Abstract
Aim
Liver cirrhosis is a consequence of chronic liver disease, and it may be caused by multiple influences of both genetic and environmental factors. Family with sequence similarity 13 member A (FAM13A) has been previously associated with lung function in several lung diseases, including chronic obstructive pulmonary disease, asthma, lung cancer, and pulmonary fibrosis. The aim of this study was to explore whether FAM13A polymorphisms confer susceptibility to liver cirrhosis.
Methods
FAM13A expression was evaluated in liver cirrhosis tissues by immunohistochemistry staining. The relationship between FAM13A gene polymorphism and liver cirrhosis was determined by association analysis. The genotypes were assessed in the Agena MassARRAY platform. Statistical analysis was performed using chi‐squared test/Fisher's exact test, genetic model analysis, and haplotype analysis.
Results
The results showed that the expression of FAM13A is obvious higher in the liver cirrhosis tissue cells than in the normal liver tissue cells. Moreover, association analysis results indicated that the minor allele “A” of rs3017895 was positively associated with high risk of liver cirrhosis in the allele model by the chi‐squared test (OR = 1.32, 95%CI = 1.03–1.68, p = 0.028). Logistic regression analyses revealed that the risk of liver cirrhosis was significantly higher in subjects with the G/A‐G/G genotype of rs3017895 than those with A/A genotype under the dominant model and log additive model, and the T/A‐A/A genotype of rs1059122 was positively associated with higher liver cirrhosis than T/T genotype based on dominant model respectively. In addition, haplotype analysis showed that the G‐A haplotype of rs3017895‐rs1059122 of the FAM13A gene significantly increased the risk of liver cirrhosis.
Conclusion
Our findings demonstrated that the high expression of FAM13A may be associated with an increased risk of liver cirrhosis.
Keywords: FAM13A, genetics polymorphism, immunohistochemistry, liver cirrhosis, SNP
1. INTRODUCTION
Liver cirrhosis caused by long‐term or repeated damage to liver parenchyma by various factors represents the main complication of chronic liver disease, which leads progressive liver failure, eventually to hepatocarcinoma (Chang et al., 2015; Schuppan & Afdhal, 2008). Liver cirrhosis is a severe public health problem worldwide, which is correlated with higher morbidity and mortality worldwide (Chung, Jo, Chung, & Kim, 2018). The pathogenesis of liver cirrhosis remains unclear; although the common risk factors, such as diet and physical inactivity for NAFLD, excessive alcohol intake for alcoholic liver disease (ALD), or infection for chronic viral hepatitis, can be identified in the majority of patients, there is a small percentage of patients with no identifiable risk factors (Xiong, Liu, & Zhang, 2018; Zocco et al., 2009).
Several studies have shown that the etiology and pathogenesis of Cirrhosis were likely to comprise a multifactorial disorder resulting from environmental and genetic factors and their interaction (Ramos‐Lopez, Martinez‐Lopez, Roman, Fierro, & Panduro, 2015). Genetic factors play a key role in determining the inter‐individual susceptibility toward liver diseases, including the cirrhosis (He, Deng, & Luo, 2015; Sheneef et al., 2017). For example, several study showed that Cytochrome certain gene polymorphisms was a molecular genetic marker of liver cirrhosis and hepatocellular carcinoma progression, such as CYP2E1, ADH2, TGF‐β, and interleukin family related genes (IL‐10, IL‐6, IL‐28B, and IL‐1; Cichoz‐Lach, Partycka, Nesina, Celiński, & Słomka, 2006; Guo, Jin, & Sun, 2015; Wu, Zeng, Gong, Chen, & Chen, 2013).
The biological function of the FAM13A (Family with sequence similarity 13 member A, OMIMl: 613299) gene is poorly understood. Researches revealed that the FAM13A gene has a putative role in signal transduction, which points out the potential importance of the FAM13A in diseases (Cohen et al., 2004). Recent genome‐wide association studies reveal that the FAM13A gene was associated with a variety of lung diseases, including chronic obstructive pulmonary disease, asthma, lung cancer, and pulmonary fibrosis (Corvol, Hodges, Drumm, & Guillot, 2014; Hawkins & Mora, 2017). However, no studies have investigated the association between genetic variants in FAM13A and the risk of liver cirrhosis. Therefore, we performed a case–control study to analyze the association between the FAM13A and the risk of liver cirrhosis.
