Abstract
Aim
Osteonecrosis of the femoral head (ONFH) refers to bony changes caused by osteocyte death under the effects of complicated factors, which is caused by genetic factors and certain risk factors. Our study aimed to explore whether IL1R1/IL1R2 polymorphisms influenced ONFH risk in the Chinese Han population.
Methods
We selected 286 patients and 441 controls, with 11 single‐nucleotide polymorphisms in IL1R1 and IL1R2 gene were successfully genotyped, and evaluated the associations using the chi‐squared test, Fisher's exact test, T test, and genetic model analyses. Odds ratios and 95% confidence intervals (CIs) were calculated using unconditional logistic regression.
Results
In the allele model, rs11674595 in IL1R2 was associated with increasing the risk of ONFH, the rs10490571 and rs3917225 in IL1R1 gene were associated with an increased risk of ONFH, respectively. In the genetic model, the rs11674595 in IL1R2 gene was associated with an increased risk of ONFH in the codominant model, dominant model, and log‐additive model, respectively. The rs10490571 and rs3917225 in IL1R1 gene conferred an increased risk of ONFH in the codominant model, dominant model, and log‐additive model, respectively. We found none of the haplotypes in the IL1R2 gene was significantly associated with theONFH risk.
Conclusion
Our findings have demonstrated that the rs11674595 (IL1R2), rs10490571, and rs3917225 (IL1R1) were significantly associated with increasing the ONFH risk in the Chinese Han population.
Keywords: case–control study, Chinese Han population, genetic polymorphism, IL1R1, IL1R2, ONFH
1. INTRODUCTION
Nontraumatic osteonecrosis of the femoral head (ONFH), known as avascular necrosis, is a refractory and progressive disease and caused by osteocyte death (Mankin, 1992; Yu et al., 2016). However, the specific pathogenesis of ONFH has not completely stated. ONFH is believed to be a multifactorial disease that is associated in some cases with both genetic factors and certain risk factors (Björkman et al., 2004; Du et al., 2016; Su et al., 2016). These risk factors include corticosteroid use, alcohol intake, smoking, and various chronic diseases (renal disease, hematological disease, inflammatory bowel disease (IBD), postorgan transplantation, and hypertension) (Zheng et al., 2014).
Previous study has reported that the immune system is substantially involved in the regulation of bone homeostasis;furthermore, ONFH may be caused by disruption of the immune system via lipopolysaccharide activated toll‐like receptor 4 (TLR4) signaling (Okazaki et al., 2009). Therefore, abnormal immune responses may contribute to the pathogenesis of ONFH by impacting bone remodeling. Interleukin (IL)‐1 is a primary proinflammatory cytokine, and it could stimulate the expression of genes that associated with inflammation and immunity (Strand & Kavanaugh, 2004). Previous studies have also indicated the association between immune‐related genes and various bone disease. For example, the IL‐1 gene cluster was related to increase the risk of ankylosing spondylitis (Maksymowych et al., 2006); inhibiting the IL‐1 could reduce cartilage damage in rheumatoid arthritis (Strand & Kavanaugh, 2004); and the recently discovered IL‐33 as an IL‐1 cytokine family member has been proved to be specifically released from osteonecrotic bones (Saidi et al., 2011).
Interleukin‐1 receptor, type 1 (IL1R1; OMIM: 147810) and IL‐1 receptor, type 2 (IL1R2; OMIM: 147811) are cytokine receptor that belongs to the IL‐1 receptor family, which is an important mediator involving in many cytokines induced by immune and inflammatory responses (Sims & Dower, 1994). Study showed that IL1R1 and IL1R2 genes could regulate the cell metabolism, and the response of immune inflammatory induced by many cytokines (Dinarello, 1994; Rock, Hardiman, Timans, Kastelein, & Bazan, 1998). Moreover, the epidemiological studies have been manifested that ONFH was impacted by hereditary factors. Therefore, the IL1R1 and IL1R2 genes may be associated with ONFH.
To identify the associations between ONFH and susceptibility loci in previous studies, we conduct a case–control study and identify the relationship between ONFH and the susceptible single‐nucleotide polymorphisms (SNPs) in the IL1R1 and IL1R2 gene to further clarify their potential roles in ONFH risk in the Chinese population.
