Abstract
Over the past three decades, extracellular vesicles (EVs) have arisen as important mediators of intercellular communication that are involved in the transmission of biological signals between cells to regulate various biological processes. EVs are largely responsible for intercellular communication through the delivery of bioactive molecules, such as proteins, messenger RNAs (mRNAs), microRNAs (miRNAs), DNAs, lipids, and metabolites. EVs released from cancer cells play a significant role in signal transduction between cancer cells and the surrounding cells, which contributes to the formation of tumors and metastasis in the tumor microenvironment. In addition, EVs released from cancer cells migrate to blood vessels and flow into various biological fluids, including blood and urine. EVs and EV-loaded functional cargoes, including proteins and miRNAs, found in these biological fluids are important biomarkers for cancer diagnosis. Therefore, EV proteomics greatly contributes to the understanding of carcinogenesis and tumor progression and is critical for the development of biomarkers for the early diagnosis of cancer. To explore the potential use of EVs as a gateway to understanding cancer biology and to develop cancer biomarkers, we discuss the mass spectrometric identification and characterization of EV proteins from different cancers. Information provided in this review may help in understanding recent progress regarding EV biology and the potential roles of EVs as new noninvasive biomarkers and therapeutic targets.
Subject terms: Cancer, Proteomics
Cancer: Protein-filled vesicles offer therapeutic and diagnostic targets
Tumor cells release tiny membrane-encapsulated packages known as extracellular vesicles containing proteins which could serve as prognostic disease biomarkers or therapeutic targets. Kwang Pyo Kim and colleagues from Kyung Hee University in Yongin, South Korea, review the use of mass spectrometry to profile the diversity of proteins found in these tumor-derived packages. The proteins found in these vesicles help mediate communication between cancer cells and their surrounding tissues. Different tumor types share many of these proteins in common, but there are differences in the protein profile related to cancer-associated biological processes such as metastasis and cell proliferation. Tests based on the proteins contained in these vesicles could help clinicians better identify, diagnose and treat specific cancers, although large, multicenter studies are needed to validate such strategies.
Introduction
Extracellular vesicles (EVs) are membrane-surrounded vesicles released by numerous cell types into the extracellular microenvironment1–3. EVs are involved in cell–cell communication, coagulation, inflammation, immune response modulation, and disease progression2,4–7. Although EVs vary in size, biological function, and components, their significance in cancer progression and the potential use of EV molecules as novel cancer biomarkers has gradually increased. Cancer cells actively release EVs into neighboring tissues, and these EVs play dynamic roles in cancer progression and metastasis, invasion, angiogenesis, tumorigenesis, and immune modulation8–10. EVs released by cancer cells are usually chosen as a gateway in the search for biomarkers for a specific cancer type. Recent results pertaining to EV-cargo molecules, including proteins and miRNAs, are summarized in EVpedia (http://evpedia.info), an integrated and comprehensive database of EVs11.
The main focus of this review is proteome profiling of EVs using mass spectrometry (MS)-based proteomic approaches. We discuss the mass spectral characterization of isolated EV proteins from different cancers and the use of these proteins as predictive cancer biomarkers. Additionally, we summarize the key characteristics of enriched proteins in cancer-associated EVs as potential therapeutic targets and provide novel information on their roles in cancer development and progression. Information provided in this review may help in understanding recent progress regarding EV biology and the prospective roles of EVs as new noninvasive biomarkers and therapeutic targets, as well as emerging therapeutic opportunities and associated challenges.
Classification of EVs
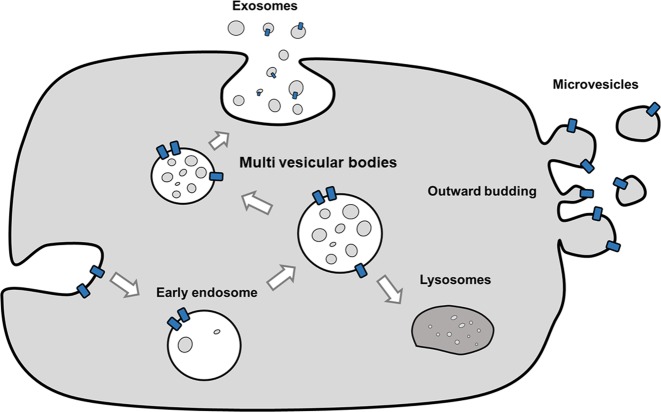
EVs are small spherical vesicles that are secreted into the extracellular milieu by many cell types. The term “EV” was invented by the International Society of Extracellular Vesicles (ISEV) and is used to define all phospholipid bilayer-bound vesicles that are secreted by cells into the extracellular microenvironment, regardless of the differences in biogenesis, size, and composition12,13. The roles of EVs in different physiological and pathological processes have made them a novel field of research. EVs are categorized into several subtypes based on their size, density, shape, subcellular origin, function, and molecular cargo14. The four major subtypes of EVs are exosomes, microvesicles, apoptotic bodies, and oncosomes (Table 1 and Fig. 1). Exosomes are 30–200-nm-sized homogeneous membrane vesicles, and they form through the endosomal trafficking pathway5,15,16. Exosomes contain late endosomal markers, even though biochemically indistinguishable vesicles can bud directly from the plasma membrane16,17. They play critical roles in cell–cell communications, such as that occurring during the regulation of cell and tissue homeostasis, as well as in pathological conditions18. Microvesicles are 100–1000-nm-sized heterogeneous membrane vesicles that originate via outward budding and the fission of the plasma membrane due to dynamic interactions during phospholipid redistribution. Phospholipid distribution is controlled by aminophospholipid translocases16,18–24 and cytoskeletal protein contraction. Microvesicles are released mostly under cellular stress or in pathological processes18. Like exosomes, microvesicles transfer bioactive molecules into target cells. Apoptotic bodies (> 1 µm) are released by cells that undergo the apoptosis process or programmed cell death18,24, and they can be characterized by cellular organelles and DNA. Finally, the vesicles named “oncosomes” are much larger than most other EV types characterized to date (1–10 μm). Owing to their unusual size, large oncosomes might have unique properties in vivo and would provide novel opportunities for tumor profiling25.
