Abstract
Tomato fruit are especially susceptible to chilling injury (CI) when continuously exposed to temperatures below 12 °C. In this study, integrative comparative analyses of transcriptomics and metabolomics data were performed to uncover the regulatory network in CI tomato fruit. Metabolite profiling analysis found that 7 amino acids, 27 organic acids, 16 of sugars and 22 other compounds had a significantly different content while transcriptomics data showed 1735 differentially expressed genes (DEGs) were down-regulated and 1369 were up-regulated in cold-stored fruit. We found that the contents of citrate, cis-aconitate and succinate were increased, which were consistent with the expression of ATP-citrate synthase (ACS) and isocitrate dehydrogenase (IDH) genes in cold-treated tomato fruit. Cold stress promotes the expression of ACS and IDH which may increase the synthesis of citrate, cis-aconitate and succinate. Alanine and leucine had increased contents, which may result from alanine aminotransferase (ALT) and branched-chain amino acid aminotransferase (BcAT)’s high expression levels, respectively. Overall the transcriptomics and metabolomics data in our study explain the molecular mechanisms of the chilling injury and expands our understanding of the complex regulatory mechanisms of a metabolic network in response to chilling injury in tomato fruit.
Introduction
Low-temperature storage is one of the most effective methods to maintain the nutrients and reduce postharvest decay of fruits and vegetables. However, it is risky to expose tropical and subtropical species to low temperatures, because it can induce the production of a physiological disorder known as chilling injury (CI). CI disorders can cause a reduction in postharvest quality and heavy economic losses1.
Tomato, originating from tropical region, is the second-most important vegetable in the world. Tomato fruit are especially susceptible to CI when continuously exposed to temperatures below 12 °C2. Chilled tomato fruit symptoms include skin pitting, water-soaking, diseases caused by pathogen, and failure to develop full color3,4. CI causes the changes of lipid composition and alterations of conformation and structure in cell membrane, resulting in a decrease of its fluidity and permeability. CI also induces an over-production of reactive oxygen species (ROS) and thus oxidative stress in storage fruit4. The ion leakage, malondialdehyde (MDA), proline content, and activities of antioxidative enzymes such as catalase (CAT) and peroxidase (POD) can act as indices to reflect physiological state of plant exposure to CI stress5.
Omics-based approaches have been used to understand the complex global biological mechanisms underlying various plant stress responses6. A transcriptomic analysis of tomato fruit treated with 6 °C for 48 h found that 38 genes were up-regulated, but only one gene coding a dehydrin was involved in cold-stress genes7. An expression analysis of tomato fruit with CI visual symptom after a long period of cold storage (4 weeks at 3 °C) indicated the alterations of genes involved in cell wall modifications, carotenoid biosynthesis, ethylene biosynthesis and signaling8. RNA-seq was used to identify differentially expressed genes in tomato fruit after the heat shock, cold treatment and subsequent ripening9. The chilling treatment down-regulated the genes related with photosynthesis, metabolism of cell wall, lipid and ethylene, and up-regulated the genes for trehalose synthesis and DOF and MYB transcription factors. On the other hand, the heat shock induced tolerance to CI including the up-regulation of genes involved in heat stress (HSTFs and HSPs), detoxification (GSTs), and sugars metabolism (TPPs and aldose 1-epimerase)9.
Proteomic studies in CI tomato fruit revealed that defensive mechanisms were linked to the uncoupling of photosynthetic processes (ATP synthase) and protein degradation machinery (26S proteosome)10. Besides, proteomic analysis of cold storage tomato fruit also indicated that the CI tolerance mechanisms were related to the accumulation of heat shock proteins (HSPs), molecular chaperones (GR-RBP), late embryogenesis abundant (LEA) proteins, and antioxidant enzymes (TPxI)11. The heat-shock induced chilling tolerance included altering levels of fruit metabolites such as arabinose, fructose-6-phosphate, valine and shikimic acid12. Volatile analysis of tomato fruit found that cold storage reduced the production of alcohol, aldehyde, ester, ketone, terpene and acid volatile compounds, and heat shock treatment prior to chilling exposure alleviated the suppression of the key volatile compounds13.
Integration of transcriptomic and metabolic profiling data can give important insights into gene-regulatory and metabolic events associated with plant growth and development processes. Recently, a number of studies have begun to integrate the multi-omics data sets, and some of them focused on the development and ripening of tomato fruit14–16. In this study, integrative comparative analysis of transcriptomics and metabolomics data from cold treatment tomato fruit was conducted to gain a broader systems perspective and to identify distinct molecular regulatory response during cold storage.
Results
Cold storage changes the metabolites profiles in tomato fruit
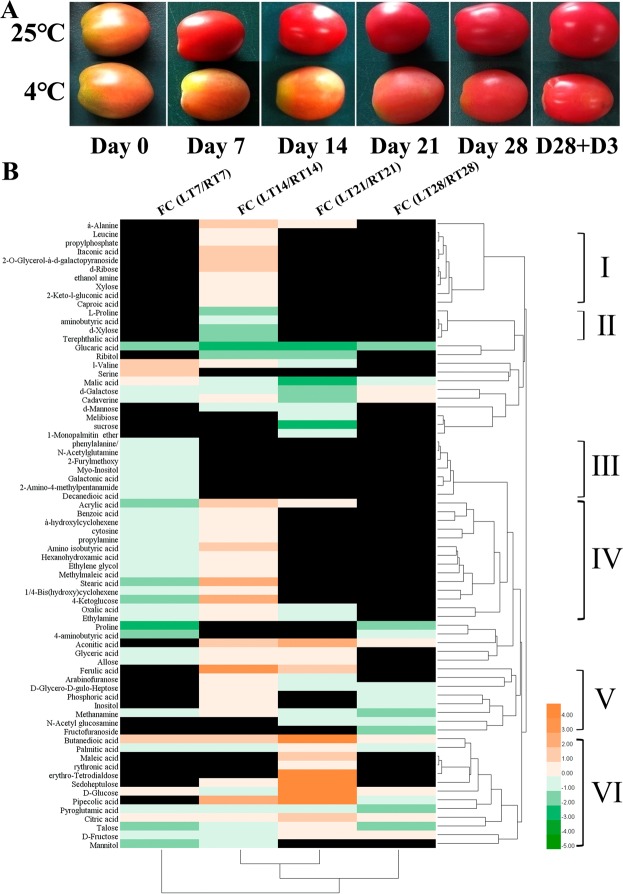
Cherry tomato fruit in the breaker developmental stage were stored at low temperature (4 °C) and room temperature (25 °C) for 28 days. The tomato fruit exhibited typical chilling injury symptoms. Tomato fruit stored at 4 °C (Fig. 1A) delayed the maturation process and 50% of the tomato fruit exhibited skin pitting when storage was continued at 25 °C for 3 days, mimicking the 28 days 4 °C shelf selling storage condition.
Figure 1.
