Abstract
The combined phenotype of thrombocytopenia accompanied by intellectual disability in patients with a de novo heterozygous mutation, i.e., p.Tyr64Cys in CDC42, signifies a clinically recognizable novel syndrome that has been eponymized as “Takenouchi-Kosaki syndrome” (OMIM #616737). In the present study, a detailed phenotypic analysis performed for a total of five patients with Takenouchi-Kosaki syndrome revealed that intellectual disability, macrothrombocytopenia, camptodactyly, structural brain abnormalities with sensorineural deafness, hypothyroidism, and frequent infections comprise the cardinal features of this condition. A morphologic analysis of platelets derived from three affected individuals was performed using electron microscopy. The platelets of the three patients were large and spherical in shape. Furthermore, platelet α-granules were decreased, while vacuoles were increased. We further performed a functional analysis of p.Tyr64Cys in CDC42 through CRISPR/Cas9-mediated gene editing in a Caenorhabditis elegans model. This functional analysis suggested that the mutant allele has hypomorphic effects. Takenouchi-Kosaki syndrome is clinically recognizable by the combined phenotype of intellectual disability, macrothrombocytopenia, camptodactyly, structural brain abnormalities with sensorineural deafness, hypothyroidism, and frequent infections as well as the identification of a heterozygous de novo mutation in CDC42, i.e., p.Tyr64Cys.
Introduction
Recently, we reported two unrelated individuals with thrombocytopenia accompanied by intellectual disability and a de novo mutation in CDC42, a critical molecule in the regulation of the cell cycle and the formation of the actin cytoskeleton1,2. Both patients shared intellectual disability, distinctive facial features, macrothrombocytopenia (increased platelet size and decreased platelet count), camptodactyly, and structural brain abnormalities with sensorineural hearing loss. Through whole exome analyses, we identified the exact same de novo mutation in CDC42, i.e., p.Tyr64Cys, in both patients. The characteristic hematologic feature, i.e., macrothrombocytopenia, was compatible with existing evidence of a similar platelet phenotype in a model organism (Cdc42 homozygous knockout mice)3. The identification of multiple unrelated patients with similar phenotypes and exactly the same de novo CDC42 mutation enabled us to confirm that these patients represent a new syndromic form of thrombocytopenia, which was eponymized as “Takenouchi-Kosaki syndrome” (OMIM #616737). Later, Motokawa et al. reported a third patient with this condition4, and we recently evaluated a fourth patient.
The cardinal features of Takenouchi-Kosaki syndrome include intellectual disability accompanied by macrothrombocytopenia. However, the detailed clinical characteristics and mechanistic basis of the macrothrombocytopenia observed in these individuals remain to be elucidated. In the present study, we sought to delineate the pathogenetic basis of the macrothrombocytopenia observed in Takenouchi-Kosaki syndrome and performed a functional analysis of p.Tyr64Cys in cdc-42 using CRISPR/Cas9-mediated gene editing and Caenorhabditis elegans.
Results
Phenotypic spectrum of Takenouchi-Kosaki syndrome
A total of five patients with macrothrombocytopenia, intellectual disability, and a de novo heterozygous mutation in CDC42, i.e., p.Tyr64Cys, have been identified. Four of these patients were reported previously1,2,4,5. We have summarized the clinical characteristics of these five patients with Takenouchi-Kosaki syndrome in a tabular form (Table 1). The shared features among the five patients were as follows: macrothrombocytopenia (present in 5/5 cases), intellectual disability (5/5), structural brain abnormalities with sensorineural deafness (5/5), camptodactyly (4/5), hypothyroidism (3/5), frequent infections (3/5), and lymph edema (2/5). All five individuals exhibited macrothrombocytopenia; however, none of the five patients exhibited bleeding diathesis. All five patients had moderate to severe intellectual disability. Structural brain abnormalities and sensorineural deafness were present in all five patients who underwent neuroimaging studies. The patterns of brain abnormalities were rather nonspecific, with various degrees of ventriculomegaly and cerebellar atrophy. Camptodactyly was present in four female patients who were all in their teens or older. Hypothyroidism was present in three patients. Three patients had a history of recurrent bacterial infections: patient 1 had a history of fulminant streptococcus infection, and patient 3 had recurrent upper respiratory infections. Four patients (patients 1, 2, 3, and 4) had decreased counts of CD19-positive cells. Abnormalities in the lymphatic system were noted in two patients: patients 1 and 2 had lymph edema in their lower extremities, and patient 2 had protein-losing enteropathy.
Table 1.