Table 1.
Brief classification of extracellular vesicles
| EV subtype | Diameter | Biogenesis | Markers | References |
|---|---|---|---|---|
| Exosomes | 30–200 nm | Released from multivesicular bodies within the endosomal network | Membrane transport and fusion proteins (annexins, GTPases, and flotillin), tetraspanins (CD9, CD63, CD81, and CD82), heat-shock proteins (Hsc70 and Hsp90), proteins involved in MVB biogenesis (Alix and Tsg101), lipid-related proteins and phospholipases, ESCRT, and MHC | 4, 5, 13– 15 |
| Microvesicles | 100–1000 nm | Produced by direct budding from the cell membrane | Selectins, integrins (B1), metalloprotease surface phosphatidylserine, vesicle-associated membrane protein 3, CD34, CD40, CD45, glycophorin, or blood group antigens | 16– 22, 25, 58 |
| Apoptotic bodies | >1 µm | Released only by cells undergoing apoptosis or programmed cell death (apoptosis fragments) | Surface phosphatidylserinehistones, calnexin, cytochrome C, annexin V, C3b, and TSP | 4, 14– 22 |
| Oncosomes | 1–10 µm | Non-apoptotic plasma membrane blebs shed by “ameboid” migrating tumor cells or from tumors | Cav-1, ARF6, Myr, Akt1, and HB‑EGF | 23 |
Fig. 1.
Biogenesis of four major subtypes of extracellular vesicles
EVs contain proteins, lipids, metabolites, and RNAs. However, the mechanisms by which these components enter EVs remain obscure. EVs are shed from almost all cell types and are present in biological fluids and conditioned cell culture media. EVs are involved in cell–cell communication, coagulation, inflammation, immune response modulation, and disease progression4–7. The functional roles of EVs in intercellular communication have made them of major interest in many scientific fields. The biomolecular composition of EVs could play a significant role in disease progression in several neurodegenerative diseases as well as in cancer.
MS in EV proteome analyses
EV proteome analysis is a novel approach and is part of the growing interest in proteomics cancer research. Over the past three decades, many proteomics studies performed on EVs have elucidated their diverse roles. Large-scale proteomics datasets and protein-interaction networks have established significant relationships between EV proteins, which improves the understanding of vesicle biogenesis and pathophysiological roles24,26,27. Proteomic studies on EVs from different origins have also suggested a controlled protein-sorting mechanism and the random packaging of EV proteins from various cell types that contain common vesicular proteins. Furthermore, proteomic studies of EVs have produced a high-throughput vesicular proteome dataset from various cell types and body fluids28. Since EVs are normally isolated in small amounts, better sensitivity is required for their analysis. Liquid chromatography (nanoscale or ultra-high performance)–electrospray ionization tandem mass spectrometry (LC/ESI–MS/MS) is the most popular and versatile analytical technique to study the molecular contents of EVs. In particular, nano-ESI–MS/MS provides high sensitivity and resolution, allowing the detection, identification, characterization, and quantification of thousands of proteins from even a single EV sample. Similar to other biological fields, LC–MS/MS-based technological platforms have become the most popular fundamental tools for elucidating the structural and functional architecture of EVs. The fragment ions from ESI (positive- and negative-ion) tandem MS experiments provide the composition, unambiguous structural characterization, and proper identification of proteins present in various biological samples. Due to the high sensitivity and small initial sample volumes required for MS, MS-based proteomic analysis has increased the understanding of EV protein content. Several investigators26,29–32 have used ESI tandem MS experiments in combination with chromatographic methods (HPLC, UHPLC, UPLC, and nano LC) to profile and structurally characterize proteins in various cancer cells, tissues, biofluids, and biological samples, which have been summarized in Table 2.
Table 2.
