Abstract
Several microRNAs (miRNAs) have been linked to chronic kidney disease (CKD) mortality, cardiovascular (CV) complications and kidney disease progression. However, their association with clinical outcomes remains poorly evaluated. We used real-time qPCR to measure serum levels of miR-126 and miR-223 in a large cohort of 601 CKD patients (CKD stage G1 to G5 patients or on renal replacement therapy – CKD G5D) from Ghent University Hospital and 31 healthy controls. All-cause mortality and cardiovascular and renal events were registered as endpoints over a 6 year follow-up period. miR-126 levels were significantly lower from CKD stage G2 on, compared to controls. The serum levels of miR-223 were significantly lower from CKD stage G3B on. When considering overall mortality, patients with levels of either miR-126 or miR-223 below the median had a lower survival rate. Similar results were observed for CV and renal events. The observed link between the two miRNAs’ seric levels and mortality, cardiovascular events or renal events in CKD appears to depend on eGFR. However, this does not preclude their potential role in the pathophysiology of CKD. In conclusion, CKD is associated with a decrease in circulating miR-223 and miR-126 levels.
Introduction
Chronic kidney disease (CKD) is a major public health burden worldwide. This condition, and end-stage kidney disease in particular, is frequently associated with disorders of mineral and bone metabolism (CKD-MBD), and an increased cardiovascular risk in which progressive vascular calcification leads to a high cardiovascular mortality rate1,2. The identification of prognostic biomarkers is therefore of the utmost relevance to CKD patients.
MicroRNAs (miRNAs) are short, single-stranded, non-coding RNAs that downregulate gene expression through translational repression or degradation of messenger RNA (mRNA) by binding to the mRNA’s 3’-untranslated region3. miRNAs are highly conserved; the first miRNA was discovered in 1993 in Caenorhabditis elegans and the first human miRNAs were identified in 20004. About 3000 miRNAs have now been identified in the human genome3. Their biogenesis and mode of action have been extensively reviewed elsewhere5.
Dysregulation of miRNAs has been linked to the pathophysiology of many diseases, including kidney and cardiovascular diseases5–7. Our team reported that miR-126 and -223 levels were altered in the aortic wall and serum in murine models of CKD8, and in the microcirculation of the brain of CKD mice9. Human targets of these miRNAs are shown in Supplementary Table 1.
miR-223 is considered to be a hematopoietic factor10 with a preponderant role in granulopoiesis11,12. It regulates cholesterol homeostasis and cardiac glucose metabolism, but it is also known to be an onco-miR13–15. In vascular disease, circulating miR-223 is able to penetrate the vascular smooth muscle cells (VSMCs) and act as an endocrine signal to regulate cell proliferation, migration and apoptosis16,17. In vitro, exposure of human VSMCs to pathological levels of inorganic phosphate (Pi) increases miR-223 expression as well as cell migration18. Finally, Ulbing et al. reported in a limited cohort of 140 patients, in CKD stages G3 to G5 or after kidney transplantation, that expression levels of miR-223-3p were lower in stage G4 and G5 CKD patients than in healthy controls19.
miR-126 is a pro-angiogenic miRNA present in endothelial cells20, and is highly expressed in vessels21. This miRNA has an important role in vascular dysfunction, since it enhances endothelial proliferation and endothelialization of large vessels, which in turn attenuates atherosclerosis. In a murine model of CKD, miR-126 is overexpressed in the aorta, along with the upregulation of SDF-18, a chemokine protein playing an important role in angiogenesis recruiting endothelial progenitor cells from the bone marrow. During tissue repair, in a mouse model of atherosclerosis, endothelial cells release miR-126 in apoptotic bodies, inducing SDF-1-dependent vascular repair22. Furthermore, miR-126 overexpression in bone marrow cells has recently been shown to promote vascular integrity following kidney injury by contributing to the recovery of the kidney microvasculature23. Plasma levels of miR-126 are abnormally low in type 2 diabetes24, and low plasma levels are associated with a poor prognosis in renal cell carcinoma25,26.
Recent studies have shown that miRNAs in blood27 and other body fluids28 are remarkably stable. They are protected from endogenous RNase activity by their binding to Argonaut proteins or their uptake in exosomes29 or microvesicles30. They are also transported by nucleophosmin 131, endothelial apoptotic bodies22 and high density lipoprotein (HDL)32. In view of the minimally invasive nature of blood and body fluid sampling and the stability of miRNAs, they may be useful as diagnostic and/or prognostic biomarkers.
The objectives of the present study were (i) to study the expression of circulating miR-126 and miR-223 in a cohort of 601 CKD patients and 31 healthy controls, and (ii) to evaluate the link between these miRNAs and cardiovascular and all-cause mortality. The secondary objective was to evaluate the association between the initial expression of miR-126 and miR-223 in serum and renal events and the decline of kidney function.
Results
Characteristics of the study population
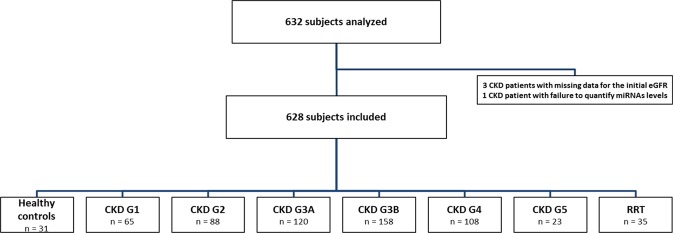
A total of 632 subjects, 601 CKD patients and 31 healthy controls were enrolled of which 628 were included in the final analysis (Fig. 1). Three CKD patients had missing data for the initial eGFR and, in one CKD patient, miRNA quantification in serum was impossible. The patients included in the analysis were divided into seven CKD groups, including patients on dialysis (22 patients on hemodialysis and 13 patients on peritoneal dialysis) and healthy controls. The clinical and biochemical characteristics of the study population are summarized in Table 1 and Supplementary Table 2.
Figure 1.
Study flow chart.
Table 1.
