Abstract
Over 60% of the modern distribution range of brown bears falls within Russia, yet palaeoecological data from the region remain scarce. Complete modern Russian brown bear mitogenomes are abundant in the published literature, yet examples of their ancient counterparts are absent. Similarly, there is only limited stable isotopic data of prehistoric brown bears from the region. We used ancient DNA and stable carbon (δ13C) and nitrogen (δ15N) isotopes retrieved from five Pleistocene Yakutian brown bears (one Middle Pleistocene and four Late Pleistocene), to elucidate the evolutionary history and palaeoecology of the species in the region. We were able to reconstruct the complete mitogenome of one of the Late Pleistocene specimens, but we were unable to assign it to any of the previously published brown bear mitogenome clades. A subsequent analysis of published mtDNA control region sequences, which included sequences of extinct clades from other geographic regions, assigned the ancient Yakutian bear to the extinct clade 3c; a clade previously identified from Late Quaternary specimens from Eastern Beringia and Northern Spain. Our analyses of stable isotopes showed relatively high δ15N values in the Pleistocene Yakutian brown bears, suggesting a more carnivorous diet than contemporary brown bears from Eastern Beringia.
Introduction
The brown bear (Ursus arctos) has a broad Holarctic distribution, spanning Eurasia and North America1. Brown bears are also found on the islands of northeast Asia, including Hokkaido (Japan) and the southern Kuril Islands (Russia)1. Until the 20th century, brown bears were common in most parts of Eurasia and in the western part of North America down to northern Mexico2,3. However, anthropogenic encroachment over the past century has resulted in reduced and fragmented brown bear populations4. Due to their extensive distribution, brown bears have become a model organism for studying biogeographic patterns across Eurasia and North America. Consequently, phylogeographic patterns of European brown bears were included in Hewitt’s seminal models of glacial contraction and postglacial expansion5,6.
Since the publication of the first brown bear phylogeographic study in 19947, many studies have further characterized the phylogeny and phylogeography of the species using mitochondrial DNA (mtDNA)8–12. Based on patterns of variation in present-day brown bears, seven main extant clades have been described (Fig. 1). Clade 1 includes bears from western European populations7. Clade 2a is found only on the Admiralty, Baranof, and Chichagof (ABC) islands of Alaska. Clade 3a is the most widely distributed, and spans from Eastern Europe to western Alaska1,13. Clade 3b is found in both eastern Alaska and in brown bears from eastern Hokkaido (Japan), the Russian Far East, and southern Siberia. Clade 4 is most common amongst brown bears of North America, but has also been identified in Southern Hokkaido (Japan)1,13. Less widespread brown bear clades have been identified in Asia, including clade 5 in Tibet1,14. Other less widely distributed brown bear clades (not included in our analyses and therefore not shown in Fig. 1) have been found in North Africa, the Middle East15,16, and the Himalayas17. The mtDNA of extant polar bears (Ursus maritimus), clade 2b, is embedded within brown bears and is most closely related to clade 2a, the ABC brown bears18.
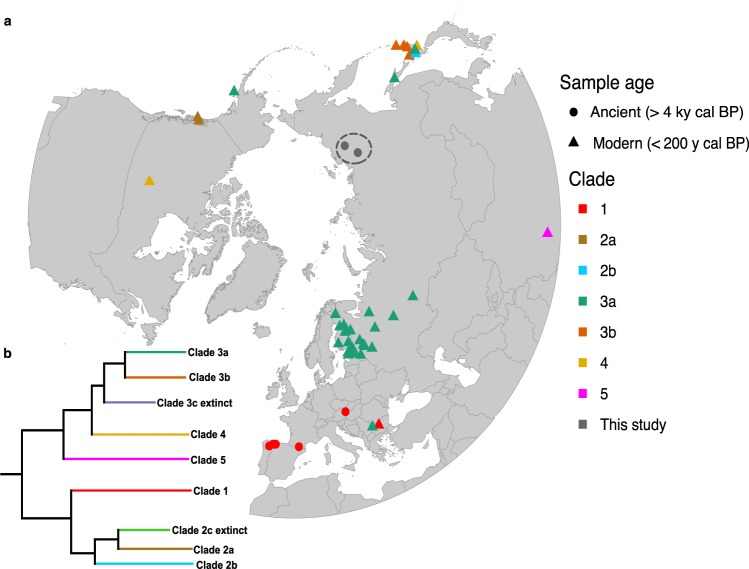
Figure 1.
(a) Map showing the sample location of the brown bears analysed using mtDNA, including information on what clades they belong to; ancient samples (dots), modern samples (triangles). The five Yakutian specimens investigated using ancient DNA and stable isotopes (grey dots) are indicated by a dashed circle. Our mitogenome study included 129 sequences (published n = 128, this study n = 1). (b) Schematic phylogeny of brown bear mitogenome clades, adapted from13. Extinct clades 2c and 3c are not shown on the map.
Fragments of the mitochondrial control region (CR) retrieved from ancient brown bear remains have been instrumental in developing our current understanding of the past biogeography of the species (e.g. refs.10,12,19). Ancient DNA analyses have documented shifting patterns of genetic diversity and structure, including the loss of genetic lineages during the Late Quaternary (such as clade 3c10,19,20). Such findings are indicative of more complex phylogeographic scenarios than initially suggested for the species by Taberlet and Bouvet7 (e.g. refs.10,12,19,21).
In this study, we extracted mitochondrial DNA of three Pleistocene subfossil brown bears collected in Yakutia, North-East Siberia, and assessed δ13C and δ15N of an additional two (totalling five) specimens. North-East Siberia is underrepresented in the large body of literature published on the species so far. Currently, there are no complete ancient mitochondrial sequences available in NCBI from the region, despite Russia covering >60% of the distribution range of brown bears3. Analysing our genetic data with the full panel of published brown bear mitogenomes, we reconstructed the maternal phylogeny of the species, to gain insight into the phylogenetic relationships among ancient Siberian brown bears and their modern contemporaries. To further elucidate the relationship between our Yakutian sequence and contemporary and extirpated brown bear populations, we included an analysis of the mtDNA CR, which included a larger temporal range of modern and ancient sequences.
Stable isotope data from Yakutian brown bears are limited22, compared with brown bears from other regions, or with other coeval large carnivores, limiting our understanding of their palaeoecology. We therefore compiled published isotopic data of Late Pleistocene megaherbivores and carnivores from North-East Siberia and Eastern Beringia, and compared them to the δ13C and δ15N isotopic compositions of our five Yakutian specimens.
