Abstract
RNA is the messenger molecule that conveys information from the genome and allows the production of biomolecules required for life in a responsive and regulated way. Most genes are able to produce multiple mRNA products in response to different internal or external environmental signals, in different tissues and organs, and at specific times in development or later life. This fine tuning of gene expression is dependent on the coordinated effects of a large and intricate set of regulatory machinery, which together orchestrate the genomic output at each locus and ensure that each gene is expressed at the right amount, at the right time and in the correct location. This complexity of control, and the requirement for both sequence elements and the entities that bind them, results in multiple points at which errors may occur. Errors of RNA biology are common and found in association with both rare, single gene disorders, but also more common, chronic diseases. Fortunately, complexity also brings opportunity. The existence of many regulatory steps also offers multiple levels of potential therapeutic intervention which can be exploited. In this review, I will outline the specific points at which coding RNAs may be regulated, indicate potential means of intervention at each stage, and outline with examples some of the progress that has been made in this area. Finally, I will outline some of the remaining challenges with the delivery of RNA-based therapeutics but indicate why there are reasons for optimism.
Keywords: mRNA processing, RNA editing, RNA export, RNA therapeutics, ncRNA, splicing, RNA epitranscriptomics, therapeutics
Introduction
The fundamental importance of RNA not only as a messenger molecule, but as a regulator of genes in its own right is increasingly being recognized. The production of mature messenger RNA (mRNA) is dependent on a plethora of processing and regulatory steps involving a complicated repertoire of sequence elements, RNA binding proteins and other regulatory RNA species. Given the complexity of the regulatory machinery, defects in non-coding regions of genes and regulatory genomic regions are common in genetic disease, being present in up to 50% of cases (Yang et al., 2013; Beaulieu et al., 2014) and are also the most common site of genetic variation conferring susceptibility to common, complex disease (Manolio et al., 2008). There is, however, a silver lining. The complexity that causes errors in gene expression or mRNA processing to be such a common occurrence, also provides multiple and differential points of potential therapeutic intervention. Over the past decade, there have been a number of examples, where the specifics of RNA regulatory machinery have been harnessed to produce novel therapeutics that are now in phase III clinical trials [e.g., Patisiran for Familial amyloid polyneuropathy (Rizk and Tuzmen, 2017), Custirsen for prostate cancer (Edwards et al., 2017) and AGS-003 for renal cell carcinoma (Figlin, 2015)]. This review aims to explore the potential for intervention in mRNA processing or post-transcriptional regulation with selected examples for future therapeutic benefit.
The Lifecycle of a Coding RNA
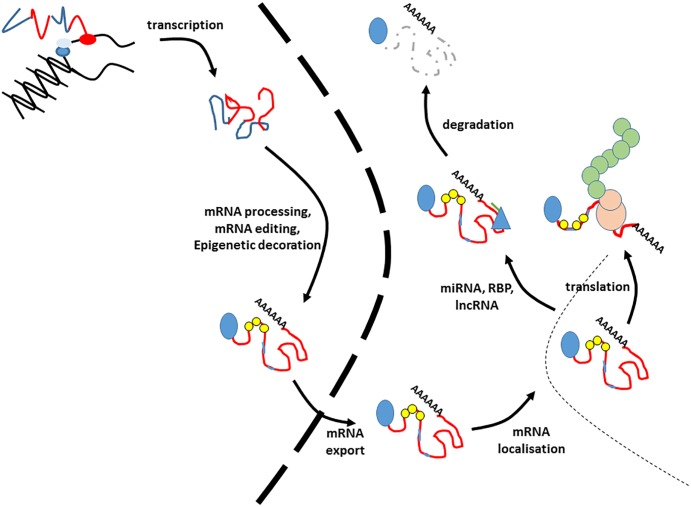
The processes involved in the production of a mature mRNA, and its subsequent fate are multifaceted and complicated (Figure 1). The life of an RNA molecule starts upon transcription, which is controlled by tissue specific promoters and enhancers. The immature primary RNA transcript (heterogeneous nuclear RNA (hnRNA) or pre-mRNA) then undergoes a series of modifications that involve the addition of the 5′ cap structure, removal of the intronic sequences by constitutive or alternative splicing and 3′ end processing events that include the addition of the poly-A tail (Chen et al., 2017; Sperling, 2017; Zhang and Tjian, 2018). These processes are not a linear pipeline and occur co-transcriptionally (Beyer and Osheim, 1988; Bentley, 2002). Newly processed RNA may also undergo RNA editing, which is mostly A to G or A to I substitution in humans (Chen, 2013). RNAs may also undergo epitranscriptomic decoration, whereby different RNA modifications such as methylation of adenosine residues (m6A) may be added. Such modifications are added by a series of RNA readers, writers and erasers (Helm and Motorin, 2017). Mature mRNAs are then exported from the nucleus to the cytoplasm. This is an active and regulated process, and one of the primary safeguards against the translation of aberrant mRNAs (Williams et al., 2018). The spatial and temporal expression of newly exported RNAs can also controlled at the level of specific localization within the cell. This can be passive, or an active process involving transport on cytoskeletal tracks (Suter, 2018). Gene expression can also be controlled at the level of translation. This can occur by virtue of selective degradation of specific RNAs by mRNA surveillance pathways such as nonsense-mediated decay, no-go decay and non-stop decay (Harigaya and Parker, 2010; Klauer and van Hoof, 2012; Lejeune, 2017), or it can be by regulation of the rate of translation itself (Gorgoni et al., 2014). The half-life of any given mRNA is then determined by a number of RNA decay pathways, most of which involve successive decapping and deadenylation of RNA molecules, which then renders them susceptible to exonucleases (Wahle and Winkler, 2013; Borbolis and Syntichaki, 2015). Finally, the fate of the RNA may also be influenced by the action of both short and long non-coding RNAs and RNA binding proteins which can result in degradation or translational blocking (Fukao et al., 2015; Iadevaia and Gerber, 2015; Fukao and Fujiwara, 2017).