Summary of biomarker candidate proteins in extracellular vesicles from different cancers
| Cancer type | Cancer-specific EV proteins | Isolation of EVs | Characterization | Sample source | References |
|---|---|---|---|---|---|
| Bladder cancer | EDIL-3 | UC | WB, TEM | TCC, T24, SV-HUC, and urine | Beckham et al.59 |
| UC | TEM, NTA | TCCSUP, T24, UMUC3, RT3 SVHUC, and urine | Silvers et al.60 | ||
| ITGB1, ITGA6, CD36, CD44, CD73, CD10, MUC1, BSG, and 5T4 | UC | NTA, TEM, and WB | HT1376, urine | Welton et al.34 | |
| UC | NTA, TEM, and WB | T24, FL3, and SLT4 | Jeppesen et al.61 | ||
| Colon cancer | HGS | ExoQuick | NTA, TEM, and WB | HCT116 | Sun et al.62 |
| Clstn1, VCP, and RuVB-like1 (O-GlcNAcylation) | UC | TEM, WB | CCD841, HT29, SW480, and SW620 | Chaiyawat et al.63 | |
| F2 | BDG | NTA, TEM, and WB | SW480, SW620 | Schillaci et al.64 | |
| YWHAZ | UC | HCT116, patient colon tumor | Hillary et al.65 | ||
| ACACA | UC | Citrus-limon, SW480 | Raimondo et al.66 | ||
| UC | NTA, WB | SW620 | Guo et al.67 | ||
| EPCAM-CLDN7 and TNIK-RAP2A | BDG | NTA, TEM, and WB | SW480, SW620 | Ji et al.53 | |
| BDG | NTA, TEM, and WB | Patient tumor | Choi et al.26 | ||
| BDG | NTA, TEM, and WB | HT-29 | Choi et al.27 | ||
| GPA33, CDH17, CEA, EpCAM, PCNA, EGFR, MUC13, MINK1, KRT18, MAPK4, CLDN (1, 3, and 7), CEP55, EFNB1, and EFNB2 | TEM, WB | LIM1215 cells, urine, mast and cells | Suresh et al.33 | ||
| UC | NTA, TEM, and WB | Dks-8, DLD-1, and DKO-1 | Demory et al.68 | ||
| BDG | NTA, TEM, and WB | SW480, SW620 | Choi et al.69 | ||
| DKK4 and DNMT3A | UC | TEM, WB | SW480, SW480APC | Lim et al.70 | |
| MAC2BP, ALIX, 14–3–3 isoforms, PFN1, CALU, and IL-8 | UC | TEM | LIM1215 cells | Ji et al.71 | |
| Prostate cancer | UC | DLS, TEM | pc3-HSP27, HEK-293 | Rauschenberger et al.72 | |
| UC | PC3, DU145, VCaP, LNCaP, C4–2, and RWPE-1 | Hosseini-Beheshti et al.73 | |||
| THBS1, GSN, and ITGB1 | UC | WB | LNCaP | Soekmadji et al.74 | |
| ITGB4 and VCL | UC, CD9 antibody magnetic beads | NTA, WB, and TEM | PC-3 | Kawakami et al.75 | |
| CD9 | UC | TEM, WB | LNCaP, DUCaP PCa cells, and plasma | Soekmadji et al.76 | |
| UC | TEM, WB, and NTA | DU145 Tax-Sen, DU145 Tax-Res | Kharaziha et al.77 | ||
| CD151 and CDCP1 | UC | PC-3 | Sandvig et al.78 | ||
| PDCD6IP, FASN, XPO1, and ENO1 | UC, SG | TEM, WB | PNT2C2, RWPE1, PC346C, and VCaP | Duijvesz et al.30 | |
| UC | Osteoblasts | Bilen et al.79 | |||
| Lung cancer | AKT and ERK1/2 | UC | WB | H3255, H1650 | Van et al.80 |
| AKT1, GSK3B, EIF4E, MTOR, RELA, and RAS | BDG | TEM, WB | PC9, PC9R | Choi et al.81 | |
| ALLIX, TSG101, CD3, EGFR, SRC, KRAS, and NRP1 | PEG precipitation, UC, and BDG | TEM, WB | A54, HCC827, and HBEC | Clark et al.39 | |
| P53 and EGFR | qEV | TRPS, TEM, and WB | 30KTp53/EGFR | Lobb et al.82 | |
| HCC | ExoQuick | TEM, WB | Hep3B, 97 H, and LM3 | Zhang et al.83 | |
| RRAS, CD44, CDC42, and CLND3 | UC | WB | HKCI-C3, HKCI-8, MHCC97L and MIHA | He et al.84 | |
| UC | NTA, TEM, and WB | HepG2 | Wang et al.43 | ||
| UC | WB, EM | Huh7.5.1 Huh7-ET | Ramakrishnaiah et al.85 | ||
| Breast cancer | UC | TEM | Plasma, bone metastasis explant-conditioned media, and pleural effusion | Tucker et al.86 | |
| vn96 affinity capture of EV | WB and TEM | SKBR3, MCF-7, and MCF-10a | Griffiths et al.87 | ||
| Free-flow electrophoresis | SKBR3 (hypoxia, normoxia) | Thomas et al.88 | |||
| UC | TEM, WB, and NTA | MCF-7, MDA-MB-231 | Harris et al.32 | ||
| DEL-1 | Plasma, MDA-MB-231 | Moon et al.89 | |||
| EDIL3 | BDG | NTA, TEM, and WB | MCF-7, MDA-MB-231 | Lee et al.29 | |
| IL-6, TNFa, GCSF, and CCL2 | UC | TEM, WB, and FC | MCF10A, MDA-MB-231, and MCF7 | Chow et al.90 | |
| POSTN | UC, SG | NTA, TEM, and WB | MCF7, MDA-MB-231, 67NR, 4T1, and plasma | Vardaki et al.91 | |
| UC | TEM, WB, and NTA | cal51 TNBC | Kavanagh et al.92 | ||
| UC | TEM, WB | MDA-MB-231 cells | Palazzolo et al.37 | ||
| MTDH and CP | UC | TEM, WB, and NTA | 4T1, 4T1.2, 67NR, and 66cl4 | Gangoda et al.93 | |
| UC | TEM, DLS, and WB | VCaP | Domenyuk et al.94 | ||
| Ovarian cancer | UC | TEM, WB | OVCAR3, OVCAR433, OVCAR5, and SKOV3 | Sinha et al.95 | |
| UC | WB, TEM | SKOV3, OVM | Escrevente et al.96 | ||
| G6PD and TKT | UC | WB, TEM | OVCA429, HO8910PM | Yi et al.97 | |
| UC | TEM, WB | OVCAR-3, IGROV1 | Liang et al.41 | ||
| UC | TEM, WB | SKOV3, CAOV3, and HUVEC | Yi et al.98 | ||
| Pancreatic cancer | EGFR | UC | WB | BxPC3, MiaPaca2, and Panc1 | Adamczyk et al.99 |
| ZIP4 | SBI ExoQuick‐TC Kit | TEM, WB | PC‐1.0 (highly malignant), PC‐1 (moderately malignant) | Jin et al.100 | |
| CEACAMs and ECM proteins | UC | TEM | Pancreatic duct fluid | Zheng et al.101 | |