Results and Discussion
We investigated five Pleistocene (one Middle Pleistocene and four Late Pleistocene) subfossil brown bears from Yakutia, one with an infinite 14C age (CGG_1_0200005) and the other four undated (CGG_1_0200006, CGG_1_0200007, DGI-1, and DGI-4). Morphological assessment of the individuals indicated that four of the five specimens, CGG_1_0200005, CGG_1_0200006, DGI-1, and DGI-4 were exceptionally large individuals (Fig. 2; Supplementary Tables 1–3).
Figure 2.
(a) Bivariate plot of the skull zygomatic width (in mm) and maximal length (in mm) of three of the ancient Yakutian bears from this study (green dots), and modern brown bears (Ursus arctos L.) from Siberia and the Russian Far East (triangles). Modern brown bears measurements are represented as mean ± SD (see Supplementary Table 1 for full details). (b) Plot representing the claw length along the outer curvature (in mm) of the ancient Yakutian bear (DGI-4) (green dot) and modern bears from Eurasia and North America (triangles). Lines represent an interval connecting minimal and maximal length values (see Supplementary Table 2).
mtDNA assembly
Three of the individuals (CGG_1_0200005, CGG_1_0200006, and CGG_1_0200007) were subjected to DNA analysis. To reconstruct mitochondrial genomes, we isolated total DNA using ancient DNA methodologies and transformed it into sequencing libraries. One library of each of the three samples was pooled with other samples and shotgun sequenced on the Illumina HiSeq. 2500 platform. Based on mapping to the polar bear reference genome (GCF_000687225.1), the endogenous content of the three samples ranged from 0.06% to 10% (Table 1). The endogenous content of 10% in sample CGG_1_0200006 indicated good preservation, suggesting nuclear work on this Late Pleistocene specimen may be possible in future studies.
Table 1.
Basic statistics derived from read processing and mapping.
| Sample | Total reads | Nuclear mapping | Endogenous content | mtDNA mapping | Depth |
|---|---|---|---|---|---|
| CGG_1_0200005 | 7,322,879 | 20,897 | 0.29% | 92 | 0.3x |
| CGG_1_0200006 | 164,297,287 | 16,563,819 | 10% | 18,612 | 69x |
| CGG_1_0200007 | 7,097,255 | 4,282 | 0.06% | 44 | 0.12x |
Mapping of the reads to the brown bear mtDNA reference genome (NC_003427.1) varied substantially between specimens: CGG_1_0200005 yielded 92 unique reads; CGG_1_0200006 18,612 unique reads; and CGG_1_0200007 44 unique reads (Table 1). Based on the number of reads after filtering, we were able to assemble the complete mitogenome (defined as >90% of the genome, covered to at least 3x read depth) of CGG_1_0200006 at 69x. The assembled mitogenome was 17,022 bp, with some gaps in the repetitive region of the CR; a region that is difficult to assemble for ancient brown bear specimens, due to low coverage or ambiguous read alignment23. The low number of unique sequencing reads derived from CGG_1_0200005 and CGG_1_0200007 were not sufficient to assemble robust mitogenomes; thus, these two samples were not subjected to further DNA analysis. Ancient damage pattern estimates from mapDamage show that all our samples have terminal base substitutions, which authenticate the sequenced fragments as ancient (Supplementary Fig. 1).
Ancient Yakutian mitogenome shifts clade 3 divergence estimate back > 25 ky
The mitogenome from CGG_1_0200006 was combined with 128 full mitochondrial genome sequences downloaded from NCBI: 17 ancient brown bears (41,201–4,635 years cal BP), 101 modern brown, and 10 polar bears; the mitochondrial lineage of the latter groups within the diversity of brown bears and hence was included in our analysis. MrBayes v3.2.624 and BEAST v.1.8.225 were used to estimate phylogenetic relationships. Both analyses yielded the same topology, and placed our Late Pleistocene Yakutian specimen as a sister lineage to clade 3b (as defined in Hirata et al. 2013) (Fig. 3; Supplementary Fig. 2). We ran BEAST v.1.8.225 using a tip-dating approach to perform the molecular dating of CGG_1_0200006, which had not been radiocarbon dated, and to estimate clade divergence times. For CGG_1_0200006, this approach gave a mean age estimate of 61,826 years BP [HPD 95%: 27,866–96,783 years BP].
Figure 3.
Tip-calibrated phylogeny of the brown bear mitogenomes analysed using BEAST. Branch labels indicate posterior clade probabilities >0.85. Tip label postfixes indicate sample age. Tips are coloured based on clade, and tip symbols indicate sample age. Nodes are centred on the median estimated divergence time, and red bars indicate the 95% HPD. Clade 3a was collapsed to facilitate visualization. Information on the specimens included in clade 3a can be found in Supplementary Table 3.
The lack of specimens representative of clade 3c in the mitogenome dataset (Fig. 3; Supplementary Fig. 2) meant that CGG_1_0200006 could not be assigned to a known clade, nor be confirmed as a novel lineage. To mitigate this, we generated a haplotype network of 225 mtDNA CR sequences representing clades 3a, 3b, 3c, and 4 from modern and ancient brown bears (Supplementary Table 4). We used PopART v.1.726 to construct a median-joining network of the overlapping region of 270 bp of the CR, with clade 4 sequences as an outgroup.
We find that CGG_1_0200006 grouped with extinct clade 3c brown bears (Fig. 4). Clade 3c has been identified in ancient Alaskan and Iberian brown bears dated >35 ky cal BP10,19,20. Our data provide further evidence that this clade was indeed widespread across the northern hemisphere during the Late Pleistocene.
Figure 4.
Clade 3 mtDNA CR haplotype network of 225 published sequences, indicating the location of the Yakutian specimen sequenced in this study (CGG_1_0200006). Specimen information listed in Supplementary Table 4. Circles are sized relative to haplotype frequency. Dashes indicate single-nucleotide mutations. The map represents the range of the clade 3 lineages (3a, 3b and 3c). Ranges were drawn based on13. Analysis was run with Clade 4 as an outgroup.
We estimated a divergence time of clade 3c from clades 3a/b of 123 ky BP [HPD 95%: 90–164] ky BP. This predates previous estimates based on short fragments of mtDNA CR sequences by >25 ky13,27. Other studies of brown bear have estimated various time to the most recent common ancestor (tMRCA) for other clades, in particular influenced by the length of the alignment (single mtDNA genes versus whole mitogenomes) and the method of calibration (e.g. tip-dating, fossil calibration)1,13,28.
