(6), 853–864. https://doi.org/10.1039/c3ib20293c . 10.1039/c3ib20293chttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3701114&tool=pmcentrez&rendertype=abstract. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3701114&tool=pmcentrez&rendertype=abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold BC, Park S-J, Ponce P, Roozeboom C, Powell C, Goodman MB, & Pruitt BL (2011). Caenorhabditis elegans body mechanics are regulated by body wall muscle tone. Biophysical Journal, 100(8), 1977–1985. 10.1016/j.bpj.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placet V, & Delobelle P (2015). Mechanical properties of bulk polydimethylsiloxane for microfluidics over a large range of frequencies and aging times. Journal of Micromechanics and Microengineering, 25(3), 035009 https://doi.org/10.1088/0960-1317/25/3/035009 . 10.1088/0960-1317/25/3/035009http://stacks.iop.org/0960-1317/25/i=3/a=035009?key=crossref.00d31edba197a446ede889fb24eb269f. http://stacks.iop.org/0960-1317/25/i=3/a=035009?key=crossref.00d31edba197a446ede889fb24eb269f . [DOI] [Google Scholar]
- Polacheck WJ, Li R, Uzel SGM, & Kamm RD (2013). Microfluidic platforms for mechanobiology. Lab on a Chip, 13(12), 2252–2267. 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto DA, Rouse TM, San-Miguel A, & Lu H (2016). Microfluidic platforms for quantitative biology studies in model organisms Microfluidic methods for molecular biology (pp. 1–18). Cham: Springer International Publishing; 10.1007/978-3-31930019-1_1. [DOI] [Google Scholar]
- Rhee SW, Taylor AM, Tu CH, Cribbs DH, Cotman CW, & Jeon NL (2005). Patterned cell culture inside microfluidic devices. Lab on a Chip, 5(1), 102 https://doi.org/10.1039/b403091e . 10.1039/b403091ehttp://xlink.rsc.org/?DOI=b403091e. http://xlink.rsc.org/?DOI=b403091e . [DOI] [PubMed] [Google Scholar]
- San-Miguel A, & Lu H (2013). Microfluidics as a tool for C. elegans research. WormBook, 1–19. https://doi.org/10.1895/wormbook.1.162.1 . 10.1895/wormbook.1.162.1http://www.wormbook.org/chapters/www_microfluidics/microfluidics.html. http://www.wormbook.org/chapters/www_microfluidics/microfluidics.html . [DOI] [PMC free article] [PubMed]
- Schoen I, Pruitt BL, & Vogel V (2013). The Yin-Yang of rigidity sensing: How forces and mechanical properties pegulate the cellular response to materials. Annual Review of Materials Research, 43(1), 589–618. 10.1146/annurev-matsci-062910-100407. [DOI] [Google Scholar]
- Shao J, Wu L, Wu J, Zheng Y, Zhao H, Jin Q, & Zhao J (2009). Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab on a Chip, 9(21), 3118 https://doi.org/10.1039/b909312e . 10.1039/b909312ehttp://xlink.rsc.org/?DOI=b909312e. http://xlink.rsc.org/?DOI=b909312e . [DOI] [PubMed] [Google Scholar]
- Siedlik MJ, Varner VD, & Nelson CM (2016). Pushing, pulling, and squeezing our way to understanding mechanotransduction. Methods, 94, 4–12. 10.1016/j.ymeth.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kishore L, & Kaur N (2014). Diabetic peripheral neuropathy: Current perspective and future directions. Pharmacological Research, 80, 21–35. 10.1016/j.phrs.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Sivagnanam V, & Gijs MAM (2013). Exploring living multicellular organisms, organs, and tissues using microfluidic systems. Chemical Reviews, 113(5), 3214–3247. https://doi.org/10.1021/cr200432q . http://pubs.acs.org/doi/abs/10.1021/cr200432q . 10.1021/cr200432qhttp://pubs.acs.org/doi/abs/10.1021/cr200432qhttp://pubs.acs.org/doi/10.1021/cr200432q. http://pubs.acs.org/doi/abs/10.1021/cr200432q . http://pubs.acs.org/doi/10.1021/cr200432q . [DOI] [PubMed] [Google Scholar]
