Abstract
A previously identified enhancer 10 kb upstream of the Aggrecan (Acan) gene (UE) can drive cartilage specific reporter expression in vivo. Here, we report that the paralogous transcription factors PAX1 and PAX9 differentially drive UE, depending on the presence or absence of SOX9-driven transactivation. In the developing vertebral column, PAX1/9 expression was inversely correlated with Acan expression. Moreover, PAX1/9 was co-expressed with SOX9/5/6 in the intervertebral mesenchyme and the inner annulus fibrosus (AF), and with SOX9 in the outer AF. Significant Acan upregulation was observed during chondrification of Pax1-silenced AF cells, while, Acan was significantly downregulated by persistent expression of Pax1 in cartilage. Deletion of UE using CRISPR/Cas9 resulted in ~30% and ~40% reduction of Acan expression in cartilage and the AF, respectively. In the UE, PAX1/9 acts as weak transactivators through a PAX1/9-binding site partially overlapped with a SOX9-binding site. In the presence of SOX9, which otherwise drives robust Acan expression along with SOX5/6, PAX1/9 competes with SOX9 for occupancy of the binding site, resulting in reduced transactivation of Acan. Coimmunoprecipitation revealed the physical interaction of Pax1 with SOX9. Thus, transactivation of the UE is differentially regulated by concerted action of PAX1/9, SOX9, and SOX5/6 in a context-dependent manner.
Introduction
Aggrecan (ACAN) is a chondroitin sulfate proteoglycan that is abundantly present in cartilage and the intervertebral discs (IVDs)1. Due to its hydrophilic properties, ACAN provides cartilage and the IVDs with a high osmotic swelling pressure to resist compressive loads. Especially in the vertebral column, the IVDs between vertebral bodies (VBs) with cartilaginous endplate at their top and bottom portions serve as cushions and allow various vertebral movements such as extension, flexion, and torsion2. The shock-absorbing gel of the nucleus pulposus (NP), containing abundant cartilaginous proteoglycans, is surrounded by the collagen-rich annulus fibrosus (AF). The inner AF (IAF) consists of several layers of fibrocartilage, while the outer AF (OAF) is a fibrous tissue containing highly organized fibers composed mainly of type I collagen, allowing it to resist tensile loads. Cartilaginous proteoglycans and type II collagen in the IAF decrease gradually closer to the OAF, while type I collagen content increases3.
Acan is induced and upregulated during chondrogenic differentiation and subsequent chondrocyte maturation. Traditional studies using transgenic reporter mice or zebrafish revealed that Acan transcription is redundantly regulated by several enhancers with or without the Sry-related HMG box (SOX) transcription factors4,5. Among them, the enhancer 10 kb upstream of the Acan gene (UE) can drive cartilage-specific reporter gene expression in transgenic mice and zebrafish, and its transactivation is mainly regulated by SOX9, SOX5, and SOX64,5. The homeobox transcription factor SHOX interacts with SOX5 and SOX6 to activate the UE6.
The paralogous transcription factors PAX1 and PAX9, containing highly conserved paired box DNA-binding domains, are coexpressed with SOX9 in the sclerotome that gives rise to the axial skeleton7; however, their expression is later restricted to the AF of the IVD8. The complete lack of VBs and IVDs in Pax1/9 double-null mice suggests their indispensable roles in medioventral sclerotomal differentiation9. PAX1 acts as either a positive or negative regulator of Acan expression. PAX1 stimulates Acan expression via NKX3.2 to promote the early chondrogenesis of presomitic mesoderm explants10,11, while persistent expression of Pax1 in chondrocytes downregulates Acan expression to result in dedifferentiation of chondrocytes12. These stage-specific actions of PAX1 in Acan expression and the restricted expression of PAX1/9 in the AF of IVDs suggest that PAX1/9 is also involved in Acan transcriptional regulation, but the molecular mechanism remains largely unknown.
Focusing on the UE, the upstream Acan enhancer, here, we demonstrate the fine-tuning of Acan expression in the UE by PAX1/9. Contribution of the UE to endogenous Acan transcription in vivo was evaluated by CRISPR/Cas9 mediated enhancer deletion. Through a PAX1/9-binding site that partially overlaps with a SOX9-binding site in the UE, PAX1/9 competes with SOX9 for occupancy of the binding site that results in a reduction in Acan transactivation, but in the absence of SOX9, PAX1/9 weakly transactivates the UE. These results suggest that PAX1/9 differentially drives the Acan UE, depending on the presence or absence of SOX9.
Results
Inverse correlation between PAX1/9 and ACAN expression in the IVDs
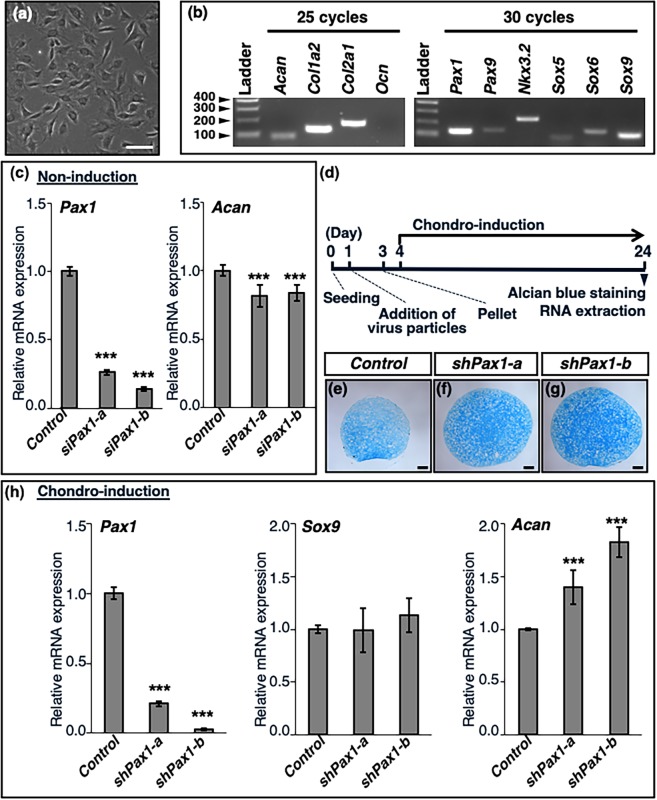
We analyzed the expression of PAX1, PAX9, SOX5, SOX6, SOX9, and ACAN in the developing mouse vertebral column from embryonic day (E) 13.5 to E16.5 by immunostaining (Fig. 1b–h,n–r) and in situ hybridization (Fig. 1j,k,s). At E13.5, cartilaginous proteoglycans that were detected by metachromatic staining with toluidine blue were abundantly present in the cartilaginous anlagen of VBs and were also present at a low level in the intervertebral mesenchyme (IM) (Fig. 1a). PAX1 was expressed in the IM and in the perichondrial tissues of VBs (Fig. 1b). PAX9 was co-localized with PAX1 in the IM and was also found in a portion of chondrocytes in the VBs (Fig. 1c,d). SOX9, SOX5, SOX6, and ACAN were distributed in the cartilaginous VBs (Fig. 1e–h). In the IM, PAX1 was co-localized with these molecules (Fig. 1e–h). At E14.5, the IVD was formed in association with the transition of the notochord into the NP. In the cross-sections of an IVD, the NP was surrounded by the AF, which is comprised of the IAF and the OAF (Fig. 1i). Pax1 was strongly expressed in the OAF but weakly expressed in the IAF (Fig. 1j). In contrast, Acan was expressed in the IAF but only weakly expressed in the OAF (Fig. 1k). In the sagittal section of the vertebral column of an embryo at E16.5, significant accumulation of proteoglycans was detected in the cartilage and the IAF, while their accumulation in the OAF was nearly undetectable (Fig. 1l,m). At E16.5, PAX1 and PAX9 were present mainly in the OAF, but were also detected in a part of the IAF (Fig. 1n,o). Although SOX5 and SOX6 in the OAF were detectable only in the boundary to the IAF, SOX9 protein was weakly expressed throughout the entire OAF (Fig. 1p–r). Acan was expressed at a high level in the cartilage, and its expression gradually decreased from the IAF towards the OAF, which expressed PAX 1 and PAX9 (Fig. 1n,o,s). Thus, in the IVD, PAX1/9 expression is inversely correlated with ACAN expression and co-expressed with SOX9 in the OAF and with SOX9/5/6 in the IAF.
Figure 1.