Central Asia is accepted as the central point of origin and diversification of brown bears2, and colonization of other areas from this region would lead to the diversification of lineages. During the glacial periods of the Pleistocene, the decline in ocean levels created a land bridge between Eurasia and North America29. A previous study investigating several biogeographic scenarios for brown bear colonization of North America suggested the entrance of several brown bear clades into North America was initiated around 70 ky BP13. This is supported by our results, which imply that divergence of clade 3c took place at the beginning of Marine Isotope Stage 5 (MIS5), during an interglacial, where temperatures were higher than today30 (Fig. 5). Due to the contraction of the ice and improved climatic conditions, brown bears could have expanded across Asia and possibly Europe. The oldest radiocarbon date of clade 3c brown bears in Alaska is as infinite date of >59 ky BP10, and our divergence estimate supports that at the end of MIS 5 or the onset of MIS 4, when climates cooled, East Asian clade 3c brown bears may have crossed Beringia across the exposed Bering Land Bridge13. At present, only one extinct clade 3c brown bear individual has been recorded in Europe, so we can only speculate as to whether this reflects an as yet undiscovered clade 3c population, or vagrant bears that moved westwards12.
Figure 5.
Curve of air temperature variation relative to the present (°C) through time30. Marine isotopic stadials (MIS) according to Lisiecki & Raymo90. Minimum age of the oldest record of a clade 3c brown bear of America (>59 ky BP, blue); Yakutian clade 3c bear (CGG_1_0200006) from this study (61,826 years BP [HPD 95%: 27,866–96,783 years BP], green); estimated clade 3c divergence time (123 ky BP [HPD 95%: 90–164] ky BP, pink).
Isotopic signatures differ in Eastern and Western Beringia
The stable isotopic compositions (δ13C and δ15N) of the five Yakutian brown bears were measured and compared to other Beringian megafauna predating the Last Glacial Maximum (LGM; only samples >25 ky cal BP) (Fig. 6). At that time, the taxonomic composition of mammoth steppe mammalian communities was similar from Europe to Eastern Beringia31. Species included large predators such as felids (e.g. lions (Panthera spelaea/Panthera atrox) spanning Europe to Alaska and scimitar cats (Homotherium serum) in Eastern Beringia), bears (e.g. brown bears across the mammoth steppe and short-faced bears (Arctodus simus) in Eastern Beringia), and canids (e.g. wolves (Canis lupus) across the entire region and hyenas (Crocuta crocuta) in western Eurasia)32. Wide-ranging mammoth steppe megaherbivores included woolly mammoths (Mammuthus primigenius), reindeer (Rangifer tarandus), bison (Bison sp.), horses (Equus ferus), caprine bovids (Bootherium cavifrons in Beringia, and Rupicapra rupicapra in Europe), and Eurasian woolly rhinoceros (Coelodonta antiquitatis).
Figure 6.
Bivariate plot of the stable carbon and nitrogen isotope values of the five Yakutian brown bears (green dots), and other pre-LGM megafauna from Western Beringia (left) and Eastern Beringia (right). Except the five bears analysed in this study, species are represented as mean ± 1 SD (see Supplementary Table 5 for full details) and the shaded areas represent the convex hulls91, which are defined as the minimum area polygon encompassing all points in bivariate space. Horse silhouette: Mercedes Yrayzoz (vectorized by T. Michael Keesey), license CC-BY-3.0 (https://creativecommons.org/licenses/by/3.0/). Scimitar cat and mammoth silhouettes: Sergiodlarosa (original images have been modified), license CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/).
We compiled the isotopic signatures of various megafauna species from Western and Eastern Beringia, including the five Yakutian brown bears from this study (Fig. 6; Supplementary Table 5). Despite the potential age difference among the Western Beringia brown bear samples, our δ13C and δ15N analysis indicate that the carnivorous diet in brown bear from the region was sustained through time, despite climatic and environmental fluctuations33.
Our results also reflect differences between the isotopic signatures of megaherbivores of Western and Eastern Beringia (Fig. 6), in agreement with previous work31,34. Megaherbivores from Western Beringia have higher δ15N (difference from +1 to +2.7‰) and lower δ13C (difference from 0 to −1.1‰) compared to Eastern Beringia (See Supplementary Table 6). The general offset in all species likely reflects environmental differences between the two regions. Soil δ15N is a product of nitrogen-transforming microorganisms and is proportional to the total soil organic nitrogen, average annual temperatures and precipitation35; soil and plant δ15N are strongly correlated36 and herbivore δ15N reflects the nitrogen isotope composition of the plants consumed37,38. In Europe, regions with higher development of permafrost during the LGM yield the lowest collagen δ15N in large herbivores during the post-LGM period, around 23–14 kyr cal BP39,40. This is comparable to contemporary patterns in which δ15N in surface soils rise with increasing distance from a glacier front, due to an increase in soil development41. The glaciers in the Brooks and Alaska ranges—in the north and south of Eastern Beringia, respectively42-could be responsible for the low δ15N in this region relative to Western Beringia, where the distance to the glacial front in Western Eurasia was larger during and after the LGM43. In addition, other climatic conditions, such as less aridity in Alaska, have been suggested33,44.
Isotopic signatures suggest Pleistocene brown bears from Western Beringia had carnivorous diets
Based on interspecific comparative palaeoecological analyses and paleoenvironmental data, we suggest that the high δ15N values in the Yakutian brown bears reflects a highly carnivorous diet. Pre-LGM clade 3c brown bears from Eastern Beringia10 had lower δ15N (presumably less carnivorous) compared to our Yakutian clade 3c brown bear (CGG_1_020000006), suggesting that carnivorous dietary habits were not linked to a mitochondrial haplogroup, but rather to palaeoecological factors.
In herbivores, interspecific differences in isotopic values have been linked to dietary specialization, and to interspecific differences in how food is digested34,39,44–46. Thus, differences exist among the hindgut fermenter horse relative to coeval ruminants, including bison. Reindeer has a diet strongly biased towards lichens that are 13C-enriched relative to vascular C3-plants47. Mammoth δ15N values are significantly higher than those of any other herbivore species31. In this way, despite the absolute differences in isotopic values of Western and Eastern Beringia (Fig. 6), the isotopic offset between megaherbivore species is preserved on both sides of Beringia.