2. MATERIALS AND METHODS
2.1. Subject recruitment and ethics committee statement
Two hundred and sixty primary liver cirrhosis patients and 384 controls were enrolled in this study, all of whom were genetically unrelated Han Chinese. All participants were recruited from Haikou People's Hospital. All primary liver cirrhosis patients were diagnosed histopathologically or based on the specific morphological criteria of liver cirrhosis with ultrasound, computed tomography, or magnetic resonance imaging. Control group were age‐ and gender‐matched healthy subjects without liver cirrhosis or other diseases. All subjects were examined without history of cancer or other diseases.
Informed consents were obtained from participants, and all of them were agree to this study. The use of human tissue and the protocol in this study were strictly conformed to the principles expressed in the Declaration of Helsinki, and this study was carried out with approval from the ethics committee of the Haikou People's Hospital.
2.2. Immunohistochemistry
Specimens obtained from surgical resection were fixed in 10% formalin prior to being processed in paraffin. Hematoxylin–eosin‐stained sections were reviewed by a pathologist, and a representative section was selected for immunohistochemical analysis. The sections were stained within 5 days of cutting using an Autostainer Link48 (Dako, USA) in strict accordance with the manufacturer's instructions. Immunohistochemical staining was performed using an EnVisionTM HRP‐polymer anti‐mouse IHC Kit (K8002; Dake BioTECH, USA, Inc) according to the manufacturer's guidelines. The primary antibodies specific to FAM13A (anti‐FAM13A, 1:500; HPA038109) were obtained from Sigma‐Aldrich (St. Louis, MO, USA). Finally, we observed the images of the scanned tissue slices through Aperio ImageScope (Version 11.1.2.752).
2.3. SNP selection and genotyping
Peripheral blood samples were collected in an anti‐coagulation tube and stored at −80°C until detection before subjects had received other therapies. Based on the manufacturer's instructions of the GoldMag‐Mini Purification Kit (GoldMag Co.Ltd. Xi'an city, China), genomic DNA was isolated from blood leukocytes samples. DNA concentrations were measured using the NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
A total of two SNPs at the 3′‐UTR of FAM13A were selected at a minor allele frequency >5% in the 1,000 Genomes Project (https://www.internationalgenome.org/). The primers were designed online (https://agenacx.com/online-tools/). The Agena Bioscience platform (https://www.agenabio.com) based on the matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) primer extension assay was used to genotype two SNPs (rs3017895 and rs1059122).
2.4. Statistical analysis
SPSS 19.0 (SPSS, Chicago, IL, USA) was used to perform statistical analyses. Genotyping results were output by Agena Bioscience TYPER version software 4.0. The Pearson's chi‐squared test and independent‐samples Student's t test were applied to assess the differences in the distribution of demographic characteristics between cases and controls. Fisher's exact tests for Hardy–Weinberg equilibrium (HWE) were performed by comparing the observed and expected genotype frequencies to calculate the genotype frequencies among the controls. Odds ratios (OR) and 95% confidence intervals (CI) were calculated to estimate the association between FAM13A gene and the risk of Cirrhosis using unconditional logistic regression analysis with or without adjustment for age and gender. The SNP effects were fitted under four models of inheritance: codominant, dominant, recessive, and a log‐additive model by PLINK software (Version 1.07). Haplotype construction and genetic association of polymorphism loci were assessed using the Haploview software package (version 4.2) and the SHEsis software (https://analysis.bio-x.cn/myAnalysis.php). All p values of statistical tests were two‐sided, and p < 0.05 was considered as statistically significant.
3. RESULT
3.1. High expression of FAM13A in the primary liver cirrhosis
As shown in Figure 1, we observed the morphological observation of normal liver tissue cells and liver cirrhosis tissue cells by hematoxylin–eosin (HE) staining showed that there are obvious differences in morphology between liver cirrhosis tissue cells and normal liver tissue cells under the electron microscope (10×), and the size and shape of liver cirrhosis tissue cells are inconsistent, and the volume of nucleus increased (Figure 1a–b). Representative photomicrographs of staining intensity of FAM13A expression in liver cirrhosis tissue cells are shown in Figure 1c–d. Compared with Figure 1c, FAM13A expression is obvious enhanced in liver cirrhosis tissue cells (Figure 1d).