FIGURE 1.
The lifecycle of an RNA. This figure illustrates the lifecycle of an mRNA. These processes are co-transcriptional, so the precise order of events is illustrative. Blue lines in the transcript refer to introns and untranslated regions, whilst exons are indicted by red lines. The 5′ cap is indicated by a blue circle. Small yellow circles indicate epitranscriptomic decoration, whilst pale blue lines within the exons refer to RNA editing events. The nuclear envelope is indicated by a large dashed line. RNA binding proteins modifying stability are given by blue triangles, and miRNAs by green lines. The translating ribosome is indicated by beige circles. Nascent polypeptide is given by green interlocked circles. Degraded RNA is indicated by a gray dashed line.
Potential Points of Therapeutic Intervention
Knowledge of the processes by which mature mRNAs are expressed, processed and regulated opens up the possibility of targeting the molecule with specific interventions for future therapeutic benefit.
Therapeutic Modulation of Transcription
Therapeutic modulation of gene activity can be achieved through several mechanisms which include triplex-forming oligonucleotides (TPOs) synthetic polyamides (SPs) and artificial transcription factors (ATFs) (Uil et al., 2003). These approaches work by altering the expression level of a gene, rather than restoring its sequence per se. TPOs and SPs work by binding the major and minor groove, respectively, of the genomic DNA in specific regions of the gene, with the consequence of modulating gene activity at the level of transcription. This can be achieved by using steric hindrance to block transcription elongation for down-regulation of gene activity or conversely, blocking access to naturally occurring repressor molecules to bring about gene activation. ATFs are custom molecules designed with DNA binding domains specific to the gene in question, coupled to a trans-regulatory domain to produce the desired activity. Although there have been some promising in vitro studies, such as reactivation of the EPB41L3 gene, usually silenced by methylation, to promote tumor suppression in breast, ovarian, and cervical cell lines (Huisman et al., 2015), they have not yet reached prominence in the clinic.
Therapeutic Modification of Splicing
RNA splicing is controlled by a complex interplay between ribonucleoprotein complexes and sequence elements in the pre-mRNA. The splicing process consists of two phosphodiester transfer reactions; the first being an interaction between the 5′ splice site and the branch site, and the second comprising cleavage at the 3′ splice site, and joining of the released exons. This occurs due to the action of a family of small nuclear ribonucleoproteins (snRNPs) named U1, U2, U4, U5, and U6, which together with a battery of approximately 80 other ancillary proteins form the core spliceosome and orchestrate the splicing process (Will and Luhrmann, 2011). The spliceosome is a dynamic machine that undergoes structural remodeling and conformational change to bring about the excision of introns and the joining of introns (Makarov et al., 2002). This machinery is necessary but sometimes not sufficient for splice site usage to occur; 98% of the genome produces multiple RNA transcripts in a process termed alternative splicing (Pan et al., 2008). The precise nature of transcripts produced under different circumstances is under tight spatial and temporal regulation. This is facilitated by the combinatorial control of a series of splice site activators and inhibitor proteins that together determine whether or not a given splicing event occurs in a given circumstance. Serine Arginine rich proteins (SRSF) splicing factors usually (but not exclusively) promote splice site usage, whereas heterogeneous nuclear ribonucleoproteins (hnRNPs) usually (but not exclusively) promote splice site silencing, as well as having roles in nuclear export and other aspects of RNA metabolism (Smith and Valcarcel, 2000; Cartegni et al., 2002). Splicing defects can arise from single base pair changes to the core and regulatory sequence elements, but can also arise from insertion or deletion events and frameshifts, or from activation of cryptic splice sites by other sequence changes. Similarly, changes occurring in exon and intron splicing enhancer and silencer elements can elicit dysregulation of splicing patterns of specific genes (Blencowe, 2000). Dysregulation of the splicing regulatory machinery by cellular stress has been reported in more complex phenotypes such as cellular senescence (Holly et al., 2013; Latorre et al., 2017) and altered global alternative splicing profiles are a key characteristics of many complex diseases such as dementia, cancer and type 2 diabetes (Tollervey et al., 2011; Berson et al., 2012; Cnop et al., 2014; Love et al., 2015; Lu et al., 2015). The complexity of splicing regulation offers several points of potential intervention.