This is not the case for wolves and brown bears, which implies that dissimilarities between these taxa are driven by dietary differences, rather than environmental variation impacting the entire food web. The difference between Western and Eastern Beringia for the brown bear is +4.6‰ for δ15N and −0.5‰ for δ13C, and in the case of the wolf is +4.1‰ for δ15N and −0.8‰ for δ13C (Supplementary Table 6). The other carnivore that is present on both sides is the lion, but the difference between regions is akin to the pattern seen in megaherbivores (+2.9‰ for δ15N and −0.2‰ for δ13C) (Supplementary Table 6). The overlap in the isotopic distribution among lions, wolves, and brown bears in Eastern Beringia is small, while in Western Beringia it is noticeable (Fig. 6). In Pleistocene Europe, wolves and brown bears competed with hyenas and lions; Pleistocene wolves exhibited lower δ15N values than contemporary cave hyenas and cave lions, despite their overlap with some cave lion individuals48. A similar scenario might have occurred in Eastern Beringia, but with more predators with which to compete, besides lions: scimitar cats and short-faced bears, which occupied a higher trophic position than wolves and brown bears. The reduced competition with other predators in Western Beringia may have provided brown bears and wolves with the opportunity to predate larger herbivores, rather than scavenging.
The typically high δ15N values in pre-LGM brown bears from North Western and Central Europe have also been attributed to competition with vegetarian cave bears (Ursus spelaeus)32,49,50. Cave bears have been reported in Western Beringia51,52, but not in Eastern Beringia. In Western Beringia, such competition may have contributed to a more carnivorous diet in brown bears. This may in turn have enabled exceptionally large body sizes, as in four of our specimens; body size in extant brown bears is correlated with meat availability53. Such a large size would have been advantageous in competition with the other large predators of the Western Beringia mammoth steppe32.
Conclusions
This study presents the first combined ancient DNA and stable isotope (δ13C and δ15N) investigation of Pleistocene brown bears from Yakutia. Using shotgun sequencing, we generated a complete mitogenome sequence (69x) that is distinct from any previously published brown bear mitogenome clades. Phylogenetic comparisons to extant and extinct brown bear sequences assigned this individual to the extinct clade 3c, which has previously been found in Eastern Beringia and Spain. Thus, this mitochondrial genome represents the first available full mtDNA for clade 3c. This mitogenome allowed us to generate a new estimation of the tMRCA of clade 3c to the other clade 3 bears, predating previous estimates by >25 ky and supporting the diversification of this clade in Western Beringia, followed by the colonization of North America across the Bering Land Bridge. Stable isotope analyses show high δ15N values in pre-LGM brown bears from Yakutia, indicating that these bears were more carnivorous than brown bears in Eastern Beringia. This could reflect reduced competition among carnivores in the region, enabling brown bears to predate larger herbivores, and possibly also competition with vegetarian cave bears present in Western Beringia, but absent in Eastern Beringia.
Importantly, our study raises a phylogenetic database issue. The lack of complete mitogenomes for some clades might bias phylogenetic inferences. Thus, if possible, we suggest performing complementary analyses using single genes/partial sequences (e.g. partial control region, cytochrome b) largely available through NBCI, whenever there is a reduced representation of age, geography, and/or lineages at the full mitogenome level. Also, to further assess evolutionary processes and to test phylogeographic hypotheses, extended ancient DNA studies—in particular from Pleistocene specimens—derived from underrepresented geographic areas are needed. Moreover, investigations that combine ancient DNA and isotopic analyses may lead to a more complete understanding of the evolutionary ecology of phylogeographic changes in Pleistocene terrestrial large mammals, and we suggest they be performed more systematically.
Materials and Methods
Samples
Five subfossil bear samples were excavated during various paleontological surveys in Yakutia from 2005 through 2016 (see Fig. 1 and Supplementary Tables 1, 2 for precise locations) as incomplete skeletal remains. For three samples, some cranial material was recovered and, for the remaining two specimens, an upper canine fragment and a claw were recovered.
The largest brown bear skull from the Uyandina River, CGG_1_0200005, belonged to an adult male, and was radiocarbon dated at the University of Groningen (see section below). Morphological measurements of this skull exceed the corresponding parameters not only for other ancient and modern bear skulls from Yakutia, but also the maximal sizes of the largest representatives of modern brown bear subspecies from Eurasia (U. a. beringianus, Amur River region, Russian Far East and U. a. piscator, Kamtchatka Peninsula)54 (Supplementary Table 1).
The second skull, CGG_1_0200006, was of smaller size and morphological identification assigned it to an adult female (Supplementary Table 1). The degree of preservation of the skulls, as well as the bone remains of accompanying fauna, suggest that this taphocenosis could have been formed during the MIS 3 (ca. 60–27 kyr cal BP) the time of the greatest warming during second half of the Late Pleistocene, when special conditions were created for the death and burial of mammoth fauna.
The third specimen, CGG_1_0200007, represents a fragment of brown bear skull, cerebral cortex, that could not be used to assess the size of this individual (Supplementary Table 1). The fragment, that was dark-brown in colour and strongly fossilized, was found at the Ulakhan Sullar outcrop, Adycha River, Verkhoyansk district55.
The tooth sample (DGI-1) from the Deputatsky village that belonged to a large brown bear, was stratigraphically assigned to the Late Pleistocene. Unfortunately, it was not possible to perform direct measurements of the skull of this bear (Supplementary Table 1).
Finally, measurements of the claw sample (DGI-4) found in 2016 at the Novosibirsk Islands Archipelago (the extreme north of Yakutia), indicate that it was a very large specimen compared to modern brown bear claws from Yakutia and other regions, as well as modern polar bear claws (Supplementary Information; Supplementary Table 2). Based on stratigraphy, this bear sample was assigned to the Upper Pleistocene.
Radiocarbon dating and stable isotope data
The lower jaw bone of CGG_1_0200005 was radiocarbon dated at the University of Groningen, based on an acid-based collagen extracted fraction56. The collagen was 14C dated by Accelerator Mass Spectrometry57, yielding an infinite age for the 14C method: >45 ky BP (GrA-66592). This is expressed in conventional 14C years, and corresponds to an age >48,000 years58. CGG_1_0200005 and CGG_1_0200006 were excavated from the same geological layer, thus CGG_1_0200006 is assumed to be from the same period as CGG_1_0200005, although this specimen was not radiocarbon dated. CGG_1_0200007 was not radiocarbon dated either, but initial superficial collagen-based assessment at the Russian Academy of Sciences (on the extent of skeletal fossilisation, as per59) indicated a likely temporal origin for this sample in the late Middle Pleistocene (ca. ~120 to 200 ky BP).
