Abstract
Background
Paired‐box gene 1 (PAX1), a member of the PAX family, plays a role in pattern formation during embryogenesis, and might be essential for development of the vertebral column.
Methods
PAX1 is silenced by methylation in several cancers and is considered a tumor suppressor gene. Our previous studies reported PAX1 as hypermethylated in cervical cancer tissues, thereby suggesting it as a potential screening marker. Recently, an increasing number of studies have confirmed PAX1 methylation as a promising biomarker in cervical cancer based on its excellent discriminatory ability between high‐grade cervical lesions and normal tissues, resulting in a reduced necessity for referral for colposcopy and biopsy. Additionally, PAX1 is also hypermethylated in other tumors, including those associated with epithelial ovarian cancer, esophageal squamous cell carcinoma, head and neck squamous cell carcinoma, and endometrial carcinoma, and shows relatively good sensitivity and specificity for the detection of these tumors.
Results
This review summarizes reports of PAX1 methylation and its promising role in cancer screening, especially that associated with cervical cancer.
Conclusion
According to current evidence, combined testing for human papillomavirus and PAX1 methylation analysis represents an efficacious cervical cancer‐screening protocol.
Keywords: cancer screening, cervical cancer, human papillomavirus, methylation, PAX1
1. INTRODUCTION
PAX1 (OMIM: 167,411) gene, a member of the PAX family located on chromosome 20p11.2, is essential to regulate fetal development. (Schnittger et al., 1992). Members of the PAX family typically contain a paired‐box domain and a paired‐type homeodomain, which are essential during fetal development and play critical roles during development of the vertebral column (Wallin et al., 1994). PAX1 plays a role in sclerotome differentiation and interacts with homeobox (HOX) genes, which play a prominent role in normal fetal development and controlling cell proliferation (Cillo, Cantile, Faiella, & Boncinelli, 2001). Substitution or deletion of this gene in mice produces variants associated with vertebral malformations and Klippel–Feil syndrome (Hol et al., 1996; McGaughran, Oates, Donnai, Read, & Tassabehji, 2003). Methylation of PAX1 promoter is an important epigenetic regulation associating to the development and the metastasis of the tumor. PAX1 gene in cervical and ovarian cancer is silenced by methylation and is considered as a tumor suppressor gene (Chang et al., 2014; Hassan, Hafez, Kamel, & Zekri, 2017; Kan et al., 2014; Kong, Du, Wang, Yang, & Zhang, 2015; Su et al., 2009).
2. PAX1 METHYLATION AND CERVICAL CANCER
2.1. Challenges in the diagnosis and treatment of cervical cancer
Cervical cancer is the second most common carcinoma among women worldwide (Torre et al., 2015) and has a long pre‐invasive phase. During cervical cancer development, normal cervical cells gradually develop into precancerous lesions; however, cervical cancer can also evolve from pre‐existing noninvasive premalignant lesions referred to as cervical intraepithelial neoplasias (CINs) that range in severity from CIN1 (mild dysplasia) to CIN2/3 (moderate/severe dysplasia/carcinoma) and which can be maintained over several years (Fabrizii, Moinfar, Jelinek, Karperien, & Ahammer, 2014; Rakotomahenina, Garrigue, Marty, & Brun, 2014). Cervical cancer has a well‐defined CIN process and can be identified and treated before malignancy formation (Jones, 2010; Wentzensen et al., 2013). Given that a long developmental process from each stage of CIN to cervical cancer, early diagnosis and treatment of CIN can effectively prevent cancer from happening.
Infection with human papillomavirus (HPV) represents a primary risk factor leading to cervical cancer (Bosch, Lorincz, Munoz, Meijer, & Shah, 2002; de Silva, Mendis, & Perera, 1999; Helmerhorst, 2000; Kaufman, Adam, Icenogle, Lawson, et al., 1997; Nessa, Rashid, E‐Ferdous, & Chowdhury, 2013; Schiffman & Castle, 2003; Wentzensen et al., 2013; Zielinski et al., 2001). HPV test is the most common screening method for cervical cancer for it high sensitivity; however, HPV test is not recommended for screening purposes because of its low specificity. Moreover, low positive‐predictive values of HPV‐positive testing results have been obtained, even in the presence of clinically relevant lesions along with Papanicolaou (Pap) smear and ThinPrep cytology tests (Cox et al., 1995; Cuzick, 2010; Dane, Batmaz, Dane, & Cetin, 2009; Kaufman, Adam, Icenogle, & Reeves, 1997; Nessa et al., 2013). Additionally, most HPV infections are subclinical, transient, and noncancerous. Evidence suggests that only persistent HPV infections are associated with precancerous lesions, as a positive HPV result might lead to overinterpretation of minor cellular abnormalities, redundant anxiety, and additional testing (Tjalma & Depuydt, 2014), which limit HPV testing as a diagnostic factor for cervical cancer. Therefore, identification of novel and accurate biomarkers for cervical cancer screening remains necessary.
For diagnosis of cervical cancer, suspicious cervical lesions will be initially evaluated by colposcopy in clinical practice, and, if necessary, biopsy samples will be taken for further histopathologic examination (Massad et al., 2013). However, it remains a challenge to choose personalized treatments and follow‐up strategies for biopsy confirmed patients with CINs. Because most CIN1 patients will regress to normal without intervention, and even high‐grade lesions (CIN2/3) exhibit a substantial rate of regression, only a small percentage of dysplasia progresses (Jones, 2010; McCredie et al., 2008; Wentzensen et al., 2013). For patients with naturally regressing CINs, unnecessary surgery can cause adverse effects, such cervical dysfunction, which can result in recurrent spontaneous abortion during subsequent pregnancies (Bjorge, Skare, Bjorge, Trope, & Lonnberg, 2016; Jakobsson & Bruinsma, 2008; Song, Seong, & Kim, 2016), whereas for patients with CINs destined to progress, medical treatment, and follow‐up are needed to prevent cervical malignancy. Therefore, reliable biomarkers are needed to assess the risk of CIN progression, reduce unnecessary referral for colposcopy and biopsy, and avoid overtreatment of patients desiring to preserve fertility.