Clinical features in patients with Takenouchi-Kosaki syndrome.
| Patient # | 1 | 2 | 3 | 4 | 5 | Frequency |
|---|---|---|---|---|---|---|
| Age/Sex | 18 y/female | 22 y/female | 12 y/female | 15 y/female | 4 y/male | |
| Macrothrombocytopenia | Present | Present | Present | Present | Present | 5/5 |
| Intellectual disability | Present | Present | Present | Present | Present | 5/5 |
| Structural brain abnormalities and sensorineural deafness | Present | Present | Present | Present | Present | 5/5 |
| Camptodactyly | Present | Present | Present | Present | Absent | 4/5 |
| Frequent infections | Present | Absent | Present | Present | Absent | 3/5 |
| Hypothyroidism | Absent | Absent | Present | Present | Present | 3/5 |
| Lymph edema | Present | Present | Absent | Absent | Absent | 2/5 |
| Heterozygous de novo mutation in CDC42 | p.Tyr64Cys | p.Tyr64Cys | p.Tyr64Cys | p.Tyr64Cys | p.Tyr64Cys | |
| References | Takenouchi et al.1 | Takenouchi et al.2 | Motokawa et al.4 | Martinelli et al.5 | Unpublished data |
Genotype
All five patients had a single de novo amino acid substitution change in CDC42, i.e., c.191A>G p.Tyr64Cys. The details of the genetic analyses performed for patients 1–4 have been published elsewhere1,2,4,5. An exome analysis was performed for patient 5 as part of routine clinical practice, and this patient is not of Japanese descent. The p.Tyr64 is located within the switch II domain of CDC426. An in silico analysis of p.Tyr64Cys in CDC42 showed that this amino acid residue is highly conserved among species, with a high Combined Annotation Dependent Depletion score7 of 23.4 (phred). According to the list of post-transcriptional modification sites8, p.Tyr64 in CDC42 is a target site of phosphorylation. According to recently proposed criteria for pathogenicity, the variant was interpreted as “pathogenic”9. p.Tyr64Cys was absent in the largest database of 2,049 normal Japanese individuals determined using whole-genome sequencing (Japanese Genome Variation Database)10, the exome sequencing of 1,208 normal Japanese individuals (Human Genetic Variation Database)11, and the 1,000 Genome12.
Electron microscopy investigation of platelets in patients
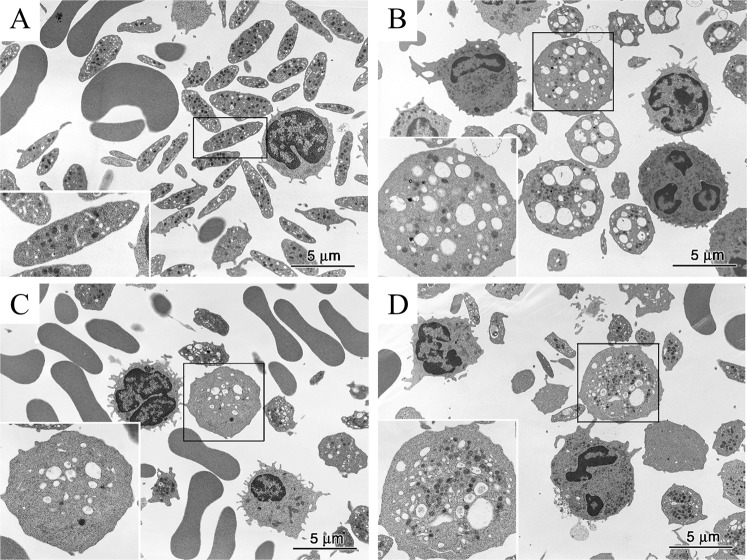
In healthy controls, platelets are smaller than red blood cells and white blood cells and have a disk-shaped appearance and a diameter of long and short axes of 3.5 ± 0.7 μm and 1.4 ± 0.5 μm, respectively (Fig. 1A). α-Granules, dense granules, and mitochondria were abundant in the cytoplasm of the platelets, and microtubules are present at the marginal ends13. Structures that appear to be small vacuoles in normal platelets were actually channels of the open canalicular system derived from the invagination of cell membranes. In the three patients with Takenouchi-Kosaki syndrome, electron microscopic images showed the presence of platelets that were larger in size and had a spherical shape (Fig. 1B–D). A violin plot of the estimated cross-sectional area of the platelets showed a positive skew, indicating that a substantial proportion of the platelets in patients 1–3 could be regarded as macrothrombocytes, although a large proportion of the platelets remained relatively unchanged in size (Fig. 2). By counting the numbers of macrothrombocytes (platelets with a cross-sectional area exceeding 12.6 μm2 and having a spherical shape), the ratio of macrothrombocytes to normal-sized platelets was shown to be significantly higher in patients 1–3, compared with the control (P < 0.001, Fisher’s exact test). In terms of platelet morphology, long and short axes of platelets from the three patients were 3.9 ± 1.1 × 3.2 ± 1.3 μm, 4.1 ± 1.5 × 2.9 ± 1.6 μm, 4.1 ± 1.9 × 2.8 ± 1.4 μm, respectively. The ratio of normal discoid shaped platelets was significantly decreased in patients 1–3 (P < 0.001, Fisher’s exact test) (Table 2). As for the organelles within the platelets, the number of α-granules was slightly reduced and the number of vacuoles with a diameter of approximately 0.5 μm or more and that were clearly different from those of the open canalicular system was increased. Consistent with these observations, our quantitative measurements showed that the number of α-granules per estimated cross-sectional area was decreased in patients (P < 0.001, Dunnett’s test) (Fig. 3). The large platelets with α-granules depletion were reminiscent of those in patients with Gray platelet syndrome14. Large vacuoles were particularly evident in platelets from patient 1 and appeared to be increased in number in patients 2 and 3; however, the numbers per area were to too small for statistical analysis. In platelets from three of the patients, interspersed microtubules were observed.