2. MATERIALS AND METHODS
2.1. Ethics statement
This investigation was conducted in accordance with the ethical standards of the Declaration of Helsinki and following national and international guidelines. The study protocol was approved by the ethics committee of the Zhengzhou Traditional Chinese Medicine Traumatology Hospital. Written informed consent was obtained from all participants after a full explanation of the study. The experimental protocol was implemented in accordance with the approved guidelines.
2.2. Subjects
All subjects were members of Chinese Han population living in the Henan Province of China. The cases were recruited from Zhengzhou Traditional Chinese Medicine Traumatology Hospital, China. ONFH was diagnosed by examining osteonecrosis in anteroposterior and frog view X‐rays of both hips and/or magnetic resonance imaging. The ONFH patients with other direct trauma, chronic diseases (such as cardiovascular diseases, congenital diseases, human immunodeficiency virus infection, diabetes mellitus, renal dysfunction, and cancer), corticosteroids, alcohol, and familial hereditary diseases were excluded. Individuals in the control group had no ONFH disease. We recruited subjects without consideration of age and gender.
2.3. SNP selection and genotyping
We selected these SNPs on the basis of their allele frequencies, location, and disease relevance through public HapMap databases. All 11 SNPs had minor allele frequencies >5% in the 1,000 genome (http://www.internationalgenome.org/). Blood samples were collected in EDTA tubes and stored at −80°C after centrifugation at 17,528 g for 10 min. Genomic DNA was extracted from peripheral blood samples using a genomic DNA purification kit (GoldMag, Xi'an, China). We used NanoDrop 2000 (Thermo Scientific, Waltham, MA) to measure the DNA concentration. The primers for amplification and extension reactions were designed with Agena MassARRAY Assay Design 3.0 Software (Gabriel, Ziaugra, & Tabbaa, 2009). We used Agena MassARRAY RS1000 to perform the SNP genotyping with the agreement of the manufacturer, and we used Agena Typer 4.0 software for data management and analysis (Gabriel et al., 2009; Thomas et al., 2007).
2.4. Statistical analysis
Microsoft Excel (Microsoft, Redmond, WA) and SPSS Statistics (version 17.0, SPSS, Chicago, IL) were used for statistical analyses. All p‐values were two‐tailed, and p < 0.05 was considered to be statistically significant. SNP genotype frequencies in the case and control groups were calculated by chi‐squared test, and the Hardy–Weinberg equilibrium (HWE) values were used to check the genotype frequency of the control group. Unconditional logistic regression analysis was used to examine the odds ratios (ORs) and 95% CIs in order to assess the association between SNPs and ONFH (Bland & Altman, 2000). Four models (codominant, dominant, recessive, and log‐additive) were used to test the association between SNPs and ONFH (Sole, Guino, Valls, Iniesta, & Moreno, 2006). Finally, the Haploview software package (version 4.2) and SHEsis software platform (http://www.nhgg.org/analysis/) were used to estimate pairwise linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci.
3. RESULT
3.1. Characteristics of the participants
This study involved 727 subjects, including 286 patients (173 males and 113 females; age at diagnosis: 41.83 ± 13.11 years) and 441 healthy controls (265 males and 176 females; age: 44.60 ± 11.55 years). The stroke cases and controls were matched by sex, but there was a significant difference in age between stroke cases and controls (p < 0.05) (Table 1).
Table 1.
General characteristics the of this study population
| Variables | Cases (n = 286) | % | Controls (n = 441) | % | p‐value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 173 | 60.50 | 265 | 60.10 | >0.05 |
| Female | 113 | 39.50 | 176 | 39.90 | |
| Age, year (mean ± SD) | 41.83 ± 13.11 | 44.60 ± 11.55 | <0.05 | ||
p‐values were calculated from two‐sided chi‐squared test/Fisher's exact test.
p ≤ 0.05 was statistically significant.