2.2. Relationships between PAX1 methylation and cervical cancer screening
Epigenetic studies demonstrate that DNA methylation could be a symbolic event of carcinogenesis. Several kinds of DNA methylation are reported as strongly associated with CINs and cervical cancer, including those in sex‐determining region Y‐box 1 (SOX1; OMIM: 602148), PAX1, LIM homeobox transcription factor 1A (OMIM: 600298), NK6 transcription factor‐related locus 1 (OMIM: 602563), and Wilms tumor 1 (OMIM: 607102) (Chang et al., 2014; Lai et al., 2008; Lim et al., 2010; Lorincz, 2016; Vasiljevic, Scibior‐Bentkowska, Brentnall, Cuzick, & Lorincz, 2014). Among these genes, multiple studies confirmed PAX1 methylation as the most highly correlated with CIN progression and cervical carcinogenesis (Chang et al., 2014; Chao et al., 2013; Chen et al., 2016; Huang et al., 2010; Kan et al., 2014; Lai et al., 2010, 2014; Luan et al., 2017; Tian et al., 2017; Xu et al., 2015).
In 2008, Lai et al. (2008) first reported that PAX1 was abnormally methylated in association with cervical cancer, and the PAX1 gene was silenced by hyper methylation and low expressed in these biopsies of cervical cancer (Lai et al., 2008). Several studies found that PAX1 methylation increased along with increased disease grade in the following order: PAX1 methylation in squamous cell carcinoma (SCC) > high‐grade squamous intraepithelial lesion (HSIL) > low‐grade squamous intraepithelial lesion (LSIL) > normal tissue (Lai et al., 2008; Lim et al., 2010; Xu et al., 2015). Detection of high‐grade cervical lesions in patients with atypical squamous cells of undetermined significance (ASCUS) remains a challenge in the screening and diagnosis of cervical cancer. PAX1 methylation demonstrated better performance as a marker than results of a high‐risk HPV‐DNA test for the detection of high‐grade lesions (CIN2+) in ASCUS cases; however, PAX1 methylation allows for the screening out of a majority of low‐grade ASCUS cases (Li et al., 2015; Wang, 2014). A result from 443 cervical scraping samples showed that PAX1 detection alone had a sensitivity and specificity of 86% and 85%, respectively, for the detection of CIN3+ lesions, whereas when used as a co‐test with the Pap test, the sensitivity and specificity were 89% and 83%, respectively (Kan et al., 2014). Additionally, our previous studies found a significant association between methylated PAX1 and CIN3+ or worse in combination with HPV16/18, with sensitivities and specificities of methylated PAX1 with HPV16/18 for CIN3+ detection at 89.2% and 76.0%, respectively (Liou et al., 2016), whereas dual methylation testing for PAX1/zinc protein finger 582 (OMIM: 615600) combined with HPV‐16/18 genotyping resulted in 100% identification of carcinoma in situ or SCC (Tian et al., 2017). Meta‐analyses also supported the utility of PAX1 methylation as an auxiliary biomarker in cervical cancer screening. One meta‐analysis reviewed 1,385 subjects with various stages of CIN and normal cervical pathology, finding that the sensitivity and specificity of PAX1 methylation in CIN3+ vs. normal samples were 0.77 and 0.92, respectively (Nikolaidis et al., 2015). Additionally, 15 individual studies showed that single PAX1 methylation allowed the accurate differential diagnosis of cervical cancer/HSIL patients from normal individuals with a sensitivity of 0.80 and a specificity of 0.89 (Kong et al., 2015).
These data suggested the efficacy of PAX1 methylation as a biomarker for cervical cancer screening, and that it plays a guiding role in triage management of LSIL, HSIL, and SCC patients, as well as displays higher accuracy than single HPV‐DNA testing. These findings suggest that incorporating PAX1‐methylation detection into current cervical cancer‐screening protocols (Figure 1) will promote the accurate screening of women requiring treatment, reduce unnecessary referrals for colposcopy and biopsy, and ease the burden on patients and medical resources.
Figure 1.