Figure 1.
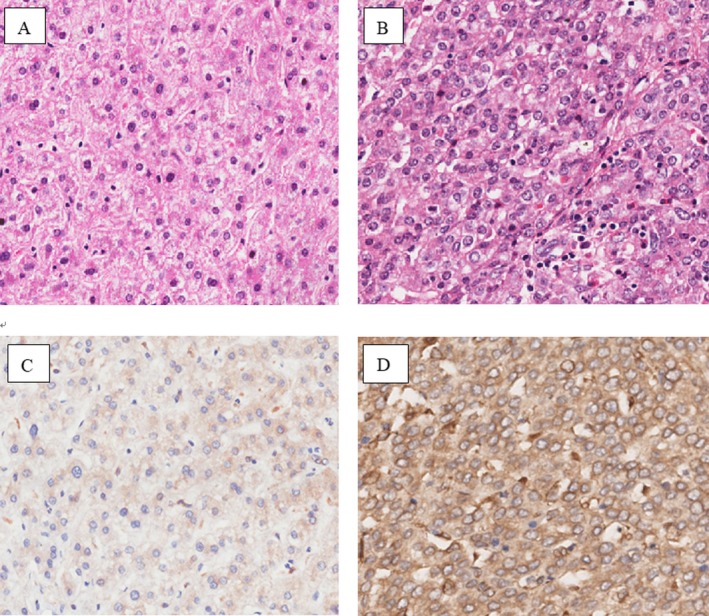
Morphological observation of normal liver tissue (a) and cirrhotic tissue (b), and the expression of FAM13A in normal liver tissue (c) and cirrhotic liver tissue (d)
3.2. Characteristics of study participants
Basic clinical characteristics of 260 cirrhosis patients and 384 disease‐free controls in this study were provided in Table 1. In patients’ characteristics, age showed no significant difference between case (51.14 ± 11.55) and controls (51.16 ± 11.49) (p = 0.722). And, there was no significant difference in gender distribution (case: male 172, female 88; control: male 278, female 106; p = 0.123).
Table 1.
The characteristic of case and control
| Variable | Case | Control | p |
|---|---|---|---|
| Age (year, SD) | 51.14 ± 11.55 | 51.16 ± 11.49 | 0.722a |
| Gender | |||
| Male | 172 (66.15%) | 278 (72.4%) | 0.123b |
| Female | 88 (33.85%) | 106 (27.6%) | |
p values were calculated by Student's t tests;
p values were calculated from two‐sided chi‐squared tests.
3.3. Association between FAM13A SNPs and the risk of cirrhosis
Rs3017895 and rs1059122 in the 3′‐UTR of FAM13A were selected. Position, alleles and minor allele frequency of these two SNPs were showed in Table 2. In control groups, two SNPs were in line with HWE (p = 0.435 for rs3017895, p = 0.179 for rs1059122). Pearson's chi‐squared test was used to assess the association between SNP variants and the risk of liver cirrhosis. The minor allele “A” of rs3017895 was associated with significant higher the risk of liver cirrhosis (OR = 1.315, 95%CI = 1.03–1.68, p = 0.028).
Table 2.
Basic information of candidate SNPs and minor allele frequency between cases and controls
| SNP rs# | Chromosome | Position | Role | Alleles A/B | Gene(s) | MAF | p ‐HWE | OR | 95%CI | p a | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||
| rs3017895 | 4 | 89649491 | 3′‐UTR | A/T | FAM13A | 0.322 | 0.266 | 0.435 | 1.32 | 1.03–1.68 | 0.028 * |
| rs1059122 | 4 | 89647424 | 3′‐UTR | G/A | FAM13A | 0.523 | 0.473 | 0.179 | 1.22 | 0.97–1.53 | 0.081 |
Alleles A/B: Minor/major alleles; CI: confidence interval; HWE: Hardy–Weinberg equilibrium; MAF, minor allele frequency; OR: odds ratio; SNP: single‐nucleotide polymorphism.