For specimens CGG_1_0200005, CGG_1_0200006, and CGG_1_0200007, collagen was extracted from powdered bone samples by demineralizing with 0.5 M HCl at room temperature for 4 h under constant motion. Samples were then rinsed to neutrality and treated with 0.1 M NaOH at room temperature for successive 20 min treatments until no colour change was observed in the solution. Samples were rinsed to neutrality, solubilized in 10−3 M HCl at 75 °C for 36 h, and then the solution containing the collagen was transferred to a glass vial and lyophilized. Stable carbon and nitrogen isotopic compositions were measured at Trent University, using a Nu Horizon (Nu Instruments, Wrexham, UK) continuous-flow isotope ratio mass spectrometer coupled to a EuroEA 3000 (Eurovector, Milan, Italy) elemental analyser. Analytical uncertainty was determined to be ±0.20‰ for both δ13C and δ15N60. Analysed samples were only included in plots and interpretations if they were characterized by acceptable atomic C:N ratios (2.9–3.6) and minimums for wt% C, wt% N, and wt% collagen61. The 14C dated jaw bone also produced stable isotope ratios. These were very similar to those obtained from the skulls.
Further stable isotope analyses were conducted in Tübingen, where the collagen was extracted for all five brown bear specimens, with a similar protocol62. For the claw sample (DGI-4), a pretreatment with 2:1 chloroform:methanol was applied to remove possible fat contaminants63. The stable carbon and nitrogen isotopic compositions were performed in duplicate at the Institute of Environmental Science and Technology (ICTA, Barcelona) using a Thermo Flash 1112 (Thermo Scientific VC) elemental analyzer coupled to a Thermo Delta V Advantage mass spectrometer with a Conflo III interface. This measures the ratios of 13C/12C and 15N/14N relative to a standard (V-PDB for carbon and AIR for nitrogen). The international laboratory standard, IAEA 600 (caffeine), was used. Analytical uncertainty was determined to be ±0.20‰ for both δ13C and δ15N, based on multiple measurements of collagen extracted from modern bones of camel (Camelus dromedarius) and elk (Alces alces). The isotopic values of the tooth sample (DGI-1) were converted into equivalent δ13C and δ15N of collagen (δ13Ccoll and δ15Ncoll) taking into consideration the mean differences observed between collagen and bone in brown bear (δ13Cbone = δ13Cdentine − 0.67; δ15Nbone = δ15Ndentine − 1.47)32. The δ13C value of the claw sample (DGI-4) was transformed into equivalent δ13C values of collagen (δ13Ccoll) considering the mean enrichment of +2‰ observed between collagen and keratin64.
In order to contextualize the isotopic data of the five brown bears from this study, 426 additional isotopic records from Eastern and Western Beringia were compiled from the literature10,22,34,44,65–69. All data is pre-LGM and have a radiocarbon or stratigraphic dating. The dataset consisted of megaherbivores that were present on both sides of the Bering Strait: mammoth (Mammuthus primigenius), horse (Equus sp.), bison (Bison sp.), and reindeer (Rangifer tarandus), in addition to all available carnivores: brown bear, wolf (Canis sp.), cave lion (Panthera spelaea), scimitar cat (Homotherium serum), and short-faced bear (Arctodus simus). See Supplementary Table 5. Bivariate analyses for δ13C and δ15N were conducted in Past3 v3.2070.
Ancient DNA extraction and amplification
We drilled 50–70 mg of bone powder from each specimen and extracted DNA using a modified version of the protocol from71: samples were incubated overnight with the extraction buffer at 42 °C instead of at 37 °C, the bone powder was pelleted out of suspension, and the supernatant concentrated down to 150–200 μl for each sample using 30 kDa Amicon centrifugal filter unit (Millipore). Binding buffer was added 13x to the concentrated supernatant and DNA was purified with MinElute columns (Qiagen), following the manufacturer’s instructions with the exception of a 15-minute incubation at 37 °C during the elution step. DNA extracts were transformed into sequencing libraries in 25 μl reactions following the protocol from72 with the following modifications: the initial DNA fragmentation was not performed; MinElute kit (Qiagen) was used for the purification steps. DNA libraries were indexed using KAPA HiFi uracil + premix (KAPA Biosystems). The number of cycles for index PCRs was determined from qPCR analysis. The resulting libraries were quantified on an Agilent 2100 Bioanalyser, pooled at equimolar concentration and sequenced on an Illumina HiSeq 2500 SR 80 basepairs (bp).
Data processing
Sequencing reads processing (adapter trimming with AdapterRemoval v2.2.273), mapping (read alignment, PCR duplicate removal, and indel realignment), and damage analyses (mapDamage v2.0.674) were performed using the PALEOMIX v1.2.13.1 pipeline75. BWA-aln v0.7.1576 with disabled seed was used for mapping, following the authors’ recommendation. Reads shorter than 30 bp were discarded during adaptor trimming. Reads were mapped to the polar bear nuclear reference to estimate the endogenous content of our libraries (GCF_000687225.177), and to the mitochondrial brown bear reference (NC_003427.178). GATK v4.0.4.079 was used to perform indel realignment within the PALEOMIX v1.2.13.1 pipeline74. ANGSD v0.91980 was used to generate mitochondrial genome consensus sequences from the unique reads BAM files. Base calling was performed under highest effective depth (EBD) at each site, with EBD being the product of the mapping quality and scores for the base under consideration81.
Phylogenetic and network analysis
We included consensus sequences covering >90% of the brown bear mitochondrial reference genome (NC_003427.166) in the phylogenetic analysis. Consensus sequences were combined with 128 published mitochondrial genomes of ancient brown bears (n = 17, spanning from 41,201 to 4,635 years cal BP), modern brown (n = 101), and polar bears (n = 10) using MAFFT v7.39282 (see Supplementary Table 3). A repetitive region of the control region was excluded from the alignment due to ambiguous alignment and missing data in the NCBI sequences. The final size of the alignment (including transfer RNA –tRNAs-, ribosomal RNA –rRNAs-, protein coding-genes, and partial control region) was 15,928 bp. PartitionFinder v1.1.183 was used to determine the partition scheme and substitution model for the mitogenome alignment using the Bayesian Information Criterion and the greedy algorithm. PartitionFinder v1.1.183 suggested three partitions for our data: rRNAs and tRNAs; 1st and 2nd codon protein-coding genes; 3rd codon protein-codon genes; and partial control region. The best models suggested by the Bayesian Information Criterion are HKY + I model for the rRNAs and tRNAs; HKY + I for the 1st and 2nd codon partition; the HKY + G for the 3rd codon partition; and HKY + G + I for the partial control region.
We constructed a Bayesian phylogeny using MrBayes v3.2.624 with two runs and four chains of 1 × 107 Markov chain Monte Carlo (MCMC) generations and sampling every 1 × 103 generations with a 25% burn-in. Convergence was assessed with Tracer v1.684. Trees were summarized with the majority-rule consensus approach, using posterior probability as a measure of clade support.