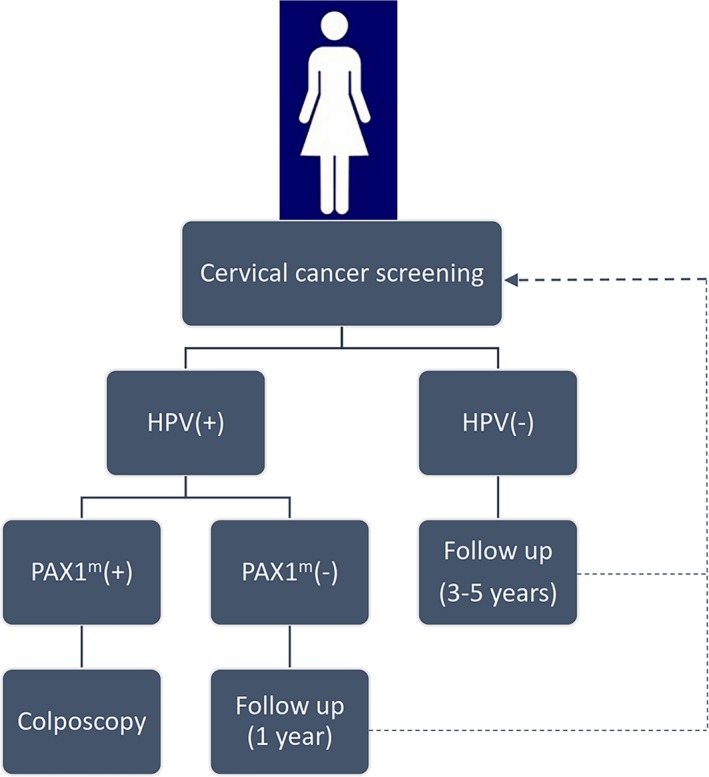
Combined HPV testing and PAX1‐methylation detection as a cervical cancer‐screening protocol. During cervical cancer screening for women, HPV‐positive results accompanied by PAX1‐methylation analysis allow accurate determination of the necessity for biopsy during colposcopy during diagnosis of CIN3+ lesions. (PAX1: NCBI Refseq: NM_006192.4, GRCh38)
3. PAX1 METHYLATION IN OTHER TUMORS
Aside from cervical cancer, PAX1 also displays hypermethylation in other tumors and offers great promise as a marker for cancer detection (Table 1). Detection of PAX1‐methylation levels in oral scrapings or oral swabs indicated that PAX1‐methylation levels and positive rates increased along with disease severity (SCC > precancerous lesions > normal oral mucosa) (Cheng et al., 2017, 2016; Huang et al., 2014), but decreased following cancer excision. However, these levels increased again at subsequent sites of recurrence in some cases at ~3‐ to ~4‐months prior to recurrence (Cheng et al., 2016). These studies suggest PAX1 methylation as an effective biomarker for oral cancer detection and the prediction of oral cancer recurrence.
Table 1.
PAX1 # gene methylation is a promising biomarker for cancer screening
| Cancer | Samples | PAX1 gene methylation | Clinical application |
|---|---|---|---|
| Cervical cancer | Cervical exfoliated cells | PAX1 hypermethylation in cervical high‐grade lesions (CIN3+and worse) |
Biomarker for
|
| Oral cancer | Oral scrapings/oral swabs | PAX1 hypermethylation in tumor samples, methylation levels decreased after cancer excision and increased again 3–4 months before cancer recurrence |
Biomarker for
|
| Esophageal squamous cell carcinoma | Tissues | PAX1 hypermethylation in tumor tissues | Biomarker for cancer detection |
| Colorectal cancer | Tissues | PAX1 hypermethylation in tumor tissues | Biomarker for cancer detection |
| Head and neck squamous cell carcinoma | tissues | PAX1 hypermethylation in tumor tissues | Biomarker for cancer detection |
| Epithelial ovarian cancer | Tissues | PAX1 hypermethylation in HPV16/18‐infected EOC tissues | / |
NCBI Refseq: NM_006192.4, GRCh38.
In epithelial ovarian cancer (EOC), PAX1 was significantly hypermethylated in HPV16/18‐infected EOC tissues (Hassan et al., 2017). Another study showed that levels of PAX1 methylation were significantly higher in esophageal squamous cell carcinoma (Huang, Wang, et al., 2017), colorectal cancer (Huang, Tan, et al., 2017), and head and neck squamous cell carcinoma (Guerrero‐Preston et al., 2014), and the PAX1 protein levels were lower in endometrial carcinoma (Liu et al., 2016). Moreover, detection of PAX1 methylation displayed relatively good sensitivity and specificity for the diagnosis of these tumors and holds great promise for tumor screening or as a prognostic marker.
4. POTENTIAL MECHANISMS OF PAX1 METHYLATION
In cancer development, hypermethylation of promoter regions containing CpG islands can inactivate tumor suppressor genes, thereby affecting genes associated with the cell cycle, DNA repair, cell–cell interactions, apoptosis, and angiogenesis (Herman & Baylin, 2003). In most vertebrates, PAX1 and PAX9 exhibit similar expression patterns and functions that belong to a highly conserved family of PAX genes, encode highly conserved transcription factors, and play roles in pattern formation during vertebrate embryogenesis (Paixao‐Cortes, Salzano, & Bortolini, 2015). Studies show that PAX genes promote cell proliferation, cell‐lineage specification, migration, and survival, and roles in tissue development and cellular differentiation in embryos (Dahl, Koseki, & Balling, 1997). In most cases, as fetal development progresses, PAX expression attenuates; however, in some tissues, PAX expression either persists into adult life or increases to exert functions, such as protection against stress‐induced cell death (Cai et al., 2005). Studies PAX1 and PAX9 mutants show that PAX1 can compensate for the loss of PAX9, although not vice versa (Wilm, Dahl, Peters, Balling, & Imai, 1998). Little is known about the functional role of PAX1 in cancer biology. Liu et al. (2016) identified PAX1 protein levels as potential histopathology biomarkers for the differential diagnosis of malignant and premalignant endometrial lesions (Liu et al., 2016). Another study reported PAX1 expression‐inducing tumor formation following subcutaneous injection of cultured cells expressing PAX1 into nude mice (Maulbecker & Gruss, 1993). Additionally, other studies showed that methylation of CpG islands in the PAX1 promoter region regulates cervical