3.2. The associations between IL1R1 and IL1R2 SNPs and ONFH
Eleven SNPs in IL1R1 and IL1R2 were analyzed in this study. Allele frequencies and basic information for all SNPs are shown in Table 2. One SNP (rs12712127) was excluded for significant deviation from HWE (p < 0.05). We used the chi‐squared test to assess the risk of gene polymorphism in the allele model, while rs11674595 in IL1R2 was significantly associated with increasing the ONFH risk (rs11674595, OR = 1.37, 95% CI = 1.07–1.75, p = 0.012), the rs10490571 and rs3917225 in IL1R1 gene were associated with an increased risk of ONFH (rs10490571, OR = 1.36, 95% CI = 1.04–1.77, p = 0.025; rs3917225, OR = 1.35, 95% CI = 1.09–1.68, p = 0.006), respectively.
Table 2.
Allele frequencies in cases and controls and odds ratio estimates for ONFH risk
| SNP | Gene(s) | Band | Alleles A/B | MAF | p‐HWE | OR (95% CI) | p‐value | |
|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||
| rs11674595 | IL1R2 | 2q11.2 | C/T | 0.267 | 0.211 | 0.250 | 1.37 (1.07–1.75) | 0.012 a |
| rs4851527 | IL1R2 | 2q11.2 | A/G | 0.259 | 0.289 | 0.486 | 0.86 (0.68–1.09) | 0.214 |
| rs719250 | IL1R2 | 2q11.2 | T/G | 0.333 | 0.317 | 0.912 | 1.07 (0.86–1.35) | 0.533 |
| rs3218896 | IL1R2 | 2q11.2 | C/T | 0.173 | 0.150 | 0.351 | 1.18 (0.89–1.57) | 0.252 |
| rs3218977 | IL1R2 | 2q11.2 | G/A | 0.263 | 0.251 | 1.000 | 1.07 (0.84–1.36) | 0.592 |
| rs2072472 | IL1R2 | 2q11.2 | G/A | 0.208 | 0.200 | 0.766 | 1.05 (0.81–1.37) | 0.696 |
| rs10490571 | IL1R1 | 2q12.1 | T/C | 0.215 | 0.168 | 0.061 | 1.35 (1.04–1.77) | 0.025 a |
| rs12712127 | IL1R1 | 2q12.1 | G/A | 0.290 | 0.215 | 0.000 | 1.50 (1.17–1.91) | 0.001 |
| rs956730 | IL1R1 | 2q12.1 | A/G | 0.248 | 0.250 | 0.016 | 0.99 (0.78–1.26) | 0.940 |
| rs3917225 | IL1R1 | 2q12.1 | T/C | 0.419 | 0.348 | 0.074 | 1.35 (1.09–1.68) | 0.006 a |
| rs3917318 | IL1R1 | 2q12.1 | G/A | 0.498 | 0.497 | 0.045 | 1.01 (0.82–1.24) | 0.951 |
SNP: single‐nucleotide polymorphism; MAF: minor allele frequency; HWE: Hardy–Weinberg equilibrium; OR: odds ratio; CI: confidence interval.
In bold, p < 0.05 indicates statistical significance
In bold, p‐HWE < 0.05 be excluded.
3.3. Associations between genotype frequencies and osteonecrosis risk
As is shown in Table 3, we examined whether the minor allele for each SNP compared to the wild‐type allele represented a risk factor in the genetic model. Our analyses showed that the rs11674595 in IL1R2 gene was associated with a 1.49‐fold increase the risk of ONFH in the codominant model (adjusted, OR = 1.49, 95% CI = 1.09–2.03, p = 0.033 for the “C/T” genotype), 1.40‐fold increase the risk of ONFH in the dominant model (adjusted, OR = 1.50, 95% CI = 1.10–2.03, p = 0.009 for the “C/T‐C/C” genotype), and 1.40‐fold increase the risk of ONFH in the log‐additive model (adjusted OR = 1.40, 95% CI = 1.08–1.81, p = 0.006), respectively. The rs10490571 in IL1R1 gene was associated with a 1.49‐fold increase the risk of ONFH in the codominant model (adjusted, OR = 1.69, 95% CI = 1.22–2.34, p = 0.006 for the “C/T” genotype), 1.57‐fold increase the risk of ONFH in the dominant model (adjusted, OR = 1.57, 95% CI = 1.15–2.16, p = 0.005 for the “C/T‐T/T” genotype), and 1.34‐fold increase the risk of ONFH in the log‐additive model (adjusted OR = 1.34, 95% CI = 1.03–1.75, p = 0.031), respectively. The rs3917225 in IL1R1 gene was associated with a 1.50‐fold and 1.65‐fold increase the risk of ONFH in the codominant model (adjusted, OR = 1.50, 95% CI = 1.07–2.09, p = 0.022 for the “A/G” genotype; OR = 1.65, 95% CI = 1.06–2.58, p = 0.022 for the “G/G” genotype), respectively. The rs3917225 was associated with a 1.54‐fold increase the risk of ONFH in the dominant model (adjusted, OR = 1.57, 95% CI = 1.13–2.10, p = 0.006 for the “A/G‐G/G” genotype) and 1.32‐fold increase the risk of ONFH in the log‐additive model (adjusted OR = 1.32, 95% CI = 1.07–1.64, p = 0.009), respectively.