Differential expression of PAX1/9 and SOX9/5/6 in the intervertebral disc. (a–h) Sagittal sections of the thoracic vertebrae of a mouse embryo at E13.5 were processed for toluidine blue staining (a) and immunostaining for PAX1 (b,d,e–h), PAX9 (c,d), SOX5 (e), SOX6 (f), SOX9 (g), or ACAN (h). Red arrowheads in (a–h) indicate the IM that is positive for PAX1. (i–k) Cross sections of the thoracic IVD of a mouse embryo at E14.5 were processed for toluidine blue staining (i) or in situ hybridization for Pax1 (j) and Acan (k). Black arrowheads in (i–k) indicate the OAF of the IVD. (l–s) Sagittal sections of the thoracic vertebrae of a mouse embryo at E16.5 were processed for toluidine blue staining (l,m); immunostaining for PAX1 (n), PAX9 (o), SOX5 (p), SOX6 (q), or SOX9 (r); and in situ hybridization for Acan (s). The boxed region in (l) is shown at a higher magnification in (m). Panels shown in (n–s) correspond to the area of (m). Dotted line in (m–s) encloses the OAF. Ventral is to the right in (a–h,l–s) or the bottom in (i–k). Ca, cartilage; IAF, inner annulus fibrosus; IM, intervertebral mesenchyme; Nc, notochord; NP, nucleus pulposus; OAF, outer annulus fibrosus; VB, vertebral body. Scale bars, 100 μm.
Upregulation of Acan in Pax1-silenced AF cells during chondrification
In the IVD, an inverse correlation between Pax1/9 and Acan expression was observed (Fig. 1h,j,k,n,s). Despite the abundant expression of SOX9 (Fig. 1g,r), the level of Acan in the OAF expressing high levels of PAX1/9 (Fig. 1d,n,o) was very low (Fig. 1k,s). We isolated AF cells from the IVDs of rat tails (Fig. 2a) and checked the expression of the genes encoding extracellular matrix proteins (Acan, Col1a2, and Col2a1) and transcription factors (Pax1, Pax9, Nkx3.2, Sox5, Sox6, and Sox9) (Fig. 2b), all of which have been reported to be expressed in the AF2,8,13. Osteocalcin (Ocn), used as a negative control, was confirmed to be undetectable (Fig. 2b). Since the expression level of Pax1 was much higher than that of Pax9, we silenced Pax1 in AF cells under non-inductive and chondro-inductive conditions. In AF cells, the level of Pax1 was decreased to 26% or 14% of that in the control by siPax1-a or siPax1-b, respectively (Fig. 2c). Upon knockdown of Pax1 by lipofection of the siPax1 oligonucleotides, the mRNA levels of Acan were slightly but significantly decreased compared with those in the control (Fig. 2c). To investigate how Pax1 affects the expression of Acan and proteoglycan accumulation during chondrogenic differentiation, AF cells infected with shRNA lentivirus were pelleted and maintained in chondrogenic differentiation medium containing l-ascorbic acid, dexamethasone, and recombinant human TGF-β1 (Fig. 2d). In the AF cells cultured under chondro-inductive conditions, the expression level of Pax1 decreased by 21%, whereas that of Sox5, Sox6, or Sox9 increased more than five-fold compared to those in the AF cells cultured under non-inductive conditions (Supplementary Fig. 1). Infection with shPax1-a or shPax1-b lentivirus resulted in increased accumulation of proteoglycans in the AF pellets as compared to levels in the control (Fig. 2e–g). The level of Pax1 was decreased to 21% or 3% of that in the control by infection with shPax1-a or shkPax1-b lentivirus, respectively (Fig. 2h). Although no significant changes were detected in the level of Sox9, the level of Acan was significantly increased to 140% or 187% of that in the control by infection with shPax1-a or shPax1-b lentivirus, respectively (Fig. 2h). No significant increase in endogenous Pax9 expression was detected in the pellets of AF cells infected with shPax1-a or shPax1-b lentivirus (Supplementary Fig. 2), suggesting that a compensatory increase in endogenous Pax9 does not occur upon knock-down of Pax1 in AF cells. Taken together, these results indicate that PAX1 stimulates Acan expression in AF cells under non-inductive conditions but inhibits its expression under chondro-inductive conditions.
Figure 2.
Enhanced chondrogenesis in pellets of AF cells by knockdown of Pax1. (a) AF cells isolated from 4-week-old Wistar rats were seeded at a density of 8 × 104 cells/well in a 12-well plate. Morphology of AF cells cultured in DMEM containing 10% FBS at day 1 is shown. (b) Total RNA was extracted from AF cells maintained in a culture dish for 96 h and then processed for RT-PCR analysis of expression of ECM molecules (Acan, Col1a2, Col2a1, Ocn) and transcription factors (Pax1, Pax9, Nkx3.2, Sox5, Sox6, Sox9). Full-length gels are presented in Supplementary Fig. 7. (c) AF cells were seeded at a density of 4 × 104 cells/well in a 24-well plate. At 24 h after inoculation, the cells were transfected with non-targeting siRNA (control) or Pax1 siRNAs (siPax1-a or siPax1-b) by lipofection. Total RNA was extracted from the cells at 48 h after lipofection, and the expression levels of Pax1 or Acan were examined by qRT-PCR. (d) Experimental schedule of chondro-induction in pelleted cultures of AF cells. AF cells were seeded at a density of 1 × 105 cells/well in 6-well plates and then infected with shRNA lentiviruses on day 1. After trypsinization on day 3, cell pellets were prepared from 1.5 × 105 cells/pellet and cultured in chondrogenic differentiation medium for another 21 days. (e–g) Sections of cell pellets infected with shRNA lentiviruses were stained with alcian blue. Increased accumulation of alcian blue-stained extracellular matrix was observed in the pellets of AF cells infected with shPax1-a or shPax1-b lentivirus (f,g) compared to that in cells infected with control lentivirus (e). (h) Total RNA was extracted from pellets on day 24. Relative expression levels of Pax1, Sox9, and Acan were examined by qRT-PCR analysis. qRT-PCR data (c,h) represent the average of three independent experiments. The relative expression of each gene is normalized to that in the control and reported as mean ± s.d. ***P < 0.001 versus control. Scale bars in (a) and (e–g), 100 μm.
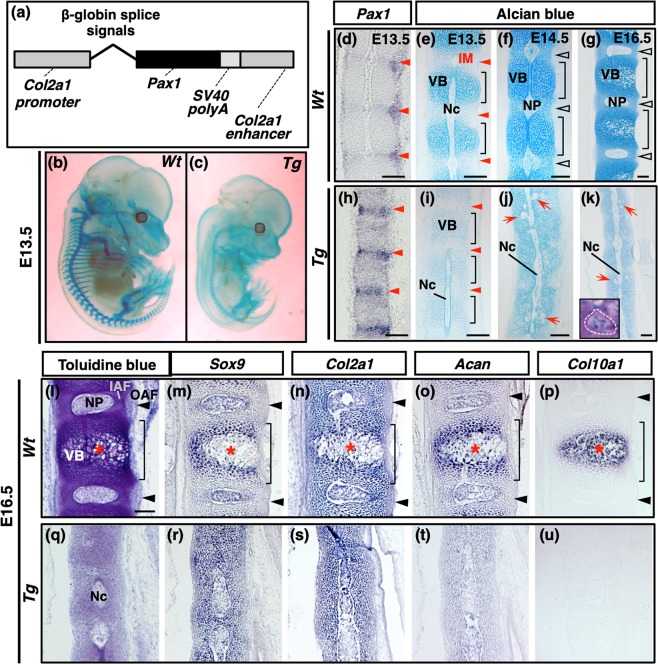
Downregulation of Acan in Col2a1-Pax1 transgenic (Tg) mouse embryos
We previously reported that overexpression of Pax1 resulted in downregulation of cartilage marker genes including Acan in the forelimbs of chicken embryos and cultured chondrocytes12. To examine the effects of persistent expression of Pax1 in cartilage, we analyzed Col2a1-Pax1 Tg mouse embryos that overexpress Pax1 under the control of Col2a1 enhancer/promoter (Fig. 3a). Whole mount alcian blue staining revealed that Col2a1-Pax1 Tg mouse embryos were smaller and their cartilaginous tissues were weakly stained with alcian blue as compared to wildtype (Wt) littermates (Fig. 3b,c and Supplementary Fig. 3a–d). At E16.5, mineralized areas stained with alizarin red were detectable both in the membranous and the endochondral bones of wild type embryos, whereas the endochondral bones were not mineralized in Col2a1-Pax1 Tg embryos (Supplementary Fig. 3e–h). In the sagittal sections of the vertebral column of a wt embryo at E13.5, the notochord was observed as a rod-like structure running in the developing vertebral column in which the metameric pattern of VB and IM was established (Fig. 3d,e). From E14.5 onwards, the nucleus pulposus derived from the notochord was formed in the middle region of each IVD in wt embryos (Fig. 3f,g). In a Col2a1-Pax1 Tg embryo, Pax1 was expressed in the VBs as well as the IM, which resulted in significant reduction of proteoglycan accumulation in the cartilaginous molds (Fig. 3h–k). Although the metameric pattern in the vertebral column of a Col2a1-Pax1 Tg embryo was observed by E13.5, such pattern was abolished with failure of IVD formation from E14.5 onwards (Fig. 3i–k). The vertebral column was ectopically bent at E14.5, and the notochord remained as a rod-like structure without forming the nucleus pulposus at E16.5 in Col2a1-Pax1 Tg embryos (Fig. 3i–k and Supplementary Fig. 3c). At E14.5 and E16.5, persistent notochordal remnants were found in the vertebral column of Col2a1-Pax1 Tg embryos (arrows in Fig. 3j,k). In the sagittal sections of the vertebral column of wild type embryos at E16.5, Sox9, Col2a1, and Acan were expressed both in the IVDs and the VBs, but these transcripts were downregulated in the hypertrophic chondrocytes expressing Col10a1 (Fig. 3l–p). In a Col2a1-Pax1 Tg embryo, arrested chondrocyte maturation was manifested by lack of Col10a1 expression as well as uniform expression of Sox9 and Col2a1 (Fig. 3q–u). Although the expression of Sox9 and Col2a1 in the cartilage was detected at a high level in a Col2a1-Pax1 Tg embryos, Acan expression was detected at a lower level than that in the cartilage of wild type embryos (Fig. 3r–t).