Bold highlights the value of p and OR (95%CI) with statistical significance.
p values were calculated using two‐sided chi‐squared test (the major allele of each SNP was a reference allele).
p ≤ 0.05 indicates statistical significance.
After adjustment for age and gender, four genotypes model of FAM13A polymorphisms were shown in Table 3. There was a significant differences between the groups. Dominant model analyses revealed that the risk of liver cirrhosis was significantly higher in subjects with the G/A‐G/G genotype of rs3017895 than those with A/A genotype (OR = 1.42, 95%CI = 1.03–1.95, p = 0.03), and there was also a significant difference under the log‐additive model (OR =1.31, 95%CI = 1.03–1.67, p = 0.03). And the T/A‐A/A genotype of rs1059122 was positively associated with higher liver cirrhosis under dominant model than T/T genotype (OR = 1.48, 95%CI = 1.03–2.15, p = 0.034).
Table 3.
Association between candidate SNPs and the risk of cirrhosis under genotype models
| SNP | Model | Genotype | Genotype frequency | OR (95% CI) | p‐Value | AIC | BIC | |
|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||
| rs3017895 | Codominant | A/A | 119 (46%) | 210 (54.7%) | 1 | 0.084 | 868 | 881.4 |
| G/A | 113 (43.6%) | 144 (37.5%) | 1.38 (0.99–1.93) | |||||
| G/G | 27 (10.4%) | 30 (7.8%) | 1.59 (0.90–2.80) | |||||
| Dominant | A/A | 119 (46%) | 210 (54.7%) | 1 | 0.03 * | 866.2 | 875.1 | |
| G/A‐G/G | 140 (54%) | 174 (45.3%) | 1.42 (1.03–1.95) | |||||
| Recessive | A/A‐G/A | 232 (89.6%) | 354 (92.2%) | 1 | 0.26 | 869.6 | 878.6 | |
| G/G | 27 (10.4%) | 30 (7.8%) | 1.37 (0.80–2.37) | |||||
| Log‐additive | — | — | — | 1.31 (1.03–1.67) | 0.03 * | 866.2 | 875.2 | |
| rs1059122 | Codominant | T/T | 57 (22%) | 111 (29.5%) | 1 | 0.1 | 860.1 | 873.5 |
| T/A | 133 (51.4%) | 174 (46.3%) | 1.49 (1.01–2.20) | |||||
| A/A | 69 (26.6%) | 91 (24.2%) | 1.48 (0.94–2.31) | |||||
| Dominant | T/T | 57 (22%) | 111 (29.5%) | 1 | 0.034 * | 858.1 | 867 | |
| T/A‐A/A | 202 (78%) | 265 (70.5%) | 1.48 (1.03–2.15) | |||||
| Recessive | T/T‐T/A | 190 (73.4%) | 285 (75.8%) | 1 | 0.49 | 862.1 | 871 | |
| A/A | 69 (26.6%) | 91 (24.2%) | 1.14 (0.79–1.63) | |||||
| Log‐additive | — | — | — | 1.21 (0.97–1.51) | 0.086 | 859.7 | 868.6 | |
AIC: Akaike's Information criterion; BIC: Bayesian Information criterion; CI: confidence interval; OR: odds ratios.
Bold highlights the value of p and OR (95%CI) with statistical significance.
p values were calculated from Wald's test adjusted for age and sex.
p ≤ 0.05 indicates statistical significance.
3.4. Haplotype association
Finally, allele frequency data from all subjects were used to do the linkage disequilibrium (LD) block, and we found a strong LD between rs3017895 and rs1059122 with D′ = 0.98 (Figure 2). The results of the association between haplotypes and the liver cirrhosis risk were shown in Table 4. There were three haplotypes “A‐T,” “G‐A,” and “A‐A”; however, only the “G‐A” haplotype was significantly associated with the liver cirrhosis risk by Pearson's chi‐squared test (p = 0.021). After unconditional logistic regression analysis adjustment for age and gender, the “G‐A” haplotype was remained significantly increasing the liver cirrhosis risk (OR = 1.34, 95%CI = 1.04–1.73, p = 0.026).
Figure 2.