Table 3.
Relationships between IL1R2 and IL1R1polymorphism and ONFH risk
| SNP | Model | Genotype | Control | Case | Before adjusted | After adjusted | AIC | BIC | ||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p avalue | OR (95% CI) | p bvalue | |||||||
| rs11674595 | Codominant | T/T | 269 (61.3%) | 147 (51.4%) | 1 | 0.03 | 1 | 0.033 | 971.5 | 985.3 |
| C/T | 155 (35.3%) | 125 (43.7%) | 1.48 (1.08–2.01) | 1.49 (1.09–2.03) | ||||||
| C/C | 15 (3.4%) | 14 (4.9%) | 1.71 (0.80–3.64) | 1.60 (0.75–3.42) | ||||||
| Dominant | T/T | 269 (61.3%) | 147 (51.4%) | 1 | 0.009 | 1 | 0.009 | 969.6 | 978.8 | |
| C/T‐C/C | 170 (38.7%) | 139 (48.6%) | 1.50 (1.11‐2.02) | 1.50 (1.10–2.03) | ||||||
| Recessive | T/T‐C/T | 424 (96.6%) | 272 (95.1%) | 1 | 0.33 | 1 | 0.420 | 975.6 | 984.7 | |
| C/C | 15 (3.4%) | 14 (4.9%) | 1.45 (0.69–3.06) | 1.36 (0.64–2.88) | ||||||
| Log‐additive | – | – | – | 1.41 (1.09–1.83) | 0.01 | 1.40 (1.08–1.81) | 0.012 | 969.8 | 979 | |
| rs10490571 | Codominant | C/C | 310 (70.5%) | 172 (60.1%) | 1 | 0.005 | 1 | 0.006 | 969.1 | 982.9 |
| C/T | 112 (25.4%) | 105 (36.7%) | 1.69 (1.22–2.34) | 1.69 (1.22–2.34) | ||||||
| T/T | 18 (4.1%) | 9 (3.1%) | 0.90 (0.40–2.05) | 0.88 (0.39–2.01) | ||||||
| Dominant | C/C | 310 (70.5%) | 172 (60.1%) | 1 | 0.004 | 1 | 0.005 | 969.3 | 978.5 | |
| C/T‐T/T | 130 (29.6%) | 114 (39.9%) | 1.58 (1.16–2.16) | 1.57 (1.15–2.16) | ||||||
| Recessive | C/C‐C/T | 422 (95.9%) | 277 (96.8%) | 1 | 0.51 | 1 | 0.470 | 977.1 | 986.3 | |
| T/T | 18 (4.1%) | 9 (3.1%) | 0.76 (0.34–1.72) | 0.74 (0.33–1.69) | ||||||
| Log‐additive | – | – | – | 1.35 (1.03–1.76) | 0.027 | 1.34 (1.03–1.75) | 0.031 | 972.6 | 981.8 | |
| rs3917225 | Codominant | A/A | 196 (44.5%) | 98 (34.4%) | 1 | 0.02 | 1 | 0.022 | 969.9 | 983.6 |
| A/G | 182 (41.4%) | 135 (47.4%) | 1.48 (1.07–2.06) | 1.50 (1.07–2.09) | ||||||
| G/G | 62 (14.1%) | 52 (18.2%) | 1.68 (1.08–2.61) | 1.65 (1.06–2.58) | ||||||
| Dominant | A/A | 196 (44.5%) | 98 (34.4%) | 1 | 0.006 | 1 | 0.007 | 968.2 | 977.4 | |
| A/G‐G/G | 244 (55.5%) | 187 (65.6%) | 1.53 (1.13–2.09) | 1.54 (1.13–2.10) | ||||||
| Recessive | A/A‐A/G | 378 (85.9%) | 233 (81.8%) | 1 | 0.14 | 1 | 0.170 | 973.4 | 982.6 | |
| G/G | 62 (14.1%) | 52 (18.2%) | 1.36 (0.91–2.04) | 1.33 (0.89–2.00) | ||||||
| Log‐additive | – | – | – | 1.33 (1.08–1.64) | 0.008 | 1.32 (1.07–1.64) | 0.009 | 968.6 | 977.8 | |
SNP: single‐nucleotide polymorphism; OR: odds ratio; 95% CI: 95% confidence interval.