Moderation of the Core Spliceosome
The global dysregulation of splicing patterns that occur in complex disease may be addressed by targeting the core spliceosome. There are several compounds of bacterial origin that affect the function of the SF3B component of the U2 snRNP, which are showing promise as anti-cancer agents by causing stalling of the cell cycle at the G1/S or G2/M checkpoints (Nakajima et al., 1996). Although these approaches show promise, to date most remain some distance from the clinic.
Moderation of Splicing Regulation
It may be possible to globally restore splicing patterns by targeting the splicing regulatory proteins themselves. This could be done at the level of mRNA expression, or at the level of activation or cellular localization. Splicing factor expression has recently been described to be negatively regulated at the mRNA level in senescent primary human dermal fibroblasts by the constitutive activation of the ERK and AKT pathways. Targeted inhibition of either ERK or AKT, as well as gene knock down of their effector genes FOXO1 and ETV6 was associated with restoration of splicing factor expression and rescue from cellular senescence (Latorre et al., 2018). Similarly, splicing factor activity and localization is controlled at the protein level by the action of a series of kinases and phosphatases including SRPK1, SRPK2, CLK1 - CLK4, DYRK1-2, PIM1-2, and PRP4. The action of these regulators ensures the correct localization of splicing factors for action at the correct time and in the correct place. Several small molecule inhibitors of SRPK1 or SRPK2 are in development currently and show promise as anti-cancer agents for prostate malignancy in humans (Mavrou et al., 2015; Bates et al., 2017). Similarly, CLK protein kinase inhibitors have been demonstrated to suppress cell growth in human mammary tumor cell lines (Araki et al., 2015).
Moderation of Splice Site Choice
If monogenic disease is due to dysregulated splicing, in some cases it may be possible to correct or reverse the defect by restoration of correct splicing patterns. There are several means of accomplishing this, including antisense oligonucleotides (AONs), or steric hindrance agents such as morpholino oligonucleotides or similar to occlude specific splicing regulatory sequences. This potential of this approach is best exemplified by novel treatments for spinal muscular atrophy (SMA) and Duchenne Muscular dystrophy (DMD) for which therapies for manipulation of splicing have been developed and are now licensed for clinical use. SMA is characterized by progressive neuromuscular disorder caused by mutations in the Survival Motor Neuron (SMN1) gene (Lefebvre et al., 1995; Lorson et al., 1999). These are often deletion events. The human genome contains a second SMN gene, SMN2, which due to the presence of a single C-to-T transition at codon 280 which disrupts a splicing enhancer site produces an unstable SMN transcript lacking exon 7 (SMNΔ7). This transcript is present at only 10% of SMN1 levels (Lorson et al., 1999) but has potential to compensate for mutation-related reduced activity of SMN1. This has formed the basis for a novel therapeutic strategy whereby an AON (Nusinersen) has been designed to influence splicing patterns of SMN2. Nusinersen targets the N1 (ISS-N1) motif in SMN2, and promotes the inclusion of exon 7 and increases levels of compensatory SMN2. Several clinical trials have now been undertaken (Parente and Corti, 2018) and Nusinersen, also known as Spinraza, has now been approved by both US and EU regulatory authorities for clinical use.