neoplasia (Chen et al., 2016; Kan et al., 2014), resulting in their characterization of PAX1 as a tumor suppressor gene. Although PAX1 functions have been hypothesized based on phenotypic outcomes associated with knockout models, PAX1 molecular functions and target genes remain largely unknown. Using mutation mouse models, studies have identified that PAX1 regulates epithelial cell death and proliferation during thymus and parathyroid organogenesis (Su, Ellis, Napier, Lee, & Manley, 2001). Studies investigating PAX1 reactivation in cervical cancer cell lines suggest that this can occur through curcumin and resveratrol administration through their effect on histone deacetylase accompanied by the downregulation of ubiquitin‐like with PHD and RING finger domains 1 (OMIM: 607990), which regulates both DNA methylation and histone acetylation (Parashar & Capalash, 2016). Moreover, PAX1 methylation levels decrease along with increases in its mRNA expression after silencing of DNA methyltransferase 1 (DNMT1; OMIM: 126375), which plays a significant role in maintaining DNA methylation status and regulating the expression of tumor suppressor genes (Zhang et al., 2011). Similar results showed that curcumin, resveratrol, and DNMT1 influence PAX1 activity and might represent effective targets for treatment of cervical cancer (Parashar, Parashar, & Capalash, 2017). PAX1 methylation is also associated with NOTCH1 mutation and the Hedgehog pathway, which is regulated by HOX transcription factors and enhancer of split 1 (OMIM: 139605) (Bolos, Grego‐Bessa, & de la Pompa, 2007; Forastiere, Koch, Trotti, & Sidransky, 2001; Guerrero‐Preston et al., 2014; Koop et al., 2010; Landsman, Parent, & Hebrok, 2011; Mammucari et al., 2005; Manley & Capecchi, 1995; Mill et al., 2003; Sang, Roberts, & Coller, 2010; Schubert et al., 2005; Wall et al., 2009). Loss of ΝOTCH1 function due to mutation or the methylation‐dependent silencing of downstream genes, such as PAX1, likely abrogates normal cell differentiation (Guerrero‐Preston et al., 2014).
5. DISCUSSION AND PROSPECTS
PAX genes encode a family of nine transcription factors that act as cell‐lineage‐specific regulators of the tissues where they are normally expressed and are now recognized as important factors in cancer progression. Additionally, these factors might play previously unrecognized fundamental roles in balancing proliferation and differentiation signals. Numerous studies have demonstrated that PAX1 methylation plays an important role in the progression of cancers and contributes significantly to the sensitivity and specificity of cancer screening, especially for cervical cancer. In scrapings for cervical cancer, analyses indicated that PAX1 is silenced by hypermethylation. Moreover, PAX1 methylation plays a guiding role in the triage management of normal tissues, as well as CIN2, CIN3, LSIL, HSIL, and SCC patients. Although HPV testing is appealing for cervical cancer diagnosis, it cannot distinguish whether or not an HPV‐positive result is associated with a clinically relevant lesion. Furthermore, these test results can be subject to overinterpretation and causing unnecessary panic. PAX1 methylation represents a novel biomarker that exhibits increased specific and accuracy for cervical cancer screening and diagnosis. There is increasing evidence that testing for methylated genes can replace cytology as a reflex test for HPV‐positive women, and interim clinical guidance approves the use of such tests as an appropriate triage tool for HPV (Huh et al., 2015; Luttmer et al., 2016).
Here, we propose a screening strategy for cervical cancer that combines using the HPV testing and PAX1‐methylation analysis as triage tests according to current evidence. In HPV‐positive patients, the detection of PAX1 methylation is necessary for diagnosis of CIN3+ lesions (Figure 1) and will greatly benefit accurate cervical cancer screening, identify women that require treatment, and reduce unnecessary referrals for colposcopy and biopsy. However, additional standardization and large‐scale clinical studies are needed to evaluate the efficacy of PAX1 methylation for cervical cancer screening and early detection. Additionally, further studies targeting the specific mechanisms associated with methylation‐induced alterations in cellular activity are required to provide additional evidence supporting the clinical use of PAX1 methylation as a screening tool.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
ACKNOWLEDGMENTS
This work was supported by Hunan Provincial Science and Technology Plan of China (2018JJ2598,2017JJ3424).