| UC, SG | EM | SOJ-6, BxPC-3, MiaPaCa-2, and Panc-1 | Ristorcelli et al.36 | ||
| MYOF | UC | DLS, TEM, and WB | MDA-MB-231, MDA-MB-468, BT-549, Hs 578 T, MCF7, MCF-10A, ZR-75–1, BT-474, SK-BR-3, and CFPAC-1 | Blomme et al.102 | |
| BDG | TEM, WB | Panc1, BxPc3, MiaPaca2, and HPSC | Klein-Scory et al.103 | ||
| MIF | UC | NTA | PKCY, PAN02 | Costa-Silva et al.31 | |
| UC, SG | TEM, WB | Panc02, Panc02-H7 cells | Yu et al.104 | ||
| CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE, and HIST2H2BF | UC | WB | 13 human PDAC, 2 non-neoplastic cell lines | Castillo et al.105 | |
| PLEC | UC, ExoQuick-TC | DLS, TEM, and WB | PDAC, C6 glioma cells, and HPDE | Shin et al.106 | |
| WNT5B | BDG, SG | EM | CHO, Caco-2 cells | Harada et al.46 | |
| CCA | UC | TEM, WB | Human bile, H69 cell line | Chaiyadet et al.107 | |
| S100A6, LUM, LCP1, YWHAZ, and VIM | UC | TEM | Hamster liver tissue, KKU055 | Khoontawad et al.108 | |
| UC | TEM, WB | KKU-100, KKU-M213, and H69 | Dutta et al.42 | ||
| Blood cancer | MARCKS | UC, SG | CM, FACS | K562, LAMA84 | Taverna et al.109 |
| VCP | UC | TEM | U937, Mec1 | Bosque et al.56 | |
| UC | WB, FC | Primary CLL cells | Paggetti et al.110 | ||
| DNMT1 and HELLS | UC | NTA, FC | Molm-14, HL-60, and OP9 cells | Huan et al.111 | |
| MHC-1, MHC-2, HSC70, HSP90, and ICMA-1 | UC | TEM, WB | Raji cells | Yao et al.40 | |
| Oral cancer | ExoQuick | NTA, TEM | HUVEC, SCC15 | Andrade et al.112 | |
| NAP1 | Ultrafiltration | NTA, TEM, WB, and CM | CAL 27, SCC-25 | Wang et al.44 | |
| HSP90 | UC | TEM, NTA | HSC-3, HSC-3-M3 | Ono et al.113 |
WB western blotting, TEM transmission electron microscopy, NTA nanoparticle tracking analysis, DLS dynamic light scattering, TRPS tunable resistive pulse sensing, FC flow cytometry, CM confocal microscopy
EV proteomes in various cancers and biomarker discovery
Proteomic analysis of EVs has revealed significant changes in protein expression under various physiological and pathological conditions26,29,30. Characterization of these proteomic profiles may be useful in understanding disease pathogenesis and assisting in the discovery of new biomarkers for different diseases. The secretion of EVs from several types of tumor cells is a significant means of conditioning and altering the tumor microenvironment by malignant cells31,32. Multiple studies have reported that the secretion of EVs from cancer cells contributes to angiogenesis, metastasis, tumor formation, and disease progression2,10,31,32. EVs are more attractive sources of biomarkers because of their biological consequences and relatively noninvasive accessibility in a wide range of biological fluids. EVs have been studied in relation to numerous cancers, such as colorectal27,33, bladder34, prostate35, pancreatic36, breast37, gastric38, lung39, blood40, ovarian41, cholangiocarcinoma42, hepatocellular carcinoma43, and oral squamous cell carcinoma44 (Table 2), as well as cardiovascular diseases45 and malignancies of the central nervous system21. The proteomic analysis of EVs, specifically the analysis of their protein composition, may be helpful for further understanding the mechanisms of their biogenesis and their functional roles. Molecular communication between cancer cells and their stromal microenvironment is a key factor for cancer progression46,47. In conjunction with typical secretory pathways, it was proposed that these small membranous vesicles are alternate mediators of intercellular communication19. EVs carry an effector-rich proteome with the ability to control different functional properties of the recipient cell48. The protein composition of EVs from different sources was studied previously by using MS30,49–54, providing a robust basis for the identification of biomarker proteins in EVs for the purpose of quality control research. A thorough understanding of the protein composition of EV subtypes and the extent to which EV composition reflects the source cell composition is essential for further development of diagnostics and therapeutics. Although EVs are secreted by almost all cell types, some available data suggest the enhanced release of EVs under pathological conditions, such as cancer55. It is reasonable to expect that these vesicles may also play key roles in tumorigenesis since they can facilitate distant intercellular communication. Tumor-derived EVs typically carry tumor antigens, and functional proteins can be transferred to recipient cells through EVs23,54,56. A better understanding of the molecular bases underlying cancer invasion and metastasis is necessary to develop effective targets for therapy.