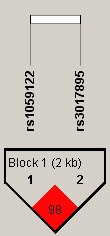
Linkage disequilibrium (LD) plots containing five SNPs from FAM13A
Table 4.
The haplotype of two SNPs (rs3017895 and rs1059122) in FAM13A and the liver cirrhosis risk
| rs3017895 | rs1059122 | Haplotype frequency | p a | OR (95%CI) | p b | ||
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| 1 | A | T | 0.476 | 0.522 | 0.106 | 1 | ‐ |
| 2 | G | A | 0.322 | 0.263 | 0.021 * | 1.34 (1.04–1.73) | 0.026 * |
| 3 | A | A | 0.202 | 0.212 | 0.643 | 1.03 (0.77–1.38) | 0.83 |
Bold highlights the value of p and OR (95%CI) with statistical significance.
p values were calculated by two side chi‐squared test;
p values were calculated by Wald test adjusted by gender and age;
p < 0.05 indicates statistical significance.
4. DISCUSSION
Liver cirrhosis occurs as a consequence of many chronic liver diseases that are prevalent worldwide. Several studies showed that the etiology and pathogenesis of Cirrhosis were likely to comprise a multifactorial disorder resulting from environmental and genetic factors and their interaction. In the present case–control study, we first used immunohistochemistry (IHC) to detect the expression of the FAM13A gene in normal liver tissue and cirrhotic tissue. We found that the expression level of FAM13A in liver cirrhosis tissues was significantly higher than the normal tissues. We predicted that this gene may be a risky gene for liver cirrhosis. Subsequently, two SNPs in this gene are screened for association analysis. The results demonstrate that FAM13A genetic polymorphisms are associated with cirrhosis risk, which indicate that the FAM13A gene may play an important role in the risk of liver cirrhosis in the Chinese population.
The FAM13A gene, mapped to chromosome 4q22, contains 25 exons spanning about 332 kb, is an important switch component of the cellular pathways controlling cell cycle and proliferation (Cohen et al., 2004; Jin et al., 2015). FAM13A is a modifier gene of cystic fibrosis lung phenotype regulating rhoa activity, actin cytoskeleton dynamics, and epithelial–mesenchymal transition (Corvol et al., 2018). The FAM13A gene was participated in the Wnt pathway, which is an important part in regulating the adult tissue homeostasis (Zhang et al., 2008). However, a dysregulation of the Wnt pathway could cause grave consequences like tumourigenesis and other severe diseases, which points out the potential importance of the FAM13A in diseases (Kikuchi, 2000; Krishnamurthy & Kurzrock, 2018). One study reported the FAM13A interacts with PP2A and β‐catenin to regulate β‐catenin protein stability it could promote cell proliferation, at the same time, the result also showed FAM13A determines susceptibility to emphysema by regulating β‐catenin signaling (Jiang et al., 2016). Recent genome‐wide association studies revealed that the FAM13A gene was associated with human lung function and a variety of lung diseases, including chronic obstructive pulmonary disease, asthma, lung cancer, and pulmonary fibrosis (Eisenhut et al., 2016; Hirano et al., 2017; Wang et al., 2013). In the meanwhile, Eisenhut et al. (2017) denoted FAM13A was associated with non‐small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. Hirano et al. (2017) reported the polymorphism of FAM13A gene had a significant association with the susceptibility to idiopathic pulmonary fibrosis, with severity of lung function impairment and with poor prognosis. However, studies about the FAM13A and other diseases have not been reported, including liver cirrhosis. Therefore, it was necessary to find genetic markers to predict people who were susceptible to liver diseases.
In our study, we investigated two SNPs in FAM13A, including rs3017895 and rs1059122, and we find the SNPs in FAM13A gene were associated with an increased the risk liver cirrhosis. As far as we know, we are the first to report the association between FAM13A polymorphisms and liver cirrhosis risk, but the results identified here should be confirmed in further studies. However, some potential limitations of our current study should be considered when decipher the results. A limited number of studies has been conducted on the FAM13A gene expression to identify a potential novel biomarker for liver cirrhosis. Further functional studies and larger population‐based prospective studies are required in order to understand the genetic factors underlying liver cirrhosis in the subsequent research.