Fang C, Wang S‐Y, Liou Y‐L, Chen M‐H, Ouyang W, Duan K‐M. The promising role of PAX1 (aliases: HUP48, OFC2) gene methylation in cancer screening. Mol Genet Genomic Med. 2019;7:e506 10.1002/mgg3.506
REFERENCES
- Bjorge, T. , Skare, G. B. , Bjorge, L. , Trope, A. , & Lonnberg, S. (2016). Adverse pregnancy outcomes after treatment for cervical intraepithelial neoplasia. Obstetrics and Gynecology, 128(6), 1265–1273. 10.1097/aog.0000000000001777. [DOI] [PubMed] [Google Scholar]
- Bolos, V. , Grego‐Bessa, J. , & de la Pompa, J. L. (2007). Notch signaling in development and cancer. Endocrine Reviews, 28(3), 339–363. 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Bosch, F. X. , Lorincz, A. , Munoz, N. , Meijer, C. J. , & Shah, K. V. (2002). The causal relation between human papillomavirus and cervical cancer. Journal of Clinical Pathology, 55(4), 244–265. 10.1136/jcp.55.4.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , Dmitrieva, N. I. , Ferraris, J. D. , Brooks, H. L. , van Balkom, B. W. , & Burg, M. (2005). Pax2 expression occurs in renal medullary epithelial cells in vivo and in cell culture, is osmoregulated, and promotes osmotic tolerance. Proceedings of the National Academy of Sciences of USA, 102(2), 503–508. 10.1073/pnas.0408840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. C. , Huang, R. L. , Wang, H. C. , Liao, Y. P. , Yu, M. H. , & Lai, H. C. (2014). High methylation rate of LMX1A, NKX6‐1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. International Journal of Gynecological Cancer, 24(2), 201–209. 10.1097/igc.0000000000000054. [DOI] [PubMed] [Google Scholar]
- Chao, T. K. , Ke, F. Y. , Liao, Y. P. , Wang, H. C. , Yu, C. P. , & Lai, H. C. (2013). Triage of cervical cytological diagnoses of atypical squamous cells by DNA methylation of paired boxed gene 1 (PAX1). Diagnostic Cytopathology, 41(1), 41–46. 10.1002/dc.21758. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Cui, Z. , Xiao, Z. , Hu, M. , Jiang, C. , Lin, Y. , & Chen, Y. (2016). PAX1 and SOX1 methylation as an initial screening method for cervical cancer: A meta‐analysis of individual studies in Asians. Annals of Translational Medicine, 4(19), 365 10.21037/atm.2016.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S. J. , Chang, C. F. , Ko, H. H. , Lee, J. J. , Chen, H. M. , Wang, H. J. , … Chiang, C. P. (2017). Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head and Neck, 40(2), 355–368. 10.1002/hed.24958. [DOI] [PubMed] [Google Scholar]
- Cheng, S. J. , Chang, C. F. , Lee, J. J. , Chen, H. M. , Wang, H. J. , Liou, Y. L. , … Chiang, C. P. (2016). Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncology, 62, 34–43. 10.1016/j.oraloncology.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Cillo, C. , Cantile, M. , Faiella, A. , & Boncinelli, E. (2001). Homeobox genes in normal and malignant cells. Journal of Cellular Physiology, 188(2), 161–169. 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- Cox, J. T. , Lorincz, A. T. , Schiffman, M. H. , Sherman, M. E. , Cullen, A. , & Kurman, R. J. (1995). Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. American Journal of Obstetrics and Gynecology, 172(3), 946–954. 10.1016/0002-9378(95)90026-8 [DOI] [PubMed] [Google Scholar]
- Cuzick, J. (2010). Long‐term follow‐up in cancer prevention trials (It ain't over 'til it's over). Cancer Prevention Research, 3(6), 689–691. 10.1158/1940-6207.capr-10-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, E. , Koseki, H. , & Balling, R. (1997). Pax genes and organogenesis. BioEssays, 19(9), 755–765. 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Dane, C. , Batmaz, G. , Dane, B. , & Cetin, A. (2009). Screening properties of human papillomavirus testing for predicting cervical intraepithelial neoplasia in atypical squamous cells of undetermined significance and low‐grade squamous intraepithelial lesion smears: A prospective study. Annals of Diagnostic Pathology, 13(2), 73–77. 10.1016/j.anndiagpath.2008.12.001. [DOI] [PubMed] [Google Scholar]
- de Silva, N. R. , Mendis, L. , & Perera, N. (1999). Human papillomavirus in carcinogenesis of cervical cancer. Ceylon Medical Journal, 44(1), 45–47. [PubMed] [Google Scholar]
- Fabrizii, M. , Moinfar, F. , Jelinek, H. F. , Karperien, A. , & Ahammer, H. (2014). Fractal analysis of cervical intraepithelial neoplasia. PLoS ONE, 9(10), e108457 10.1371/journal.pone.0108457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiere, A. , Koch, W. , Trotti, A. , & Sidransky, D. (2001). Head and neck cancer. New England Journal of Medicine, 345(26), 1890–1900. 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Guerrero‐Preston, R. , Michailidi, C. , Marchionni, L. , Pickering, C. R. , Frederick, M. J. , Myers, J. N. , … Sidransky, D. (2014). Key tumor suppressor genes inactivated by "greater promoter" methylation and somatic mutations in head and neck cancer. Epigenetics, 9(7), 1031–1046. 10.4161/epi.29025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, Z. K. , Hafez, M. M. , Kamel, M. M. , & Zekri, A. R. (2017). Human papillomavirus genotypes and methylation of CADM1, PAX1, MAL and ADCYAP1 genes in epithelial ovarian cancer patients. Asian Pacific Journal of Cancer Prevention, 18(1), 169–176. 10.22034/apjcp.2017.18.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst, T. J. (2000). Human papillomavirus and etiology of cervical cancer: Concept of carcinogenesis. Nederlands Tijdschrift Voor Geneeskunde, 144(39), 1891–1892. [PubMed] [Google Scholar]