Figure 3.
Impaired vertebral column formation in transgenic mouse embryos overexpressing Pax1 in the Col2a1 expressing region. (a) A schematic illustration of the transgene for generation of transgenic mice overexpressing Pax1 under the control of Col2a1 enhancer/promoter (Col2a1-Pax1 Tg). (b,c) Lateral views of E13.5 Col2a1-Pax1 Tg founder embryos stained with alcian blue are shown. Cartilaginous vertebrae, ribs, and limbs are stained with alcian blue in Wt (b), whereas accumulation of alcian blue-stainable matrix in those cartilaginous elements is significantly reduced in Tg (c). (d–u) Sagittal sections of thoracic vertebrae of Wt (d–g,l–p) or Tg (h–k,q–u) embryos at E13.5 (d,e,h,i), E14.5 (f,j), and E16.5 (g,k–u) are shown. The sections were processed for in situ hybridization of Pax1 (d,h), Sox9 (m,r), Col2a1 (n,s), Acan (o,t) or Col10a1 (p,u) and for alcian blue staining (e-g,i–k) or toluidine blue staining (l,q). Brackets in (e–g,i) and red arrowheads in (d,e,h,i) indicate the VB and the IM, respectively. Hollowed arrowheads in (f,g) and black arrowheads in (l–p) indicate the IVD and the OAF, respectively. Arrows in (j,k) indicate remnants of the notochord. The inset in (k) shows a magnified image of a toluidine blue stained notochordal remnant that is enclosed with a dotted line. Asterisks in (l–p) indicate the zone of hypertrophic chondrocytes. Ventral is to the right. IAF, inner annulus fibrosus; IM, intervertebral mesenchyme; Nc, notochord; NP, nucleus pulposus; OAF, outer annulus fibrosus; VB, vertebral body. Scale bars, 100 μm.
Thus, persistent expression of Pax1 in the developing cartilage inhibits Acan expression and proteoglycan accumulation, suggesting that downregulation of Pax1 is necessary for the proper progression of chondrocyte maturation during vertebral column development.
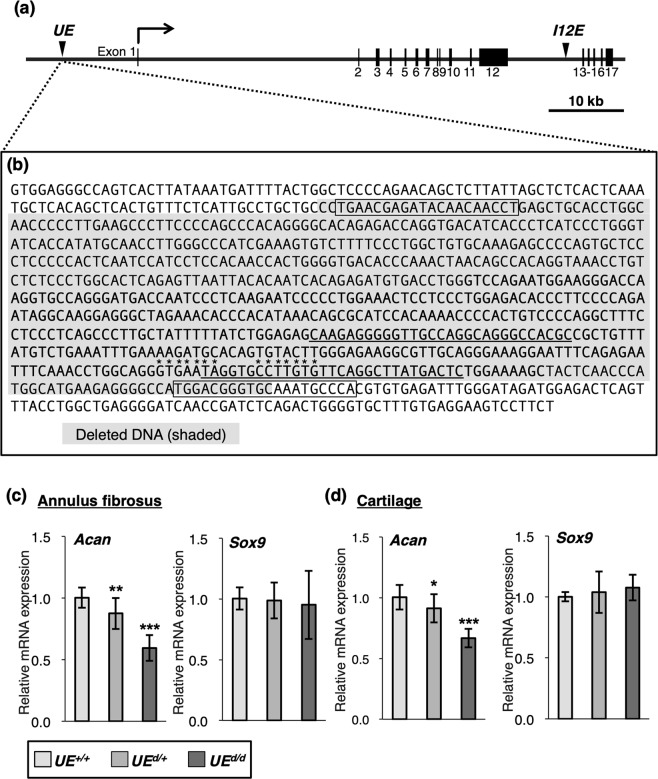
Decreased endogenous Acan expression in the AF and the costal cartilage by in vivo deletion of the genome region containing the upstream Acan enhancer
Han and Lefebvre5 have previously identified the upstream Acan enhancer (UE) harboring SOX9- and SOX5/6-binding sites as a major cis-regulatory element for Acan expression (Fig. 4a). We used MatInspector software (Genomatix) to identify two potential binding sites for PAX2/5/8 (Fig. 4b). We then generated mutant mice lacking the SOX9-binding sites of the enhancer using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) to evaluate its contribution to endogenous Acan expression (Fig. 4a,b). We established a mouse line with a deletion of a 695-bp region containing 359 bp of the UE (Fig. 4b). The expression level of Acan in the AF of the IVD was significantly reduced by 12% in heterozygous UEd/+ mice and by 41% in homozygous UEd/d mice (Fig. 4c) compared to that in wildtype UE+/+ mice. In the costal cartilage, the Acan expression level was decreased by 9% in heterozygous UEd/+ mice and by 33% in homozygous UEd/d mice compared to that in wildtype UE+/+ mice (Fig. 4d). The in vivo deletion of the UE-containing region did not significantly affect Sox9 expression in either tissue (Fig. 4c,d). These results suggest that the UE is a major enhancer that regulates Acan expression in the AF of the IVD and the costal cartilage.
Figure 4.
In vivo deletion of the Acan upstream enhancer. (a) Genomic structure of the mouse Acan gene, consisting of 17 exons (black boxes). Black arrowheads indicate the positions of the upstream enhancer (UE), located approximately 10 kb upstream of the transcription start site, and of the I12 enhancer (I12E) located in intron 12. (b) The mouse DNA sequence surrounding the UE. The sequence of the UE is shown in bold letters. Two potential binding sites for PAX2/5/8, predicted by MatInspector software (Genomatix), are underlined. The shaded letters indicate the region of a deletion mutation produced by the CRISPR/Cas system. The boxed letters indicate the sgRNA target sequence. The bases comprising the SOX9-binding sites are indicated with asterisks. (c,d) Total RNA was extracted from the AF (c) and the costal cartilage (d) of UE+/+, UEd/+, or UEd/d male mice at 3 weeks after birth. Relative expression levels of Acan and Sox9 were examined by qRT-PCR analysis. qRT-PCR data represent the average of 4 mice in each group. The relative expression of each gene is normalized to that of UE+/+ and reported as mean ± s.d. *P < 0.05 versus UE+/+, **P < 0.01 versus UE+/+, ***P < 0.001 versus UE+/+.