- Sofela S, Sahloul S, Rafeie M, Kwon T, Han J, Warkiani ME, & Song Y-A (2018). High-throughput sorting of eggs for synchronization of C. elegans in a microfluidic spiral chip. Lab on a Chip, 18(4), 679–687. https://doi.org/10.1039/C7LC00998D . 10.1039/C7LC00998Dhttp://xlink.rsc.org/?DOI=C7LC00998D. http://xlink.rsc.org/?DOI=C7LC00998D . [DOI] [PubMed] [Google Scholar]
- Sonnenfeld R, Hansma PK, Drake B, Albrecht TR, Quate CF, & Hansma PK (1986). Atomic-resolution microscopy in water. Science (New York, NY), 232(4747), 211–213. https://doi.org/10.1126/science.232.4747.211 . 10.1126/science.232.4747.211http://www.ncbi.nlm.nih.gov/pubmed/17780805. http://www.ncbi.nlm.nih.gov/pubmed/17780805 . [DOI] [PubMed] [Google Scholar]
- Spill F, Reynolds DS, Kamm RD, & Zaman MH (2016). Impact of the physical microenvironment on tumor progression and metastasis. Current Opinion in Biotechnology, 40, 41–48. https://doi.org/10.1016/J.COPBIO.2016.02.007 . 10.1016/J.COPBIO.2016.02.007https://www.sciencedirect.com/science/article/pii/S0958166916300301. https://www.sciencedirect.com/science/article/pii/S0958166916300301 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strähle U, Scholz S, Geisler R, Greiner P, Hollert H, Rastegar S,…Braunbeck T (2012). Zebrafish embryos as an alternative to animal experiments—A commentary on the definition of the onset of protected life stages in animal welfare regulations. Reproductive Toxicology, 33(2), 128–132. 10.1016/j.reprotox.2011.06.121. [DOI] [PubMed] [Google Scholar]
- Thompson DW (1942). On growth and form. Cambridge Univ. Press. [Google Scholar]
- Ugur B, Chen K, & Bellen HJ (2016). Drosophila tools and assays for the study of human diseases. Disease Models & Mechanisms, 9(3), 235–244. https://doi.org/10.1242/dmm.023762 . http://www.ncbi.nlm.nih.gov/pubmed/26935102 . 10.1242/dmm.023762http://www.ncbi.nlm.nih.gov/pubmed/26935102http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4833332. http://www.ncbi.nlm.nih.gov/pubmed/26935102 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4833332 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung WL, Mutafopulos K, Spink P, Rambach RW, Franke T, & Weitz DA (2017). Enhanced surface acoustic wave cell sorting by 3D microfluidic-chip design. Lab on a Chip, 17(23), 4059–4069. https://doi.org/10.1039/C7LC00715A . 10.1039/C7LC00715Ahttp://xlink.rsc.org/?DOI=C7LC00715A. http://xlink.rsc.org/?DOI=C7LC00715A . [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, & Bitter W (2004). A star with stripes: Zebrafish as an infection model. Trends in Microbiology, 12(10), 451–457. https://doi.org/10.1016/J.TIM.2004.08.001 . 10.1016/J.TIM.2004.08.001https://www.sciencedirect.com/science/article/pii/S0966842X04001805. https://www.sciencedirect.com/science/article/pii/S0966842X04001805 . [DOI] [PubMed] [Google Scholar]