5. CONCLUSION
Our results indicate that the expression level of FAM13A in liver cirrhosis tissues was significantly higher than the normal tissues. The polymorphisms of rs3017895 and the rs1059122 in FAM13A are associated with an increased risk of liver cirrhosis.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to report.
ACKNOWLEDGMENT
This research was supported by Key Research and Development Program of Hainan Province (ZDYF2018174), Major Science and Technology Program of Hainan Province (ZDKJ2017007), and Natural Science Foundation of Hainan Province grants (817379). We thank all of the patients and individuals for their participation. We thank all of the physicians and nurses of Haikou Hospital for their offers of liver cirrhosis blood samples.
Zhang Y, Wang S, Wang C, Xiao J, Zhang S, Zhou H. High expression of FAM13A was associated with increasing the liver cirrhosis risk. Mol Genet Genomic Med. 2019;7:e543 10.1002/mgg3.543
REFERENCES
- Chang, P. E. , Wong, G. W. , Li, J. W. , Lui, H. F. , Chow, W. C. , & Tan, C. K. (2015). Epidemiology and clinical evolution of liver cirrhosis in Singapore. Annals of the Academy of Medicine Singapore, 44(6), 218. [PubMed] [Google Scholar]
- Chung, W. , Jo, C. , Chung, W. J. , & Kim, D. J. (2018). Liver cirrhosis and cancer: Comparison of mortality. Hepatology International, 12(3), 1–8. 10.1007/s12072-018-9850-5 [DOI] [PubMed] [Google Scholar]
- Cichoz‐Lach, H. , Partycka, J. , Nesina, I. , Celiński, K. , & Słomka, M. (2006). The influence of genetic polymorphism of CYP2E1 on the development of alcohol liver cirrhosis. Wiadomosci Lekarskie, 59(11–12), 757–761. [PubMed] [Google Scholar]
- Cohen, M. , Reichenstein, M. , Wind, E. V. D. , Heon‐Lee, J. , Shani, M. , Lewin, H. A. , … Seroussi, E. (2004). Cloning and characterization of FAM13A1—A gene near a milk protein QTL on BTA6: Evidence for population‐wide linkage disequilibrium in Israeli Holsteins ☆. Genomics, 84(2), 374–383. 10.1016/j.ygeno.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Corvol, H. , Hodges, C. A. , Drumm, M. L. , & Guillot, L. (2014). Moving beyond genetics: Is FAM13A a major biological contributor in lung physiology and chronic lung diseases? Journal of Medical Genetics, 51(10), 646. [DOI] [PubMed] [Google Scholar]
- Corvol, H. , Rousselet, N. , Thompson, K. E. , Berdah, L. , Cottin, G. , Foussigniere, T. , … Guillot, L. (2018). FAM13A is a modifier gene of cystic fibrosis lung phenotype regulating rhoa activity, actin cytoskeleton dynamics and epithelial‐mesenchymal transition. Journal of Cystic Fibrosis, 17(2), 190–203. 10.1016/j.jcf.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Eisenhut, F. , Heim, L. , Trump, S. , Mittler, S. , Sopel, N. , Andreev, K. , … Springel, R. (2016). FAM13A is associated with non‐small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. Oncoimmunology, 6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut, F. , Heim, L. , Trump, S. , Mittler, S. , Sopel, N. , Andreev, K. , … Warnecke, C. (2017). FAM13A is associated with non‐small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. OncoImmunology, 6(1), e1256526 10.1080/2162402x.2016.1256526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, P. F. , Jin, J. , & Sun, X. (2015). Influence of IL10 gene polymorphisms on the severity of liver fibrosis and susceptibility to liver cirrhosis in HBV/HCV‐infected patients. Infection Genetics & Evolution, 30, 89–95. 10.1016/j.meegid.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Hawkins, G. A. , & Mora, A. L. (2017). FAM13A, a fatty acid oxidation switch in mitochondria. Friend or foe in chronic obstructive pulmonary disease pathogenesis? American Journal of Respiratory Cell & Molecular Biology, 56(6), 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Deng, T. , & Luo, H. S. (2015). Genetic polymorphism in alcohol dehydrogenase 2 (ADH2) gene and alcoholic liver cirrhosis risk. International Journal of Clinical & Experimental Medicine, 8(5), 7786. [PMC free article] [PubMed] [Google Scholar]