Tomato storage and heatmap analysis of differential metabolites at low temperature (LT) and room temperature (RT). (A) Tomatoes were stored for 7 days, 14 days, 21 days, 28 days at 25 °C and 4 °C and then stored at 4 °C to 25 °C for 3 days mimicking shelf storage; (B) Fold change (FC) at LT and RT was shown and the blocks colored in red means the up-regulated metabolites and the down-regulated metabolites were colored in green. The blocks colored in black means no significant difference at that time point. Group numbers from I to VI were marked manually by cluster analysis results of the metabolites.
To investigate metabolic changes during cold storage, metabolite profiling was analyzed by using gas chromatography–mass spectrometry (GC-MS) method. For the analysis, the tomato fruit were chosen after 0, 7, 14, 21 and 28 days of low temperature treatment. Cold-stored fruit with significantly different metabolites content, including 7 amino acids, 27 organic acids, 16 sugars and 22 other compounds (Fig. 1B, Supplementary Table S1) from each storage stage were selected for further analysis. Cluster analysis showed these metabolites were divided into 6 groups based on their content pattern. In group I metabolites were induced at 14 days of treatment, while in group II the metabolites showed an opposite pattern that was suppressed at 14 days of treatment. The metabolites were inhibited at 7 days in group III. The content was suppressed at 7 days and induced at 14 days of treatment for metabolites in group IV. Most of the metabolites in group V and VI had significantly different contents during long term storage of 14, 21 and 28 days, and the metabolites were significantly induced at 21 days in group VI. It is interesting that the contents of butanedioic acid and citrate were continually induced by cold temperature at 7, 14, 21 and 28 days. However, glucaric acid and pyroglutamic acid showed an opposite pattern of gradually decreasing at 7, 14, 21 and 28 days cold treatment.
Cold treatment entails extensive transcriptome reprogramming
RNA-Seq was performed to obtain a global view of the transcriptome of cold-treated tomato fruit from 7, 14, 21 and 28 days of low temperature treatment. The differentially expressed genes (DEGs) between cold-stored fruit and room temperature stored fruit in each time stage were analyzed to identify the genes that respond to cold stress. A Venn diagram showed that 1735 DEGs were down-regulated and 1369 were up-regulated at each of the five storage stages, which were selected for further analysis (Fig. 2A). These DEGs were useful indicators to identify candidate genes for further in-depth analyses by qRT-PCR. Twenty-two out of 23 genes involved in sugar, amino acids and lipids metabolism, except Solyc02g088680.1 (UDP-glucose 6-dehydrogenase 2), showed similar expression patterns by RNA-Seq assay (Fig. 2B, Supplementary Table S2). The results also indicated that the data from the RNA-Seq were reproducible and reliable.
Figure 2.
Venn diagram and qRT-PCR validation of DEGs. (A) Left figure stands for the down-regulated genes while the right one stands for the up-regulated genes at 4 stages; (B) QRT-PCR validation of randomly selected genes related with sugar, organic acid, amino acid and lipid metabolome.
The DEGs were used for Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Interestingly, GO and KEGG analysis indicated a different metabolism network may exist after cold treatment since there were no coinciding GO and KEGG term for down and up-regulated DEGs. GO analysis identified 4 significant terms related with chloroplast and 11 DEGs related with photosynthesis that were up-regulated, which indicated that cold treatment tomato fruit still maintain the function of chloroplast (Table 1, Supplementary Table S3). GO terms was significantly enriched for down-regulated DEGs, integral component of membrane (212 DEGs), plasma membrane (47 DEGs), cell wall organization (14 DEGs), cell wall biogenesis (7 DEGs) as well as plant-type cell wall (13 DEGs) (Table 2, Supplementary Table S4) indicating that cold stress potentially affects membrane and cell wall functioning. Moreover, 24 DEGs involving in calcium binding and 8 DEGs related with calcium-dependent kinase activity were down-regulated, which showed that cold stress decreased the calcium-dependent signal transductions process (Table 2, Supplementary Table S4).
Table 1.
GO and KEGG enrichment for up-regulated DEGs.
| Category | Term | Name | Count | P-Value |
|---|---|---|---|---|
| Biological Process | GO:0006270 | DNA replication initiation | 6 | 0.00207 |
| Biological Process | GO:0052696 | flavonoid glucuronidation | 16 | 0.002376 |
| Biological Process | GO:0015979 | Photosynthesis | 11 | 0.002461 |
| Biological Process | GO:0009813 | flavonoid biosynthetic process | 16 | 0.004199 |
| Biological Process | GO:0009735 | response to cytokinin | 6 | 0.005635 |
| Biological Process | GO:0000103 | sulfate assimilation | 7 | 0.015144 |
| Biological Process | GO:0006541 | glutamine metabolic process | 5 | 0.020492 |
| Biological Process | GO:0009693 | ethylene biosynthetic process | 4 | 0.021628 |
| Biological Process | GO:0009773 | photosynthetic electron transport in photosystem I | 3 | 0.041851 |
| Cellular Component | GO:0009535 | chloroplast thylakoid membrane | 33 | 1.17E-09 |
| Cellular Component | GO:0009570 | chloroplast stroma | 31 | 1.45E-07 |
| Cellular Component | GO:0009941 | chloroplast envelope | 28 | 1.54E-07 |
| Cellular Component | GO:0009507 | Chloroplast | 43 | 9.46E-06 |
| Cellular Component | GO:0009654 | photosystem II oxygen evolving complex | 7 | 2.85E-04 |
| Cellular Component | GO:0042555 | MCM complex | 4 | 0.001528 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | 19 | 0.003011 |
| Cellular Component | GO:0009579 | Thylakoid | 6 | 0.003862 |
| Cellular Component | GO:0019898 | extrinsic component of membrane | 6 | 0.004843 |
| Cellular Component | GO:0010319 | Stromule | 4 | 0.025165 |
| Cellular Component | GO:0005829 | Cytosol | 39 | 0.025596 |
| Molecular Function | GO:0003678 | DNA helicase activity | 4 | 0.001548 |
| Molecular Function | GO:0080043 | quercetin 3-O-glucosyltransferase activity | 13 | 0.004606 |
| Molecular Function | GO:0080044 | quercetin 7-O-glucosyltransferase activity | 13 | 0.004606 |
| Molecular Function | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | 7 | 0.013155 |
| Molecular Function | GO:0003824 | catalytic activity | 14 | 0.018301 |
| Molecular Function | GO:0030170 | pyridoxal phosphate binding | 12 | 0.042889 |
| KEGG_PATHWAY | sly01200 | Carbon metabolism | 34 | 5.41E-06 |
| KEGG_PATHWAY | sly00710 | Carbon fixation in photosynthetic organisms | 16 | 1.23E-05 |
| KEGG_PATHWAY | sly00195 | Photosynthesis | 15 | 1.24E-04 |
| KEGG_PATHWAY | sly01100 | Metabolic pathways | 137 | 1.58E-04 |
| KEGG_PATHWAY | sly01130 | Biosynthesis of antibiotics | 40 | 0.001247 |
| KEGG_PATHWAY | sly01230 | Biosynthesis of amino acids | 23 | 0.006324 |
| KEGG_PATHWAY | sly03030 | DNA replication | 8 | 0.012285 |
| KEGG_PATHWAY | sly00030 | Pentose phosphate pathway | 8 | 0.018663 |
| KEGG_PATHWAY | sly01110 | Biosynthesis of secondary metabolites | 76 | 0.027292 |
| KEGG_PATHWAY | sly00920 | Sulfur metabolism | 6 | 0.039347 |
Table 2.