p‐values were calculated from unconditional logistic regression analysis.
p‐values were calculated by unconditional logistic regression analysis with adjustments for age and gender.
The bold values and p ≤ 0.05 indicate statistical significance.
3.4. Associations between haplotype analyses and ONFH risk
Linkage disequilibrium and haplotype analyses of the SNPs in the case and control samples were further studied. Haplotype analysis revealed two blocks in the IL1R2 gene (Figure 1). Although the three SNPs (rs4851527, rs719250, and rs3218896) and the two SNPs (rs3218977 and rs2072472) in the IL1R2 gene have showed strong linkage, but the result for the IL1R2 haplotype was not found to be associated with a risk of ONFH, because the p‐value has no statistical difference (Table 4). In addition, we have not found any association between IL1R1 haplotype and the risk of ONFH.
Figure 1.
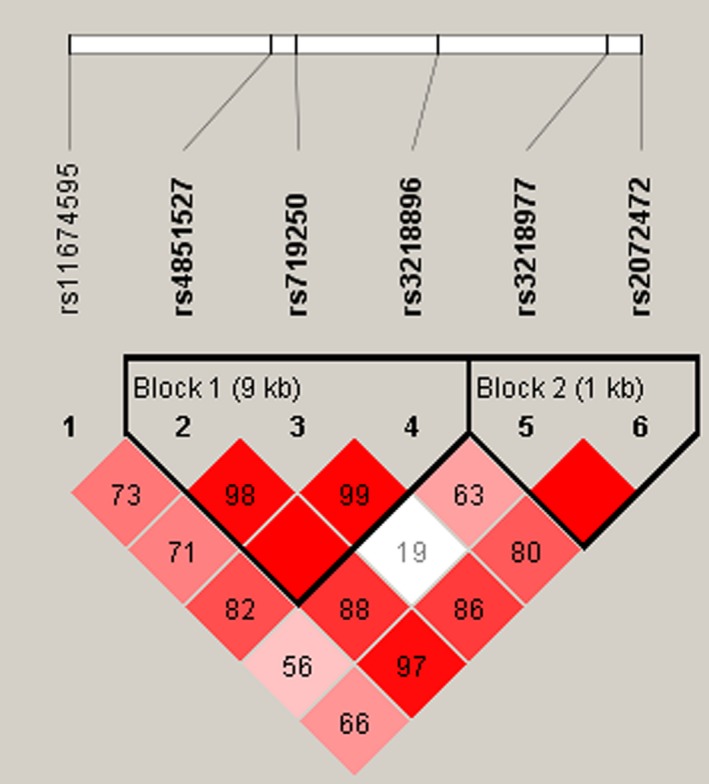
Linkage disequilibrium plots containing four SNPs from IL1‐R2
Table 4.