- Hirano, C. , Ohshimo, S. , Horimasu, Y. , Iwamoto, H. , Fujitaka, K. , Hamada, H. , … Kohno, N. (2017). FAM13A polymorphism as a prognostic factor in patients with idiopathic pulmonary fibrosis. Respiratory Medicine, 123, 105–109. 10.1016/j.rmed.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Jiang, Z. , Lao, T. , Qiu, W. , Polverino, F. , Gupta, K. , Guo, F. , … Zhou, X. (2016). A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of beta‐catenin. American Journal of Respiratory Cell and Molecular Biology, 194(2), 185–197. 10.1164/rccm.201505-0999OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. , Chung, J. W. , Mei, W. , Strack, S. , He, C. , Lau, G. W. , & Yang, J. (2015). Regulation of nuclear‐cytoplasmic shuttling and function of Family with sequence similarity 13, member A (Fam13a), by B56‐containing PP2As and Akt. Molecular Biology of the Cell, 26(6), 1160–1173. 10.1091/mbc.E14-08-1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, A. (2000). Regulation of beta‐catenin signaling in the Wnt pathway. Biochemical & Biophysical Research Communications, 268(2), 243–248. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, N. , & Kurzrock, R. (2018). Targeting the Wnt/beta‐catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treatment Reviews, 62, 50–60. 10.1016/j.ctrv.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos‐Lopez, O. , Martinez‐Lopez, E. , Roman, S. , Fierro, N. A. , & Panduro, A. (2015). Genetic, metabolic and environmental factors involved in the development of liver cirrhosis in Mexico. World Journal of Gastroenterology, 21(41), 11552 10.3748/wjg.v21.i41.11552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan, D. , & Afdhal, N. H. (2008). Liver cirrhosis. Lancet, 371(9615), 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheneef, A. , Esmat, M. M. , Mohammad, A. N. , Mahmoud, A. A. , Moghazy, H. M. , & Noureldin, A. K. (2017). Interleukin‐10 and interferon gamma gene polymorphisms and hepatitis C virus‐related liver cirrhosis risk. Journal of Interferon & Cytokine Research, 37(4), 175–180. 10.1089/jir.2016.0106 [DOI] [PubMed] [Google Scholar]
- Wang, B. , Liang, B. , Yang, J. , Xiao, J. , Ma, C. , Xu, S. , … Liu, H. (2013). Association of FAM13A polymorphisms with COPD and COPD‐related phenotypes in Han Chinese. Clinical Biochemistry, 46(16–17), 1683–1688. 10.1016/j.clinbiochem.2013.07.013 [DOI] [PubMed] [Google Scholar]
- Wu, X. D. , Zeng, K. , Gong, C. S. , Chen, J. , & Chen, Y. Q. (2013). Transforming growth factor‐beta genetic polymorphisms on development of liver cirrhosis in a meta‐analysis. Molecular Biology Reports, 40(1), 535–543. 10.1007/s11033-012-2090-1 [DOI] [PubMed] [Google Scholar]
- Xiong, J. P. , Liu, H. Y. , & Zhang, Y. X. (2018). Analysis of risk factors for portal vein thrombosis in liver cirrhosis patients. Chinese Hepatology, 23(3), 206-209. [Google Scholar]
- Zhang, Y. , Goss, A. M. , Cohen, E. D. , Kadzik, R. , Lepore, J. J. , Muthukumaraswamy, K. , … Parmacek, M. S. (2008). A Gata6‐Wnt pathway required for epithelial stem cell development and airway regeneration. Nature Genetics, 40(7), 862–870. 10.1038/ng.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocco, M. A. , Di, S. E. , De, C. R. , Novi, M. , Ainora, M. E. , Ponziani, F. , … Flore, R. (2009). Thrombotic risk factors in patients with liver cirrhosis: Correlation with MELD scoring system and portal vein thrombosis development. Journal of Hepatology, 51(4), 682–689. 10.1016/j.jhep.2009.03.013 [DOI] [PubMed] [Google Scholar]