Baseline characteristics of the cohort participants overall and by group (CKD patients, patients on RRT, and healthy controls).
| n | Cohort | Healthy | CKD G1 | CKD G2 | CKD G3A | CKD G3B | CKD G4 | CKD G5 | CKD G5D | P value* |
|---|---|---|---|---|---|---|---|---|---|---|
| 628 | 31 | 65 | 88 | 120 | 158 | 108 | 23 | 35 | ||
| Age, years | 64 [49–74] | 30 [25–58]¥ | 38 [28–50] | 54 [43–62] | 66 [56–73] | 71 [64–78] | 73 [63–81] | 72 [57–78] | 66 [62–75] | p < 0.05 |
| Male, n (%) | 365 (58) | 13 (42) | 32 (49) | 50 (57) | 68 (57) | 96 (61) | 72 (67) | 11 (48) | 23 (66) | p = 0.26 |
| Caucasian, n (%) | 619 (99) | 31 (100) | 63 (97) | 85 (97) | 118 (98) | 158 (100) | 106 (98) | 23 (100) | 35 (100) | p = 0.36 |
| BMI, kg/m² | 27 [23–31] | 22 [20–23]¥ | 25 [22–28] | 27 [23–30] | 27 [24–31] | 29 [25–32] | 28 [24–32] | 26 [24–29] | 26 [23–32] | p < 0.05 |
| Mean blood pressure, mmHg | 97 [90–105] | 93 [86–99] | 95 [89–101] | 99 [90–106] | 96 [89–105] | 98 [93–106] | 97 [90–107] | 99 [91–112] | 92 [76–97] | p < 0.05 |
| Hypertension, n (%) | 537 (86) | 4 (13)¥ | 48 (73) | 71 (81) | 110 (92) | 153 (97) | 106 (98) | 22 (96) | 23 (66) | p < 0.05 |
| Diabetes, n (%) | 182 (29) | 0 (0)¥ | 9 (14) | 21 (24) | 28 (23) | 65 (41) | 42 (39) | 9 (39) | 8 (3) | p < 0.05 |
| Cardiovascular history, n (%) | 53 (9) | 0 (0)¥ | 1 (2) | 4 (5) | 6 (5) | 20 (13) | 18 (17) | 2 (9) | 2 (6) | p < 0.05 |
| Etiology, n (%) | n = 597 | |||||||||
| Nephroangiosclerosis | 105 (17.5) | 1 (2) | 10 (11) | 26 (22) | 31 (20) | 29 (27) | 4 (17) | 4 (11) | ||
| Diabetic nephropathy | 93 (15.5) | 4 (6) | 9 (10) | 11 (9) | 42 (27) | 20 (18) | 2 (9) | 5 (14) | ||
| ADPKD/Alport | 41 (7) | 8 (12) | 12 (14) | 7 (6) | 6 (4) | 4 (4) | 2 (9) | 2 (6) | ||
| Unique kidney | 48 (8) | 1 (2) | 8 (9) | 14 (12) | 14 (9) | 10 (9) | 1 (4) | 0 (0) | ||
| Lupus/vasculitis | 39 (6.5) | 14 (21) | 9 (10) | 10 (8) | 4 (2) | 1 (1) | 1 (4) | 0 (0) | ||
| Membranous nephropathy | 8 (1) | 0 (0) | 4 (5) | 2 (2) | 1 (1) | 1 (1) | 0 (0) | 0 (0) | ||
| IgA nephropathy | 27 (4.5) | 9 (14) | 4 (5) | 4 (3) | 4 (2) | 5 (5) | 1 (4) | 0 (0) | ||
| Urologic/reflux nephropathy | 17 (3) | 1 (2) | 3 (3) | 3 (3) | 5 (3) | 1 (1) | 2 (9) | 2 (6) | ||
| Tubulo-interstitial nephritis | 40 (7) | 6 (9) | 3 (3) | 11 (9) | 5 (3) | 8 (7) | 4 (17) | 2 (6) | ||
| Other glomerulonephritis | 39 (6.5) | 10 (15) | 8 (9) | 2 (2) | 7 (4) | 5 (5) | 1 (4) | 6 (17) | ||
| Renovascular nephropathy | 10 (2) | 2 (3) | 1 (1) | 0 (0) | 4 (2) | 3 (3) | 0 (0) | 0 (0) | ||
| Non-recovery from AKI | 18 (3) | 1 (2) | 5 (6) | 4 (3) | 7 (4) | 1 (1) | 1 (4) | 0 (0) | ||
| Toxic | 33 (5.5) | 0 (0) | 2 (2) | 10 (8) | 11 (7) | 9 (8) | 1 (4) | 0 (0) | ||
| Other or unknown | 79 (13) | 8 (12) | 10 (11) | 16 (13) | 17 (11) | 11 (10) | 3 (13) | 14 (40) | ||
| Medications, n (%) | ||||||||||
| ACEIs and/or ARBs | 401 (64) | 2 (6.5)¥ | 43 (66) | 57 (65) | 84 (70) | 111 (70) | 78 (72) | 13 (56) | 13 (37) | p < 0.05 |
| Diuretics | 243 (39) | 3 (9.7)¥ | 12 (18) | 30 (34) | 37 (31) | 74 (47) | 66 (61) | 12 (52) | 9 (26) | p < 0.05 |
| Calcium channel blockers | 226 (36) | 1 (3.2)¥ | 10 (15) | 24 (27) | 44 (37) | 74 (47) | 56 (52) | 11 (48) | 6 (17) | p < 0.05 |
| Beta-blockers | 280 (45) | 2 (6.5)¥ | 9 (14) | 22 (25) | 60 (50) | 100 (63) | 70 (65) | 6 (26) | 11 (31) | p < 0.05 |
| Other antihypertensive drugs | 83 (13) | 0 (0.0)¥ | 2 (3) | 60 (7) | 12 (10) | 28 (18) | 25 (23) | 6 (26) | 4 (11) | p < 0.05 |
| Statins | 340 (54) | 1 (3.2)¥ | 18 (28) | 39 (44) | 68 (57) | 112 (71) | 76 (70) | 16 (70) | 10 (29) | p < 0.05 |
BMI: body mass index; ADPKD: autosomal dominant polycystic kidney disease; ACEI: angiotensin-converting-enzyme inhibitor; ARB: angiotensin II receptor blocker.
Continuous variables are expressed as the median [IQR] and binary variables are expressed as the number (%).
*The p value is for the comparison between CKD groups. Healthy controls vs CKD patients: ¥p < 0.001.
The median age was 64 years, 58.1% of the participants were male, 29% had diabetes mellitus, and 85.5% had hypertension. The groups differed with regard to age; controls, CKD G1 and G2 groups being significantly younger than CKD G4 and G5 groups. There were no statistically significant intergroup differences in gender or ethnicity. The mean arterial pressure was higher in the CKD G2 to G5 groups than in the other groups. Prevalence of diabetes was higher in the CKD G3B, CKD G4 and CKD G5 groups.
We observed statistically significant intergroup differences for serum phosphate, C-reactive protein (CRP), hemoglobin, glucose and lipid parameters and percentage of patients with significant proteinuria (>200 mg/L) (Supplementary Table 2).