GO and KEGG enrichment for down-regulated DEGs.
| Category | Term | Name | Count | P-Value |
|---|---|---|---|---|
| Biological Process | GO:0046777 | protein autophosphorylation | 9 | 0.001982 |
| Biological Process | GO:0016132 | brassinosteroid biosynthetic process | 7 | 0.003056 |
| Biological Process | GO:0018105 | peptidyl-serine phosphorylation | 10 | 0.004176 |
| Biological Process | GO:0071555 | cell wall organization | 14 | 0.004704 |
| Biological Process | GO:0042546 | cell wall biogenesis | 7 | 0.006139 |
| Biological Process | GO:0010411 | xyloglucan metabolic process | 7 | 0.006139 |
| Biological Process | GO:0009738 | abscisic acid-activated signaling pathway | 10 | 0.00645 |
| Biological Process | GO:0009408 | response to heat | 5 | 0.016273 |
| Biological Process | GO:0035556 | intracellular signal transduction | 13 | 0.019121 |
| Biological Process | GO:0032259 | Methylation | 8 | 0.023571 |
| Biological Process | GO:0009833 | plant-type primary cell wall biogenesis | 4 | 0.028812 |
| Cellular Component | GO:0005802 | trans-Golgi network | 21 | 8.92E-08 |
| Cellular Component | GO:0005768 | Endosome | 17 | 2.66E-05 |
| Cellular Component | GO:0016021 | integral component of membrane | 212 | 0.001422 |
| Cellular Component | GO:0009505 | plant-type cell wall | 13 | 0.011292 |
| Cellular Component | GO:0005886 | plasma membrane | 47 | 0.012016 |
| Cellular Component | GO:0005794 | Golgi apparatus | 17 | 0.021236 |
| Cellular Component | GO:0009506 | Plasmodesma | 14 | 0.046915 |
| Molecular Function | GO:0005509 | calcium ion binding | 24 | 4.16E-04 |
| Molecular Function | GO:0005516 | calmodulin binding | 8 | 0.002049 |
| Molecular Function | GO:0004683 | calmodulin-dependent protein kinase activity | 8 | 0.002049 |
| Molecular Function | GO:0009931 | calcium-dependent protein serine/threonine kinase activity | 8 | 0.002049 |
| Molecular Function | GO:0005524 | ATP binding | 96 | 0.004468 |
| Molecular Function | GO:0016762 | xyloglucan:xyloglucosyl transferase activity | 7 | 0.006891 |
| Molecular Function | GO:0016790 | thiolester hydrolase activity | 3 | 0.016552 |
| Molecular Function | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity | 9 | 0.033368 |
| Molecular Function | GO:0016759 | cellulose synthase activity | 4 | 0.037445 |
| Molecular Function | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | 5 | 0.044504 |
| Molecular Function | GO:0003700 | transcription factor activity, sequence-specific DNA binding | 34 | 0.047623 |
| KEGG_PATHWAY | sly04626 | Plant-pathogen interaction | 25 | 6.73E-06 |
| KEGG_PATHWAY | sly00330 | Arginine and proline metabolism | 9 | 0.013333 |
| KEGG_PATHWAY | sly00905 | Brassinosteroid biosynthesis | 4 | 0.029545 |
| KEGG_PATHWAY | sly00062 | Fatty acid elongation | 6 | 0.031366 |
| KEGG_PATHWAY | sly00520 | Amino sugar and nucleotide sugar metabolism | 13 | 0.039295 |
KEGG annotation suggested that photosynthesis pathways (15 DEGs), the biosynthesis of amino acids (23 DEGs) and the pentose phosphate pathway (8 DEGs) were induced after cold treatment in tomato fruit (Supplementary Fig. S1). On the other hand, fatty acid elongation (6 DEGs), arginine and proline metabolism (9 DEGs) and amino sugar and nucleotide sugar metabolism (13 DEGs) were significantly depressed in cold stress (Supplementary Fig. S1).
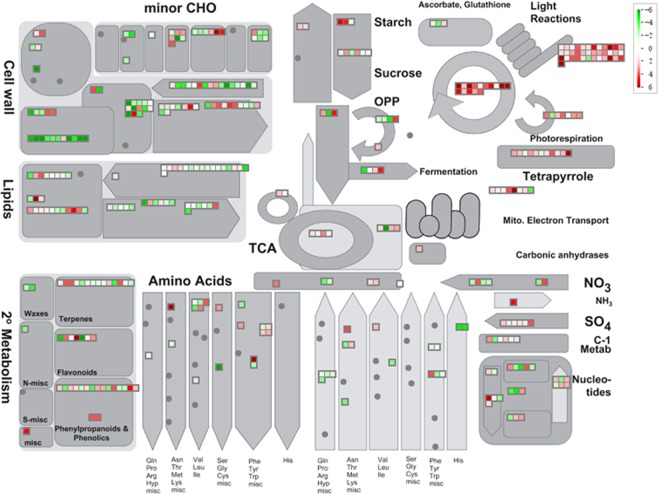
To have a broader view of all DEGs, Mapman analysis was used for the metabolism overview (Fig. 3, Supplementary Table S5). Similar to the GO and KEGG analysis, genes related to cell wall and lipid metabolism have decreased expression levels, whereas genes involved in Calvin cycle and light reactions pathway have increased expression levels after cold treatment in tomato fruit.
Figure 3.
Metabolism network overview of all DEGs by Mapman. The block colored in red and green means the up-regulated genes and down-regulated genes, respectively. The higher the intensity of the color, the greater the difference.
Sugar and organic acid metabolism analysis of cold-stored tomato fruit
Based on transcriptome and metabolome data, a schematic sugar and organic acid metabolism network was built with differential metabolites and DEGs (Fig. 4, Supplementary Table S6). In total, 13 metabolites and 27 DEGs were involved in the network. In the TCA cycle, the contents of citrate, butanedioic acid, cis-aconitate and succinate were increased, which were consistent with the expression of ATP-citrate synthase (ACS, EC: 2.3.3.8) and isocitrate dehydrogenase (IDH, EC: 1.1.1.42) genes at all four cold treatment stages in tomato fruit. The result indicates cold stress promote the expression of ACS and IDH genes, thus may increase the synthesis of citrate, butanedioic acid, cis-aconitate and succinate. Besides, malate content showed another pattern: increasing in cold-stored fruit at 7 days and decreasing thereafter. Oxalic acid content was relatively lower in cold-treated fruit at 7 and 21 days storage and higher at 14 days storage.