Transactivation of the upstream enhancer and the intragenic region regulated by PAX1/9, SOX9, and SOX5/6
To test whether Acan is a direct target gene of PAX1 or PAX9, we performed dual luciferase reporter assays using human embryonic kidney (HEK) 293T cells, primary chondrocytes, and NIH3T3 cells. One copy or four tandem repeats of 359 bp of the mouse UE (1xUE-Luc and 4xUE-Luc) was inserted into the pGL4.23[luc2/minP] vector, containing a minimal promoter and a luciferase (luc) reporter gene. Co-expression of Pax1, Pax9, or Sox9 with 1xUE-Luc in HEK293T cells resulted in a 2.7-, 2.1-, or 3.8-fold increase in luc activity compared to that with the pcDNA3 empty vector (Fig. 5a). The luc activity of 4xUE-Luc was significantly increased by Sox9 (12.8-fold), while luc activity was increased 2.2- or 1.9-fold when cells were co-expressed with Pax1 or Pax9 (Fig. 5a). In the presence of Sox9 and Sox5/6, the luc activity of 4xUE-Luc in HEK293T cells increased to 71-fold, but it decreased to 30% or 7% upon co-expression with Pax1 or Pax9, respectively (Fig. 5b). We also identified a genomic region containing a potential PAX2-binding site in intron 12 (I12E) of the mouse Acan gene (Supplementary Fig. 4a,b). Gel shift assays revealed that PAX1 and PAX9, but not SOX9, bound to the predicted PAX2-binding site in I12E (Supplementary Fig. 4c–e). When co-expressed with Pax1, the luc activity of I12E (I12E-Luc) was significantly increased in HEK293T cells (Fig. 5c). In chondrocytes expressing Sox9 and Sox5/6 at high levels, the luc activities of 1xUE-Luc and 4xUE-Luc increased to 19- and 52-fold that of the pGL4.23[luc2/minP] empty vector, respectively (Supplementary Fig. 5). As expected, the luc activity of 1xUE-Luc co-expressed with Pax1 and Pax9 decreased to 54% and 21%, respectively. Similarly, the luc activity of 4xUE-Luc co-expressed with Pax1 and Pax9 decreased to 29% and 15%, respectively (Fig. 5d). In contrast, when the I12E fragment with the PAX1/9-binding site not overlapped with a SOX9-binding site was tested, PAX1 and PAX9 acted as transactivators in chondrocytes (Fig. 5e). Interestingly, in the previously identified Nkx3.2 promoter region (Nkx3.2-P), which contains a PAX1/PAX9-binding site that partially overlaps with a SOX9-binding site10,14, SOX9-induced transactivation was significantly decreased by co-expression with PAX1 or PAX9 (Fig. 5f). Taken together, these results suggest that PAX1 and PAX9 act as weak transactivators in the transcriptional regulation of Acan and Nkx3.2. However, SOX9- or SOX9/5/6-induced transactivation of Acan and Nkx3.2 expression is attenuated by PAX1 and PAX9, as the PAX1/9-binding sites partially overlap with the SOX9-binding sites.
Figure 5.
Transactivation of Acan enhancers by PAX1/PAX9 and SOX9/SOX5/SOX6. (a–f) Dual luciferase assays were performed with HEK293T cells (a–c), chondrocytes (d,e), or NIH3T3 cells (f). Cells were co-transfected with reporter plasmids (pGL4.74 [hRluc/TK] and pGL4.23[luc2/minP] or pGL3-Basic vectors) and expression plasmids (pcDNA3 vectors). The pGL4.23[luc2/minP] and pGL3-Basic vectors used for the assays were pGL4.23[luc2/minP]-1xUE-Luc (1xUE-Luc), pGL4.23[luc2/minP]-4xUE-Luc (4xUE-Luc), or pGL4.23[luc2/minP]-I12E-Luc (I12E-Luc), and pGL3-Basic-Nkx3.2-P-Luc (Nkx3.2-P-Luc). The expression plasmids used were pcDNA3 empty vector (pcDNA3), pcDNA3-hSox5 (Sox5), pcDNA3-hSox6 (Sox6), pcDNA3-mSox9 (Sox9), pcDNA3-FLAG-Pax1 (Pax1), or pcDNA3-FLAG-Pax9 (Pax9). For the co-transfection of pcDNA3 vectors in (b), 10 ng each of Sox5, Sox6, and Sox9, and 20 ng of pcDNA3, Pax1, or Pax9 were used. For the co-transfection of pcDNA3 vectors in (f), 25 ng each of Pax1 and Pax9 (the fourth bar from the left), or 25 ng of Sox9 and 25 ng of pcDNA3, Pax1, or Pax9 (from the fifth to the seventh bars from the left), or 12.5 ng each of Pax1 and Pax9 (the right end bar) were used. The firefly and Renilla luciferase activities were measured 24 h after transfection. Values were normalized using a pGL4.74[hRluc/TK] vector and are presented as fold induction relative to pcDNA3 co-transfected with reporter plasmids. Graphs show a representative experiment out of at least three. Each bar represents the average of three independent transfections (means ± s.d.). *P < 0.05 versus pcDNA3, **P < 0.01 versus pcDNA3, ***P < 0.001 versus pcDNA3 (a,c,d,e,f), pcDNA3 + Sox5 + Sox6 + Sox9 (b), or pcDNA3 + Sox9 (f).
Competitive binding of PAX1/9 and SOX9 in the UE
A potential binding site for PAX2/5/8 was found near the previously identified SOX9-binding sites located from 317 to 334 in the UE (Figs 4b and 6a). Chromatin immunoprecipitation (ChIP) assays revealed that PAX1 directly binds to the UE and I12E of Acan (Acan-UE and Acan-I12E) and the promoter region of Nkx3.2 (Nkx3.2-P), as shown in Fig. 6b. We then performed gel shift assays to determine whether PAX1 and PAX9 directly bind to the predicted binding site that partially overlaps with the SOX9-binding sites found in the 359-bp UE. We tested three DNA fragments corresponding to the sequences at 1–130, 118–259, and 249–359 and identified the specific binding of PAX1-containing nuclear extracts to the 111-bp of the 3′-fragment of the UE (Fig. 6c). This specific binding competed with the non-labelled 45-bp oligonucleotide encoding the sequence at 315–359, but it was not affected by the non-labelled 249–290 or 281–325 oligonucleotides (Fig. 6d). We also confirmed the specific binding of PAX1 and PAX9 to the predicted binding site found in intron 12 (Supplementary Fig. 4c,d). In the UE, PAX1 bound the biotin-labelled 55-bp oligonucleotide (wt) corresponding to the sequence at 301–355 in the UE, which was consistent with the in-silico prediction of the potential PAX2/5/8-binding site at 322–350 (Fig. 4b, Supplementary Fig. 6a). Gel shift assays with mutated oligonucleotides revealed that PAX1 bound the 334–351 sequence in the UE, partially overlapping with the SOX9-binding site (Supplementary Fig. 6a). We also confirmed the binding of PAX9 to the UE by detection of the specific binding of PAX9 protein to the wt oligonucleotide and m3 oligonucleotide (containing a mutated SOX9-binding site), but not to the m7 oligonucleotide (containing a mutated PAX1-binding site), as was observed for PAX1 (Supplementary Fig. 6b).
Figure 6.
Binding of PAX1 to the 3′ region of the UE. (a) A 359-bp DNA sequence of the mouse UE. The SOX9-binding sites are underlined. (b) ChIP assays on extracts from AF cells. After immunoprecipitation of the cross-linked extracts with anti-PAX1 antibody, anti-SOX9 antibody, or normal IgG antibody, the DNA was subjected to PCR with primers that amplify a 116-bp fragment of the UE, 216-bp fragment of the I12E, 135-bp fragment of Nkx3.2-P, or 328-bp fragment of the Col2a1-E. Full-length gels are presented in Supplementary Fig. 7. (c,d) Gel shift assays were performed using 20 fmol of biotin-labelled dsDNA probes, 20 pmol of unlabeled dsDNA fragments, and nuclear extracts (N.E.) of HEK293T cells transfected with pcDNA3 empty vector (N.E. Control) or pcDNA3-FLAG-Pax1 (N.E. FLAG-PAX1). The biotin-labelled and unlabeled DNA fragments correspond to positions 1–130, 118–259, and 249–359 of the UE. A shifted band was caused by interactions between FLAG-PAX1 and the biotin-labelled 249–359 of UE. The shifted band in (c) disappeared in the presence of the unlabeled 249–359 oligonucleotide or anti-FLAG antibody (Antibody). The shifted band in (d) disappeared in the presence of the unlabeled 315–359 oligonucleotide but remained detectable in the presence of the unlabeled 249–290 or 281–325 oligonucleotide. (e,f) Gel shift assays were performed using biotin-labelled oligonucleotides shown in Supplementary Table 1 and nuclear extracts (N.E.) of HEK293T cells transfected with pCAG empty vector (N.E Control), pCAG-Sox9 (N.E. SOX9), or pCAG-Pax1 (N.E. PAX1). For the assays in (e,f), 100 and 5 fmol of the biotin-labelled oligonucleotides were used, respectively. A total of 1.5 μL of N.E. was used for each binding reaction in (f). A shifted band is shown with an arrow. Non-specific bindings are shown with open arrowheads. s, shifted band.