Figure 1.
Electron micrographs of platelets from normal subjects and three patients. (A) Normal platelets have a disk-like appearance with a diameter of approximately 3.5 μm. The inset shows an enlarged platelet in the boxed region in which platelet granules and an open canalicular system are visible. (B–D) Some typical platelets of three patients are larger and spherical in shape. In these platelets, the number of α-granules is slightly decreased and the number of vacuoles with a diameter of approximately 0.5 μm and that are notably different from those belonging to the open canalicular system is increased. The insets show enlarged images of the boxed regions.
Figure 2.
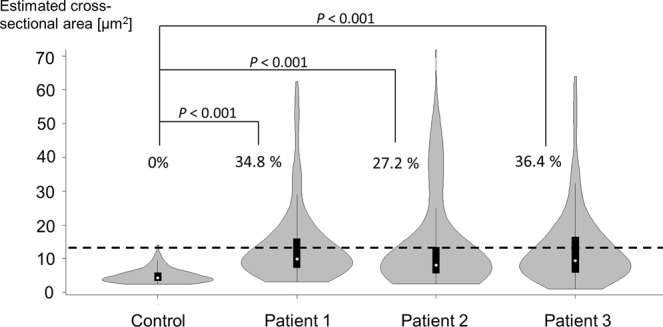
Skewed distribution of platelet size in patients. The vertical axis represents the estimated cross-sectional area [μm2]. The horizontal dotted line represents the cut-off value (12.6 μm2) of the estimated cross-sectional area for macrothrombocytes. Note that the distributions of platelet size were skewed positively (upwards) in patients 1–3. Macrothrombocytes comprised none (1/120) of the platelets in the control, 34.8% (47/135) of the platelets in patient 1, 27.2% (24/88) of the platelets in patient 2, and 36.4% (51/140) of the platelets in patient 3.
Table 2.
Platelet shape in the control and the three patients.
| Ratio of normal discoid-shaped platelets | P value | |
|---|---|---|
| Control | 93% (111/120) | — |
| Patient 1 | 20% (27/135) | <0.001 |
| Patient 2 | 34% (30/88) | <0.001 |
| Patient 3 | 34% (48/140) | <0.001 |
Figure 3.
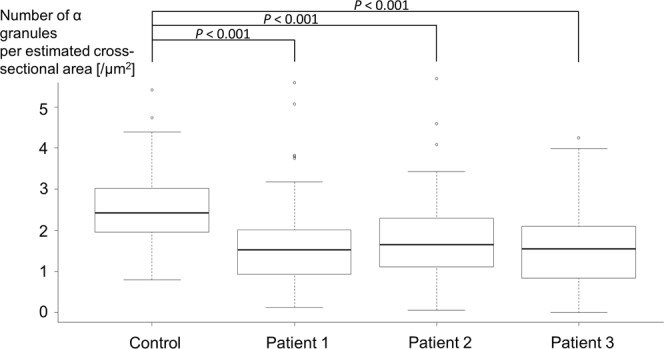
Decreased α-granule density in patients’ platelets. The vertical axis represents the number of α-granules per estimated cross-sectional area [μm2]. Note that the α-granule density is significantly lower in all three patients than in the control.
Functional analysis using C. elegans
We generated a model organism of cdc-42 p.Tyr64Cys using C. elegans for the following reasons: C. elegans can be grown less expensively and can be frozen and then thawed and revived when needed. Along with its fully mapped genome and extensively studied cell fate specification, many gene-editing and analytic methods have been established for C. elegans and are commonly available. C. elegans also has an ortholog of human CDC42, namely cdc-42, and the nucleotide sequence in the vicinity of the p.Tyr64Cys mutation is highly conserved between CDC42 in humans and cdc-42 in C. elegans. Using the CRISPR/Cas9-mediated gene editing methods15, we successfully generated worms carrying one or two alleles of the missense mutation, [cdc-42p.Tyr64Cys/+] and [cdc-42p.Tyr64Cys], i.e., tm9602/+ strain and tm9602 strain respectively, from the standard N2 background.
Validation of knock-in model generated through gene editing
We first validated the knock-in model by confirming that a protruding vulva and mild morphological changes in gonads were observed in approximately 30% of all the knock-in worms homozygous for the p.Tyr64Cys mutant allele and in approximately 10% of the worms that were heterozygous for the p.Tyr64Cys mutant allele (data not shown). These morphological changes were compatible with reduction-of-function of cdc-42 rather than the loss-of-function of cdc-42 in prior reports16–18.