- Vining KH, & Mooney DJ (2017). Mechanical forces direct stem cell behaviour in development and regeneration. Nature Reviews Molecular Cell Biology, 18(12), 728–742. https://doi.org/10.1038/nrm.2017.108 . 10.1038/nrm.2017.108http://www.nature.com/doifinder/10.1038/nrm.2017.108. http://www.nature.com/doifinder/10.1038/nrm.2017.108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M, Butler D, Haj AE, Bodle JC, Loboa EG, & Banes AJ (2017). Key developments that impacted the field of mechanobiology and mechanotransduction. Journal of Orthopaedic Research, 36, 20–22. 10.1002/jor.23707. [DOI] [PubMed] [Google Scholar]
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P,…Hartl FU (2015). Widespread proteome remodeling and aggregation in aging C. elegans. Cell, 161(4), 919–932. https://doi.org/10.1016/j.cell.2015.03.032 . https://www.sciencedirect.com/science/article/pii/S0092867415003207 . 10.1016/j.cell.2015.03.032https://www.sciencedirect.com/science/article/pii/S0092867415003207http://linkinghub.elsevier.com/retrieve/pii/S0092867415003207. https://www.sciencedirect.com/science/article/pii/S0092867415003207 . http://linkinghub.elsevier.com/retrieve/pii/S0092867415003207 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JHC, & Thampatty BP (2006). An introductory review of cell mechanobiology. Biomechanics and Modeling in Mechanobiology, 5(1), 1–16. 10.1007/s10237-005-0012-z. [DOI] [PubMed] [Google Scholar]
- Wang Z, Volinsky AA, & Gallant ND (2014). Crosslinking effect on polydimethylsiloxane elastic modulus measured by custom-built compression instrument. Journal of Applied Polymer Science, 131(22), 41050 https://doi.org/10.1002/app.41050 . 10.1002/app.41050http://doi.wiley.com/10.1002/app.41050. http://doi.wiley.com/10.1002/app.41050 . [DOI] [Google Scholar]
- Warkiani ME, Wu L, Tay AKP, & Han J (2015). Large-volume microfluidic cell sorting for biomedical applications. Annual Review of Biomedical Engineering, 17(1), 1–34. https://doi.org/10.1146/annurev-bioeng-071114-040818 . 10.1146/annurev-bioeng-071114-040818http://www.annualreviews.org/doi/10.1146/annurev-bioeng-071114-040818. http://www.annualreviews.org/doi/10.1146/annurev-bioeng-071114-040818 . [DOI] [PubMed] [Google Scholar]
- Watanabe K, Chiu H, Pfeiffer BD, Wong AM, Hoopfer ED, Rubin GM, & Anderson DJ (2017). A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron, 95(5), 1112–1128. https://doi.org/10.1016/j.neuron.2017.08.017 . http://www.ncbi.nlm.nih.gov/pubmed/28858617 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5588916 . 10.1016/j.neuron.2017.08.017http://www.ncbi.nlm.nih.gov/pubmed/28858617http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5588916http://linkinghub.elsevier.com/retrieve/pii/S0896627317307328. http://www.ncbi.nlm.nih.gov/pubmed/28858617 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5588916 . http://linkinghub.elsevier.com/retrieve/pii/S0896627317307328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE,…Yang J (2015). Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nature Cell Biology, 17(5), 678–688. https://doi.org/10.1038/ncb3157 . 10.1038/ncb3157http://www.nature.com/articles/ncb3157. http://www.nature.com/articles/ncb3157 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Po MD, Hulme E, Chen S, Liu X, Kwok SW,…Samuel AD (2012). Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron, 76(4), 750–761. https://doi.org/10.1016/j.neuron.2012.08.039 . 10.1016/j.neuron.2012.08.039http://linkinghub.elsevier.com/retrieve/pii/S0896627312008057. http://linkinghub.elsevier.com/retrieve/pii/S0896627312008057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2000). The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Univ. of Oregon Press. [Google Scholar]
- Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH,… Daridon A (2003). Microfluidic device for single-cell analysis. Analytical Chemistry, 75(14), 3581–3586. https://doi.org/10.1021/ac0340758 . http://pubs.acs.org/doi/abs/10.1021/ac0340758 . 10.1021/ac0340758http://pubs.acs.org/doi/abs/10.1021/ac0340758https://pubs.acs.org/doi/abs/10.1021/ac0340758. http://pubs.acs.org/doi/abs/10.1021/ac0340758 . https://pubs.acs.org/doi/abs/10.1021/ac0340758 . [DOI] [PubMed] [Google Scholar]
- Whitesides GM (2006). The origins and the future of microfluidics. Nature, 442(7101), 368–373. https://doi.org/10.1038/nature05058 . 10.1038/nature05058http://www.nature.com/articles/nature05058. http://www.nature.com/articles/nature05058 . [DOI] [PubMed] [Google Scholar]
- Wirtz D, Konstantopoulos K, & Searson PC (2011). The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nature Reviews Cancer, 11(7), 512–522. https://doi.org/10.1038/nrc3080 . http://www.nature.com/articles/nrc3080 . http://www.nature.com/nrc/journal/v11/n7/abs/nrc3080.html%5Cn . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3262453&tool=pmcentrez&rendertype=abstract . 10.1038/nrc3080http://www.nature.com/articles/nrc3080http://www.nature.com/nrc/journal/v11/n7/abs/nrc3080.html%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3262453&tool=pmcentrez&rendertype=abstracthttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1360529. http://www.nature.com/articles/nrc3080 . http://www.nature.com/nrc/journal/v11/n7/abs/nrc3080.html%5Cn . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3262453&tool=pmcentrez&rendertype=abstract . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1360529 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, & Rockman HA (2006). Drosophila as a model for the identification of genes causing adult human heart disease. Proceedings of the National Academy of Sciences of the United States of America, 103(5), 1394–1399. https://doi.org/10.1073/pnas.0507359103 . 10.1073/pnas.0507359103http://www.ncbi.nlm.nih.gov/pubmed/16432241. http://www.ncbi.nlm.nih.gov/pubmed/16432241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J (2010a). The classic: On the inner architecture of bones and its importance for bone growth. 1870. Clinical Orthopaedics and Related Research, 468(4), 1056–1065. 10.1007/s11999-010-1239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J (2010b). The classic: On the theory of fracture healing. 1873. Clinical Orthopaedics and Related Research, 468(4), 1052–1055. 10.1007/s11999-010-1240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J (2011). The classic: On the significance of the architecture of the spongy substance for the question of bone growth: A preliminary publication. 1869. Clinical Orthopaedics and Related Research, 469(11), 3077–3078. https://doi.org/10.1007/s11999-011-2041-5 . http://www.ncbi.nlm.nih.gov/pubmed/21866420 . 10.1007/s11999-011-2041-5http://www.ncbi.nlm.nih.gov/pubmed/21866420http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3183176. http://www.ncbi.nlm.nih.gov/pubmed/21866420 . http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3183176 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- WormBase, n.d. http://www.wormbase.org.
- WormBook, n.d. http://www.wormbook.org/.