The binding site for PAX1 partially overlapped with that for SOX9, raising the possibility that PAX1 interferes with the binding of SOX9 in the UE. To test this possibility, we performed gel shift assays using nuclear extracts from HEK293T cells expressing PAX1 or SOX9. In the presence of both PAX1 and SOX9 with biotin-labelled wt oligonucleotide, containing the two SOX9-binding sites and a PAX1/9-binding site, two specific bands were detected (lane 2 in Fig. 6e). When biotin-labelled m3 oligonucleotide, containing a mutated SOX9-binding site, or m7 oligonucleotide, containing a mutated PAX1/9-binding site, was used, specific shift bands for PAX1 and SOX9 were detected as PAX1/m3 and SOX9/m7 complexes, respectively (lanes 3 and 4 in Fig. 6e). A shift band for SOX9 (lane 2 in Fig. 6f) was detected in the presence of the wt oligonucleotide. When incubated with both SOX9 and PAX1, the intensity of the shift band for SOX9 was reduced, and instead the shift band for PAX1 was detected (lanes 3 and 4 in Fig. 6f). These data suggest that PAX1 and SOX9 compete for the partially overlapping binding sites in the UE.
A previous study demonstrated that SOX9 directly interacted with RUNX2 to repress its activity15. To explore the possibility of a physical interaction between PAX1 and SOX9, we performed coimmunoprecipitation assays using the nuclear extracts of HEK293T cells transfected with pcDNA3-Sox9 and pcDNA3-FLAG-Pax1. When the nuclear extracts were immunoprecipitated with anti-FLAG antibody, SOX9 was detected in the immunoprecipitates of the nuclear extracts containing FLAG-PAX1 and SOX9, and the nuclear extracts containing FLAG-RUNX2 and SOX9 (Fig. 7). These results suggest that the physical interactions between PAX1 and SOX9 interfere with each other to reduce their transcription activities. Thus, the extent of SOX9-driven transactivation of the UE is influenced by the presence or absence of PAX1.
Figure 7.

Physical interaction between FLAG-PAX1 and SOX9. Coimmunoprecipitation assays were performed with (lanes 1, 2) or without (lane 3) nuclear extracts. The nuclear extracts prepared from HEK293T cells co-transfected with pcDNA3-Sox9 and pcDNA3-FLAG-Runx2 or pcDNA3-FLAG-Pax1 were immunoprecipitated with the anti-FLAG antibody. The immunoprecipitates (lanes 1–3) and the nuclear extracts (lanes 4, 5) were subjected to immunoblotting with the anti-Sox9 antibody. The band representing the SOX9 protein is indicated by the arrowhead on the right. Lane 1, immunoprecipitates of FLAG-RUNX2 and SOX9; lane 2, immunoprecipitates of FLAG-PAX1 and SOX9; lane 3, immunoprecipitates of the buffer for nuclear extraction; lane 4, nuclear extracts of the cells expressing FLAG-RUNX2 and SOX9; lane 5, nuclear extracts of the cells expressing FLAG-PAX1 and SOX9.
Discussion
In this study, our CRISPR/Cas9 mediated enhancer deletion in vivo demonstrated for the first time that the upstream Acan enhancer, UE, accounts for ~40% and ~30% of endogenous Acan expression in the AF and costal cartilage, respectively. In the developing vertebral column, we found an inverse correlation between PAX1/9 and Acan expression. PAX1/9 acted as weak transactivators of both the intragenic (intron 12) and the UE. However, in the presence of SOX9, which directs modest but robust transcriptional activation together with SOX5/6, PAX1 and SOX9 physically interacted with each other and competed for occupancy of their partially overlapping binding sites within the UE (Fig. 8a), thus resulting in reduced transactivation (Fig. 8b). These results suggest that unique combinations of PAX1/9, SOX9, and SOX5/6 contribute to the establishment of the graded expression and accumulation of ACAN.
Figure 8.
Differential Acan expression achieved by unique combinations of PAX1/PAX9, SOX9, and SOX5/SOX6. (a) The mouse UE sequence from 301 to 355 is shown. Underlined and boxed letters in the sequence are the SOX9-binding sites and the PAX1-binding site, respectively, as revealed by gel shift assays in Supplementary Fig. 6. (b) A proposed model for the transcriptional regulation of Acan mediated by the interaction of PAX1/9, SOX9, and SOX5/6 with the Acan enhancers is shown. During vertebral column development, UE weakly activates Acan transcription in the OAF (I), which expresses PAX1/9 and SOX9; moderately in the IAF (II), which expresses PAX1/9 and SOX9/5/6; and strongly in the cartilaginous VB (III), which expresses SOX9/5/6. In parallel, I12E activates Acan transcription weakly in the OAF and IAF, which express PAX1/9. Details are discussed in the text. Ca, cartilage; IAF, inner annulus fibrosus; IM, intervertebral mesenchyme; IVD, intervertebral disc, Nc, notochord; NP, nucleus pulposus; OAF, outer annulus fibrosus; VB, vertebral body. Scale bar, 100 μm.
During vertebral column development, the formation of IVDs is associated with the physical transition of the rod-like notochord into nuclei pulposi16. No residual notochordal tissue normally remains in the vertebral body. In the Col2a1-Pax1 Tg embryos, the notochord remained as a rod-like structure without forming the nucleus pulposus nor IVDs. This was similarly observed in the vertebral column of Col2a1−/− mouse embryos17. It has been proposed that swelling pressure exerted by the cartilaginous vertebral bodies serves to push notochord cells into the intervertebral regions during the transition of the notochord into nuclei pulposi4. Our results suggest that persistent expression of Pax1 in Col2a1-Pax1 Tg downregulates Acan expression to induce dedifferentiation of chondrocytes, reduced matrix synthesis and accumulation in cartilage, the notochord, and the notochord sheath, thus resulting in a failure of the normal transition of the notochord into nuclei pulposi.
The silencing of Pax1 in AF cells isolated from the IVD of 4-week-old rats resulted in the slight downregulation of Acan expression. This is in accordance with the previous observation that the loss of Pax1 causes a decrease in Acan expression8. However, significant accumulation of proteoglycan and upregulation of Acan gene expression were observed in Pax1-silenced AF cell pellets under chondro-inductive conditions, indicating that PAX1 acts as a negative regulator of Acan expression during chondrification. This is consistent with our previous and present findings that Acan expression is markedly downregulated in chondrocytes upon forced expression of Pax1 in chicks12 and transgenic mice. Since there were no significant changes in Sox9 mRNA levels, and no increase in those of Pax9, in the control vs. the Pax1-silenced AF pellets under chondrogenic conditions, reduced Pax1 expression could ultimately lead to augmented transcriptional activity of SOX trio (SOX5, 6, 9) complexes, which drive robust Acan expression.
When two transcription factors compete for shared DNA binding sites, robust transactivation can be blocked by enhancer or promoter occupancy18. This mode of regulation has been proposed for several combinations of transcription factors, such as HMX1 and NKX2.5, PAX3 and MITF, and SP1 and SP319–21. Such antagonistic regulation can be mediated not only by a combination of a transcriptional activator and repressor, but also by a combination of activators, such as SP1 and SP321. We found that PAX1/9 competes with SOX9 for occupancy of the shared binding sites within the UE (Fig. 8a). In HEK293T cells, where neither SOX9 nor SOX5/6 is expressed, PAX1/9 and SOX9 transactivated the Acan upstream enhancer (4xUE-Luc) ~2-fold and ~12-fold, respectively. When co-expressed with SOX9 and SOX5/6, the luc activity of 4xUE-Luc was ~70-fold greater than that of the control. Either PAX1 or PAX9 can attenuate this transactivation driven by SOX9 and SOX5/6 through decreased binding of SOX9 to the shared binding site. Consistent with these observations, luc activity of 1xUE-Luc or 4xUE-Luc decreased when PAX1 or PAX9 was expressed in chondrocytes which express SOX9 and SOX5/6 endogenously at high levels. Both PAX1 and PAX9 compete with SOX9 for occupancy of the partially overlapping binding sites, resulting in reduced transactivation of Acan in chondrocytes. This suggests that a unique molecular mechanism operates in non-hyaline cartilaginous tissues such as the AF, in which weak and strong transactivators cooperatively contribute to the fine-tuning of Acan expression by changing their relative expression ratios.