Analysis of apoptotic cell clearance in gonads and embryos
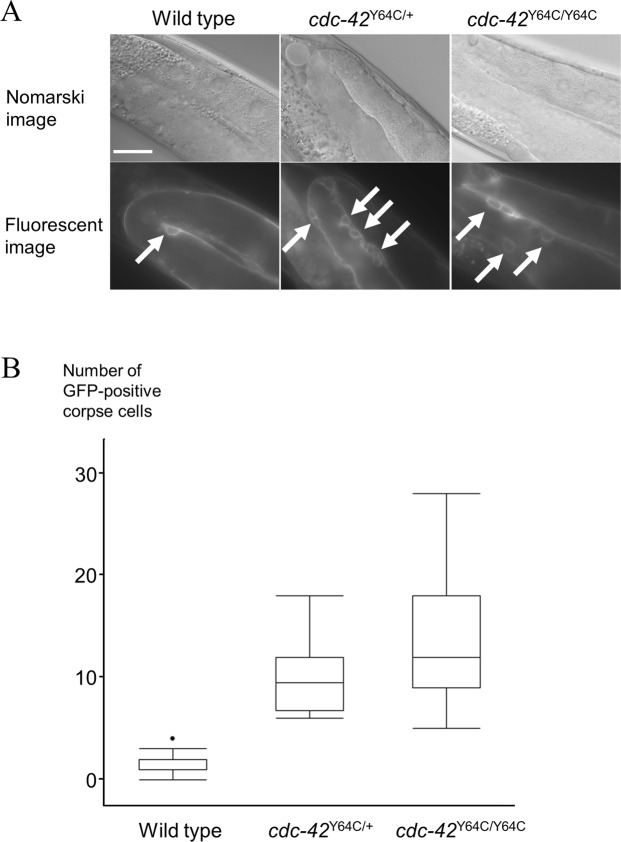
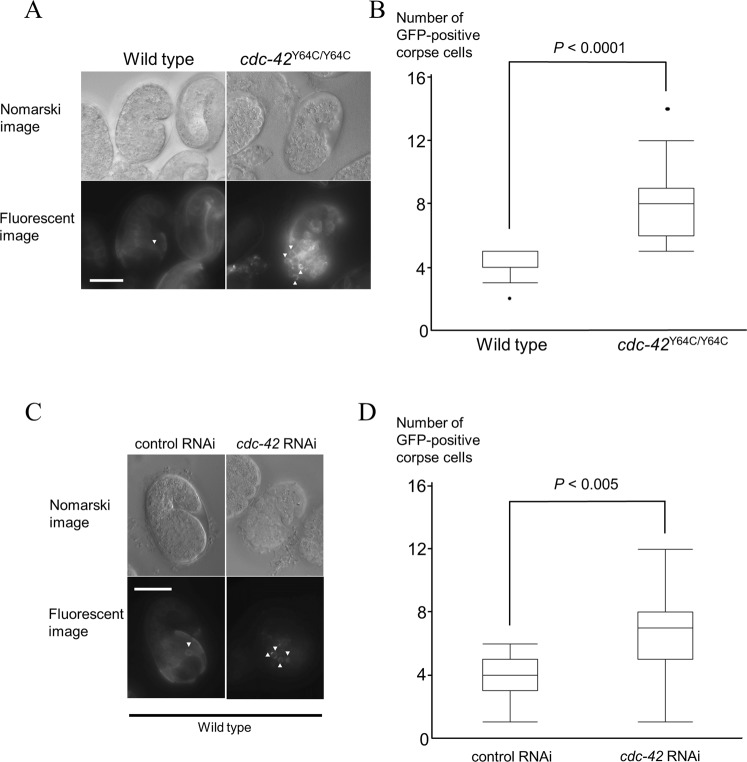
We opted to analyze the number of corpse cells as a functional consequence of mutant cdc-42 p.Tyr64Cys because the involvement and roles of cdc-42 in the process of apoptosis have been well established in previous studies of C. elegans, and corpse cells in gonads and eggs can be readily visualized and are countable under microscopy. The numbers of corpse cells derived from gonadal cells in the gonads were increased in heterozygous (tm9602/+) and homozygous (tm9602) worms in a gene dosage-dependent manner (P < 0.05, nonparametric max3 test) (Fig. 4). Furthermore, the number of corpse cells derived from embryonic cells was also increased in the homozygous (tm9602) strain, compared with wild-type worms (P < 0.0001, Student’s t-test). Hence, this increase in the number of corpse cells was ascribed to the decreased clearance of apoptotic cells in cdc-42 knockout worms19.
Figure 4.
p.Tyr64Cys mutation of CDC-42 affects apoptotic cell clearance in gonads. (A) Nomarski and fluorescent images (x100) of gonads in L4-stage hermaphrodites in a wild type worm (left), a knock-in model of heterozygous p.Tyr64Cys-mutated cdc-42 (cdc-42Y64C/+, middle), and a knock-in model of homozygous p.Tyr64Cys-mutated cdc-42 (cdc-42Y64C/Y64C, right). The white arrows indicate GFP-positive corpse cells. Scale bar = 20 µm. (B) Bar chart showing the average number of corpse cells expressing GFP in wild type, cdc-42Y64C/+, and cdc-42Y64C/Y64C L4-stage hermaphrodites (n = 15–36). The error bars indicate the standard error of the mean.