We also constructed a phylogeny in BEAST v.1.8.225 using the ancient dated sequences to estimate a mutation rate across the mitochondrial genome and divergence dates of the lineages. BEAST was also used to estimate the age of CGG_1_0200006. The alignment used in the BEAST analysis was a reduced version of the alignment analysed with MrBayes, as we did not include a published ancient undated sample (see Supplementary Table 3). Phylogenetic age estimation was performed using a tip-dating method85. The prior posterior distribution of the tip date of the undated sample was set between zero (present day) and one million years, following the analyses of reference22. Substitution rate priors for all the partitions were assigned an independent mutation rate prior according to86. A cross-validation method was used to estimate the date of samples with known age23. A date-randomization test was performed to test for the temporal signal in the data. TipDatingBeast v1.0–887 was used to generate 20 date-randomized BEAST xml files and compare the parameter estimates of the real dataset with the date-randomized datasets. Our date-randomization test was passed, indicating that there is a temporal signal in our dataset (Supplementary Fig. 3).
All BEAST analyses were run in two independent MCMC chains of 1 × 108 generations each, sampling trees and model parameters every 1 × 103 generations. Tracer v1.684 was used to combine and inspect the results of each run and to determine the convergence of each parameter, all of which had ESS values >200. We identified the Maximum Clade Credibility (MCC) tree in TreeAnnotator v1.8.0, and visualized and graphically edited the MCC tree using ggtree88.
To further evaluate the phylogenetic placement of the brown bear mtDNA generated in this study, we compiled a dataset of 225 published control region (CR) sequences for clades 3a, 3b, 3c and 4 (Supplementary Table 4). The consensus sequence for CGG_1_0200006 was aligned to the sequences in the CR dataset using MAFFT v7.39282. The alignment was trimmed down to 270 bp, which allowed for the incorporation of sequences from the extinct clade 3c10. A haplotype network was generated using the median-joining algorithm89 implemented in the program PopART v.1.726.
Accession number
Complete mitogenome for the specimen CGG_1_0200006 is stored in the NCBI GenBank with accession number MH255807.
Supplementary information
Acknowledgements
This work was supported by the Villum Fonden Young Investigator Programme, grant No. 13151.
Author Contributions
E.D.L. conceived and designed the research. G.G.B. provided the bone samples. G.G.B. and G.F.B. conducted initial morphological analyses and contributed to the discussion. A.R.I. and E.C.T. performed D.N.A. extraction, sequencing libraries preparation, and bioinformatics analysis. J.P. conducted the radiocarbon analysis. H.B. and P.S. conducted the stable isotope analyses. A.G.V. compiled the control region and stable isotope databases, and stable isotope data interpretation. A.R.I. wrote the manuscript with input from all the authors, especially from E.D.L. and A.G.V. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alba Rey-Iglesia, Email: ardelaiglesia@snm.ku.dk.
Eline D. Lorenzen, Email: elinelorenzen@snm.ku.dk
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40168-7.
References
- 1.Hirata D, et al. Molecular phylogeography of the brown bear (Ursus arctos) in Northeastern Asia based on analyses of complete mitochondrial DNA sequences. Mol. Biol. Evol. 2013;30:1644–1652. doi: 10.1093/molbev/mst077. [DOI] [PubMed] [Google Scholar]
- 2.Kurtén, B. Pleistocene mammals of Europe (Aldine, Chicago, 1968).
- 3.McLellan, B. N., Proctor, M. F., Huber, D. & Michel, S. Ursus arctos (amended version of 2017 assessment). The IUCN Red List of Threatened Species 2017: e.T41688A121229971, 10.2305/IUCN.UK.2017-3.RLTS.T41688A121229971.en (2017).
- 4.Swenson JE, Taberlet P, Bellemain E. Genetics and conservation of European brown bears Ursus arctos. Mammal Rev. 2011;41:87–98. [Google Scholar]
- 5.Hewitt G. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999;68:87–112. [Google Scholar]
- 6.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 7.Taberlet P, Bouvet J. Mitochondrial DNA polymophism, phylogeography, and conservation genetics of the brown bear Ursus arctos in Europe. Proc. R. Soc. Lond. B. 1994;255:195–200. doi: 10.1098/rspb.1994.0028. [DOI] [PubMed] [Google Scholar]
- 8.Matsuhashi T, Masuda R, Mano T, Yoshida MC. Microevolution of the mitochondrial DNA control region in the Japanese brown bear (Ursus arctos) population. Mol. Biol. Evol. 1999;16:676–84. doi: 10.1093/oxfordjournals.molbev.a026150. [DOI] [PubMed] [Google Scholar]
- 9.Leonard JA, Wayne RK, Cooper A. Population genetics of Ice Age brown bears. Proc. Natl. Acad. Sci. USA. 2000;97:1651–1654. doi: 10.1073/pnas.040453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene Population Extinctions in Beringian Brown Bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- 11.Zachos FE, Otto M, Unici R, Lorenzini R, Hartl GB. Evidence of a phylogeographic break in the Romanian brown bear (Ursus arctos) population from the Carpathians. Mamm. Biol. 2008;73:93–101. [Google Scholar]
- 12.García-Vázquez, A., Pinto Llona, A. C. & Grandal-d’Anglade, A. Post-glacial colonization of Western Europe brown bears from a cryptic Atlantic refugium out of the Iberian Peninsula. Hist. Biol. 1–13 In press (2017).
- 13.Davison J, et al. Late-Quaternary biogeographic scenarios for the brown bear (Ursus arctos), a wild mammal model species. Quat. Sci. Rev. 2011;30:418–430. [Google Scholar]
- 14.Guskov VY, Sheremeteva IN, Seredkin IV, Kryukov AP. Mitochondrial cytochrome b gene variation in brown bear (Ursus arctos Linnaeus, 1758) from southern part of Russian Far East. Russ. J. Genet. 2013;49:1213–1218. [PubMed] [Google Scholar]
- 15.Calvignac S, Hughes S, Hänni C. Genetic diversity of endangered brown bear (Ursus arctos) populations at the crossroads of Europe, Asia and Africa. Divers. Distributions. 2009;15:742–750. [Google Scholar]
- 16.Calvignac S, et al. Ancient DNA evidence for the loss of a highly divergent brown bear clade during historical times. Mol. Ecol. 2008;17:1962–1970. doi: 10.1111/j.1365-294X.2008.03631.x. [DOI] [PubMed] [Google Scholar]
- 17.Lan, T. et al. Evolutionary history of enigmatic bears in the Tibetan Plateau–Himalaya region and the identity of the yeti. Proc. R. Soc. B. 284 (2017). [DOI] [PMC free article] [PubMed]
- 18.Edwards CJ, et al. Ancient Hybridization and an Irish Origin for the Modern Polar Bear Matriline. Curr. Biol. 2011;21:1251–1258. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdiosera CE, et al. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos) Proc. Natl. Acad. Sci. USA. 2008;105:5123–5128. doi: 10.1073/pnas.0712223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray, S.C.E. Mitochondrial DNA analysis of the evolution and genetic diversity of ancient and extinct bears (University of Adelaide, 2010).