The Acan UE was identified as an evolutionarily conserved genomic region that drives cartilage-specific reporter gene expression in transgenic mice5. Later, Hu et al. identified multiple Acan enhancers, whose activities were analyzed in zebrafish22. In these studies, transgenic reporter assays using a single copy or tandem copies of the candidate enhancer regions were performed to evaluate the transactivation potentials and tissue specificities of these enhancers. Accumulating evidence has suggested that the pervasive presence of multiple enhancers with similar activities near the same gene confers phenotypic robustness against loss-of-function mutations in individual enhancers23. Deletion of an individual enhancer would cause no noticeable change, even though it drives reporter gene expression in transgenic animals23. However, our in vivo deletion analysis of the Acan UE demonstrated that ~40% of the endogenous Acan expression in the AF is driven by this enhancer. Considering that even subtle changes in Acan expression levels could cause degeneration of the IVD or osteoarthritis, the Acan UE, the activity of which is finely tuned by combinations of strong and weak transactivators, is considered to be important in maintaining vertebral column integrity in the postnatal IVD.
Our study revealed a novel PAX1/9-binding motif, 5′ GTTCAGGCTTATGACTCT 3′, which contains the core sequence of a PAX2-binding motif, 5′ TCANGCNTGA 3′24. In the mouse Acan UE, the PAX1-binding site partially overlapped with one of the SOX9-binding sites. Similar PAX1/9-binding motifs that partially overlap with SOX9-binding sites are also present in the Acan UE in other mammals (data not shown). Interestingly, it has been reported that a PAX1/9-binding site partially overlaps with the SOX9-binding site in the mouse Nkx3.2 promoter14. In Pax1/9-deficient mice, Nkx3.2 expression is undetectable in the sclerotome10. During early chondrogenesis, PAX1/9 induces the expression of Nkx3.2, which stimulates Acan expression in explants of the chick presomitic mesoderm10. In contrast, persistent expression of Pax1 and Nkx3.2 in chick chondrocytes results in significant downregulation of Acan12. PAX1/9 physically interacts with the Nkx3.2 promoter to transactivate it10. Transactivation of the Nkx3.2 promoter by SOX9 is significantly suppressed by PAX1 or PAX9, suggesting a common mechanism in which SOX9-driven transactivation is modulated by PAX1 or PAX9 through competition for overlapping binding sites and physical interactions.
The OAF is a fibrous tissue that still expresses Acan at a low level. It is of interest that the OAF predominantly expresses not only Pax1/Pax9, but also Scx, which was also reported to be a transactivator of Acan25. In Pax1/Pax9 double-null mice, IVD is completely missing9. Loss of Scx also causes defective formation of the OAF during development26. We previously reported that the conditional inactivation of Sox9 in SCX+/SOX9+ cells caused defective formation of the IAF but expansion of the OAF27, suggesting that the SCX+/SOX9+ cell population contributes to the formation of the AF. IAF formation is delayed in Sox5−/−/Sox6−/− mice, in which Acan expression is almost undetectable4. ACAN content increases from the PAX1+/SOX9+ OAF towards the PAX1+/SOX9+/SOX5+/SOX6+ IAF and the central SOX9+/SOX5+/SOX6+ NP. It is apparent that Acan expression in the IAF and the NP is predominantly regulated by SOX9 and SOX5/6. In contrast, predominant PAX1/9 and SCX expression in the OAF and IVD defects of mice lacking these genes suggest the importance of these weak transactivators in transcriptional regulation of Acan in the OAF. Since the UE accounts for ~40% of endogenous Acan expression, it is reasonable to speculate that PAX1/9 regulates Acan expression directly through other Acan enhancers or indirectly by affecting the expression of downstream genes that regulate Acan expression. So far, we have revealed that the SOX9-driven transactivation in the Nkx3.2 promoter is modulated by PAX1/9 through competition for overlapping binding sites and physical interactions. Genome wide identification of Acan enhancers and ChIP Seq studies will reveal whether a similar mechanism is found in other enhancers to regulate Acan transactivation.
Methods
Animals
C57BL/6 mice and Wistar rats were purchased from Shimizu Laboratory Supplies (Kyoto, Japan). Fertile White Leghorn chicken eggs were obtained from Takeuchi Farm (Nara, Japan). All animal experimental procedures used in this study were approved by the Animal Care Committee of the Institute for Frontier Life and Medical Sciences, Kyoto University and conformed to institutional guidelines for the study of vertebrates.
Generation of transgenic mice
To generate the Col2a1-Pax1 construct, mouse Pax1 cDNA was amplified by PCR using the primers listed in Supplementary Table 2 and inserted into the NotI site of the pCol2a1 vector containing the promoter and intronic enhancer of the rat Col2a1 gene28,29. The purified Col2a1-Pax1 transgene was injected into the pronuclei of fertilized eggs from C57BL/6× C3H F1 mice. Injected founder embryos were dissected and analyzed from E12.5 to E16.5. Transgene positive embryos were identified by PCR using the primers listed in Supplementary Table 2 and genomic DNA extracted from the yolk sac.
Generation of mouse lines by CRISPR/Cas9-mediated gene editing
For CRISPR/Cas9-mediated gene editing, crisprRNAs (crRNAs) were designed using the CRISPRdirect website30 and chemically synthesized and purified by Fasmac (Kanagawa, Japan). The crRNAs, trans-activating crRNA, and Cas9 protein were microinjected into the cytoplasm or pronuclei of fertilized eggs obtained from C57BL/6 mice. Injected eggs were transferred into the oviducts of pseudopregnant surrogate ICR female mice. Deletions in genomic DNA were identified by direct sequencing of the PCR products amplified using the primers listed in Supplementary Table 2. Mouse lines were established from founder mice carrying deletions in targeted DNA sites.
Tissue sections
For the preparation of frozen sections of embryonic tissues, mouse embryos were fixed in 4% paraformaldehyde dissolved in PBS (PFA/PBS) for 3 h, infiltrated with 18% sucrose/PBS, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Japan), cut at a thickness of 8 μm, and mounted onto glass slides.
Histological staining
For alcian blue or toluidine blue staining of frozen sections, the sections were fixed with 4% PFA/PBS for 5 min and stained with 1% alcian blue 8GX (Sigma, St. Louis, MO, USA) at pH 1 or 0.05% toluidine blue O (Sigma) at pH 4 for 5 min. For whole mount alcian blue staining, embryos were fixed in 5% trichloroacetic acid for 3 h and washed with distilled water. The embryos were then stained with 0.1% alcian blue 8GX (Sigma) in 70% ethanol containing 0.1% HCl for 3 h and destained in 70% ethanol containing 0.1% HCl. After dehydrating twice in 100% ethanol for 1 h, the samples were cleared and stored in methyl salicylate.
Immunostaining
Sections for the detection of PAX1, PAX9, SOX5, and SOX6 were boiled in 10 mM sodium citrate at pH 6 for 10 min. Sections for the detection of ACAN were pretreated with 0.2 U/mL chondroitinase ABC (Sigma) at 37 °C for 30 min. After blocking with 3.2% skim milk/PBS, the sections were incubated with primary antibodies for 16 h, washed, and then incubated with appropriate secondary antibodies conjugated with Alexa Fluor 488 or 594 purchased from Thermo Fisher Scientific (Waltham, MA, USA). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma). The primary antibodies used were anti-ACAN (Merck Millipore, Burlington, MA, USA, AB1031; 1:500), anti-PAX1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA, sc-7744; 1:500), anti-PAX9 (Active Motif, Carlsbad, CA, USA, 61077; 1:500), anti-SOX5 (Abcam, Cambridge, UK, ab94396; 1:500), anti-SOX6 (Abcam, ab30455; 1:500), and anti-SOX9 (Merck Millipore, AB5535; 1:800). The images were captured under a Leica DMRXA microscope equipped with a Leica DC500 camera (Leica Microsystems).
In situ hybridization
For RNA probes, the complementary DNAs (cDNAs) for Acan, type II collagen (Col2a1), and Pax1 were amplified by reverse transcription-polymerase chain reaction (RT-PCR) based on sequence information available in GenBank (Acan, L07049; Col2a1, NM031163; Pax1, NM008780). Mouse Sox9 and Col10a1 cDNAs were generously given by Dr. Gerd Scherer (University of Freiburg)28 and Dr. Bjorn R Olsen (Harvard Medical School)31, respectively. The RNA probes were transcribed from the linearized plasmids with a digoxigenin (DIG) RNA labelling kit (Roche, Mannheim, Germany). The frozen sections were fixed in 4% PFA/PBS for 10 min, washed with PBS, and treated with 10 μg/mL proteinase K (Thermo Fisher Scientific) for 15 min. After post-fixation with 4% PFA/PBS for 10 min, the sections were carbethoxylated twice in 0.1% DEPC/PBS and hybridized with DIG-labelled RNA probes diluted in 50% formaldehyde/5 × SSC containing 40 μg/mL salmon sperm DNA at 55 °C for 16 h. To detect DIG-labelled RNA probes, immunological detection was performed with anti-DIG-AP Fab fragment (Roche) and BM purple (Roche).