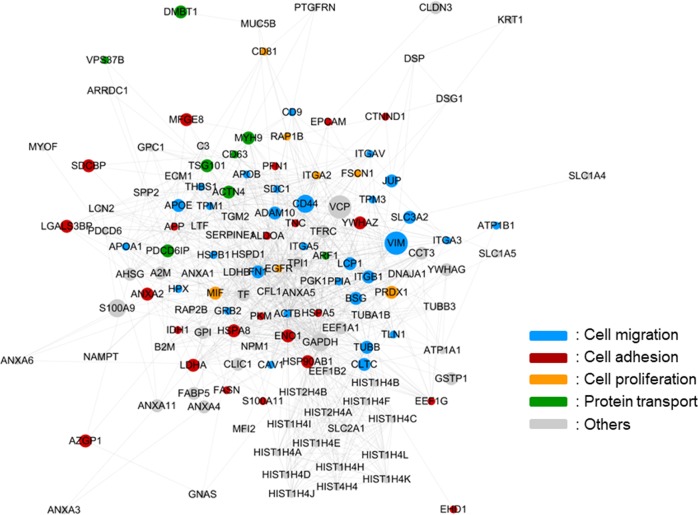
EV proteins from many cancers have similar biological processes and functions. To understand the functions of differentially expressed proteins (DEPs) in cancer, we performed gene ontology analysis on a variety of DEPs57. As expected, the EV–DEPs from different cancer types were implicated in similar biological processes, such as cell adhesion, migration, and transport. Considering that EVs are potential metastasis factors, those proteins appear to be relevant for cancer metastasis or cancer cell proliferation. Of the 12 different cancers evaluated, we observed that DEPs that overlapped more than five times were primarily related to cancer metastasis or cancer cell proliferation, and many of the DEPs had strong interactions with each other (Fig. 2). Even though the selection of these DEPs from different cancers was biased, the roles of EVs in different cancers focused mainly on cell adhesion and cell migration.
Fig. 2.
Protein–protein interaction network of differentially expressed extracellular vesicle proteins in cancer cell-derived EVs
Conclusions
In this review, we summarized different EV studies to discuss the potential of EVs in cancer treatment. All studies discussed in this review indicated that the specific protein composition of various EVs has high potential for identifying different cancers. The majority of these studies revealed the relationship of cancer with changes in the protein contents of various body fluids. Moreover, we have highlighted the emerging roles of EVs in cancer, specifically their role in metastasis, which opens the possibility of the rapid translation of EV research for clinical applications in diagnosis, prognosis, and treatment. Ultimately, the majority of the investigations discussed in this review need further verification in large-cohort, multicenter clinical studies. In the future, highly reliable EV proteome data could be combined with well-developed current popular genomic and other “omics-” studies to provide extended knowledge of EVs from the perspective of systems biology approaches.
Future perspectives
There are many perspectives on the potential contribution of EV research for the development of cancer therapeutics and diagnosis. EVs could play key roles in intercellular communication during cancer development, which may offer new therapeutic strategies for various cancers. EV protein composition in different body fluids reveals the overall condition of the patient and is also useful for screening the efficacy and toxicity of anticancer treatments. Additionally, EVs could be used as cancer vaccines and drug delivery components. Moreover, the inhibition of intercellular communication through EVs might provide opportunities to suppress tumor progression. In the near future, clinical applications of EVs could contribute to cancer management and treatment. However, before EV-targeted therapy can be applied in cancer, the identification of cancer-specific genes or molecules that are crucial for EV communication is necessary.
Acknowledgements
This work was supported by a grant from Kyung Hee University in 2017 (KHU-20171191) and the GRRC program of Gyeonggi province [GRRC-kyunghee2018(B03)].