RNA interference
To distinguish between the dominant negative mechanism and the hypomorphic mechanism, we analyzed the numbers of corpse cells in cdc-42 knockdown worms mediated by a cdc-42-specific RNAi clone. The application of a cdc-42-specific RNAi clone resulted in an increased number of corpse cells (P < 0.005, Student’s t-test), similar to the tm9602 strains (Fig. 5).
Figure 5.
Both the p.Tyr64Cys mutation of CDC-42 and cdc-42-specific RNA interference affect apoptotic cell clearance in embryonic cells. (A) Nomarski and fluorescent images (x100) of comma-stage eggs in a wild type (left) and a knock-in model of homozygous p.Tyr64Cys-mutated cdc-42 (cdc-42Y64C/Y64C; right). The white arrow heads indicate GFP-positive corpse cells. (B) Bar chart showing the average number of corpse cells expressing GFP in wild type and cdc-42Y64C/Y64C comma-stage eggs (n = 12–36). (C) Nomarski and fluorescent images (x100) of comma-stage eggs treated with empty-vector (L4440) RNAi (left) and cdc-42-specific RNAi (right). The white arrows indicate GFP-positive corpse cells. (D) Bar chart showing the average number of corpse cells expressing GFP in comma-stage eggs treated with L4440 (RNAi) and cdc-42 (RNAi) (n = 12–22). The error bars indicate the standard error of the mean. Scale bar = 20 µm.
Discussion
In the present report, we showed that Takenouchi-Kosaki syndrome is a clinically recognizable condition characterized by a combination of macrothrombocytopenia, intellectual disability, sensorineural hearing loss with structural brain abnormalities, camptodactyly, and frequent infections and decreased CD19-positive cells. An electron microscopy investigation of platelets demonstrated a markedly increased size of platelets accompanied by an increase in vacuoles. We then successfully generated a knock-in model of cdc-42 mutated C. elegans carrying this p.Tyr64Cys mutation through CRISPR/Cas9-mediated gene editing. The knock-in model exhibited delays in the clearance of corpse cells both in the gonads and embryonic cells, and these findings were reproduced using RNAi in a gene dosage-dependent fashion. The present results suggest that the mechanistic basis of macrothrombocytopenia in Takenouchi-Kosaki syndrome involves a hypomorphic effect exerted by the p.Tyr64Cys mutation in CDC42.
Following our initial observation of two patients with Takenouchi-Kosaki syndrome, Martinelli et al. recently reported a series of patients with de novo mutations in CDC42 and various phenotypes. In their report, one patient (subject #3) had the p.Tyr64Cys mutation in CDC42 and exhibited macrothrombocytopenia, intellectual disability, hypothyroidism, camptodactyly, and recurrent infections5, all of which were all compatible with the phenotypes of our previously reported patients. Their functional analysis showed that the p.Tyr64Cys mutation was associated with the largest reduction, by far, in GTPase activity in an in vitro assay; this finding is compatible with the notion that patients with p.Tyr64Cys represent the most prototypic and severe end of the clinical spectrum.
The characteristic platelet phenotype described in this report provides a specific diagnostic clue to Takenouchi-Kosaki syndrome. Despite the presence of giant platelets, none of the patients exhibited a clinically overt bleeding tendency. The electron microscopy findings for the platelets from the presently reported patients, such as the increased number of vacuoles, were compatible with those obtained in double knockout mice (Cdc42 and Rac1)20. We would like to emphasize that macrothrombocytopenia, which is readily detectable during routine clinical laboratory examinations, is a useful diagnostic clue in the differential diagnosis of patients with intellectual disabilities. From the perspective of future research directions, further functional and morphological analyses of platelets, i.e., expression analysis of surface markers, proplatelet formation assays and analyses of tubulin organization and action cytoskeleton of platelets in comparison to prior studies3,20–22, would provide further insight into the mechanistic basis of macrothrombocytopenia in Takenouchi-Kosaki syndrome.
We showed that the mechanistic basis of Takenouchi-Kosaki syndrome is likely to involve a hypomorphic effect derived from the mutant allele. Through an in vivo analysis of mutant C. elegans, we observed the abnormal clearance of corpse cells using two complementary methods and confirmed that the mutant allele is hypomorphic. First, we assessed the clearance of corpse cells in the gonads, which occurs as a physiological apoptotic process. The number of corpse cells in both the mutant worms and the null-mutant worms increased in a gene dose-dependent manner. This observation was compatible with the notion that p.Tyr64Cys represents a hypomorphic allele. Second, we evaluated the clearance of corpse cells in worms during the embryonic stage. The self-fertilization of C. elegans results in diploid embryos. Embryos at the comma stage exhibit corpse cell clearance as a physiological apoptotic process. Similar to the findings for the gonads, an increased number of corpse cells was observed at this stage of worm development. Furthermore, the increase in the number of corpse cells was reproduced in the gonads of worms with one mutated allele. The increase in the number of corpse cells was reproduced using RNAi. Given that RNAi specifically suppresses the expression of cdc-42, this reproducibility using RNAi was compatible with the notion that p.Tyr64Cys represents a hypomorphic allele, rather than a neomorphic or antimorphic allele.