Haplotype analysis results in this study
| Block | SNPs | Haplotype | Freq | Before adjusted | After adjusted | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p‐value | OR (95% CI) | p a‐value | ||||
| Block 1 | rs4851527/rs719250/rs3218896 | GCT | 0.4011 | 1 | – | 1 | – |
| ACT | 0.2749 | 0.88 (0.67–1.14) | 0.33 | 0.87 (0.67–1.14) | 0.32 | ||
| GTT | 0.1638 | 0.93 (0.68–1.27) | 0.65 | 0.93 (0.68–1.28) | 0.68 | ||
| GTC | 0.1583 | 1.13 (0.82–1.54) | 0.45 | 1.13 (0.82–1.55) | 0.45 | ||
| Block 2 | rs3218977/rs2072472 | AA | 0.5411 | 1 | – | 1 | – |
| GA | 0.2555 | 1.09 (0.85–1.41) | 0.51 | 1.08 (0.84–1.40) | 0.55 | ||
| AG | 0.2035 | 1.09 (0.82–1.43) | 0.56 | 1.07 (0.81–1.42) | 0.63 | ||
SNP: single‐nucleotide polymorphism; OR: odds ratio; CI: confidence interval; pa: Adjusted by gender and age.
4. DISCUSSION
Genetic studies have provided insight into numerous diseases, including ONFH. In the present case–control study, we investigated the associations between 11 SNPs in IL1R1 and IL1R2 gene risk of ONFH. We demonstrated that IL1R2 and IL1R1 genetic polymorphisms were associated with ONFH risk in Chinese Han population. Our results indicate that the rs11674595 (in the IL1R2), the rs10490571, and rs3917225 (in the IL1R1) were associated with an increased risk of ONFH. These results suggest that polymorphisms in IL1R2 and IL1R1 genes may play an important role in the risk of ONFH in the Han Chinese population.
The biological activity of the multifunctional cytokine IL‐1 is mediated by its receptors (Lukens, Gross, & Thirumala‐Devi, 2012). The IL1R1 and IL1R2 genes are cytokine receptors that belong to the IL‐1 receptor family, which is an important mediator involved in many cytokine‐induced responses (Sims & Dower, 1994). IL1R1 (on chromosome 2q12) is an important mediator involved in many cytokine‐induced immune and inflammatory responses (Dinarello, 1998). Recently, some studies revealed that the expression of IL1R1 was observably increased in several bone disease. For example, Latiano et al. (2013) reported that the rs13015714 and rs2058660 in IL1R1 could increase the risk of IBD; Kouhia et al. (2010) indicated that four SNPs (rs1465325, rs956730, rs3917225, and rs2287047) in the IL1R1 gene provided evidence for association with hand osteoarthritis. Another study involving the association between five SNPs polymorphisms in IL1R1 (rs10490571, rs12712127, rs956730, rs3917225, and rs3917318) and osteoarthritis risk, the result found that rs3917225 in IL1R1 was associated with increasing the risk of knee OA (Na et al., 2017; Smith et al., 2004). However, we have not found any evidence for the role of heredity between IL1R1 and ONFH susceptibility in previous study. Therefore, our study fully discusses the relationship between IL1R1 and ONFH risk. In our case–control study, we found that rs10490571 and rs3917225 were associated with an increased risk of ONFH.
IL1R2 (on 2q11.2) is a molecular decoy that traps IL‐1β and does not initiate subsequent signaling events, thereby suppressing an inflammatory response. Many studies reported that the IL1R2 as a risk factor in some disease, such as IgA nephropathy (Xie et al., 2017). The other study reported that epithelial IL1R2 acted as a homeostatic regulator during remission of ulcerative colitis (Mora‐Buch et al., 2016), and another suggested that IL1R2 was an important regulator of arthritis by acting specifically on macrophages as a decoy receptor for IL‐1 (Shimizu et al., 2015). However, previous studies based on the IL1R2 gene polymorphisms were rare, until now, there has no study reported the association between IL1R2 and the ONFH risk. For this study, the rs11674595 in IL1R2 showed an increased risk of ONFH. Hence, IL1R2 gene may play an important function in affecting ONFH.
To sum up, we provide new evidence for the association between IL1R1 and IL1R2 variant and ONFH risk in Han Chinese population for the first time, which may provide new data to facilitate earlier diagnosis and promote early prevention, and shed light on the new candidate genes and new ideas for the study. Nevertheless, there are limitations that need to be noticed. Our current research is fundamental, and further functional studies and larger population‐based prospective studies are required in order to understand the genetic factors underlying ONFH.