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Perez-Hernandez D, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013;288:11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell. Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J. Proteom. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 6.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Conde I, Shrimpton CN, Thiagarajan P, Lopez JA. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106:1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- 8.Andaloussi EL, Mager S, Breakefield I, X. O., Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug. Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 9.Valenti R, et al. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66:9290–9298. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 10.Kim CW, et al. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002;62:6312–6317. [PubMed] [Google Scholar]
- 11.Kim DK, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 13.O’Driscoll L. Expanding on exosomes and ectosomes in cancer. N. Engl. J. Med. 2015;372:2359–2362. doi: 10.1056/NEJMcibr1503100. [DOI] [PubMed] [Google Scholar]
- 14.Jang SC, et al. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small. 2015;11:456–461. doi: 10.1002/smll.201401803. [DOI] [PubMed] [Google Scholar]
- 15.Gould Stephen J., Raposo Graça. As we wait: coping with an imperfect nomenclature for extracellular vesicles. Journal of Extracellular Vesicles. 2013;2(1):20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bobrie Angélique, Colombo Marina, Krumeich Sophie, Raposo Graça, Théry Clotilde. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. Journal of Extracellular Vesicles. 2012;1(1):18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell. Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell. Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 21.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyorgy B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Zembala M. Tumour-derived microvesicles modulate biological activity of human monocytes. Immunol. Lett. 2007;113:76–82. doi: 10.1016/j.imlet.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 25.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell. Dev. Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi DS, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11:2745–2751. doi: 10.1002/pmic.201100022. [DOI] [PubMed] [Google Scholar]
- 27.Choi DS, et al. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J. Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 28.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 29.Lee JE, et al. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J. Proteom. 2016;131:17–28. doi: 10.1016/j.jprot.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Duijvesz D, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris DA, et al. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS One. 2015;10:e0117495. doi: 10.1371/journal.pone.0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathivanan S, et al. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteom. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welton JL, et al. Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteom. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen FH, et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol. Cell. Proteom. 2009;8:1192–1205. doi: 10.1074/mcp.M800443-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ristorcelli E, et al. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 2008;22:3358–3369. doi: 10.1096/fj.07-102855. [DOI] [PubMed] [Google Scholar]
- 37.Palazzolo G, et al. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012;32:847–860. [PubMed] [Google Scholar]
- 38.Fu H, et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res. 2018;37:162. doi: 10.1186/s13046-018-0825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J. Proteom. 2016;133:161–169. doi: 10.1016/j.jprot.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Yao Y, et al. Proteomic analysis of exosomes derived from human lymphoma cells. Eur. J. Med. Res. 2015;20:8. doi: 10.1186/s40001-014-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang B, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013;80:171–182. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 42.Dutta S, et al. Proteomics profiling of cholangiocarcinoma exosomes: a potential role of oncogenic protein transferring in cancer progression. Biochim. Biophys. Acta. 2015;1852:1989–1999. doi: 10.1016/j.bbadis.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, et al. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J. Hematol. Oncol. 2018;11:82. doi: 10.1186/s13045-018-0625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, et al. Oral cancer-derived exosomal NAP1 enhances cytotoxicity of natural killer cells via the IRF-3 pathway. Oral. Oncol. 2018;76:34–41. doi: 10.1016/j.oraloncology.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Harada T, et al. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci. 2017;108:42–52. doi: 10.1111/cas.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kharmate G, Hosseini-Beheshti E, Caradec J, Chin MY, Tomlinson Guns ES. Epidermal growth factor receptor in prostate cancer derived exosomes. PLoS One. 2016;11:e0154967. doi: 10.1371/journal.pone.0154967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Kreimer S, et al. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J. Proteome Res. 2015;14:2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 50.Clark DJ, et al. Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster analysis. Anal. Chem. 2015;87:10462–10469. doi: 10.1021/acs.analchem.5b02586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT) Proteomics. 2012;12:329–338. doi: 10.1002/pmic.201100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalra H, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 53.Ji H, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 54.Graner MW, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohno S, Ishikawa A, Kuroda M. Roles of exosomes and microvesicles in disease pathogenesis. Adv. Drug Deliv. Rev. 2013;65:398–401. doi: 10.1016/j.addr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 56.Bosque A, et al. Comparative proteomics of exosomes secreted by tumoral Jurkat T cells and normal human T cell blasts unravels a potential tumorigenic role for valosin-containing protein. Oncotarget. 