- 21.Hofreiter M, et al. Lack of phylogeography in European mammals before the last glaciation. Proc. Natl. Acad. Sci. USA. 2004;101:12963–12968. doi: 10.1073/pnas.0403618101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirillova IV, et al. On the discovery of a cave lion from the Malyi Anyui River (Chukotka, Russia) Quat. Sci. Rev. 2015;117:135–151. [Google Scholar]
- 23.Fortes GG, et al. Ancient DNA reveals differences in behaviour and sociality between brown bears and extinct cave bears. Mol. Ecol. 2016;25:4907–4918. doi: 10.1111/mec.13800. [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 25.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.8. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Met. Ecol. Evol. 2015;6:1110–1116. [Google Scholar]
- 27.Korsten M, et al. Sudden expansion of a single brown bear maternal lineage across northern continental Eurasia after the last ice age: a general demographic model for mammals? Mol. Ecol. 2009;18:1963–1979. doi: 10.1111/j.1365-294x.2009.04163.x. [DOI] [PubMed] [Google Scholar]
- 28.Anijalg P, et al. Large‐scale migrations of brown bears in Eurasia and to North America during the Late Pleistocene. J. Biogeogr. 2017;1:1–12. [Google Scholar]
- 29.Hopkins DM. Sea level history in Beringia during the past 250,000 years. Quat. Res. 1973;3:520–540. [Google Scholar]
- 30.Bintanja R, van de Wal RSW, Oerlemans J. Modelled atmospheric temperatures and global sea levels over the past million years. Nature. 2005;437:125–128. doi: 10.1038/nature03975. [DOI] [PubMed] [Google Scholar]
- 31.Bocherens, H. In Advances in Mammoth Research (eds Reumer, J. W., De Vos, J. & Mol, D.) 57–76 (Deinsea, 2003).
- 32.Bocherens H. Isotopic tracking of large carnivore palaeoecology in the mammoth steppe. Quaternary Science Reviews. 2015;117:42–71. [Google Scholar]
- 33.Lozhkin AV, Anderson PM, Matrosova TV, Minyuk PS. The pollen record from El’gygytgyn Lake: implications for vegetation and climate histories of northern Chukotka since the late middle Pleistocene. J. Paleolimnol. 2007;37:135–153. [Google Scholar]
- 34.Szpak P, et al. Regional differences in bone collagen δ13C and δ15N of Pleistocene mammoths: Implications for palaeoecology of the mammoth steppe. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2010;286:88–96. [Google Scholar]
- 35.Amundson R, et al. Global patterns of the isotopic composition of soil and plant nitrogen. Global Biogeochem. Cycles. 2003;17:1031. [Google Scholar]
- 36.Craine JM, et al. Ecological interpretations of nitrogen isotope ratios of terrestrial plants and soils. Plant and Soil. 2015;396:1–26. [Google Scholar]
- 37.Murphy BP, Bowman DMJS. Kangaroo metabolism does not cause the relationship between bone collagen δ15N and water availability. Func. Ecol. 2006;20:1062–1069. [Google Scholar]
- 38.Hartman G. Are elevated δ15N values in herbivores in hot and arid environments caused by diet or animal physiology? Func. Ecol. 2011;25:122–131. [Google Scholar]
- 39.Drucker DG, Bocherens H, Billiou D. Evidence for shifting environmental conditions in Southwestern France from 33000 to 15000 years ago derived from carbon-13 and nitrogen-15 natural abundances in collagen of large herbivores. Earth Planet. Sci. Lett. 2003;216:163–173. [Google Scholar]
- 40.Drucker DG, Bridault A, Cupillard C. Environmental context of the Magdalenian settlement in the Jura Mountains using stable isotope tracking (13C, 15N, 34S) of bone collagen from reindeer (Rangifer tarandus) Quat. Int. 2012;272–273:322–332. [Google Scholar]
- 41.Hobbie EA, Jumpponen A, Trappe J. Foliar and fungal 15N:14N ratios reflect development of mycorrhizae and nitrogen supply during primary succession: Testing analytical models. Oecologia. 2005;146:258–268. doi: 10.1007/s00442-005-0208-z. [DOI] [PubMed] [Google Scholar]
- 42.Kaufman, D. S. & Manley, W. F. In Quaternary Glaciations-Extent and Chronology Part II: North America (eds Ehlers, J. & Gibbard, P. L.) 2, 9–27 (Elsevier B.V, 2004).
- 43.Svendsen JI, et al. Late Quaternary ice sheet history of northern Eurasia. Quat. Sci. Rev. 2004;23:1229–1271. [Google Scholar]
- 44.Fox-Dobbs K, Leonard JA, Koch PL. Pleistocene megafauna from eastern Beringia: Paleoecological and paleoenvironmental interpretations of stable carbon and nitrogen isotope and radiocarbon records. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2008;261:30–46. [Google Scholar]
- 45.Iacumin P, Nikolaev V, Ramigni M. C and N stable isotope measurements on Eurasian fossil mammals, 40000 to 10000 years BP: herbivore physiologies and palaeoenvironmental reconstruction. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2000;163:33–47. [Google Scholar]
- 46.Iacumin P, Davanzo S, Nikolaev V. Spatial and temporal variations in the 13C/12C and 15N/14N ratios of mammoth hairs: Palaeodiet and palaeoclimatic implications. Chem. Geol. 2006;231:16–25. [Google Scholar]
- 47.Fizet M, et al. Effect of diet, physiology and climate on carbon and nitrogen stable isotopes of collagen in a late pleistocene anthropic palaeoecosystem: Marillac, Charente, France. J. Archaeol. Sci. 1995;22:67–79. [Google Scholar]
- 48.Bocherens H, et al. Isotopic evidence for dietary ecology of cave lion (Panthera spelaea) in North-WesternEurope: prey choice, competition and implications for extinction. Quat. Int. 2011;245:249–261. [Google Scholar]
- 49.Münzel SC, et al. Pleistocene bears in the Swabian Jura (Germany): genetic replacement, ecological displacement, extinctions and survival. Quat. Int. 2011;245:225–237. [Google Scholar]
- 50.Bocherens H, et al. Reconstruction of the Gravettian food-web at Předmostí I using isotopic tracking of bone collagen. Quat. Int. 2015;359:211–228. [Google Scholar]
- 51.Sher AV, et al. The first record of “spelaeoid” bears in Arctic Siberia. Quat. Sci. Rev. 2011;30:2238–2249. [Google Scholar]
- 52.Boeskorov GG, Grigoriev SE, Baryshnikov GF. New evidence for the existence of Pleistocene cave bears in Arctic Siberia. Dokl.Biol. Sci. 2012;445:239–243. doi: 10.1134/S0012496612040060. [DOI] [PubMed] [Google Scholar]
- 53.Hilderbrand GV, et al. The importance of meat, particularly salmon, to body size, population productivity, and conservation of North American brown bears. Can. J. Zool. 1999;77:132–138. [Google Scholar]
- 54.Baryshnikov GF, Boeskorov GG. Skull of the Pleistocene brown bear (Ursus arctos) from Yakutia, Russia. Russ. J. Theriol. 2004;3:71–75. [Google Scholar]
- 55.Boeskorov, G. G. & Baryshnikov, G. F. Late Quaternary Carnivora of Yakutia [In Russian] (Nauka Publishers, 2013).