Relationship between eGFR and miRNA baseline levels
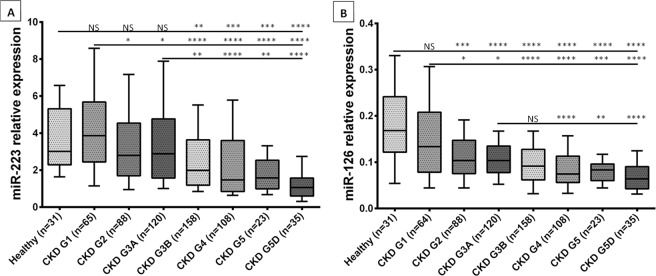
The two miRNAs were detected in all samples. The serum levels of miR-126 and miR-223 over the course of CKD are summarized in Fig. 2.
Figure 2.
Relative expression (serum levels) of miRNAs in healthy controls, patients at different CKD stages, and patients on RRT. (A) Serum levels of miR-223. (B) Serum levels of miR-126. Data are shown as the median with box and whisker plots, including the 10th and 90th percentiles. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (in a Mann-Whitney test).
The serum levels of miR-223 were significantly lower in the CKD G3B, CKD G4, CKD G5 and CKD G5D groups than in the healthy controls and the CKD G1 and CKD G3A groups. There were no statistically significant differences between the control group on one hand, and the CKD G1, CKD G2, and CKD G3A groups on the other.
When compared with controls, serum levels of miR-126 were significantly lower in the CKD G2, CKD G3A, CKD G3B, CKD G4, CKD G5 and CKD G5D groups but not in the CKD G1 group.
The correlation between serum miR-126 and miR-223 levels in the cohort is presented in Supplementary Figure 1 and is statistically significant. Serum levels of miR-126 and miR-223 were the same whatever dialysis technique (Supplementary Figure 2). It should be noted that the exogenous control used here (Cel-miR-39) had a similar Cq for all groups (Supplementary Figure 3).
Association of miR-126 and miR-223 with clinical and laboratory parameters
The results of linear regression analysis between miR-126 or miR-223 and baseline clinical and laboratory parameters are presented in Table 2.
Table 2.
Association of miR-223 and miR-126 with clinical and laboratory parameters (Spearman’s rank correlation coefficient, ρ).
| Parameters | miR-223 | miR-126 | ||
|---|---|---|---|---|
| ρ | p | ρ | p | |
| Age (n = 628) | −0.21 | p < 0.0001 | −0.28 | p < 0.0001 |
| Sex (ref:female) (n = 628) | 0.15 | p = 0.0001 | 0.01 | p = 0.78 |
| Hypertension (n = 627) | −0.05 | p = 0.19 | −0.18 | p < 0.0001 |
| MAP (n = 627) | −0.02 | p = 0.60 | −0.02 | p = 0.66 |
| BMI (n = 625) | −0.10 | p = 0.009 | −0.13 | p = 0.0008 |
| Diabetes (n = 628) | −0.08 | p = 0.04 | −0.09 | p = 0.03 |
| CRP (n = 591) | −0.03 | p = 0.43 | −0.04 | p = 0.28 |
| Serum Creatinine (n = 562) | −0.31 | p < 0.0001 | −0.23 | p < 0.0001 |
| eGFR (n = 562) | 0.30 | p < 0.0001 | 0.26 | p < 0.0001 |
| Urea (n = 549) | −0.29 | p < 0.0001 | −0.25 | p < 0.0001 |
| Calcium (n = 552) | 0.09 | p = 0.03 | −0.02 | p = 0.68 |
| Phosphate (n = 548) | −0.09 | p = 0.04 | −0.17 | p < 0.0001 |
| PTH (n = 438) | −0.26 | p < 0.0001 | −0.23 | p < 0.0001 |
| 25-OH-D (n = 330) | −0.08 | p = 0.17 | −0.09 | p = 0.12 |
| Proteinuria (n = 524) | −0.07 | p = 0.19 | −0.10 | p = 0.02 |
| Hemoglobin (n = 583) | 0.29 | p < 0.0001 | 0.24 | p < 0.0001 |
| Leukocyte count (n = 581) | 0.31 | p < 0.0001 | 0.04 | p = 0.28 |
| Platelet count (n = 580) | 0.24 | p < 0.0001 | 0.18 | p < 0.0001 |
| Glucose (n = 537) | −0.04 | p = 0.31 | −0.08 | p = 0.08 |
| Triglycerides (n = 518) | −0.03 | p = 0.44 | 0.01 | p = 0.91 |
| Cholesterol (n = 527) | 0.16 | p = 0.0002 | 0.14 | p = 0.0009 |
| HDL (n = 440) | 0.08 | p = 0.11 | 0.06 | p = 0.22 |
| LDL (n = 422) | 0.15 | p = 0.002 | 0.13 | p = 0.006 |
MAP: mean arterial pressure; BMI: body mass index; CRP: C-reactive protein; eGFR: estimated glomerular filtration rate; PTH: parathyroid hormone; 25-OH-D: 25-hydroxy vitamin D; HDL: high-density lipoprotein; LDL: low-density lipoprotein.
The healthy control group was excluded from the analysis of laboratory characteristics and the RRT group was excluded for non-interpretable laboratory parameters (serum calcium, phosphate, urea, eGFR and proteinuria), due to the dialysis.
A multiple regression analysis was used to assess the relationship between miR-223 and the parameters significantly associated with miRNA expression in a univariate analysis. Parameters independently associated with miR-223 levels were the leukocyte count, eGFR, hemoglobin and sex (Supplementary Table 3).
Similarly, we used a multiple linear regression model of the entire cohort that included the variables significantly associated with miR-126 expression in univariate analysis; only eGFR, platelet count, hemoglobin level, and age were independently associated with serum miR-126 levels (Supplementary Table 3).