Figure 4.
Schematic network of sugar and organic metabolism network. The numbers colored in red are enzyme identifier number. Differential metabolites are in green boxes while the white boxes are other pathway related metabolites. The bars with four small blocks in each DEGs or differential metabolites are the relative fold change value on 7, 14, 21 and 28 days. Orange color means up-regulated under chilling injury and green means down-regulated at that time point. The higher the intensity of the color, the greater the difference. Solid line between two metabolites means a straight reaction while a dashed line means some omission of several metabolites and genes. The enzyme identifier meaning are as follows: 1.1.1.22: UDP-glucose 6-dehydrogenase 2; 1.1.1.37: malate dehydrogenase; 1.1.1.42: isocitrate dehydrogenase; 1.2.1.12: glyceraldehyde-3-phosphate dehydrogenase GAPCP1; 1.2.1.9: NADP-dependent glyceraldehyde-3-phosphate dehydrogenase; 1.2.4.1: pyruvate dehydrogenase E1 component subunit; 2.2.1.1: transketolase; 2.3.1.12: dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex; 2.3.3.8: ATP-citrate synthase; 2.4.1.1: alpha-1,4 glucan phosphorylase L-2 isozyme; 2.4.1.13: sucrose synthase; 2.4.1.15: trehalose-phosphate synthase; 2.7.1.1: hexokinase; 2.7.1.90: pyrophosphate–fructose 6-phosphate 1-phosphotransferase subunit alpha; 2.7.2.3: phosphoglycerate kinase; 3.1.3.11: fructose-1,6-bisphosphatase; 3.1.3.12: trehalose-phosphate synthase; 3.2.1.2: beta-amylase 3; 3.2.1.21: beta-glucosidase; 3.2.1.4: endoglucanase 24-like; 4.1.2.13: fructose-bisphosphate aldolase 1; 5.1.3.1: ribulose-phosphate 3-epimerase; 5.3.1.6; probable ribose-5-phosphate isomerase 2; 5.4.2.12: probable 2-carboxy-D-arabinitol-1-phosphatase; 2.3.3.9: malate synthase; 1.1.1.95: glycerate dehydrogenase; 2.7.1.11: ATP-dependent 6-phosphofructokinase 6.
The glucose content was decreased at 14 days storage and increased at 7, 21 and 28 days storage of cold-treated fruit. The expression level of hexokinase (HXK1, EC: 2.7.1.1) was induced at all four cold treatment stages in tomato fruit. Ribose and sedoheptulose had higher contents in cold-treated fruit at 14 and 21 days storage, and transketolase (TKT, EC: 2.2.1.1), ribulose-phosphate 3-epimerase (RPE, EC: 5.1.3.1), and probable ribose-5-phosphate isomerase 2 (RPI2, EC: 5.3.1.6) genes also had higher expression levels in cold-stored fruit. The result demonstrated that cold stress increases the expression of TKT, RPE and PRI2 genes, thus may improve the accumulation of ribose and sedoheptulose in tomato fruit.
Amino acid metabolism analysis of cold-stored tomato fruit
For amino acid metabolism, 6 DEGs and 5 differential amino acids were involved in the schematic network (Fig. 5, Supplementary Table S6). Alanine and leucine had increased contents in cold-stressed fruit at 14 and 21 days storage, and alanine aminotransferase (ALT, EC: 2.6.1.2) and branched-chain amino acid aminotransferase (BcAT, EC: 2.6.1.42) had higher expression levels in cold-stored fruit. The higher expression of ALT and BcAT genes induced by cold stress may improve the synthesis of alanine and leucine in fruit, respectively. The serine content was also increased in cold stressed fruit at 7 days storage, and the expression of threonine ammonia lyase (TAL, EC: 4.3.1.19) was decreased in the fruit. The result indicates that cold stress can suppress the expression of TAL gene, thus potentially leading to an increase in the accumulation of serine in cold stress fruit. Moreover, proline content was decreased only at 14 days storage and stayed constant at 7, 21 and 28 days storage in cold-stressed fruit.
Figure 5.
Schematic network of amino acid metabolism. The enzyme identifier meaning are as follows: 2.6.1.2: alanine aminotransferase 2-like; 2.2.1.6: acetolactate synthase small subunit 1; 1.1.1.86: ketol-acid reductoisomerase; 2.6.1.42: branched-chain-amino-acid aminotransferase 5; 4.3.1.19: chloroplast threonine deaminase 1 precursor; 1.1.1.42: isocitrate dehydrogenase.
Lipid metabolism analysis of cold-stored tomato fruit
In total, two metabolites and 12 DEGs were involved in fatty acid metabolism pathway (Fig. 6, Supplementary Table S6). The contents of two saturated fatty acids, palmitic acid and stearic acid, were reduced in the cold-stressed fruit after 7 days storage. Intriguingly, stearic acid content increased after 14 days cold storage. Five fatty acids synthesis genes, including two acetyl-CoA carboxylase 1 genes (ACC1, EC: 6.3.4.14), one malonyl-CoA-acyl carrier protein transacylase gene (MCAT, EC: 2.3.1.39) and two 3-oxoacyl-[acyl-carrier-protein] reductase genes (OAR, EC: 1.1.1.100), had increased expression levels in the cold-stressed tomato fruit at all four storage stages. This result indicates that cold stress can increase the synthesis of fatty acids in tomato fruit.
Figure 6.
Schematic network of fatty acid metabolism. The enzyme identifier meaning are as follows: 1.3.3.6: acyl-coenzyme A oxidase 4; 3.1.2.22: palmitoyl-protein thioesterase 1; 6.3.4.14: biotin carboxyl carrier protein of acetyl-CoA carboxylase 1; 2.3.1.39: malonyl-CoA-acyl carrier protein transacylase; 1.1.1.100: short-chain type dehydrogenase/reductase-like; 1.1.1.330: very-long-chain 3-oxoacyl-CoA reductase 1; 1.3.1.93: very-long-chain enoyl-CoA reductase-like; 1.14.19.2: acyl-[acyl-carrier-protein] desaturase; 3.1.2.2: acyl-coenzyme A thioesterase 8; 4.2.1.17: peroxisomal fatty acid beta-oxidation multifunctional protein AIM1-like; 1.1.1.35: peroxisomal fatty acid beta-oxidation multifunctional protein AIM1-like; 3.1.2.14: oleoyl-acyl carrier protein thioesterase.