- 56.Longin R. New Method of Collagen Extraction for Radiocarbon Dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 57.van der Plicht J, Wijma S, Aerts-Bijma AT, Pertuisot MH, Meijer HAJ. The Groningen AMS facility: status report. Nuclear Instruments and Methods. 2000;B172:58–65. [Google Scholar]
- 58.Reimer PJ, et al. IntCal13 and Marine13 Radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. [Google Scholar]
- 59.Trueman CN, Martill DM. The long-term survival of bone: The role of bioerosion. Archaeometry. 2002;44:371–382. [Google Scholar]
- 60.Szpak P, Metcalfe JZ, Macdonald RA. Best Practices for Calibrating and Reporting Stable Isotope Measurements inArchaeology. J. Archaeol. Sci. Rep. 2017;13:609–616. [Google Scholar]
- 61.Ambrose SH. Preparation and characterization of bone and tooth collagen for isotopic analysis. J. Archaeol. Sci. 1990;17:431–451. [Google Scholar]
- 62.Bocherens H, et al. Paleobiological implications of the isotopic signatures (13C, 15N) of fossil mammal collagen in Scladina Cave (Sclayn, Belgium) Quat. Research. 1997;48:370–380. [Google Scholar]
- 63.Dufour E, Bocherens H, Mariotti A. Palaeodietary implications of isotopic variability in Eurasian lacustrine fish. J. Archaeol. Sci. 1999;26:627–637. [Google Scholar]
- 64.Bocherens H, Grandal-d’Anglade A, Hobson KA. Pitfalls in comparing modern hair and fossil bone collagen C and N isotopic data to reconstruct ancient diets: a case study with cave bears (Ursus spelaeus) Isotopes Environ. Health. Stud. 2014;50:291–299. doi: 10.1080/10256016.2014.890193. [DOI] [PubMed] [Google Scholar]
- 65.Matheus PE. Diet and co-ecology of Pleistocene short-faced bears and brown bears in eastern Beringia. Quat. Res. 1995;44:447–453. [Google Scholar]
- 66.Bocherens H, Pacaud G, Lazarev PA, Mariotti A. Stable isotope abundances (13C, 15N) in collagen and soft tissues from Pleistocene mammals from Yakutia: Implications for the palaeobiology of the Mammoth Steppe. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1996;126:31–44. [Google Scholar]
- 67.Iacumin P, Di Matteo A, Nikolaev V, Kuznetsova TV. Climate information from C, N and O stable isotope analyses of mammoth bones from northern Siberia. Quat. Int. 2010;212:206–212. [Google Scholar]
- 68.Leonard JA, et al. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 2007;17:1146–1150. doi: 10.1016/j.cub.2007.05.072. [DOI] [PubMed] [Google Scholar]
- 69.Mann DH, Groves P, Kunz ML, Reanier RE, Gaglioti BV. Ice-age megafauna in Arctic Alaska: extinction, invasion, survival. Quat. Sci. Rev. 2013;70:91–108. [Google Scholar]
- 70.Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST-Palaeontological statistics, www. uv. es/~pardomv/pe/2001_1/past/pastprog/past. pdf, acessado em 25 (2001).
- 71.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protoc. 5 (2010). [DOI] [PubMed]
- 73.Schubert M, Lindgreen S, Orlando L. AdapterRemovalv2: rapid adapter trimming, identification, and read merging. BMC res. notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2. 0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schubert M, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 76.Li H, Durbin R. Fast and accurate short read alignment with Burrows – Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu S, et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157:785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delisle I, Strobeck C. Conserved Primers for Rapid Sequencing of the Complete Mitochondrial Genome from Carnivores, Applied to Three Species of Bears. Mol. Biol. Evol. 2002;19:357–361. doi: 10.1093/oxfordjournals.molbev.a004090. [DOI] [PubMed] [Google Scholar]
- 79.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. (2010). [DOI] [PMC free article] [PubMed]
- 80.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Lu J, Yu J, Gibbs RA, Yu F. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013;23:833–842. doi: 10.1101/gr.146084.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 84.Rambaut, A. & Drummond, A. J. Tracer, v.1.5., http://beast.bio.ed.ac.uk/Tracer (2007).
- 85.Shapiro B, et al. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 2011;28:879–887. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rieux A, et al. Improved calibration of the human mitochondrial clock using ancient genomes. Mol. Biol. Evol. 2014;31:2780–2792. doi: 10.1093/molbev/msu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rieux A, Khatchikian CE. TipDatingBeast: An R package to assist the implementation of phylogenetic tip‐dating tests using BEAST. Mol. Ecol. Resour. 2017;17:608–613. doi: 10.1111/1755-0998.12603. [DOI] [PubMed] [Google Scholar]
- 88.Yu G, Smith DK, Zhu H, Guan Y, Lam TT. Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. [Google Scholar]
- 89.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 90.Lisiecki LE, Raymo ME. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography. 2005;20:1–17. [Google Scholar]
- 91.Layman CA, Arrington DA, Montaña CG, Post DM. Can stable isotope ratios provide for community-wide measures of trophic structure? Ecol. 2007;88:42–48. doi: 10.1890/0012-9658(2007)88[42:csirpf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.