All-cause mortality and cardiovascular outcomes
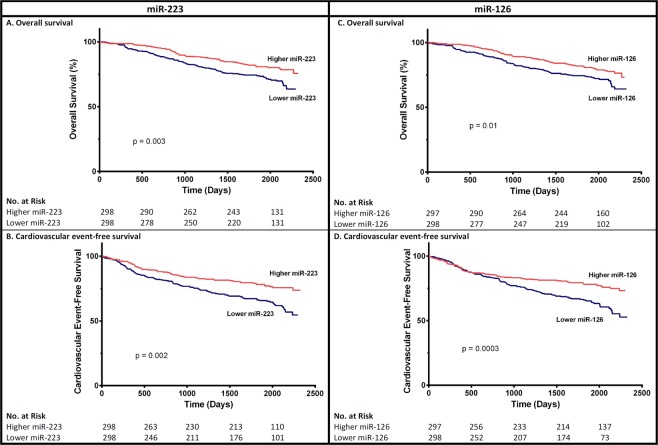
Overall, during a median follow-up period of 1948 days [1630–2146], 173 patients experienced cardiovascular events and 150 died (Table 3). Infection accounted for 25 deaths, malignant disease for 24, and cardiovascular events for 29 deaths (Table 3). Twelve patients were lost to follow-up. When considering all-cause mortality, patients with levels of miR-223 and miR-126 below the median had a lower survival rate (Fig. 3A and C). However, the associations between both miRNA levels and overall mortality were no longer significant after adjustment for baseline eGFR (Supplementary Table 4). To confirm this result, we performed a power analysis. We assume a one-sided adjusted alpha risk of 2.5% (two miRNA were studied), a power of 80% and a correlation coefficient of 0.35 between eGFR and each miRNA. With the latter hypotheses, 150 deaths could permit the detection of a HR = 1.63 between the two groups defined by the median miRNA. An HR around 1.50 is realistic for a superiority time-to-event trial, therefore our study had enough statistical power to compared overall survival according to miRNA. Sensitivity analysis using Cox models adjusted for each CKD stage or a model with the eGFR*miR interaction or combining both miRNAs levels did not reveal any significant associations between miRNA levels and mortality after adjustment for eGFR baseline either (data not shown). When we focused more specifically on cardiovascular mortality, there was no significant association with miRNA levels after adjustment for baseline eGFR (Supplementary Table 5). When considering CV events, patients with below-median levels of miR-223 and miR-126 had a lower CV event rate (Fig. 3B and D). However, theses associations were no longer significant after adjustment for baseline eGFR (Supplementary Table 6).
Table 3.
Outcomes in the CKD cohort and each CKD stage group.
| n | CKD Cohort | CKD G1 | CKD G2 | CKD G3A | CKD G3B | CKD G4 | CKD G5 | CKD G5D | p |
|---|---|---|---|---|---|---|---|---|---|
| 597 | 65 | 88 | 120 | 158 | 108 | 23 | 35 | ||
| Follow-up time (days) | 1948 [1630–2146] | 1988 [1744–2181] | 1966 [1787–2142] | 2034 [1735–2182] | 1920 [1417–2117] | 1869 [938–2130] | 1907 [775–2171] | 1839 [1135–2157] | p = 0.006 |
| Alive, n (%) | 447 (75) | 64 (98) | 79 (90) | 99 (82) | 112 (71) | 58 (54) | 13 (56) | 22 (63) | p < 0.0001 |
| Primary outcome, n (%) | 250 (42) | 4 (6) | 16 (18) | 43 (36) | 85 (54) | 65 (60) | 14 (61) | 23 (66) | p < 0.0001 |
| Death, n (%) | 150 (25) | 1 (2) | 9 (10) | 21 (17) | 46 (29) | 50 (46) | 10 (43) | 13 (37) | p < 0.0001 |
| infection | 25 (4) | 1 (2) | 0 (0) | 3 (3) | 6 (4) | 11 (10) | 1 (4) | 3 (9) | |
| malignancy | 24 (4) | 0 (0) | 0 (0) | 6 (5) | 9 (6) | 7 (6) | 1 (4) | 1 (3) | |
| cardiovascular cause | 29 (5) | 0 (0) | 3 (3) | 3 (3) | 11 (7) | 9 (8) | 1 (4) | 2 (6) | |
| refuse dialysis | 4 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) | 0 (0) | 2 (6) | |
| undetermined | 68 (11) | 0 (0) | 6 (7) | 9 (8) | 20 (13) | 21 (19) | 7 (30) | 5 (14) | |
| Cardiovascular events, n (%) | 173 (29) | 3 (5) | 11 (12) | 33 (27) | 63 (40) | 39 (36) | 8 (35) | 16 (46) | p < 0.0001 |
| atheromatous | 119 (20) | 3 (5) | 6 (7) | 27 (22) | 41 (26) | 26 (24) | 6 (26) | 10 (29) | |
| non-atheromatous | 71 (12) | 0 (0) | 6 (7) | 9 (7) | 26 (16) | 20 (19) | 3 (13) | 7 (20) | |
| Secondary outcome, n (%)* | 83 (15) | 0 (0) | 7 (8) | 6 (5) | 18 (11) | 38 (35) | 14 (61) | n.a. | p < 0.0001 |
| Renal events, n (%) | 57 (10) | 0 (0) | 3 (3) | 1 (1) | 7 (4) | 32 (30) | 14 (61) | n.a. | p < 0.0001 |
| dialysis | 49 (9) | 3 (3) | 1 (1) | 7 7 (4) | 27 (25) | 11 (48) | p < 0.0001 | ||
| transplant | 16 (3) | 0 (0) | 0 (0) | 0 (0) | 6 (6) | 5 (22) | p < 0.0001 | ||
| Renal impairment, n (%) | 45 (8) | 0 (0) | 6 (7) | 6 (5) | 13 (8) | 18 (17) | 2 (9) | n.a. | p < 0.0001 |
n.a.: not applicable.
*n = 562 (excluding CKD 5D).
Continuous variables are expressed as the median [IQR] and binary variables are expressed as the number (%).
Figure 3.
Unadjusted survival curves for primary outcomes for participants with above-median and below-median serum miR-223 (A and B) or miR-126 (C and D) levels (Log-rank test).
We further divided CV events into atheromatous and non-atheromatous events and performed the statistical analysis for each in relation with the miRNA levels. The results were globally similar, but it should be pointed out that in univariate analysis, miR-223 was associated with non-atheromatous events, but not with atheromatous events (Supplementary Tables 7 and 8).