Two other genes involved in the fatty acid elongation process (from 16* to 18*) were successfully mapped17. One gene called very-long-chain 3-oxoacyl-CoA reductase 1 (KCR, EC: 1.1.1.330) was down-regulated while another gene called very-long-chain enoyl-CoA reductase-like (ECR, EC: 1.3.1.93) were up-regulated in tomato fruit at all four cold treatment stages. Moreover, acyl-[acyl-carrier-protein] desaturase gene (ACPD, EC: 1.14.19.2) and oleoyl-acyl carrier protein thioesterase gene (OTE, EC: 3.1.2.14) were down-regulated at all four cold treatment stages. These results indicated that cold stress may inhibit the transformation of saturated fatty acid to unsaturated fatty acid in tomato fruit.
Co-expression analysis of DEGs involved in sugar, organic acid, fatty acid and amino acid metabolism
In an attempt to validate whether the DEGs in each metabolism network were “hub genes”, co-expression analysis was performed between the selected genes and other genes involved in the network. Hub genes are genes that have high centralities and play a crucial role in a network.
A total of 57 tomato gene annotations involved in sugar and organic acid, amino acid as well as fatty acid schematic pathway were selected for co-expression analysis (Supplementary Fig. S2). In the sugar and organic acid metabolism network, 7 genes were found to be significantly co-related with ACS (EC: 2.3.3.8, Solyc05g005160.2, Solyc01g059880.2) and 1 gene was significantly co-related with IDH (EC: 1.1.1.42, Solyc01g005560.2), indicating an important role of ACS in response to cold stress in tomato fruit. In amino acid metabolism, 3 genes were co-related with ALT (EC: 2.6.1.2, Solyc06g063090.2) and 1 gene was co-related with BcAT (EC: 2.6.1.42, Solyc03g043880.2). In lipids metabolism, the ACC1 (EC: 6.3.4.14, Solyc01g008330.2), MCAT (EC: 2.3.1.39, Solyc01g006980.2) and FabG (EC: 1.1.1.100, Solyc06g071910.2) were significantly co-related, indicating a cooperation of fatty acids synthesis in cold-treated tomato fruit.
Discussion
Tomato fruit are susceptible to CI, resulting in heavy economic losses. With transcriptomic and metabolic conjoint analysis, we were able to reveal some molecular mechanisms of sugar, organic acids, fatty acids, amino acids and other compounds of tomato fruit in response to CI. Even with the caveat that transcript abundance may not lead to changes in enzyme activity, and thus to alterations in metabolite levels, it is nevertheless tempting to speculate that there may be functional. Indications of low temperature increasing the concentration of citric acid are present in other cold-tolerant fruits beside tomato. For instance, banana fruit had the highest concentration of citric acid during the cool dry season harvest18. Another example is low temperature induction of the expression of citrate synthase (CS) genes and significantly increased citrate content 1.4–1.9 fold in Ponkan fruit19. In our results, cold stress increased the content of citrate in tomato fruit. These results indicated that citrate play important roles in response to cold stress in plant fruits and accumulation of citrate may be involved in fruit tolerance to cold stresses.
In the present study, malate content increased in cold-stored fruit at 7 days then decreased. A similar pattern was exhibited in myrtle fruit during cold storage20. In contrast, a decline in malate concentration during cold-storage of grape berries has already been reported to exhibit the opposite effect21. These examples support our results, indicating the important effect of malate content in cold-stored fruit.
Postharvest treatments with oxalic acid can alleviate chilling injury in sweet cherry by promoting a higher content of bioactive compounds and antioxidant activity22. Oxalic acid was also reported to have a CI alleviation function in tomato and mango through exogenous application23. In our study, tomato fruit had relatively higher content of oxalic acid at 14 days cold storage. Thus, oxalic acid may benefit the control of CI and the maintenance of fruit quality in cold storage.
The accumulation of organic acids was reported to relate to the plant response to many abiotic stresses. For example, drought treatment induced the accumulation of citrate in the leaves of cotton24. Another example is the frost injury increase in malate and titratable acidity in orange fruit25. For instance, when plants are under aluminum stress, organic acids, such as citrate, malate and so on are induced and accumulated, resulting in the elimination of aluminum toxicity26. These reports suggested that organic acid accumulation may play an important role in plant tolerance to many different abiotic stresses.
Our results indicated that the glucose content was decreased at 14 days storage and increased at 7, 21 and 28 days storage of cold-treated fruit, which resembled a similar pattern found in carambola fruit under cold treatment27. Glucose, a central carbohydrate metabolite, was identified to be positively correlated with freezing tolerance28. It was reported that a higher content of glucose tends to have a better tolerance of chilling injury in loquat fruit29. Cold stress also induced the expression of hexokinase (HXK1) gene in tomato fruit. Therefore, the HXK1 gene may be a response to cold stress and is required for glucose synthesis in tomato fruit under at low temperature.
In this study, alanine, serine, leucine and valine had increased contents in fruit at early cold treatment. Previous studies of peach fruit metabolism affected by cold treatment at 3 days and 5 days confirmed our findings that for alanine, serine and valine contents tended to increase30. Obata et al. reported that valine and leucine, belonging to branched chain amino acids (BCAAs), were generally accumulated under abiotic stress conditions, including cold stress31. BCAAs have a possible role as an alternative electron donor for the mitochondrial electron transport chain under stress. BCAAs can directly provide electrons to the electron transport chain via electron transfer flavoprotein complex, as well as indirectly feed into the Krebs cycle via their catabolic products31,32.
Branched-chain aminotransferase (BCAT) enzymes are at the interface of BCAA synthesis and catabolism33. Gonda et al. reported that the expression of the branched chain amino acid transaminas 2 (Os03g0231600), a BCAT family gene, was induced by both cold and dehydration treatment in rice34. In this study, the expression of the BCAT gene was induced, and the synthesis of leucine was improved by cold stress in tomato fruit. BCAT genes may be a new candidate gene for genetic engineering for cold stress tolerance in plants.
Proline has highly beneficial functions in plants exposed to many stress conditions including chilling injury35,36. Proline plays many roles such as being an excellent osmolyte, metal chelator, antioxidative defense molecule and signaling molecule during stress37. However, our study showed the proline content was decreased at 14 days storage of cold-treated fruit. We suggested that proline may be induced by cold stress only at the first several days in tomato fruit.
Low temperature storage at 4 °C can restrain the maturation process of tomatoes from breaker stage (Fig. 1A). However, 50% of tomatoes had skin pitting and electrolyte leakage and it was present only after transferring to room temperature for 3 days. Sharom et al. observed similar symptoms in tomatoes transferred from 5 °C to 25 °C, possibly due to the lipid phase change in membrane38.
CI has an effect on membrane permeability, including electrolyte leakage, lipid phase transitions and changes in lipid composition39–42. Our GO analysis confirmed that CI regulate genes significantly enriched in membrane related terms, including up-regulated extrinsic component of membrane, intracellular membrane-bounded organelle and down-regulated integral component of membrane, and plasma membrane. Moreover, cell wall metabolism has been found to play important roles in CI in fruits43–45. Recently, an integrative analysis reported that CI significantly regulates long non-coding RNAs targeting cell wall degradation in tomato fruit46. We found that cell wall related terms like cell wall organization, cell wall biogenesis as well as plant-type cell wall was down-regulated, indicating CI may induce cell wall degradation and thus may easily lead to skin pitting and electrolyte leakage at shelf storage.