CONFLICTS OF INTEREST
None.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (no. 81160228, 81260284, 81660378). We thank all the patients and individuals for their participation and all the physicians and nurses of the Zhengzhou Traditional Chinese Medicine Traumatology Hospital for their offers of ONFH blood samples.
An F, Wang J, Gao H, et al. Impact of IL1R1 and IL1R2 gene polymorphisms on risk of osteonecrosis of the femoral head from a case–control study. Mol Genet Genomic Med. 2019;7:e557 10.1002/mgg3.557
Contributor Information
Wanlin Liu, Email: 1009902058@qq.com.
Jianzhong Wang, Email: Jianzhongwang123@163.com.
REFERENCES
- Björkman, A. , Svensson, P. J. , Hillarp, A. , Burtscher, I. M. , Rünow, A. , & Benoni, G. (2004). Factor V leiden and prothrombin gene mutation: Risk factors for osteonecrosis of the femoral head in adults. Clinical Orthopaedics & Related Research, 425(425), 168–172. [PubMed] [Google Scholar]
- Bland, J. M. , & Altman, D. G. (2000). Statistics notes. The odds ratio. BMJ, 320(7247), 1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello, C. A. (1994). The interleukin‐1 family: 10 years of discovery. FASEB J, 8(15), 1314–1325. [PubMed] [Google Scholar]
- Dinarello, C. A. (1998). Interleukin‐1, interleukin‐1 receptors and interleukin‐1 receptor antagonist. International Reviews of Immunology, 16(5–6), 457. [DOI] [PubMed] [Google Scholar]
- Du, J. , Jin, T. , Cao, Y. , Chen, J. , Guo, Y. , Sun, M. , … Wang, J. (2016). Association between genetic polymorphisms of MMP8 and the risk of steroid‐induced osteonecrosis of the femoral head in the population of northern China. Medicine, 95(37), e4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S. , Ziaugra, L. , & Tabbaa, D. (2009). SNP genotyping using the Sequenom MassARRAY iPLEX platform. John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Kouhia, S. T. , Saarela, J. , Harilainen, A. , Tallroth, K. , Videman, T. , Kaprio, J. , … Kujala, U. M. (2010). Allelic variants of IL1R1 gene associate with severe hand osteoarthritis. BMC Medical Genetics, 11(1), 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latiano, A. , Palmieri, O. , Pastorelli, L. , Vecchi, M. , Pizarro, T. T. , Bossa, F. , … Corritore, G. (2013). Associations between genetic polymorphisms in IL‐33, IL1R1 and risk for inflammatory bowel disease. PLoS ONE, 8(4), e62144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, J. R. , Gross, J. M. , & Thirumala‐Devi, K. (2012). IL‐1 family cytokines trigger sterile inflammatory disease. Frontiers in Immunology, 3, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksymowych, W. P. , Rahman, P. , Reeve, J. P. , Gladman, D. D. , Peddle, L. , & Inman, R. D. (2006). Association of the IL1 gene cluster with susceptibility to ankylosing spondylitis: An analysis of three Canadian populations. Arthritis and Rheumatism, 54(3), 974–985. 10.1002/art.21642 [DOI] [PubMed] [Google Scholar]
- Mankin, H. J. (1992). Nontraumatic necrosis of bone (osteonecrosis). New England Journal of Medicine, 326(22), 1473–1479. 10.1056/nejm199205283262206 [DOI] [PubMed] [Google Scholar]