Renal outcomes
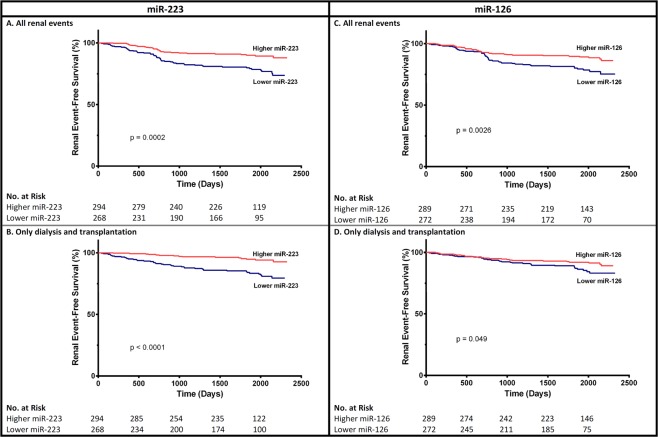
Among the 562 non-dialysis CKD patients, 57 developed need for renal replacement therapy, either dialysis or transplantation during follow-up (Table 3) and 45 experienced an accelerated loss of renal function (defined as a doubling of the serum creatinine level, or a reduction of eGFR by 30%, over two years). CKD patients stage G1 to G5 not yet on dialysis with above-median levels of miR-223 and miR-126 had a significantly better event-free survival rate (Fig. 4). However, the associations between both miRNA levels and renal events were no longer significant after adjustment for baseline eGFR (Supplementary Table 9).
Figure 4.
Unadjusted survival curves for secondary outcomes for participants with above-median and below-median serum miR-223 (A and B) or miR-126 (C and D) levels (Log-rank test).
After a median follow up of 737 days, among 458 patients with non-dialysis CKD, 134 patients were considered as fast progressors (an eGFR decrease of more than 3 mL/min/1.73 m²/year), 229 as non-progressors (an eGFR decrease of less than 1 mL/min/1.73 m²/year) and 95 as slow progressors (an eGFR decrease of between 1 and 3 mL/min/1.73 m²/year). There was no difference between the three groups with regard to the serum levels of miR-223 and miR-126 (Supplementary Figure 4).
Discussion
The present study demonstrated a statistically significant decrease in circulating miR-126 and miR-223 levels in patients with more severe stages of CKD. Levels of miR-126 were independently associated with eGFR but also with hemoglobin level, platelet count, and age. The circulating level of miR-223 was independently associated with leukocyte count and hemoglobin level, sex, and eGFR. We did not find an independent correlation between any studied miRNA and the serum level of CRP or with diabetes. The event-free survival rate was better in patients with higher circulating miRNA levels. After adjustment for eGFR, neither miR-126 nor miR-223 level were prognostic markers of all-cause mortality, cardiovascular events or renal events. We did not observe an association between the initial circulating levels of miR-126 and miR-223 and the decline of kidney function over two years.
Glomerular filtration rate estimated from the serum level of creatinine currently remains the most convenient marker of renal function for everyday practice and large epidemiologic studies. Albuminuria, proteinuria and seric urea levels are useful indicators of renal function when normalized for urinary creatinine. They are however not very sensitive when used to detect the first stages of CKD33. Several other markers have been assessed for their predictive value of mortality, cardiovascular (CV) complications and kidney disease progression without success34–40. miRNAs have recently attracted attention as biomarker candidates to assess the severity and/or the etiologies of kidney disease41,42. One of the main advantages of miRNAs is their seric stability, which makes them suitable as a non-invasive biomarker27. One can thus hope that these small RNAs could prove to be reliable enough to be useful in potential clinical practice43,44.
Quantitative PCR is a technique commonly used for accurate quantification of miRNA levels in blood samples and transplantable in a clinical laboratory. In quantitative PCR, it is important to choose an adequate control gene, especially as miRNA levels are low in the circulation. Some teams have chosen to use small endogenous RNAs, such as U6, to normalize circulating miRNAs levels but the results have proven to be unreliable due to marked variability of the expression of these RNAs according to pathological conditions45. An emerging consensus is now to spike-in in the blood sample before extraction a given quantity of exogenous miRNA (such as the synthetic Caenorhabditis elegans miR-39)46,47.
Several teams have shown that the overall expression of miRNAs blood concentration diminishes as CKD advances48,49. In a cohort of 90 CKD patients (stages G3 to G5D) Chen et al. published that miR-125b, miR-145 and miR-155 levels decline as CKD progresses50. The blood levels of cardiac miRNAs also seems to decline with eGFR51. In the present study, we observed a decrease in serum levels of miR-126 and miR-223 as renal function declined. However, proof that kidney function affects miRNA levels in plasma and urine is sparse and sometimes contradictory. Some studies were interested in the possible accumulation of RNases known in renal failure52 which could increase degradation of circulating miRNAs in the plasma, but these are protected in particular by exosomes and Argonaut proteins. In the present study, serum RNA levels after extraction did not differ as a function of the CKD stage (data not shown) – suggesting that there is no degradation, in line with other reports50.
Over the last years, we studied the role of uremic toxins in in vitro models of osteoclastogenesis and vascular calcification.5 We highlighted the roles of miR-223 and miR-126 in the trans-differentiation of VSMCs to an osteoblastic phenotype linked with vascular calcification18,53. We confirmed this finding in vivo by showing that miR-126 and miR-223 expression was enhanced in aortas of murine CKD models, while their serum levels were decreased8 which may indicate an accumulation of these two miRNAs in vascular walls. In 2016, Ulbing et al. found a decrease of seric levels of miR-223 in CKD patients that was alleviated after renal graft19. In our large cohort covering the whole spectrum of CKD, we also observed low expression - from stage G3B onwards for miR-223, and from stage G2 onwards for miR-126.
In the present study, the eGFR was the clinical feature most strongly correlated with miRNA concentration (apart from leukocyte count for miR-223 and age for miR-126). The observed associations between the miRNA level and the other features of severe kidney failure (e.g. hyperparathyroidism and hyperphosphatemia) in univariate analyses were mediated by the impairment in glomerular filtration. There was a good degree of correlation between miR-223 level on one hand and sex, leukocyte count, platelet count, and blood hemoglobin and cholesterol levels on the other. The relationship with blood cell counts has already been reported, and is related to the involvement of miR-223 in hematopoiesis and its strong expression in granulocytes, platelets and red blood cells10–12,54–56. miR-223 has been involved before in cholesterol homeostasis57, and we found accordingly a correlation with cholesterol levels using univariate analysis. In our multivariate analysis, only loss of kidney function, a low leukocyte count, a low hemoglobin level, and male sex were associated with low circulating levels of miR-223. Although miR-223 is often considered to be a marker of inflammation58, levels were not correlated with serum CRP. In univariate analyses, the circulating level of miR-126 was notably correlated with age, hemoglobin and cholesterol levels, and platelet count. The correlation with cholesterol and LDL has already been reported but the underlying mechanism have not yet been identified59. In our multivariate analysis, miR-126 level was independently associated with eGFR, platelet count, hemoglobin level, and age. The correlations with hemoglobin and eGFR have previously been described60, as have the correlations with platelet count61 and age62. Grabher et al. demonstrated that miR-126 regulates the proto-oncogene c-Myb during hematopoiesis63.