It was also reported that a higher proportion of unsaturated fatty acids produced higher tolerance to low temperature stress. Examples are found in banana, pomegranate and loquat fruit47–49. Membrane lipids from chilling-resistant plant species exhibited higher content of unsaturated fatty acids than sensitive species50–52. For instance, the decrease in lipid unsaturation was related to the induction of CI in loquat fruit47. It was also reported that a decrease in unsaturation of peel tissue occurred during the early phase of CI in chilled cucumber fruit53,54. In the present study, acyl-[acyl-carrier-protein] desaturase gene (EC: 1.14.19.2) and oleoyl-acyl carrier protein thioesterase gene (EC: 3.1.2.14) was down-regulated at all four cold treatment stages. The results indicated that cold stress may inhibit the transformation of saturated fatty acid to unsaturated fatty acid in tomato fruit, thus decreasing the contents of unsaturated fatty acids in chilled tomato fruit.
Conclusions
Our results indicate that cold stress promotes the expression of ACS and IDH genes, which may have increased the synthesis of citrate, cis-aconitate and succinate. The improved expression of the ALT and BcAT genes induced by cold stress may improve the synthesis of alanine and leucine in fruit, respectively. In this study, integrative comparative analysis of transcriptomics and metabolomics data uncovered complex regulatory network in tomato fruit under chilling injury. The DEGs found can act as the indicator genes to reflect the physiological state of plants exposed to CI stress. The DEGs can also act as candidate genes for genetic engineering to improve the fruits tolerance to chilling in further studies.
Materials and Methods
Plant material and experimental treatment
Cherry tomato (Solanum lycopersicum L. cv. Micro-Tom) was harvested from Baishiyi orchard, Chongqing, China. Ninety tomato fruit with the same developmental stage (breaker) were carefully selected for storage experiment at low temperature (4 °C) and room temperature (25 °C), separately (Fig. 1A). Tomato fruit were stored for 28 days and around 5 tomatoes from each group were collected every 7 days and stored in a −80 °C freezer for metabolome and transcriptomic analysis.
Gas chromatography–mass spectrometry (GC-MS) metabolome analysis
Six biological replicates were used for each group at each time stage in the GC-MS experiment. Tomato fruit were ground into mixed powder and for each replicate a 400 mg sample was collected and mixed with 2 ml of methanol then vortexed for 10 s before ultrasonic extraction at 70 °C. The supernatant was collected after centrifugation at 12,000 rpm. Two milliliter of water and 1 ml of chloroform was added to the test bottle and centrifuged at 14,000 rpm for 5 minutes. A 400 µl sample of supernatant was collected into the test bottle and dried in nitrogen. Later, 80 µl of a 15 mg/ml methoxyamine pyridine stock solution was added to the pellet and vortexed for 30 s. The reaction was incubated at 37 °C for 90 min. Finally, add 80 µl BSTFA with 1% TMCS to the reaction and incubate at 70 °C for 60 min for GC-MS analysis.
GC-MS analysis was performed on an Agilent 6980 GC system equipped with a 30.0 m × 0.25 mm i.d. fused-silica capillary column with 0.25 μm HP-5MS stationary phase (Agilent, Shanghai, China). The inject temperature was set at 250 °C. The column temperature was initially kept at 80 °C for 3 min and then increased from 80 °C to 280 °C at 10 °C/min, where it was held for 2 min. Helium was used as the carrier gas at a constant flow rate of 1 ml/min through the column. A split-less injection volume of 1 µl was used. The column effluent was introduced into the ion source of an Agilent 5973 mass selective detector (Agilent Technologies). The MS quadrupole temperature was set at 150 °C and the ion source temperature was 230 °C. Masses were acquired from m/z 50 to 800. The mass accuracy of the instrument was 0.1 atomic mass unit (amu). The acceleration voltage was turned on after a solvent delay of 180 s.
Transcriptomic analysis
QIAGEN RNeasy Plant mini kit (Qiagen, Hilden, Germany) was used for RNA extraction of tomato fruit. RNA-seq was performed in Shanghai Majorbio Biopharm Technology Co., Ltd described by Zhang55. RNA-seq was performed without replications, acting as indicators only to identify candidate genes which need further analyses by qRT-PCR. Strict criteria were adapted to ensure accuracy. Based on the distribution of the raw data, all sequences were of high quality. All raw data were further filtered to obtain high quality data (Supplementary Fig. S3). All clean reads were aligned to the tomato genome (ITAG 2.3, http://solgenomics.net/) using Tophat (http://tophat.cbcb.umd.edu/). To identify differentially expressed genes (DEGs), transcript abundance was normalized by FPKM (Fragments per Kilobase of exon model per Million mapped reads)56 method using R (https://www.r-project.org/). A FDR < 0.05 and |logFC| > 1 was used to determine the threshold for DEGs57.
Gene Oncology (GO) and KEGG function enrichment analysis
GO and KEGG function enrichment analysis was carried out through DAVID bioinformatics resources 6.8 (https://david.ncifcrf.gov/home.jsp)58,59. All tomato ID was converted to Entrez ID through Gene Accession Conversion Tool of DAVID before function enrichment analysis. Ease score, a modified Fisher Exact P-value, was used for gene-enrichment analysis. Ease score <0.05 was considered strongly enriched in the annotation categories and was accepted for this analysis.
Mapman network analysis
Mapman (http://mapman.gabipd.org/, version 3.6.0) was used to provide a graphical overview of tomato metabolic networks60–63. It is a user-driven tool which can display large datasets onto diagrams of metabolic pathways or biological processes. A table with tomato gene ID and log transformed data (RT/LT) was imported and visualized.
Co-expression analysis
Gene expression data of 4 storage stages (7, 14, 21 and 28 days) were selected and imported to Cytoscape (http://cytoscape.org/, version 3.4.0) for co-expression analysis. Pearson’s correlation was performed and P-value < 0.05 was considered significant.
Quantitative reverse transcription PCR (qRT-PCR) expression analysis
Total RNA was extracted using RNAprep Pure Plant Kit (Tiangen). First strand cDNA was reversed transcribed from 1 μg total RNA using the First Strand cDNA Synthesis Kit (Thermo Scientific). qRT-PCR was carried out as described previously55. The primers used are listed in Supplementary Table S2.
Ethics approval and consent to participate
The experimental research on plants carried out in this work complies with institutional, national, and international guidelines.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0400100), the Project of Chongqing Science and Technology Commission (CSTC2015JCYJA80018), the National Natural Science Foundation of China (31272165) and the National Basic Research Program of China (2013CB127106).