- Mora‐Buch, R. , Dotti, I. , Planell, N. , Calderón‐Gómez, E. , Jung, P. , Masamunt, M. C. , … Panés, J. (2016). Epithelial IL‐1R2 acts as a homeostatic regulator during remission of ulcerative colitis. Mucosal Immunology, 9(4), 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, Y. , Rui, B. , Zhao, Z. , Wei, Y. , Li, D. , Yong, W. , … Jin, T. (2017). IL1R1 gene polymorphisms are associated with knee osteoarthritis risk in the Chinese Han population. Oncotarget, 8(3), 4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, S. , Nishitani, Y. , Nagoya, S. , Kaya, M. , Yamashita, T. , & Matsumoto, H. (2009). Femoral head osteonecrosis can be caused by disruption of the systemic immune response via the toll‐like receptor 4 signalling pathway. Rheumatology (Oxford), 48(3), 227–232. 10.1093/rheumatology/ken462 [DOI] [PubMed] [Google Scholar]
- Rock, F. L. , Hardiman, G. , Timans, J. C. , Kastelein, R. A. , & Bazan, J. F. (1998). A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A, 95(2), 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi, S. , Bouri, F. , Lencel, P. , Duplomb, L. , Baud'huin, M. , Delplace, S. , … Magne, D. (2011). IL‐33 is expressed in human osteoblasts, but has no direct effect on bone remodeling. Cytokine, 53(3), 347–354. 10.1016/j.cyto.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Shimizu, K. , Nakajima, A. , Sudo, K. , Liu, Y. , Mizoroki, A. , Ikarashi, T. , … Iwakura, Y. (2015). IL‐1 receptor type 2 suppresses collagen‐induced arthritis by inhibiting IL‐1 signal on macrophages. Journal of Immunology, 194(7), 3156–3168. [DOI] [PubMed] [Google Scholar]
- Sims, J. E. , & Dower, S. K. (1994). Interleukin‐1 receptors. European Cytokine Network, 5(6), 539. [PubMed] [Google Scholar]
- Smith, A. J. P. , Keen, L. J. , Billingham, M. J. , Perry, M. J. , Elson, C. J. , Kirwan, J. R. , … Bidwell, J. L. (2004). Extended haplotypes and linkage disequilibrium in the IL1R1‐IL1A‐IL1B‐IL1RN gene cluster: Association with knee osteoarthritis. Genes & Immunity, 5(6), 451. [DOI] [PubMed] [Google Scholar]
- Sole, X. , Guino, E. , Valls, J. , Iniesta, R. , & Moreno, V. (2006). SNPStats: A web tool for the analysis of association studies. Bioinformatics, 22(15), 1928–1929. 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- Strand, V. , & Kavanaugh, A. F. (2004). The role of interleukin‐1 in bone resorption in rheumatoid arthritis. Rheumatology (Oxford), 43 (Suppl 3(6)), iii10. [DOI] [PubMed] [Google Scholar]
- Su, B. , Yu, Y. , Yang, H. , Ouyang, Y. , Feng, T. , Jin, T. , & Zhao, H. (2016). Nitric oxide of genetic variants is associated with alcohol‐induced osteonecrosis risk of the femoral head in a Han population. International Journal of Clinical & Experimental Pathology, 8(20):33770–33778. [Google Scholar]
- Thomas, R. K. , Baker, A. C. , Debiasi, R. M. , Winckler, W. , Laframboise, T. , Lin, W. M. , … Garraway, L. A. (2007). High‐throughput oncogene mutation profiling in human cancer. Nature Genetics, 39(3), 347–351. 10.1038/ng1975 [DOI] [PubMed] [Google Scholar]
- Xie, M. , Zhang, D. , Zhang, Y. , Yang, X. , Su, Y. , Wang, Y. , … Fu, K. (2017). Association of genetic polymorphisms in IL‐1R1 and IL‐1R2 genes with IgA nephropathy in the Han Chinese population. Oncotarget, 8(31), 50673–50679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Xie, Z. , Wang, J. , Chen, C. , Du, S. , Chen, P. , … Zhao, H. (2016). Single‐nucleotide polymorphisms of MMP2 in MMP/TIMP pathways associated with the risk of alcohol‐induced osteonecrosis of the femoral head in Chinese males: a case‐control study. Medicine (Baltimore), 95(49), e5407 10.1097/md.0000000000005407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C. , Zheng, Q. J. , Wang, Y. S. , Liao, J. X. , Yuan Chen, M. A. , & Zheng, X. Q. (2014). The clinical prognostic risk factors of osteonecrosis of femoral head. Guangdong Medical Journal, 35(14), 2178–2181. [Google Scholar]