2016;7:29287–29305. doi: 10.18632/oncotarget.8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Barteneva NS, et al. Circulating microparticles: square the circle. Bmc. Cell. Biol. 2013;14:23. doi: 10.1186/1471-2121-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beckham CJ, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014;192:583–592. doi: 10.1016/j.juro.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 60.Silvers CR, Miyamoto H, Messing EM, Netto GJ, Lee YF. Characterization of urinary extracellular vesicle proteins in muscle-invasive bladder cancer. Oncotarget. 2017;8:91199–91208. doi: 10.18632/oncotarget.20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeppesen DK, et al. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics. 2014;14:699–712. doi: 10.1002/pmic.201300452. [DOI] [PubMed] [Google Scholar]
- 62.Sun Y, et al. A novel TP53 pathway influences the HGS-mediated exosome formation in colorectal cancer. Sci. Rep. 2016;6:28083. doi: 10.1038/srep28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaiyawat P, et al. Elevated O-GlcNAcylation of extracellular vesicle proteins derived from metastatic colorectal cancer cells. Cancer Genom. Proteom. 2016;13:387–398. [PMC free article] [PubMed] [Google Scholar]
- 64.Schillaci O, et al. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: their emerging role in tumor heterogeneity. Sci. Rep. 2017;7:4711. doi: 10.1038/s41598-017-05002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mulvey HE, et al. Extracellular vesicle-mediated phenotype switching in malignant and non-malignant colon cells. Bmc. Cancer. 2015;15:571. doi: 10.1186/s12885-015-1568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raimondo S, et al. Label-free quantitative proteomic profiling of colon cancer cells identifies acetyl-CoA carboxylase alpha as antitumor target of Citrus limon-derived nanovesicles. J. Proteom. 2018;173:1–11. doi: 10.1016/j.jprot.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 67.Guo J, et al. Phosphoproteome characterization of human colorectal cancer SW620 cell-derived exosomes and new phosphosite discovery for C-HPP. J. Proteome Res. 2016;15:4060–4072. doi: 10.1021/acs.jproteome.6b00391. [DOI] [PubMed] [Google Scholar]
- 68.Demory Beckler M, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteom. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi Dong-Sic, Choi Do-Young, Hong BokSil, Jang SuChul, Kim Dae-Kyum, Lee Jaewook, Kim Yoon-Keun, Pyo Kim Kwang, Gho YongSong. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. Journal of Extracellular Vesicles. 2012;1(1):18704. doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim JW, et al. Restoration of full-length APC protein in SW480 colon cancer cells induces exosome-mediated secretion of DKK-4. Electrophoresis. 2012;33:1873–1880. doi: 10.1002/elps.201100687. [DOI] [PubMed] [Google Scholar]
- 71.Ji H, Greening DW, Kapp EA, Moritz RL, Simpson RJ. Secretome-based proteomics reveals sulindac-modulated proteins released from colon cancer cells. Proteom. Clin. Appl. 2009;3:433–451. doi: 10.1002/prca.200800077. [DOI] [PubMed] [Google Scholar]
- 72.Rauschenberger L, et al. Exosomal particles secreted by prostate cancer cells are potent mRNA and protein vehicles for the interference of tumor and tumor environment. Prostate. 2016;76:409–424. doi: 10.1002/pros.23132. [DOI] [PubMed] [Google Scholar]
- 73.Hosseini-Beheshti E, Pham S, Adomat H, Li N, Tomlinson Guns ES. Exosomes as biomarker enriched microvesicles: characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol. Cell. Proteom. 2012;11:863–885. doi: 10.1074/mcp.M111.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soekmadji Carolina, Rockstroh Anja, Ramm Grant A., Nelson Colleen C., Russell Pamela J. Extracellular Vesicles in the Adaptive Process of Prostate Cancer during Inhibition of Androgen Receptor Signaling by Enzalutamide. PROTEOMICS. 2017;17(23-24):1600427. doi: 10.1002/pmic.201600427. [DOI] [PubMed] [Google Scholar]
- 75.Kawakami K, et al. Integrin beta4 and vinculin contained in exosomes are potential markers for progression of prostate cancer associated with taxane-resistance. Int. J. Oncol. 2015;47:384–390. doi: 10.3892/ijo.2015.3011. [DOI] [PubMed] [Google Scholar]
- 76.Soekmadji C, et al. Modulation of paracrine signaling by CD9 positive small extracellular vesicles mediates cellular growth of androgen deprived prostate cancer. Oncotarget. 2017;8:52237–52255. doi: 10.18632/oncotarget.11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kharaziha P, et al. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget. 2015;6:21740–21754. doi: 10.18632/oncotarget.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandvig K, Llorente A. Proteomic analysis of microvesicles released by the human prostate cancer cell line PC-3. Mol. Cell. Proteom. 2012;11:M111–012914. doi: 10.1074/mcp.M111.012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bilen MA, et al. Proteomics profiling of exosomes from primary mouse osteoblasts under proliferation versus mineralization conditions and characterization of their uptake into prostate cancer cells. J. Proteome Res. 2017;16:2709–2728. doi: 10.1021/acs.jproteome.6b00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Mijn JC, et al. Analysis of AKT and ERK1/2 protein kinases in extracellular vesicles isolated from blood of patients with cancer. J. Extracell. Vesicles. 2014;3:25657. doi: 10.3402/jev.v3.25657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi DY, et al. Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics. 2014;14:1845–1856. doi: 10.1002/pmic.201400008. [DOI] [PubMed] [Google Scholar]
- 82.Lobb Richard J., Hastie Marcus L., Norris Emma L., van Amerongen Rosa, Gorman Jeffrey J., Möller Andreas. Oncogenic transformation of lung cells results in distinct exosome protein profile similar to the cell of origin. PROTEOMICS. 2017;17(23-24):1600432. doi: 10.1002/pmic.201600432. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Jing, Lu Shaohua, Zhou Ye, Meng Kun, Chen Zhipeng, Cui Yizhi, Shi Yunfeng, Wang Tong, He Qing-Yu. Motile hepatocellular carcinoma cells preferentially secret sugar metabolism regulatory proteins via exosomes. PROTEOMICS. 2017;17(13-14):1700103. doi: 10.1002/pmic.201700103. [DOI] [PubMed] [Google Scholar]