Author Contributions
Wei Deng and Wen-Fa Zhang designed the experiments and wrote the manuscript. Ze-Hao Gong and Yu-Jin Yuan drafted the manuscript and helped with figure preparation. Meng-Bo Wu helped with the qRT-PCR analysis. Ning Tang, Qiang Zhang, Ming-Jun Miao and Helen Chan performed or assisted in the bioinformatics analysis. Zheng-Guo Li, Wei Chang, Zhi Li, Helen Chan and Liang Jin guided the study and revised the manuscript. All authors read and approved the manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang Jin, Email: jinliang4002@126.com.
Wei Deng, Email: dengwei1977@cqu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41065-9.
References
- 1.Lauxmann MA, et al. Transcriptomic profiling during the post-harvest of heat-treated Dixiland Prunus persica fruits: common and distinct response to heat and cold. PloS one. 2012;7:e51052. doi: 10.1371/journal.pone.0051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vega-Garcia MO, et al. Changes in Protein Expression Associated with Chilling Injury in Tomato Fruit. J Am Soc Hortic Sci. 2010;135:83–89. doi: 10.21273/JASHS.135.1.83. [DOI] [Google Scholar]
- 3.Wang CY. Approaches to reduce chilling injury of fruits and vegetables. Hort Rev. 1993;15:63–95. [Google Scholar]
- 4.Sevillano L, Sanchez-Ballesta MT, Romojaro F, Flores FB. Physiological, hormonal and molecular mechanisms regulating chilling injury in horticultural species. Postharvest technologies applied to reduce its impact. Journal Of the Science Of Food And Agriculture. 2009;89:555–573. doi: 10.1002/jsfa.3468. [DOI] [Google Scholar]
- 5.Imahori Y, Takemura M, Bai J. Chilling-induced oxidative stress and antioxidant responses in mume (Prunus mume) fruit during low temperature storage. Postharvest Biol Tec. 2008;49:54–60. doi: 10.1016/j.postharvbio.2007.10.017. [DOI] [Google Scholar]
- 6.Mochida K, Shinozaki K. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant & cell physiology. 2011;52:2017–2038. doi: 10.1093/pcp/pcr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss J, Egea-Cortines M. Transcriptomic analysis of cold response in tomato fruits identifies dehydrin as a marker of cold stress. J Appl Genet. 2009;50:311–319. doi: 10.1007/BF03195689. [DOI] [PubMed] [Google Scholar]
- 8.Rugkong A, McQuinn R, Giovannoni JJ, Rose JKC, Watkins CB. Expression of ripening-related genes in cold-stored tomato fruit. Postharvest Biol Tec. 2011;61:1–14. doi: 10.1016/j.postharvbio.2011.02.009. [DOI] [Google Scholar]
- 9.Cruz-Mendívil AEA. Transcriptional changes associated with chilling tolerance and susceptibility in ‘Micro-Tom’tomato fruit using RNA-Seq. Postharv Biol Technol. 2015;99:141–151. doi: 10.1016/j.postharvbio.2014.08.009. [DOI] [Google Scholar]
- 10.Sanchez-Bel P, et al. Proteome changes in tomato fruits prior to visible symptoms of chilling injury are linked to defensive mechanisms, uncoupling of photosynthetic processes and protein degradation machinery. Plant & cell physiology. 2012;53:470–484. doi: 10.1093/pcp/pcr191. [DOI] [PubMed] [Google Scholar]
- 11.Page D, et al. Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta. 2010;232:483–500. doi: 10.1007/s00425-010-1184-z. [DOI] [PubMed] [Google Scholar]
- 12.Luengwilai K, Saltveit M, Beckles DM. Metabolite content of harvested Micro-Tom tomato (Solanum lycopersicum L.) fruit is altered by chilling and protective heat-shock treatments as shown by GC-MS metabolic profiling. Postharvest Biol Tec. 2012;63:116–122. doi: 10.1016/j.postharvbio.2011.05.014. [DOI] [Google Scholar]
- 13.Wang LB, et al. Suppression of volatile production in tomato fruit exposed to chilling temperature and alleviation of chilling injury by a pre-chilling heat treatment. Lwt-Food Sci Technol. 2015;62:115–121. doi: 10.1016/j.lwt.2014.12.062. [DOI] [Google Scholar]
- 14.Lee JM, et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012;70:191–204. doi: 10.1111/j.1365-313X.2011.04863.x. [DOI] [PubMed] [Google Scholar]
- 15.Osorio S, et al. Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 2012;159:1713–1729. doi: 10.1104/pp.112.199711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osorio S, et al. Systems Biology of Tomato Fruit Development: Combined Transcript, Protein, and Metabolite Analysis of Tomato Transcription Factor (nor, rin) and Ethylene Receptor (Nr) Mutants Reveals Novel Regulatory Interactions. Plant Physiology. 2011;157:405–425. doi: 10.1104/pp.111.175463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki K, et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugaud C, Alter P, Daribo MO, Brillouet JM. Comparison of the physico-chemical characteristics of a new triploid banana hybrid, FLHORBAN 920, and the Cavendish variety. J Sci Food Agr. 2009;89:407–413. doi: 10.1002/jsfa.3459. [DOI] [Google Scholar]
- 19.Lin Q, et al. Low Temperature Induced Changes in Citrate Metabolism in Ponkan (Citrus reticulata Blanco cv. Ponkan) Fruit during Maturation. PloS one. 2016;11:e0156703. doi: 10.1371/journal.pone.0156703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulas M, Fadda A, Angioni A. Effect of maturation and cold storage on the organic acid composition of myrtle fruits. J Sci Food Agric. 2013;93:37–44. doi: 10.1002/jsfa.5724. [DOI] [PubMed] [Google Scholar]
- 21.Lasko AK, Weinfeld M. The influence of temperature on malic acid metabolism in grape berries. Plant Physiol. 1975;56:370–372. doi: 10.1104/pp.56.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valero D, et al. Postharvest treatments with salicylic acid, acetylsalicylic acid or oxalic acid delayed ripening and enhanced bioactive compounds and antioxidant capacity in sweet cherry. J Agric Food Chem. 2011;59:5483–5489. doi: 10.1021/jf200873j. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Yin F, Song L, Zheng X. Alleviation of chilling injury in tomato fruit by exogenous application of oxalic acid. Food chemistry. 2016;202:125–132. doi: 10.1016/j.foodchem.2016.01.142. [DOI] [PubMed] [Google Scholar]
- 24.Timpa JD, Burke JJ, Quisenberry JE, Wendt CW. Effects of water stress on the organic Acid and carbohydrate compositions of cotton plants. Plant Physiol. 1986;82:724–728. doi: 10.1104/pp.82.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcone Ferreyra MLEA. Carbohydrate metabolism and fruit quality are affected in frost‐exposed Valencia orange fruit. Physiol Plant. 2006;128:224–236. doi: 10.1111/j.1399-3054.2006.00744.x. [DOI] [Google Scholar]
- 26.Kochian LV, Pineros MA, Liu J, Magalhaes JV. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu Rev Plant Biol. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Tello GO, Silva-Espinoza BA, Vargas-Arispuro I, Briceno-Torres BO, Martinez-Tellez MA. Effect of temperature on enzymatic and physiological factors related to chilling injury in carambola fruit (Averrhoa carambola L.) Biochem Biophys Res Commun. 2001;287:846–851. doi: 10.1006/bbrc.2001.5670. [DOI] [PubMed] [Google Scholar]