According to our results, miR-126 and miR-223 cannot be used as prognostic markers because of their eGFR-related variability. Indeed, miR-126 and miR-223 levels were not independently associated with poor survival after adjustment for eGFR. The results were similar for mortality, cardiovascular events, and renal events – none of which were predictive of a further decline in eGFR.
The present study is the first having investigated these two miRNAs as potential biomarkers in CKD. Its strengths include the large study population and the availability of data over the full range of CKD stages including a group of controls without CKD, enabling stratification by CKD stage. We also assessed a number of important, potentially confounding routine laboratory parameters with a long term follow-up. We studied for the first time in a large CKD cohort the impact of serum miRNA levels on hard outcomes. However, our study also has several limitations. Estimated GFR was used in the analysis, instead of the more accurate direct measurement of GFR. Nevertheless, we used the CKD-EPI formula which estimates GFR more accurately compared to other widely-used formulas. Another limitation is that miRNA levels were only measured at the baseline examination.
In conclusion, lower circulating levels of miR-126 and miR-223 were associated with lower eGFR in a cohort of patients with CKD at different stages. The observed link between the two miRNAs’ seric levels and mortality, cardiovascular events or renal events in CKD appears to depend on eGFR. However, this does not preclude their potential role in the pathophysiology of CKD, which will need further evaluation.
Methods
The study population
A total of 601 patients with CKD stages G1 to G5 (as defined by the Kidney Disease Outcomes Quality Initiative guidelines (KDOQI)) or on renal replacement therapy (RRT: hemodialysis or peritoneal dialysis) and 31 healthy controls were included in this single-center study at the outpatient clinic of the Section of Nephrology, Ghent University Hospital (Ghent, Belgium). Inclusion took place between January 2011 and January 2014. The exclusion criteria were pregnancy, age under 18, active infection, active malignancy or history of transplantation. Outcome parameters were monitored until June 2017 (for up to 6 years). Patients with CKD were categorized into subgroups according to their estimated glomerular filtration rate (eGFR), using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) – creatinine equation, as recommended by the KDOQI. Eight subgroups were compared, including six CKD subgroups not on dialysis (eGFR: over 90, 60–89, 45–59, 30–44, 15–29, and below 15 ml/min/1.73 m²), one CKD subgroup on RRT (stage G5D), and healthy controls. Hypertension was defined as a systolic blood pressure >140 mmHg and/or a diastolic blood pressure >90 mmHg, or the need for anti-hypertensive medication.
Ethical approval
This study was approved by the local ethical committee at Ghent University Hospital. Written informed consent was obtained from all participants. The study complied with the tenets of the Declaration of Helsinki and its amendments.
Blood samples
A sample of venous blood was collected in Venosafe serum (Terumo Europe, Leuven, Belgium) tubes. The blood was left to coagulate for 30 minutes on the bench and was then centrifuged at 2,095 × g for 10 min at room temperature (RT). The serum samples were divided into 500 μL aliquots on ice and stored at −80 °C until batch analysis.
RNA extraction
Total RNA was extracted with the miRNeasy Serum/Plasma kit (Qiagen, Germany), according to the manufacturer’s instructions. Stored serum was thawed on ice and shaken. Next, 1000 μl of Qiazol lysis reagent was added to 200 μl of serum and incubated at RT for 5 min. A fixed amount of synthetic Caenorhabditis elegans miR-39 (3.5 µL of Cel-miR-39 at 1.6 × 108 copies/µL, i.e. 1 fmol) was added as an internal control. Next, 200 μl of chloroform was added, and the sample was shaken for 15 s. After incubation for 3 min at room temperature, the samples were centrifuged for 15 min at 16,100 × g at 4 °C. The upper aqueous phase (approximately 700 μl) was transferred to a new tube, and 1000 μl of 100% ethanol was added. The RNA was isolated with a miRNeasy MinElute spin column and solutions (Qiagen, Germany), eluted from the columns with 14 μl RNase-free water, and stored at −80 °C.
The purity and concentration of the isolated RNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA).
Reverse transcription
Isolated RNA was reverse-transcribed into complementary DNA using TaqMan miRNA-specific primers and the TaqMan microRNA reverse transcription kit (Applied Biosystems, USA) on ice. Each well contained the following mixture: 2.8 μL of RNase-free water, 1.5 μL of 10X RT buffer, 1.5 μL of dNTP mix (10 mM), 0.2 μL of RNase inhibitor (20 U/μL), 1 μL of MultiScribe® RT enzyme ΜL (50 U/µL), 3 μL of 5X primers (i.e. for miR-223, miR-126 or Cel-miR-39) and 5 μL of RNA 5 ng/μL (i.e. 25 ng). The Eppendorf Mastercycler® thermocycler was used to incubate the samples at 16 °C for 30 min, at 42 °C for 30 min and then at 85 °C for 5 min. The cDNA samples were then stored at −20 °C.
Quantification of serum miRNA levels
Serum levels of miR-223 and miR-126 were quantified using TaqMan qRT-PCR. cDNA (2.4 μL) was amplified using 18 μL of SsoAdvanced™ Universal Probes Supermix (Bio-Rad Laboratories, USA), 1.8 μL of 20X Primers and 13.8 μL of RNase-free water, in a final volume of 36 μL. All reactions and analyses were performed in triplicate on CFX Connect (Bio-Rad) according to the following protocol: incubation at 95 °C for 30 seconds, then 40 cycles of 15 seconds at 95 °C, and incubation at 60 °C for 30 seconds. Exogenously added Caenorhabditis elegans-miR-39 (Cel-miR-39) was used as a spiked-in normalization control. The relative expression levels of miR-223 and miR-126 were calculated using the 2-ΔCq method, where ΔCq = Cq (miR-223 or miR-126) − Cq (cel-miR-39). The maximum accepted coefficient of variation for intra-assay replicates was set to 5%. All experiments were performed by the same person (O.F.) who was blinded for the outcomes, to avoid bias.
Outcomes
All the outcomes were collected prospectively. The primary outcomes were all-cause mortality, and cardiovascular events (fatal and non-fatal). Cardiovascular events were defined as atheromatous or non-atheromatous cardiovascular disease (CVD). Atheromatous CVD was defined as coronary artery disease (CAD), ischemic stroke or transient ischemic attack, peripheral arterial disease requiring a revascularization procedure or an amputation. Non atheromatous CVD was defined as heart failure in the absence of CAD, cardiac rhythm or conduction disorders, or aortic aneurysm.