Vector construction
The entire coding regions of mouse Pax1, mouse Pax9, human Sox5, human Sox6, and mouse Sox9 were obtained from RCAS(A)-Pax1, RCAS(A)-Pax910, pMXs-gw/hSOX5, pMXs-gw/hSOX632, and pCMV-SPORT6-Sox9 (OriGene Technologies, Rockville, USA), respectively. To construct expression plasmids, each DNA fragment was inserted into pcDNA3 or CAG-IRES-EGFP vectors. To generate N-terminal FLAG-tagged PAX1 and PAX9, each DNA fragment was amplified by PCR to add NotI sites and inserted into the pcDNA3-FLAG vector. A 359-bp UE was amplified by PCR from mouse genomic DNA with forward and reverse primers designed with BamHI and BglII sites, respectively, and then PCR products were cloned into the pCR4-TOPO vector (Thermo Fisher Scientific). Four tandem copies of UE were obtained by ligation of monomers digested with BamHI and BglII. To construct pGL4.23[luc2/minP]-1xUE-Luc or pGL4.23[luc2/minP]-4xUE-Luc, one copy or four tandem copies of UE were inserted into the pGL4.23[luc2/minP] vector (Promega, Madison, WI, USA) containing a minimal promoter and the firefly luciferase reporter gene. The 508-bp I12E or 1109-bp Nkx3.2-P were amplified by PCR from mouse genomic DNA and inserted into the pGL4.23[luc2/minP] or pGL3-Basic vector (Promega) containing the firefly luciferase reporter gene, respectively. The PCR primers for constructs are listed in Supplementary Table 2.
Cell culture and transient transfection
AF cells were isolated from the tail IVDs of 4-week-old male Wistar rats. AFs were minced and partially digested with 0.2% collagenase (Roche) and 0.1% trypsin (BD Biosciences) at 37 °C for 20 min. The tissue fragments were placed onto 10-cm cell culture dishes (BD Biosciences) coated with type I collagen (KOKEN, Tokyo, Japan), and the cultures were maintained in MF-start (TOYOBO, Osaka, Japan). At 80% confluence, the cells grown from AFs were passaged twice and grown in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) containing 10% fetal bovine serum (FBS). Chondrocytes were isolated from the distal tibiotarsal cartilage of chicken embryos at stage 41, as previously described33. Human embryonic kidney (HEK) 293T cells and NIH3T3 cells were cultured in DMEM containing 10% FBS. Transfection of plasmids into the cells was performed using Lipofectamine LTX Reagent (Thermo Fisher Scientific) or Lipofectamine 3000 Reagent (Thermo Fisher Scientific). Cell cultures were maintained in a humidified atmosphere of 5% CO2 in air.
Gene silencing experiments
Small interfering RNA (siRNA) oligonucleotide duplexes were purchased from GE Healthcare Life Sciences (Little Chalfont, UK). Pax1 was knocked down using siPax1-a (J-092982-12) and siPax1-b (J-092982-17), included in the ON-TARGET plus Rat Pax1 siRNA-Set of four (GE Healthcare, LQ-092982-02-002). For the control experiment, siGENOME non-targeting siRNA Pool number 1 (GE Healthcare, D-001206-13-05) was used. Transfection of siRNA into AF cells was performed with Lipofectamine 3000 Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For lentivirus-mediated knockdown of Pax1, lentiviral particles were produced using HIV-based lentiviral vector constructs purchased from SBI (Palo Alto, CA, USA). The short hairpin RNA (shRNA) sequences for shPax1-a and shPax1-b were created based on the sequences of J-092982-11 and J-092982-17 included in the ON-TARGET plus Rat Pax1 siRNA-Set of four (GE Healthcare, LQ-092982-02-002), respectively. For the control experiment, the non-targeting sequence (UAAGGCUAUGAAGAGAUAC) included in siGENOME non-targeting siRNA Pool no. 1 (GE Healthcare, D-001206-13-05) was used. The target sequences were connected by a loop sequence (CUUCCUGUCAGA) and restriction enzyme sites and then inserted into the lentivirus-mediated shRNA-expressing vector pGreenPuro (SBI, SI505A-1). The lentiviral particles were produced in Lenti-X 293T cells (TaKaRa Bio, Shiga, Japan) and concentrated with PEG-it Virus Precipitation Solution (SBI), according to the manufacturer’s instructions.
Chondrogenic induction
For induction of chondrogenic differentiation, aliquots of 1.5 × 105 AF cells were pelleted in 15-mL conical tubes and maintained for 21 days in 0.5 mL DMEM containing 6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL sodium selenite, 1.25 mg/mL bovine serum albumin, 50 μg/mL l-ascorbic acid, 1 μM dexamethasone, and 10 ng/mL recombinant human TGF-β1, as previously described34.
Immunoprecipitation and Immunoblotting
Nuclear extracts prepared from HEK293T cells transfected with a total of 10 μg of plasmids were incubated with 5 μg of anti-FLAG M2 antibody (Sigma, F3165) in the immunoprecipitation buffer [0.1% NP-40, 1 mM EDTA, 100 mM NaCl, 50 mM Tris-HCl (pH 7.5)] at 4 °C for 4 h with gentle rocking. Twenty-five microliters of Protein G magnetic beads (Active motif, 53033) was then added to the mixture and incubated at 4 °C for another 1 h with gentle rocking. After incubation, the beads were washed twice with the immunoprecipitation buffer and then with ice-cold PBS twice. The bound protein was eluted with 20 μL of 2 × SDS sample buffer [100 mM Tris-HCl (pH 6.8), 4% SDS, 100 mM DTT, 20% glycerol] and boiled for 5 min. The eluate and nuclear extracts were separated by SDS-PAGE and transferred onto Immobilon-P transfer membrane (Merk Millipore). The membranes were preincubated with 3.2% skim milk in Tris-buffered saline (TBS) for 30 min and incubated with anti-SOX9 antibody (Merck Millipore, AB5535; 1:2000) at 4 °C overnight. Blots were washed with TBS containing 0.05% Tween 20 and incubated with Trueblot HRP-conjugated anti-rabbit IgG (Rockland, 18-8816-31). Antibodies were detected using ImmunoStar Zeta (Wako, 291-72401) and ImageQuant LAS 4000mini (GE healthcare).
RT-PCR and quantitative RT-PCR (qRT-PCR) analysis
For RNA extraction from the costal cartilage and the AF, tissues dissected from the rib cage and the tail, respectively, were immersed in RNAlater solution (Thermo Fisher Scientific). Total RNA was extracted from AF tissues or AF cells using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The costal cartilage was frozen in liquid nitrogen and crushed using SK-mill (Tokken, Chiba, Japan), and then the total RNA was purified by RNeasy MinElute Cleanup kit (Qiagen) after phenol-chloroform extraction. One or two hundred nanograms of total RNA was used to synthesize cDNA with a PrimeScript RT reagent Kit (Takara Bio). RT-PCR was performed with Takara Ex Taq (Takara Bio) and the same primers for qRT-PCR listed in Supplementary Table 3. qRT-PCR was performed using SYBR Premix Ex Taq II (Takara Bio) on a StepOne instrument (Thermo Fisher Scientific). Relative mRNA expression was normalized to that of 18 S rRNA and calculated using the 2−ΔΔCT method.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using a ChIP-IT Express Magnetic Chromatin Immunoprecipitation kit (Active Motif) following the manufacturer’s protocol. Sheared chromatin prepared from AF cells was immunoprecipitated using anti-PAX1 (Santa Cruz Biotechnology, sc-7744), anti-SOX9 (Millipore, AB5535), or normal goat IgG (Santa Cruz Biotechnology, sc-2028) antibodies. The sequences of Acan-UE, Acan-I12E, Nkx3.2-P10, and Col2a1-E5 were detected by PCR using Takara Ex Taq (Takara Bio) and the ChIP primers listed in Supplementary Table 2. The amplification products were electrophoresed on a 2% agarose gel and visualized by ethidium bromide.