- Herman, J. G. , & Baylin, S. B. (2003). Gene silencing in cancer in association with promoter hypermethylation. New England Journal of Medicine, 349(21), 2042–2054. 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hol, F. A. , Geurds, M. P. , Chatkupt, S. , Shugart, Y. Y. , Balling, R. , Schrander‐Stumpel, C. T. , … Mariman, E. C. (1996). PAX genes and human neural tube defects: An amino acid substitution in PAX1 in a patient with spina bifida. Journal of Medical Genetics, 33(8), 655–660. 10.1136/jmg.33.8.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T. H. , Lai, H. C. , Liu, H. W. , Lin, C. J. , Wang, K. H. , Ding, D. C. , & Chu, T. Y. (2010). Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. International Journal of Gynecological Cancer, 20(4), 513–519. 10.1111/IGC.0b013e3181c7fe6e. [DOI] [PubMed] [Google Scholar]
- Huang, Y. K. , Peng, B. Y. , Wu, C. Y. , Su, C. T. , Wang, H. C. , & Lai, H. C. (2014). DNA methylation of PAX1 as a biomarker for oral squamous cell carcinoma. Clinical Oral Investigations, 18(3), 801–808. 10.1007/s00784-013-1048-6. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Tan, Z. R. , Yu, J. , Li, H. , Lv, Q. L. , Shao, Y. Y. , & Zhou, H. H. (2017). DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. Onco Targets and Therapy, 10, 4739–4751. 10.2147/ott.s143389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Wang, G. , Tang, J. , Zhuang, W. , Wang, L. P. , Liou, Y. L. , … Zhu, Y. S. (2017). DNA methylation status of PAX1 and ZNF582 in esophageal squamous cell carcinoma. International Journal of Environmental Research and Public Health, 14(2), 216 10.3390/ijerph14020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K. , Ault, K. A. , Chelmow, D. , Davey, D. D. , Goulart, R. A. , Garcia, F. A. , … Einstein, M. H. (2015). Use of primary high‐risk human papillomavirus testing for cervical cancer screening: Interim clinical guidance. Gynecologic Oncology, 136(2), 178–182. 10.1016/j.ygyno.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Jakobsson, M. , & Bruinsma, F. (2008). Adverse pregnancy outcomes after treatment for cervical intraepithelial neoplasia. BMJ, 337, a1350 10.1136/bmj.a1350. [DOI] [PubMed] [Google Scholar]
- Jones, R. W. (2010). The natural history of cervical and vulvar intraepithelial neoplasia. American Journal of Obstetrics and Gynecology, 202(3), e12–e13. 10.1016/j.ajog.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Kan, Y. Y. , Liou, Y. L. , Wang, H. J. , Chen, C. Y. , Sung, L. C. , Chang, C. F. , & Liao, C. I. (2014). PAX1 methylation as a potential biomarker for cervical cancer screening. International Journal of Gynecological Cancer, 24(5), 928–934. 10.1097/igc.0000000000000155. [DOI] [PubMed] [Google Scholar]
- Kaufman, R. H. , Adam, E. , Icenogle, J. , Lawson, H. , Lee, N. , Reeves, K. O. , … Reeves, W. C. (1997). Relevance of human papillomavirus screening in management of cervical intraepithelial neoplasia. American Journal of Obstetrics and Gynecology, 176(1 Pt 1), 87–92. 10.1016/S0002-9378(97)80017-8 [DOI] [PubMed] [Google Scholar]
- Kaufman, R. H. , Adam, E. , Icenogle, J. , & Reeves, W. C. (1997). Human papillomavirus testing as triage for atypical squamous cells of undetermined significance and low‐grade squamous intraepithelial lesions: Sensitivity, specificity, and cost‐effectiveness. American Journal of Obstetrics and Gynecology, 177(4), 930–936. 10.1016/S0002-9378(97)70296-5 [DOI] [PubMed] [Google Scholar]
- Kong, L. Y. , Du, W. , Wang, L. , Yang, Z. , & Zhang, H. S. (2015). PAX1 methylation hallmarks promising accuracy for cervical cancer screening in asians: results from a meta‐analysis. Clinical Laboratory, 61(10), 1471–1479. 10.7754/Clin.Lab.2015.150232 [DOI] [PubMed] [Google Scholar]
- Koop, D. , Holland, N. D. , Semon, M. , Alvarez, S. , de Lera, A. R. , Laudet, V. , … Schubert, M. (2010). Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: Insights into early anterior‐posterior patterning of the chordate body plan. Developmental Biology, 338(1), 98–106. 10.1016/j.ydbio.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Lai, H. C. , Lin, Y. W. , Huang, R. L. , Chung, M. T. , Wang, H. C. , Liao, Y. P. , … Yu, M. H. (2010). Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer, 116(18), 4266–4274. 10.1002/cncr.25252. [DOI] [PubMed] [Google Scholar]
- Lai, H. C. , Lin, Y. W. , Huang, T. H. , Yan, P. , Huang, R. L. , Wang, H. C. , … Yu, M. H. (2008). Identification of novel DNA methylation markers in cervical cancer. International Journal of Cancer, 123(1), 161–167. 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- Lai, H. C. , Ou, Y. C. , Chen, T. C. , Huang, H. J. , Cheng, Y. M. , Chen, C. H. , … Wang, K. L. (2014). PAX1/SOX1 DNA methylation and cervical neoplasia detection: A Taiwanese Gynecologic Oncology Group (TGOG) study. Cancer Medicine, 3(4), 1062–1074. 10.1002/cam4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman, L. , Parent, A. , & Hebrok, M. (2011). Elevated Hedgehog/Gli signaling causes beta‐cell dedifferentiation in mice. Proceedings of the National Academy of Sciences of USA, 108(41), 17010–17015. 10.1073/pnas.1105404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. R. , Wang, Z. M. , Wang, Y. H. , Wang, X. B. , Zhao, J. Q. , Xue, H. B. , & Jiang, F. G. (2015). Value of PAX1 Methylation analysis by MS‐HRM in the triage of atypical squamous cells of undetermined significance. Asian Pacific Journal of Cancer Prevention, 16(14), 5843–5846. 10.7314/APJCP.2015.16.14.5843 [DOI] [PubMed] [Google Scholar]