In conclusion, Takenouchi-Kosaki syndrome is a clinically recognizable human disease that is characterized by macrothrombocytopenia, intellectual disability, sensorineural hearing loss with structural brain abnormalities, camptodactyly, and frequent infections as well as a de novo heterozygous single amino acid substitution change in CDC42, i.e., p.Tyr64Cys. The p.Tyr64Cys mutation in CDC42 exerts a hypomorphic effect and represents the most severe and prototypic end of the CDC42 spectrum. In patients with intellectual disability, the identification of giant platelets with an increased number of vacuoles in routine laboratory testing is a diagnostic clue for Takenouchi-Kosaki syndrome.
Materials and Methods
Study subjects
Individuals with thrombocytopenia, intellectual disability, and a p.Tyr64Cys mutation in CDC42 were identified through a MEDLINE search and our local collaborative research network.
Platelet preparation and electron microscopy
The research protocol was approved by Keio University School of Medicine Ethics Committee. All methods were carried out in accordance with the World Medical Association Declaration of Helsinki. After obtaining informed consent, venous blood samples from patients 1, 2, and 3 were directly collected into respective syringes containing 3.8% sodium citrate as an anticoagulant (9:1 whole blood:anticoagulant, v/v). At each collection time, blood was also sampled from healthy 3 normal volunteers who had not received any medication for at least 10 days. The tubes containing the blood samples were allowed to settle for 2 hours at room temperature to obtain platelet-rich plasma (PRP). PRP was carefully harvested, and prostaglandin E1 (PGE1, 1 μM; Sigma-Aldrich) was added to the PRP. Platelets were sedimented by centrifugation at 900 g for 15 min and resuspended in HEPES-Tyrode’s washing solution (pH 7.4) containing 137 mM NaCl, 2.7 mM KCl, 0.4 mM NaH2PO4, 12 mM NaHCO3, 1 mM MgCl2, 5 mM HEPES, 1 μM PGE1, 0.35% bovine serum albumin (BSA), and 0.1% glucose. The suspension was kept at 37 °C for 30 min. After centrifugation at 900 g for 10 min, platelets were finally suspended in the above-mentioned HEPES-Tyrode’s solution without PGE1 and maintained at 37 °C prior to use.
Washed platelets obtained from control and 3 patients were fixed by mixing with an equal volume of 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 30 min. The fixed cells were transferred to Eppendorf tubes, then centrifuged at 3000 rpm for 3 min at 4 °C. The platelet pellets were washed 3 times in 0.1 M phosphate buffer, post-fixed with 1% osmium tetroxide for 1 hour at 4 °C, dehydrated with a graded ethanol series, and embedded in Epon 812 according to conventional methods. Thin sections were cut with a diamond knife and stained with uranyl acetate and lead citrate, then examined using a JEM-1400plus electron microscope (JEOL, Tokyo, Japan) at an accelerating voltage of 80 kV.
Quantitative measurement of size and microstructure of platelets
Using electron microscopic images, we performed quantitative measurements of the long and short axes of the platelets and counted the numbers of α-granules and vacuoles; we then compared the results of patients 1–3 with those of the control. The cross-sectional area of the platelets was estimated using the mathematical formula for the area of an ellipse, i.e., , where a represents the semi-major axis and b represents the semi-minor axis. The typical size of normal platelets ranges from 3 to 4 μm. A diameter of 4 μm corresponds to a circular area of 3.14 × 2 × 2 = 12.6 μm2. Therefore, we defined macrothrombocytes as platelets with a cross-sectional area exceeding 12.6 μm2. Vacuoles were counted only when their diameter exceeded 0.5 μm.
Strains of Caenorhabditis elegans
All strains of C. elegans were seeded with Escherichia coli OP50. All the experiments were performed at 20 °C using standard techniques23. The wild-type strain Bristol N2 was obtained from the Caenorhabditis Genetics Center (CGC, Minneapolis, MN). Strains carrying the following mutations were obtained from the trimethylpsoralen/ultraviolet-mutagenized library, as described previously24.