- 84.He M, et al. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36:1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 85.Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tucker R, Pedro A. Blood-derived non-extracellular vesicle proteins as potential biomarkers for the diagnosis of early ER + breast cancer and detection of lymph node involvement. F1000Res. 2018;7:283. doi: 10.12688/f1000research.14129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Griffiths SG, Cormier MT, Clayton A, Doucette AA. Differential proteome analysis of extracellular vesicles from breast cancer cell lines by chaperone affinity enrichment. Proteomes. 2017;5:pii: E25. doi: 10.3390/proteomes5040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas SN, et al. Exosomal proteome profiling: a potential multi-marker cellular phenotyping tool to characterize hypoxia-induced radiation resistance in breast cancer. Proteomes. 2013;1:87–108. doi: 10.3390/proteomes1020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moon PG, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin. Cancer Res. 2016;22:1757–1766. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 90.Chow A, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci. Rep. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vardaki I, et al. Periostin is identified as a putative metastatic marker in breast cancer-derived exosomes. Oncotarget. 2016;7:74966–74978. doi: 10.18632/oncotarget.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kavanagh EL, et al. Protein and chemotherapy profiling of extracellular vesicles harvested from therapeutic induced senescent triple negative breast cancer cells. Oncogenesis. 2017;6:e388. doi: 10.1038/oncsis.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gangoda Lahiru, Liem Michael, Ang Ching-Seng, Keerthikumar Shivakumar, Adda Christopher G., Parker Belinda S., Mathivanan Suresh. Proteomic Profiling of Exosomes Secreted by Breast Cancer Cells with Varying Metastatic Potential. PROTEOMICS. 2017;17(23-24):1600370. doi: 10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- 94.Domenyuk V, et al. Plasma exosome profiling of cancer patients by a next generation systems biology approach. Sci. Rep. 2017;7:42741. doi: 10.1038/srep42741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, Kislinger T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem. Biophys. Res. Commun. 2014;445:694–701. doi: 10.1016/j.bbrc.2013.12.070. [DOI] [PubMed] [Google Scholar]
- 96.Escrevente C, et al. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS One. 2013;8:e78631. doi: 10.1371/journal.pone.0078631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yi H, et al. Exosomes mediated pentose phosphate pathway in ovarian cancer metastasis: a proteomics analysis. Int. J. Clin. Exp. Pathol. 2015;8:15719–15728. [PMC free article] [PubMed] [Google Scholar]
- 98.Yi H, et al. High-grade ovarian cancer secreting effective exosomes in tumor angiogenesis. Int. J. Clin. Exp. Pathol. 2015;8:5062–5070. [PMC free article] [PubMed] [Google Scholar]
- 99.Adamczyk KA, et al. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life. Sci. 2011;89:304–312. doi: 10.1016/j.lfs.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 100.Jin H, et al. Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 2018;109:2946–2956. doi: 10.1111/cas.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng J, et al. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB. 2018;20:597–604. doi: 10.1016/j.hpb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blomme A, et al. Myoferlin is a novel exosomal protein and functional regulator of cancer-derived exosomes. Oncotarget. 2016;7:83669–83683. doi: 10.18632/oncotarget.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klein-Scory S, et al. New insights in the composition of extracellular vesicles from pancreatic cancer cells: implications for biomarkers and functions. Proteome Sci. 2014;12:50. doi: 10.1186/s12953-014-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu Z, et al. Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget. 2017;8:63461–63483. doi: 10.18632/oncotarget.18831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castillo J, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018;29:223–229. doi: 10.1093/annonc/mdx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shin SJ, et al. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2013;110:19414–19419. doi: 10.1073/pnas.1309720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chaiyadet S, et al. Carcinogenic liver fluke secretes extracellular vesicles that promote cholangiocytes to adopt a tumorigenic phenotype. J. Infect. Dis. 2015;212:1636–1645. doi: 10.1093/infdis/jiv291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khoontawad J, et al. Differential protein expression marks the transition from infection with opisthorchis viverrini to cholangiocarcinoma. Mol. Cell. Proteom. 2017;16:911–923. doi: 10.1074/mcp.M116.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taverna S, et al. Curcumin modulates chronic myelogenous leukemia exosomes composition and affects angiogenic phenotype via exosomal miR-21. Oncotarget. 2016;7:30420–30439. doi: 10.18632/oncotarget.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paggetti J, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126:1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huan J, et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29:2285–2295. doi: 10.1038/leu.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Andrade A, et al. Extracellular vesicles from oral squamous carcinoma cells display pro- and anti-angiogenic properties. Oral. Dis. 2018;24:725–731. doi: 10.1111/odi.12765. [DOI] [PubMed] [Google Scholar]
- 113.Ono K, et al. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J. Cell. Biochem. 2018;119:7350–7362. doi: 10.1002/jcb.27039. [DOI] [PubMed] [Google Scholar]