- Lim, E. H. , Ng, S. L. , Li, J. L. , Chang, A. R. , Ng, J. , Ilancheran, A. , … Tay, E. H. (2010). Cervical dysplasia: Assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2, PAX1 and TFPI2 to improve diagnostic accuracy. Gynecologic Oncology, 119(2), 225–231. 10.1016/j.ygyno.2010.07.028. [DOI] [PubMed] [Google Scholar]
- Liou, Y. L. , Zhang, T. L. , Yan, T. , Yeh, C. T. , Kang, Y. N. , Cao, L. , … Zhou, H. (2016). Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clinical Epigenetics, 8, 66 10.1186/s13148-016-0232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. C. , Lai, H. C. , Chou, Y. C. , Huang, R. L. , Yu, M. H. , Lin, C. P. , … Chao, T. K. (2016). Paired boxed gene 1 expression: A single potential biomarker for differentiating endometrial lesions associated with favorable outcomes in patients with endometrial carcinoma. Journal of Obstetrics and Gynaecology Research, 42(9), 1159–1167. 10.1111/jog.13040. [DOI] [PubMed] [Google Scholar]
- Lorincz, A. T. (2016). Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytologica, 60(6), 501–512. 10.1159/000450595. [DOI] [PubMed] [Google Scholar]
- Luan, T. , Hua, Q. , Liu, X. , Xu, P. , Gu, Y. , Qian, H. , … Li, P. (2017). PAX1 Methylation as a Potential Biomarker to Predict the Progression of Cervical Intraepithelial Neoplasia: A Meta‐analysis of Related Studies. International Journal of Gynecological Cancer, 27(7), 1480–1488. 10.1097/igc.0000000000001011. [DOI] [PubMed] [Google Scholar]
- Luttmer, R. , De Strooper, L. M. , Berkhof, J. , Snijders, P. J. , Dijkstra, M. G. , Uijterwaal, M. H. , … Meijer, C. J. (2016). Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). International Journal of Cancer, 138(4), 992–1002. 10.1002/ijc.29824. [DOI] [PubMed] [Google Scholar]
- Mammucari, C. , Tommasi di Vignano, A. , Sharov, A. A. , Neilson, J. , Havrda, M. C. , Roop, D. R. , … Dotto, G. P. (2005). Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Developmental Cell, 8(5), 665–676. 10.1016/j.devcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Manley, N. R. , & Capecchi, M. R. (1995). The role of Hoxa‐3 in mouse thymus and thyroid development. Development, 121(7), 1989–2003. [DOI] [PubMed] [Google Scholar]
- Massad, L. S. , Einstein, M. H. , Huh, W. K. , Katki, H. A. , Kinney, W. K. , Schiffman, M. , … Lawson, H. W. (2013). 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstetrics and Gynecology, 121(4), 829–846. 10.1097/AOG.0b013e3182883a34. [DOI] [PubMed] [Google Scholar]
- Maulbecker, C. C. , & Gruss, P. (1993). The oncogenic potential of Pax genes. EMBO Journal, 12(6), 2361–2367. 10.1002/j.1460-2075.1993.tb05890.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCredie, M. R. , Sharples, K. J. , Paul, C. , Baranyai, J. , Medley, G. , Jones, R. W. , & Skegg, D. C. (2008). Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: A retrospective cohort study. The Lancet Oncology, 9(5), 425–434. 10.1016/s1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- McGaughran, J. M. , Oates, A. , Donnai, D. , Read, A. P. , & Tassabehji, M. (2003). Mutations in PAX1 may be associated with Klippel‐Feil syndrome. European Journal of Human Genetics, 11(6), 468–474. 10.1038/sj.ejhg.5200987. [DOI] [PubMed] [Google Scholar]
- Mill, P. , Mo, R. , Fu, H. , Grachtchouk, M. , Kim, P. C. , Dlugosz, A. A. , & Hui, C. C. (2003). Sonic hedgehog‐dependent activation of Gli2 is essential for embryonic hair follicle development. Genes & Development, 17(2), 282–294. 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessa, A. , Rashid, M. H. U. , E‐Ferdous, N. , & Chowdhury, A. (2013). Screening for and management of high‐grade cervical intraepithelial neoplasia in Bangladesh: A cross‐sectional study comparing two protocols. Journal of Obstetrics and Gynaecology Research, 39(2), 564–571. 10.1111/j.1447-0756.2012.01998.x. [DOI] [PubMed] [Google Scholar]
- Nikolaidis, C. , Nena, E. , Panagopoulou, M. , Balgkouranidou, I. , Karaglani, M. , Chatzaki, E. , … Constantinidis, T. C. (2015). PAX1 methylation as an auxiliary biomarker for cervical cancer screening: A meta‐analysis. Cancer Epidemiology, 39(5), 682–686. 10.1016/j.canep.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Paixao‐Cortes, V. R. , Salzano, F. M. , & Bortolini, M. C. (2015). Origins and evolvability of the PAX family. Seminars in Cell & Developmental Biology, 44, 64–74. 10.1016/j.semcdb.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Parashar, G. , & Capalash, N. (2016). Promoter methylation‐independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clinical and Experimental Medicine, 16(3), 471–478. 10.1007/s10238-015-0366-1. [DOI] [PubMed] [Google Scholar]
- Parashar, G. , Parashar, N. , & Capalash, N. (2017). ‐(‐) Menthol induces reversal of promoter hypermethylation and associated up‐regulation of the FANCF gene in the SiHa cell line. Asian Pacific Journal of Cancer Prevention, 18(5), 1365–1370. 10.22034/apjcp.2017.18.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotomahenina, H. , Garrigue, I. , Marty, M. , & Brun, J. L. (2014). Prevention and screening of cervical cancer. La Revue Du Praticien, 64(6), 780–785. [PubMed] [Google Scholar]