Generation of knock-in model of C. elegans using CRISPR/Cas9-mediated gene editing
We chose to generate a knock-in model, rather than a transgenic model, of the p.Tyr64Cys mutation in C. elegans using the CRISPR/Cas9 system and one copy of the abnormal allele and one copy of the normal allele to better recapitulate the pathogenesis in humans with Takenouchi-Kosaki syndrome. A comparison of the human CDC42 protein (NP_001782.1) sequence with that for C. elegans CDC-4225 revealed a high degree of evolutionary conservation of the protein sequence flanking the p.Tyr64Cys missense mutation in humans5, and the p.Tyr64Cys corresponded to p.Tyr64Cys in C. elegans. To generate the strain with the cdc-42 missense mutation, tm9602, we used the CRISPR/Cas9 method as described previously15. First, we selected two spots of the guide sequence for cdc-42 to be used for Cas9 cleavage (#1_AGGTCACAGTAATGATCGG and #2_TTTCTTGTTTGCTTCTCCG) through “CRISPR Design” (http://crispr.mit.edu). The two sequences were inserted into a Cas9-sgRNA (single guide RNA) expression vector (pPD162) using Addgene26. The primers used to generate the sgRNAs according to the infusion method were as follows: sgRNA_#1, 5′-ACC TCC TAT TGC GAG ATG TCT TGA GGT CAC AGT AA-3′, 5′-TCT AGC TCT AAA ACC CGA TCA TTA CTG TGA CCT CAA-3′; and sgRNA_#2, 5′-TCC TAT TGC GAG ATG TCT TGT TTC TTG TTT GCT TCT-3′, 5′-GCT ATT TCT AGC TCT AAA ACC GGA GAA GCA AAC AAG AA-3′. Next, to generate the cdc-42 genome fragment plasmid (i.e., the targeting vector), pPD95.79_ cdc-42p.Tyr64Cys, a mutated cdc-42 genome, was amplified by PCR and sewing PCR using the following primers: #1; 5′-GAA ATG AAA TAA GCT TTC TCT GCG TAT CTC ACC AC-3′, 5′-GAT CAT TAC TGT CAC GGC GT-3′; #2; 5′-TAC GCC GTG ACA GTA ATG ATC G-3′, 5′-GGA GAA GCA AAC AAG GAA CA-3′, #3; 5′-ACC GAC GTG TTC CTT GTT TGC T-3′, 5′-CCA ATC CCG GGG ATC GAA ATT CTA TAC GAA ACA AT-3′. The fragment was inserted into the pPD95.79 plasmid using the EcoRI and BamHI sites. The primers used to generate the target vector according to infusion methods were as follows: 5′-TTC AGG TGA CAG TAA TGA TCG-3′, and 5′-GCC ACC GAT CAT TAC TGT CA-3′. To generate integration lines with cdc-42p.Tyr64Cys, two sgRNAs and one target vector were injected using standard C. elegans microinjection methods27 with the myo-2::Venus selection marker. Strain tm9602 was backcrossed twice with N2. To generate a worm strain carrying one allele of the missense mutation, we balanced tm9602 worms with mIn1 [mIs14dpy-10 (e128)].
Analysis of corpse cells
Since CDC-42 plays a critical role in apoptotic cell clearance19,28, we evaluated the functional state of cdc-42p.Tyr64Cys by counting corpse cells, which reflect the clearance of apoptotic cells, in both somatic cells (gonads) and embryonic cells (comma-stage eggs). Functional abnormalities in the tm9602 strain (homozygous mutant), the tm9602/+ strain (heterozygous mutant), and cdc-42 knockdown worms were studied with regard to the clearance of post-apoptotic cells by engulfment cells. The increases in corpse cell numbers derived from both gonadal cells and embryonic cells were interpreted as a quantitative measurement of the clearance function of post-apoptotic cells. To assay the number of corpse cells, we used the integrated array SmIs34, which contains ced-1p::ced-1::gfp. The transgenic strain was obtained through the Caenorhabditis Genetics Center.
RNA interference
To generate cdc-42 knockdown worms, we used RNA interference (RNAi) methods. RNAi was performed by feeding animals dsRNA-producing bacteria, as described previously29. Briefly, the RNAi clones were transformed into E. coli HT115(DE3); then, approximately 10–20 P0 animals at the early L1 stage were transferred to plates containing RNAi-bacteria grown on 100 µg/mL of ampicillin and 1 mmol/L of isopropyl-beta-D-thiogalactopyranoside (IPTG). To analyze the phenotypes of cdc-42-knockdown worms, the numbers of apoptotic corpse cells were counted in both the P0 generation at the L4 larval stage and the F1 generation at the comma-stage. We then compared the numbers of corpse cells in cdc-42-knockdown and control worms. The RNAi clone was obtained from the Ahringer RNAi library.
Microscope
Differential interference contrast and fluorescence images were obtained using a BX51 microscope equipped with a DP30BW CCD camera (Olympus Optical Co., Ltd, Tokyo, Japan).
Statistical analyses
The results are expressed as mean ± standard deviation. All the analyses were performed using R software, version 3.4.3. The Dunnett’s test was used to analyze the numbers of granules per estimated cross-sectional area. The Fisher’s exact test was used to analyze the ratio of macrothrombocytes to normal-sized platelets. Nonparametric max 3 (a procedure for testing the association between a biallelic single nucleotide polymorphism and a quantitative trait using the maximum value of the three nonparametric trend tests derived for the recessive, additive, and dominant models) was used to analyze the gene dosage effect on the number of corpse cells in gonads30. The Student’s t-test was used to compare the numbers of corpse cells in embryonic cells and the effect of RNAi. Differences with a P value of 0.05 were considered to indicate a statistical significance.
Acknowledgements
We thank Ms. Chika Kanoe and Keiko Tsukue for their technical assistance in the preparation of this article. This work was supported by a Grant-in-Aid for Scientific Research [16K09974] from the Japan Society for the Promotion of Science and the grants [JP18ek0109301 and JP18ek0109398] from Japan Agency for Medical Research and Development.