Luciferase reporter assay
HEK293T or NIH3T3 cells seeded in 96-well microtiter plates were transfected with 50 ng of the pGL4.23 construct, 50 ng of pcDNA3 expression vectors, and 1 ng of pGL4.74[hRluc/TK] vector (Promega) containing the HSV-TK promoter and Renilla luciferase. Chondrocytes seeded in 96-well microtiter plates were transfected with 100 ng of the pGL4.23 construct, 100 ng of pcDNA3 expression vectors, and 1 ng of pGL4.74[hRluc/TK] vector. After 24 h, luciferase activities were measured using the Pica gene Dual luciferase kit (Toyo ink, Tokyo, Japan) and a microplate luminometer (Centro XS3 LB960, Berthold Technologies, Bad Wildbad, Germany). Reporter construct activity was normalized by comparison with activity from the Renilla luciferase construct. All experiments were performed in triplicate.
Gel shift assay
Protein-DNA binding was assayed with the Gelshift Chemiluminescent EMSA kit (Active Motif) following the manufacturer’s protocol. For the SOX9 protein, the binding reactions were carried out in 20 μL of binding buffer containing 1 μg of poly(dG-dC)·poly(dG-dC) and 20 μg of BSA. To generate the DNA probes or competitors above 100 bp in size, double-stranded DNA (dsDNA) fragments were amplified by PCR with the biotin-labelled or unlabeled PCR primers listed in Supplementary Table 2, respectively. The biotin-labelled DNA probes less than 100 bp in size were prepared from synthesized DNA oligonucleotides using the Biotin 3′ End DNA Labeling kit (Thermo Fisher Scientific). Nuclear proteins were isolated using the Nuclear Extract Kit (Active Motif) from HEK293T cells transfected with pcDNA3 vectors to express FLAG-PAX1, FLAG-PAX9, and SOX9 protein or pCAG-IRES-EGFP vectors to express PAX1 and SOX9 protein. Five micrograms of anti-FLAG M2 monoclonal antibody (Sigma, F3165) was used for each reaction of the antibody with the nuclear protein.
Statistical analysis
P-values were calculated by t-test or one-way analysis of variance using the SPSS software package (SPSS 21.0). The data were considered statistically significant when the P value was < 0.05.
Supplementary information
Acknowledgements
The pCol2a1 vector and the entire coding cDNAs of mPax1 and mPax9 were generously gifted from Dr. Y. Yamada and Dr. K. Imai, respectively. The pMXs-gw/hSOX5 and pMXs-gw/hSOX6 vectors were generously gifted from Dr. N. Tsumaki. Mouse Sox9 and Col10a1 cDNAs were generously gifted from Dr. G. Scherer and Dr. B. R. Olsen, respectively. We thank Ms. H. Sugiyama for her valuable secretarial help. This study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 26861186, 16K10899, 24300154, 18H02966, the Frontier Development Program for Genome Editing funded by the Doctoral Program for World-leading Innovative and Smart Education, Ministry of Education, Culture, Sports, Science and Technology, and the Cooperative Research Program of the Institute for Frontier Life and Medical Sciences, Kyoto University, Japan.
Author Contributions
A.T. performed experimental design and immunostaining, knockdown, construction of the transgene, skeletal preparation, in situ hybridization, in vivo deletion of the UE, construction and luciferase assay, wrote the manuscript, performed ChIP assay and EMSA. C.S. performed construction and luciferase assays. H.W. and G.K. generated the transgenic founder mice. T.S. and T.Y. provided genome editing tools. C.K. and Y.H. approved the manuscript. C.S. conceived the project and designed the experiments, wrote and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40810-4.
References
- 1.Roughley PJ, Melching LI, Heathfield TF, Pearce RH, Mort JS. The structure and degradation of aggrecan in human intervertebral disc. Eur Spine J. 2006;15(Suppl 3):S326–332. doi: 10.1007/s00586-006-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes AJ, Benjamin M, Ralphs JR. Extracellular matrix in development of the intervertebral disc. Matrix Biol. 2001;20:107–121. doi: 10.1016/S0945-053X(01)00125-1. [DOI] [PubMed] [Google Scholar]
- 3.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 4.Smits P, Lefebvre V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003;130:1135–1148. doi: 10.1242/dev.00331. [DOI] [PubMed] [Google Scholar]
- 5.Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aza-Carmona M, et al. SHOX interacts with the chondrogenic transcription factors SOX5 and SOX6 to activate the aggrecan enhancer. Hum Mol Genet. 2011;20:1547–1559. doi: 10.1093/hmg/ddr032. [DOI] [PubMed] [Google Scholar]
- 7.Neubuser A, Koseki H, Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- 8.Sivakamasundari V, et al. A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol Open. 2017;6:187–199. doi: 10.1242/bio.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters H, et al. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigo I, Hill RE, Balling R, Munsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130:473–482. doi: 10.1242/dev.00240. [DOI] [PubMed] [Google Scholar]
- 11.Murtaugh LC, Zeng L, Chyung JH, Lassar AB. The chick transcriptional repressor Nkx3.2 acts downstream of Shh to promote BMP-dependent axial chondrogenesis. Dev Cell. 2001;1:411–422. doi: 10.1016/S1534-5807(01)00039-9. [DOI] [PubMed] [Google Scholar]
- 12.Takimoto A, Mohri H, Kokubu C, Hiraki Y, Shukunami C. Pax1 acts as a negative regulator of chondrocyte maturation. Exp Cell Res. 2013;319:3128–3139. doi: 10.1016/j.yexcr.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Tribioli C, Frasch M, Lufkin T. Bapx1: an evolutionary conserved homologue of the Drosophila bagpipe homeobox gene is expressed in splanchnic mesoderm and the embryonic skeleton. Mech Dev. 1997;65:145–162. doi: 10.1016/S0925-4773(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita S, et al. Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp Cell Res. 2009;315:2231–2240. doi: 10.1016/j.yexcr.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G, et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawson L, Harfe BD. Notochord to Nucleus Pulposus Transition. Current osteoporosis reports. 2015;13:336–341. doi: 10.1007/s11914-015-0284-x. [DOI] [PubMed] [Google Scholar]
- 17.Aszodi A, Chan D, Hunziker E, Bateman JF, Fassler R. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. The Journal of cell biology. 1998;143:1399–1412. doi: 10.1083/jcb.143.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermsen R, Tans S, ten Wolde PR. Transcriptional regulation by competing transcription factor modules. PLoS Comput Biol. 2006;2:e164. doi: 10.1371/journal.pcbi.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amendt BA, Sutherland LB, Russo AF. Transcriptional antagonism between Hmx1 and Nkx2.5 for a shared DNA-binding site. J Biol Chem. 1999;274:11635–11642. doi: 10.1074/jbc.274.17.11635. [DOI] [PubMed] [Google Scholar]
- 20.Lang D, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, Datta PK, Bagchi S. Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 2003;31:5368–5376. doi: 10.1093/nar/gkg706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu G, Codina M, Fisher S. Multiple enhancers associated with ACAN suggest highly redundant transcriptional regulation in cartilage. Matrix Biol. 2012;31:328–337. doi: 10.1016/j.matbio.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterwalder M, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose SK, Gibson W, Bullard RS, Donald CD. PAX2 oncogene negatively regulates the expression of the host defense peptide human beta defensin-1 in prostate cancer. Mol Immunol. 2009;46:1140–1148. doi: 10.1016/j.molimm.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, et al. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. J Biol Chem. 1997;272:29880–29885. doi: 10.1074/jbc.272.47.29880. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimoto Y, et al. Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci Rep. 2017;7:45010. doi: 10.1038/srep45010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimoto Y, et al. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140:2280–2288. doi: 10.1242/dev.096354. [DOI] [PubMed] [Google Scholar]
- 28.Tsumaki N, et al. Separable cis-regulatory elements that contribute to tissue- and site-specific alpha 2(XI) collagen gene expression in the embryonic mouse cartilage. The Journal of cell biology. 1996;134:1573–1582. doi: 10.1083/jcb.134.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebsbach PH, et al. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene. J Biol Chem. 1996;271:4298–4303. doi: 10.1074/jbc.271.8.4298. [DOI] [PubMed] [Google Scholar]
- 30.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apte SS, Seldin MF, Hayashi M, Olsen BR. Cloning of the human and mouse type X collagen genes and mapping of the mouse type X collagen gene to chromosome 10. Eur J Biochem. 1992;206:217–224. doi: 10.1111/j.1432-1033.1992.tb16919.x. [DOI] [PubMed] [Google Scholar]
- 32.Outani H, et al. Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS One. 2013;8:e77365. doi: 10.1371/journal.pone.0077365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298:234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 34.Takimoto A, Oro M, Hiraki Y, Shukunami C. Direct conversion of tenocytes into chondrocytes by Sox9. Exp Cell Res. 2012;318:1492–1507. doi: 10.1016/j.yexcr.2012.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.