- Sang, L. , Roberts, J. M. , & Coller, H. A. (2010). Hijacking HES1: How tumors co‐opt the anti‐differentiation strategies of quiescent cells. Trends in Molecular Medicine, 16(1), 17–26. 10.1016/j.molmed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman, M. H. , & Castle, P. (2003). Epidemiologic studies of a necessary causal risk factor: Human papillomavirus infection and cervical neoplasia. Journal of the National Cancer Institute, 95(6), E2 10.1093/jnci/95.6.E2 [DOI] [PubMed] [Google Scholar]
- Schnittger, S. , Rao, V. V. , Deutsch, U. , Gruss, P. , Balling, R. , & Hansmann, I. (1992). Pax1, a member of the paired box‐containing class of developmental control genes, is mapped to human chromosome 20p11.2 by in situ hybridization (ISH and FISH). Genomics, 14(3), 740–744. [DOI] [PubMed] [Google Scholar]
- Schubert, M. , Yu, J. K. , Holland, N. D. , Escriva, H. , Laudet, V. , & Holland, L. Z. (2005). Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development, 132(1), 61–73. 10.1242/dev.01554. [DOI] [PubMed] [Google Scholar]
- Song, T. , Seong, S. J. , & Kim, B. G. (2016). Regeneration process after cervical conization for cervical intraepithelial neoplasia. Obstetrics and Gynecology, 128(6), 1258–1264. 10.1097/aog.0000000000001755. [DOI] [PubMed] [Google Scholar]
- Su, D. , Ellis, S. , Napier, A. , Lee, K. , & Manley, N. R. (2001). Hoxa3 and pax1 regulate epithelial cell death and proliferation during thymus and parathyroid organogenesis. Developmental Biology, 236(2), 316–329. 10.1006/dbio.2001.0342. [DOI] [PubMed] [Google Scholar]
- Su, H. Y. , Lai, H. C. , Lin, Y. W. , Chou, Y. C. , Liu, C. Y. , & Yu, M. H. (2009). An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. International Journal of Cancer, 124(2), 387–393. 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- Tian, Y. , Wu, N. Y. , Liou, Y. L. , Yeh, C. T. , Cao, L. , Kang, Y. N. , … Zhang, Y. (2017). Utility of gene methylation analysis, cytological examination, and HPV‐16/18 genotyping in triage of high‐risk human papilloma virus‐positive women. Oncotarget, 8(37), 62274–62285. 10.18632/oncotarget.19459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalma, W. A. , & Depuydt, C. E. (2014). Cervical atypical glandular cells and false negative HPV testing: A dramatic reality of the wrong test at the right place. European Journal of Gynaecological Oncology, 35(2), 117–120. [PubMed] [Google Scholar]
- Torre, L. A. , Bray, F. , Siegel, R. L. , Ferlay, J. , Lortet‐Tieulent, J. , & Jemal, A. (2015). Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians, 65(2), 87–108. 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Vasiljevic, N. , Scibior‐Bentkowska, D. , Brentnall, A. R. , Cuzick, J. , & Lorincz, A. T. (2014). Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high‐risk HPV positive women. Gynecologic Oncology, 132(3), 709–714. 10.1016/j.ygyno.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, D. S. , Mears, A. J. , McNeill, B. , Mazerolle, C. , Thurig, S. , Wang, Y. , … Wallace, V. A. (2009). Progenitor cell proliferation in the retina is dependent on Notch‐independent Sonic hedgehog/Hes1 activity. Journal of Cell Biology, 184(1), 101–112. 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin, J. , Wilting, J. , Koseki, H. , Fritsch, R. , Christ, B. , & Balling, R. (1994). The role of Pax‐1 in axial skeleton development. Development, 120(5), 1109–1121. [DOI] [PubMed] [Google Scholar]
- Wang, Z. M. (2014). PAX1 methylation analysis by MS‐HRM is useful in triage of high‐grade squamous intraepithelial lesions. Asian Pacific Journal of Cancer Prevention, 15(2), 891–894. 10.7314/APJCP.2014.15.2.891 [DOI] [PubMed] [Google Scholar]
- Wentzensen, N. , Walker, J. , Schiffman, M. , Yang, H. P. , Zuna, R. E. , Dunn, S. T. , … Wang, S. S. (2013). Heterogeneity of high‐grade cervical intraepithelial neoplasia related to HPV16: Implications for natural history and management. International Journal of Cancer, 132(1), 148–154. 10.1002/ijc.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm, B. , Dahl, E. , Peters, H. , Balling, R. , & Imai, K. (1998). Targeted disruption of Pax1 defines its null phenotype and proves haploinsufficiency. Proceedings of the National Academy of Sciences of USA, 95(15), 8692–8697. 10.1073/pnas.95.15.8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Xu, L. , Yang, B. , Wang, L. , Lin, X. , & Tu, H. (2015). Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. International Journal of Clinical and Experimental Pathology, 8(2), 1674–1681. [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Chen, F. Q. , Sun, Y. H. , Zhou, S. Y. , Li, T. Y. , & Chen, R. (2011). Effects of DNMT1 silencing on malignant phenotype and methylated gene expression in cervical cancer cells. Journal of Experimental & Clinical Cancer Research, 30, 98 10.1186/1756-9966-30-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, G. D. , Snijders, P. J. , Rozendaal, L. , Voorhorst, F. J. , van der Linden, H. C. , Runsink, A. P. , … Meijer, C. J. (2001). HPV presence precedes abnormal cytology in women developing cervical cancer and signals false negative smears. British Journal of Cancer, 85(3), 398–404. 10.1054/bjoc.2001.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]