Author Contributions
T.U. performed experiments and wrote the initial manuscript. H.S. performed all the electron microscopic analyses of the platelets. N.O. collected and analyzed clinical data. T.K. collected and analyzed clinical data. A.A. collected and analyzed clinical data. B.C.O. collected and analyzed clinical data. S.Y. designed and performed experiments and analyzed the data. S.M. designed and performed experiments and analyzed the data. K.K. designed and supervised the entire project. T.T. organized the entire project, analyzed the data and edited the final manuscript. All authors critically revised and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takenouchi T, Okamoto N, Ida S, Uehara T, Kosaki K. Further evidence of a mutation in CDC42 as a cause of a recognizable syndromic form of thrombocytopenia. Am J Med Genet A. 2016;170A:852–855. doi: 10.1002/ajmg.a.37526. [DOI] [PubMed] [Google Scholar]
- 2.Takenouchi T, Kosaki R, Niizuma T, Hata K, Kosaki K. Macrothrombocytopenia and developmental delay with a de novo CDC42 mutation: Yet another locus for thrombocytopenia and developmental delay. Am J Med Genet A. 2015;167A:2822–2825. doi: 10.1002/ajmg.a.37275. [DOI] [PubMed] [Google Scholar]
- 3.Pleines I, et al. Multiple alterations of platelet functions dominated by increased secretion in mice lacking Cdc42 in platelets. Blood. 2010;115:3364–3373. doi: 10.1182/blood-2009-09-242271. [DOI] [PubMed] [Google Scholar]
- 4.Motokawa M, et al. A hot-spot mutation in CDC42 (p.Tyr64Cys) and novel phenotypes in the third patient with Takenouchi-Kosaki syndrome. J Hum Genet. 2018;63:387–390. doi: 10.1038/s10038-017-0396-5. [DOI] [PubMed] [Google Scholar]
- 5.Martinelli S, et al. Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am J Hum Genet. 2018;102:309–320. doi: 10.1016/j.ajhg.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdul-Manan N, et al. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 7.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi-Kabata Y, et al. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Human genome variation. 2015;2:15050. doi: 10.1038/hgv.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higasa K, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–553. doi: 10.1038/jhg.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki, H. et al. Immunocytochemical aspects of platelet membrane glycoproteins and adhesive proteins during activation. Progress in histochemistry and cytochemistry30, 1–106 (1996). [DOI] [PubMed]
- 14.Clauser, S. & Cramer-Borde, E. Role of platelet electron microscopy in the diagnosis of platelet disorders. Seminars in thrombosis and hemostasis35, 213–223, 10.1055/s-0029-1220329 (2009). [DOI] [PubMed]
- 15.Yoshina S, Suehiro Y, Kage-Nakadai E, Mitani S. Locus-specific integration of extrachromosomal transgenes in C. elegans with the CRISPR/Cas9 system. Biochemistry and biophysics reports. 2016;5:70–76. doi: 10.1016/j.bbrep.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmer LL, et al. A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation. Plos genetics. 2016;12:e1005786. doi: 10.1371/journal.pgen.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meighan CM, Kelly VE, Krahe EC, Gaeta A. J. alpha integrin cytoplasmic tails can rescue the loss of Rho-family GTPase signaling in the C. elegans somatic gonad. Mech Dev. 2015;136:111–122. doi: 10.1016/j.mod.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neukomm LJ, Zeng S, Frei AP, Huegli PA, Hengartner MO. Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell death and differentiation. 2014;21:845–853. doi: 10.1038/cdd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pleines I, et al. Defective tubulin organization and proplatelet formation in murine megakaryocytes lacking Rac1 and Cdc42. Blood. 2013;122:3178–3187. doi: 10.1182/blood-2013-03-487942. [DOI] [PubMed] [Google Scholar]
- 21.Dutting S, et al. A Cdc42/RhoA regulatory circuit downstream of glycoprotein Ib guides transendothelial platelet biogenesis. Nat Commun. 2017;8:15838. doi: 10.1038/ncomms15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pula G, Poole AW. Critical roles for the actin cytoskeleton and cdc42 in regulating platelet integrin alpha2beta1. Platelets. 2008;19:199–210. doi: 10.1080/09537100701777303. [DOI] [PubMed] [Google Scholar]
- 23.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Lim HH, Lim L. The CDC42 homologue from Caenorhabditis elegans. Complementation of yeast mutation. J Biol Chem. 1993;268:13280–13285. [PubMed] [Google Scholar]
- 26.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh HH, Hsu TY, Jiang HS, Wu YC. Integrin alpha PAT-2/CDC-42 signaling is required for muscle-mediated clearance of apoptotic cells in Caenorhabditis elegans. Plos genetics. 2012;8:e1002663. doi: 10.1371/journal.pgen.1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome biology. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Li Q. Nonparametric Risk and Nonparametric Odds in Quantitative Genetic Association Studies. Scientific reports. 2015;5:12105. doi: 10.1038/srep12105. [DOI] [PMC free article] [PubMed] [Google Scholar]