Renal events were defined as progression towards RRT (dialysis or renal transplantation), or a doubling of the serum creatinine level, or a reduction of eGFR by 30%, over two years (according to the FDA criteria64). The CKD G5D group was excluded from the analysis of the renal events. The variation of eGFR between inclusion and end of renal follow-up (at one or two years of follow-up according to the patients) was calculated. We also studied miRNA levels in three strata: a decrease in the eGFR <1 mL/min/1.73 m²/year (non-progressors), 1–3 mL/min/1.73 m²/year (slow progressors) or >3 mL/min/1.73 m²/year (fast progressors).
Statistical analyses
Patient characteristics and the relative expression levels of miR-223 and miR-126 are presented as median [interquartile range (IQR)] for continuous variables (because the data were not normally distributed except for hemoglobin) or as the number (frequency) for binary variables. Intergroup comparisons were performed using a Kruskal-Wallis test for continuous variables and a χ² test for binary variables. Comparisons between two groups were performed with a Mann-Whitney test. The relationships between patient characteristics and miRNA levels were explored using Spearman’s rank correlation tests. Multiple linear regression analyses were performed to investigate the association between clinical/laboratory parameters and the expression of miRNAs. The Kaplan-Meier survival curves were constructed to estimate overall mortality, cardiovascular and renal events by dividing the studied population according to the selected cut-off (the median level: 2.28 for miR-223 and 0.09 for miR-126). The log-rank test was used to compare survival curves. Univariate and multivariate analyses of outcomes were performed by building a Cox proportional hazards model based on miRNA levels. Due to non-normal distribution, the data were log-transformed. The multivariate analysis included all the parameters with a p-value < 0.05 in the univariate analysis. In all the tests, the threshold for statistical significance was set to p < 0.05. Statistical analyses were performed using GraphPad Prism® software version 6 and SPSS® software version 21. The power analysis was performed using package powerSurvEpi with software “RStudio software Version 1.0.143 – © 2009–2016 RStudio (R.3.4.0 software)” as described previously65.
Supplementary information
Acknowledgements
Part of this work was funded by a grant MIRNA from the Picardie Regional Council. Anneleen Pletinck is a post-doctoral researcher of the Research Fund Flanders (FWO). ZAM reports Receipt of grants/research supports: Amgen, Baxter, Fresenius Medical Care, GlaxoSmithKline, Merck Sharp and Dohme-Chibret, Genzyme/Sanofi, Lilly, Otsuka, and Government support for CKD REIN PROJECT AND EXPERIMENTAL PROJECTS. Receipt of honoraria or consultation fees: To charities from Amgen, Bayer, and Daichii. Other authors reported no conflict of interest.
Author Contributions
O.F., E.S., V.M.L.M., G.G., R.V., G.C., S.B., Z.A.M. and L.M. made a substantial contribution to conception and design of the study, O.F., E.S., B.B., F.V. and A.P. made a substantial contribution to data acquisition, O.F., E.S., S.L., V.M.L.M., M.D., G.G., R.V., G.C., Z.A.M. and L.M. made a substantial contribution to data analysis and interpretation; O.F., E.S., V.M.L.M., G.G., R.V., G.C., S.B., Z.A.M. and L.M. drafted the article and/or revised it for important intellectual content. All Authors approved the final version of the submitted manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
A comprehensive list of consortium members appears at the end of the paper
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ophélie Fourdinier and Eva Schepers contributed equally.
Contributor Information
Laurent Metzinger, Email: laurent.metzinger@u-picardie.fr.
on behalf of the European Uremic Toxin Work Group-EUTox:
Angel Argiles, Joachim Beige, Philippe Brunet, Gerald Cohen, Omar Abou Deif, Pieter Evenepoel, Danilo Fliser, Ivo Fridolin, Andreas Gmerek, Joachim Jankowski, Vera Jankowski, Roos Masereeuw, Harald Mischak, Alberto Ortiz, Alessandra Perna, Juan Mariano Rodriguez-Portillo, Joost Schanstra, Goce Spasovski, Dimitrios Stamatialis, Sonja Steppan, Markus Storr, Bernd G. Stegmayr, Peter Stenvinkel, Paul J. Thornalley, and Andrej Wiecek
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41101-8.
References
- 1.Vallianou NG, Mitesh S, Gkogkou A, Geladari E. Chronic Kidney Disease and Cardiovascular Disease: is there any relationship? Curr Cardiol Rev. Curr Cardiol Rev. 2019;15(1):55. doi: 10.2174/1573403X14666180711124825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rennenberg RJ, et al. Vascular calcifications as a marker of increased cardiovascular risk: a meta-analysis. Vasc Health Risk Manag. 2009;5(1):185. doi: 10.2147/vhrm.s4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rong D, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Metzinger-Le Meuth V, et al. The expanding roles of microRNAs in kidney pathophysiology. Nephrol Dial Transplant. 2019;34(1):7. doi: 10.1093/ndt/gfy140. [DOI] [PubMed] [Google Scholar]
- 6.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2010;31(11):2383. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 7.Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol. 2015;11(1):23. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 8.Taibi F, et al. Possible involvement of microRNAs in vascular damage in experimental chronic kidney disease. Biochim Biophys Acta. 2014;1842(1):88. doi: 10.1016/j.bbadis.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Metzinger-Le Meuth V, et al. microRNAs are dysregulated in the cerebral microvasculature of CKD mice. Front Biosci (Elite Ed) 2014;6:80. doi: 10.2741/e693. [DOI] [PubMed] [Google Scholar]
- 10.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 11.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 12.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Kang W, et al. Stathmin1 plays oncogenic role and is a target of microRNA-223 in gastric cancer. PLoS One. 2005;7(3):e33919. doi: 10.1371/journal.pone.0033919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138(5):763. doi: 10.1007/s00432-012-1154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang H, et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer. 2015;14:58. doi: 10.1186/s12943-015-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, et al. Downregulation of miR-223 and miR-153 mediates mechanical stretch-stimulated proliferation of venous smooth muscle cells via activation of the insulin-like growth factor-1 receptor. Arch Biochem Biophys. 2012;528(2):204. doi: 10.1016/j.abb.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Shan Z, et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J Am Coll Cardiol. 2015;65(23):2526. doi: 10.1016/j.jacc.2015.03.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangrez AY, et al. Inorganic phosphate accelerates the migration of vascular smooth muscle cells: evidence for the involvement of miR-223. PLoS One. 2012;7(10):e47807. doi: 10.1371/journal.pone.0047807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulbing M, et al. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone. 2017;95:115. doi: 10.1016/j.bone.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris TA, et al. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105(5):1516. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zernecke A, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 23.Bijkerk R, et al. Hematopoietic microRNA-126 protects against renal ischemia/reperfusion injury by promoting vascular integrity. J Am Soc Nephrol. 2014;25(8):1710. doi: 10.1681/ASN.2013060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zampetaki A, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 25.Slaby O, et al. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer. 2012;51(7):707. doi: 10.1002/gcc.21957. [DOI] [PubMed] [Google Scholar]