- 28.Hannah MA, et al. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S, Yang Z, Zheng Y. Sugar metabolism in relation to chilling tolerance of loquat fruit. Food chemistry. 2013;136:139–143. doi: 10.1016/j.foodchem.2012.07.113. [DOI] [PubMed] [Google Scholar]
- 30.Lauxmann MA, et al. Deciphering the metabolic pathways influencing heat and cold responses during post-harvest physiology of peach fruit. Plant Cell Environ. 2014;37:601–616. doi: 10.1111/pce.12181. [DOI] [PubMed] [Google Scholar]
- 31.Obata T, Fernie AR. The use of metabolomics to dissect plant responses to abiotic stresses. Cellular And Molecular Life Sciences. 2012;69:3225–3243. doi: 10.1007/s00018-012-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. Protein degradation–an alternative respiratory substrate for stressed plants. Trends in plant science. 2011;16:489–498. doi: 10.1016/j.tplants.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Maloney GS, et al. Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 2010;153:925–936. doi: 10.1104/pp.110.154922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonda I, et al. Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. Journal Of Experimental Botany. 2010;61:1111–1123. doi: 10.1093/jxb/erp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food chemistry. 2017;219:76–84. doi: 10.1016/j.foodchem.2016.09.123. [DOI] [PubMed] [Google Scholar]
- 36.Palma F, Carvajal F, Jamilena M, Garrido D. Contribution of polyamines and other related metabolites to the maintenance of zucchini fruit quality during cold storage. Plant physiology and biochemistry: PPB. 2014;82:161–171. doi: 10.1016/j.plaphy.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Hayat S, et al. Role of proline under changing environments: a review. Plant Signal Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharom M, Willemot C, Thompson JE. Chilling injury induces lipid phase changes in membranes of tomato fruit. Plant Physiology. 1994;105:305–308. doi: 10.1104/pp.105.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurdo A, Wilson J. Chilling injury and Arrhenius plots. Cryo-letters. 1980;1:231–238. [Google Scholar]
- 40.King, M. & Ludford, P. Chilling injury and electrolyte leakage in fruit of different tomato cultivars [Lycopersicon esculentum, postharvest]. Journal American Society for Horticultural Science (1983).
- 41.Autio, W. & Bramlage, W. Chilling sensitivity of tomato fruit in relation to ripening and senescence. Journal of the American Society for Horticultural Science (USA) (1986).
- 42.Lyons JM. Chilling injury in plants. Annual review of plant physiology. 1973;24:445–466. doi: 10.1146/annurev.pp.24.060173.002305. [DOI] [Google Scholar]
- 43.Manganaris G, Vicente A, Crisosto C, Labavitch J. Cell wall modifications in chilling-injured plum fruit (Prunus salicina) Postharvest Biology and Technology. 2008;48:77–83. doi: 10.1016/j.postharvbio.2007.09.017. [DOI] [Google Scholar]
- 44.Cao S, Zheng Y, Wang K, Rui H, Tang S. Effect of methyl jasmonate on cell wall modification of loquat fruit in relation to chilling injury after harvest. Food Chemistry. 2010;118:641–647. doi: 10.1016/j.foodchem.2009.05.047. [DOI] [Google Scholar]
- 45.Brummell DA, Dal Cin V, Lurie S, Crisosto CH, Labavitch JM. Cell wall metabolism during the development of chilling injury in cold-stored peach fruit: association of mealiness with arrested disassembly of cell wall pectins. Journal of Experimental Botany. 2004;55:2041–2052. doi: 10.1093/jxb/erh228. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Integrative analysis of long non-coding RNA acting as ceRNAs involved in chilling injury in tomato fruit. Gene. 2018;667:25–33. doi: 10.1016/j.gene.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 47.Cao SF, Zheng YH, Wang KT, Jin P, Rui HJ. Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chemistry. 2009;115:1458–1463. doi: 10.1016/j.foodchem.2009.01.082. [DOI] [Google Scholar]
- 48.Promyou S, Ketsa S, van Doorn WG. Hot water treatments delay cold-induced banana peel blackening. Postharvest Biol Tec. 2008;48:132–138. doi: 10.1016/j.postharvbio.2007.09.006. [DOI] [Google Scholar]
- 49.Mirdehghan SH, et al. Reduction of pomegranate chilling injury during storage after heat treatment: Role of polyamines. Postharvest Biol Tec. 2007;44:19–25. doi: 10.1016/j.postharvbio.2006.11.001. [DOI] [Google Scholar]
- 50.Wongsheree T, Ketsa S, van Doorn WG. The relationship between chilling injury and membrane damage in lemon basil (Ocimum x citriodourum) leaves. Postharvest Biol Tec. 2009;51:91–96. doi: 10.1016/j.postharvbio.2008.05.015. [DOI] [Google Scholar]
- 51.Lee SH, et al. Differential impact of low temperature on fatty acid unsaturation and lipoxygenase activity in figleaf gourd and cucumber roots. Biochem Biophys Res Commun. 2005;330:1194–1198. doi: 10.1016/j.bbrc.2005.03.098. [DOI] [PubMed] [Google Scholar]
- 52.Boonsiri K, Ketsa S, van Doorn WG. Seed browning of hot peppers during low temperature storage. Postharvest Biol Tec. 2007;45:358–365. doi: 10.1016/j.postharvbio.2007.03.014. [DOI] [Google Scholar]
- 53.Rui H, et al. Effects of heat treatment on internal browning and membrane fatty acid in loquat fruit in response to chilling stress. J Sci Food Agric. 2010;90:1557–1561. doi: 10.1002/jsfa.3993. [DOI] [PubMed] [Google Scholar]
- 54.Parkin KL, Kuo SJ. Chilling-Induced Lipid Degradation in Cucumber (Cucumis sativa L. cv Hybrid C) Fruit. Plant Physiol. 1989;90:1049–1056. doi: 10.1104/pp.90.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XL, et al. Auxin Response Gene SlARF3 Plays Multiple Roles in Tomato Development and is Involved in the Formation of Epidermal Cells and Trichomes. Plant And Cell Physiology. 2015;56:2110–2124. doi: 10.1093/pcp/pcv136. [DOI] [PubMed] [Google Scholar]
- 56.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 58.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Urbanczyk-Wochniak E, et al. Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol. 2006;60:773–792. doi: 10.1007/s11103-005-5772-4. [DOI] [PubMed] [Google Scholar]
- 61.Usadel B, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiology. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usadel B, et al. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell And Environment. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 63.Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant journal: for cell and molecular biology. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.