- Xia Y, & Whitesides GM (1998). Soft lithography. Annual Review of Materials Science, 28(1),153–184. https://doi.org/10.1146/annurev.matsci.28.1.153 . 10.1146/annurev.matsci.28.1.153http://www.annualreviews.org/doi/abs/10.1146/annurev.matsci.28.1.153. http://www.annualreviews.org/doi/abs/10.1146/annurev.matsci.28.1.153 . [DOI] [Google Scholar]
- Yang F, Chen Z, Pan J, Li X, Feng J, & Yang H (2011). An integrated microfluidic array system for evaluating toxicity and teratogenicity of drugs on embryonic zebrafish developmental dynamics. Biomicrofluidics, 5(2), 024115 10.1063/1.3605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gao C, Wang P, Zhang G-J, Chen Z, Roensch J,…Kim S (2016). Fish-ona-chip: Microfluidics for zebrafish research. Lab on a Chip, 16(7), 1106–1125. https://doi.org/10.1039/C6LC00044D . 10.1039/C6LC00044Dhttp://xlink.rsc.org/?DOI=C6LC00044D. http://xlink.rsc.org/?DOI=C6LC00044D . [DOI] [PubMed] [Google Scholar]
- Yang L, Hong T, Zhang Y, Arriola JGS, Nelms BL, Mu R, & Li D (2017). A microfluidic diode for sorting and immobilization of Caenorhabditis elegans. Biomedical Microdevices, 19(2), 38 10.1007/s10544-017-0175-2. [DOI] [PubMed] [Google Scholar]
- Yanik MF, Rohde CB, & Pardo-Martin C (2011). Technologies for micromanipulating, imaging, and phenotyping small invertebrates and vertebrates. Annual Review of Biomedical Engineering, 13(1), 185–217. 10.1146/annurev-bioeng-071910-124703. [DOI] [PubMed] [Google Scholar]
- Yi C, Li C-W, Ji S, & Yang M (2006). Microfluidics technology for manipulation and analysis of biological cells. Analytica Chimica Acta, 560(1–2), 1–23. https://doi.org/10.1016/J.ACA.2005.12.037 . 10.1016/J.ACA.2005.12.037https://www.sciencedirect.com/science/article/pii/S0003267005020945. https://www.sciencedirect.com/science/article/pii/S0003267005020945 . [DOI] [PubMed] [Google Scholar]
- Young EWK, & Beebe DJ (2010). Fundamentals of microfluidic cell culture in controlled microenvironments. Chemical Society Reviews, 39(3), 1036 https://doi.org/10.1039/b909900j . 10.1039/b909900jhttp://xlink.rsc.org/?DOI=b909900j. http://xlink.rsc.org/?DOI=b909900j . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H, Kim K, & Lee WG (2013). Cell manipulation in microfluidics. Biofabrication, 5(2), 022001 https://doi.org/10.1088/1758-5082/5/2/022001 . 10.1088/1758-5082/5/2/022001http://stacks.iop.org/1758-5090/5/i=2/a=022001?key=crossref.e52e7bcad258fe147b5a32f620a51ade. http://stacks.iop.org/1758-5090/5/i=2/a=022001?key=crossref.e52e7bcad258fe147b5a32f620a51ade . [DOI] [PubMed] [Google Scholar]
- Zeng F, Rohde CB, & Yanik MF (2008). Sub-cellular precision on-chip small-animal immobilization, multi-photon imaging and femtosecond-laser manipulation. Lab on a Chip, 8(5), 653 https://doi.org/10.1039/b804808h . 10.1039/b804808hhttp://xlink.rsc.org/?DOI=b804808h. http://xlink.rsc.org/?DOI=b804808h . [DOI] [PubMed] [Google Scholar]
- ZFIN, n.d. http://www.zfin.org.
- Zhang W, Dong X, & Liu X (2017). Switched Fuzzy-PD Control of contact forces in robotic microbiomanipulation. IEEE Transactions on Biomedical Engineering, 64(5), 1169–1177. 10.1109/TBME.2016.2594054. [DOI] [PubMed] [Google Scholar]
- Zhao W, Cheng R, Miller JR, & Mao L (2016). Label-free microfluidic manipulation of particles and cells in magnetic liquids. Advanced Functional Materials, 26(22), 3916–3932. 10.1002/adfm.201504178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Nguyen J, Wei Y, & Sun Y (2013). Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab on a Chip, 13(13), 2464 https://doi.org/10.1039/c3lc50355k . 10.1039/c3lc50355khttp://xlink.rsc.org/?DOI=c3lc50355k. http://xlink.rsc.org/?DOI=c3lc50355k . [DOI] [PubMed] [Google Scholar]