- 26.Vergho DC, et al. Impact of miR-21, miR-126 and miR-221 as prognostic factors of clear cell renal cell carcinoma with tumor thrombus of the inferior vena cava. PLoS One. 2014;9(10):e109877. doi: 10.1371/journal.pone.0109877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 30.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickers KC, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waikar SS, Betensky RA, Bonventre JV. Creatinine as the gold standard for kidney injury biomarker studies? Nephrol Dial Transplant. 2009;24(11):3263. doi: 10.1093/ndt/gfp428. [DOI] [PubMed] [Google Scholar]
- 34.Kern EF, et al. Early urinary markers of diabetic kidney disease: a nested case-control study from the Diabetes Control and Complications Trial (DCCT) Am J Kidney Dis. 2014;55(5):824. doi: 10.1053/j.ajkd.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TQ, et al. Urinary connective tissue growth factor excretion correlates with clinical markers of renal disease in a large population of type 1 diabetic patients with diabetic nephropathy. Diabetes Care. 2006;29(1):83. doi: 10.2337/diacare.29.1.83. [DOI] [PubMed] [Google Scholar]
- 36.Boes E, et al. Apolipoprotein A-IV predicts progression of chronic kidney disease: the mild to moderate kidney disease study. J Am Soc Nephrol. 2006;17(2):528. doi: 10.1681/ASN.2005070733. [DOI] [PubMed] [Google Scholar]
- 37.Bhavsar NA, Kottgen A, Coresh J, Astor BC. Neutrophil gelatinase-associated lipocalin (NGAL) and kidney injury molecule 1 (KIM-1) as predictors of incident CKD stage 3: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Kidney Dis. 2012;60(2):233. doi: 10.1053/j.ajkd.2012.02.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabbisetti VS, et al. Blood kidney injury molecule-1 is a biomarker of acute and chronic kidney injury and predicts pro gression to ESRD in type I diabetes. J Am Soc Nephrol. 2014;25(10):2177. doi: 10.1681/ASN.2013070758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fliser D, et al. Fibroblast growth factor 23 (FGF23) predicts progression of chronic kidney disease: the Mild to Moderate Kidney Disease (MMKD) Study. J Am Soc Nephrol. 2007;18(9):2600. doi: 10.1681/ASN.2006080936. [DOI] [PubMed] [Google Scholar]
- 40.Peters HP, et al. High urinary excretion of kidney injury molecule-1 is an independent predictor of end-stage renal disease in patients with IgA nephropathy. Nephrol Dial Transplant. 2007;26(11):3581. doi: 10.1093/ndt/gfr135. [DOI] [PubMed] [Google Scholar]
- 41.Nassirpour R, Raj D, Townsend R, Argyropoulos C. MicroRNA biomarkers in clinical renal disease: from diabetic nephropathy renal transplantation and beyond. Food Chem Toxicol. 2016;98(Pt A):73. doi: 10.1016/j.fct.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Etheridge A, et al. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1-2):85. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerr KF, et al. Evaluating biomarkers for prognostic enrichment of clinical trials. Clin Trials. 2014;14(6):629. doi: 10.1177/1740774517723588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang M, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 46.Roberts TC, Coenen-Stass AM, Wood MJ. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One. 2014;9(2):e89237. doi: 10.1371/journal.pone.0089237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang G, et al. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med Sci Monit. 2015;21:617. doi: 10.12659/MSM.891028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv LL, et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9(10):1021. doi: 10.7150/ijbs.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neal CS, et al. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26(11):3794. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 50.Chen NX, et al. Decreased microRNA is involved in the vascular remodeling abnormalities in chronic kidney disease (CKD) PLoS One. 2013;8(5):e64558. doi: 10.1371/journal.pone.0064558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gidlof O, et al. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118(4):217. doi: 10.1159/000328869. [DOI] [PubMed] [Google Scholar]
- 52.Karpetsky TP, Humphrey RL, Levy CC. Influence of renal insufficiency on levels of serum ribonuclease in patients with multiple myeloma. J Natl Cancer Inst. 1977;58(4):875. doi: 10.1093/jnci/58.4.875. [DOI] [PubMed] [Google Scholar]
- 53.M’Baya-Moutoula E, et al. High inorganic phosphate concentration inhibits osteoclastogenesis by modulating miR-223. Biochim Biophys Acta. 2015;1852(10 Pt A):2202. doi: 10.1016/j.bbadis.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 54.O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118(11):2960. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan JY, et al. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J Cell Mol Med. 2009;13(11–12):4551. doi: 10.1111/j.1582-4934.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M’Baya-Moutoula E, et al. A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim Biophys Acta. 2018;1864(8):2664. doi: 10.1016/j.bbadis.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Vickers KC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA. 2014;111(40):14518. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taibi F, Metzinger-Le Meuth V, Massy ZA, Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta. 2014;1842(7):1001. doi: 10.1016/j.bbadis.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Sun X, et al. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL. cholesterol. Thromb J. 2012;10(1):16. doi: 10.1186/1477-9560-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, et al. Circulating levels of inflammation-associated miR-155 and endothelial-enriched miR-126 in patients with end-stage renal disease. Braz J Med Biol Res. 2012;45(12):1308. doi: 10.1590/S0100-879X2012007500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaudewitz D, et al. Association of MicroRNAs and YRNAs With Platelet Function. Circ Res. 2016;118(3):420. doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olivieri F, et al. Age- and glycemia-related miR-126-3p levels in plasma and endothelial cells. Aging (Albany NY) 2014;6(9):771. doi: 10.18632/aging.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grabher C, et al. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2011;25(3):506. doi: 10.1038/leu.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levey AS, et al. GFR decline as an end point for clinical trials in CKD: a scientific workshop sponsored by the National Kidney Foundation and the US Food and Drug Administration. Am J Kidney Dis. 2014;64(6):821. doi: 10.1053/j.ajkd.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 65.Hsieh FY, Lavori PW. Sample-size calculations for the Cox proportional hazards regression model with nonbinary covariates. Control Clin Trials. 2000;21(6):552–60. doi: 10.1016/s0197-2456(00)00104-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.