Similar strategies have also been employed for Duchenne Muscular dystrophy, an X-linked neuromuscular disorder that affects 1:5000 newborn boys (Mendell et al., 2012), and is primarily caused by deletions, frameshift or nonsense mutations in the dystrophin (DMD) gene (Monaco et al., 1988). The majority of these mutations yield mRNAs containing premature termination codons, which trigger nonsense-mediated decay and degradation of affected DMD transcripts. Several strategies involving AONs targeted to specific splice sites have now been employed to bring about exon skipping to remove the offending exon(s) and lead to the production of a truncated, but still partially functional DMD protein (Aartsma-Rus, 2010; Niks and Aartsma-Rus, 2017). Similar approaches have been employed to modify the effects of duplication mutations in cell lines (Wein et al., 2017). Most AONs under assessment as DMD therapeutics are chemically modified 2′-O-methyl-phosphorothioate oligonucleotides (2OMePS) or phosphorodiamidate morpholino oligomers (PMOs) which can be administered systemically (Goemans et al., 2011). One of these, eteplirsen, a PMO which brings about skipping of exon 51, a hotspot for DMD mutations, has demonstrated promising results in a number of clinical trials and been designated ‘reasonably likely to predict a clinical benefit’ by the FDA (Goemans et al., 2011). Other approaches have employed ‘readthrough’ agents such as ataluren that allow bypass of the premature termination codon and are now in Phase III clinical trials (Namgoong and Bertoni, 2016).
Therapeutic Moderation of Polyadenylation
Polyadenylation is an essential step in mRNA processing, with a pivotal role in maintenance of RNA stability and management of RNA turnover. Many genes contain more than one polyadenylation site and display alternative polyadenylation, producing mRNA transcripts with novel 3′ untranslated regions. These may be differentially targeted by non-coding RNAs such as miRNAs or RNA binding proteins, or have differential translation efficiency (Elkon et al., 2013). Control of polyadenylation is mediated by a number of sequence elements such as the polyadenylation site itself, but also a series of upstream (U and UGUA rich) and downstream (U and GU rich) elements (Tian and Graber, 2012) that bind the protein complexes that orchestrate the process. These sequence elements bind the polyadenylation machinery that include the cleavage and polyadenylation specificity factors, the cleavage stimulation factors and the polyadenylate polymerase itself (Shi et al., 2009). Differential choice of polyadenylation site is linked to the proliferation and differentiation capacity of the cells; transcripts in highly proliferative cells tend to have shorter 3′UTRs (Sandberg et al., 2008). Differential use of polyadenylation sites may also have impacts on mRNA stability, mRNA export and localization, translation rates and protein localization (Tian and Graber, 2012). Patterns of alternative polyadenylation are also regulated by differential binding of RNA binding proteins; CSTF2 and CFIm subunits of the main polyadenylation machinery have been shown to have effects on relative expression of alternatively polyadenylated isoforms (Zheng and Tian, 2014). Other RNA binding proteins such as HNRNPs H and I (Katz et al., 2010), as well as CPEB1 (Bava et al., 2013) have also been associated with alternative isoform choice. RBPs such as these may in the future form the basis of therapies to influence the 3′ end processing of alternatively polyadenylated transcripts as therapeutic agents.
Therapeutic Modification of RNA Editing
RNA editing is a mechanism of generating further transcriptomic diversity and can impact the final sequence or structure of both encoded proteins and non-coding RNAs (ncRNAs) (Ganem and Lamm, 2017; Yablonovitch et al., 2017). RNA editing is an extremely common event, occurring in the many dynamically regulated mRNA transcripts and can comprise a variety of modifications, the most common of which is adenosine to inosine (A to I), which is eventually read as guanosine (Peng et al., 2012). RNA editing is especially prevalent in Small Interspersed Repetitive Elements (SINE) elements such as Alu, and also in transcripts in the brain (Jepson and Reenan, 2008; Osenberg et al., 2010). RNA editing events have been implicated in control of mRNA splicing and miRNA regulation (Farajollahi and Maas, 2010; Nishikura, 2010). RNA editing events are primarily mediated by a family of adenosine deaminases acting on RNA (ADARs), of which there are three major members; ADAR1, ADAR2, and ADAR3. The three ADARs have common functional domains, but differential structural features and some degree of site specificity (Nishikura, 2016). ADAR expression is itself regulated by transcription factors such as CREB and activated by kinases such as JNK1 (Peng et al., 2006; Yang et al., 2012). Dysfunction of ADAR1 is associated with diseases such as Aicardi-Goutières syndrome (Rice et al., 2012), with psychiatric disorders due to attenuated 5-HT2CR levels (Eran et al., 2013), and also with cancer (Ganem et al., 2017), whereas ADAR2 is linked to circadian rhythm and epilepsy (Gallo et al., 2017). Although less advanced than therapies targeting splicing defects, strategies to target ADARs to influence RNA editing are beginning to be evaluated for future clinical benefit. ADAR1 has been demonstrated to target let7, a miRNA involved in many processes including control of cell cycle (Roush and Slack, 2008). Over-expression of ADAR1 and subsequent down-regulation of Let7 has been shown to drive the self-renewal of leukemic stem cells in human blood, an observation that can be reversed by inhibition of ADAR1-mediated RNA editing (Zipeto et al., 2016). Very recently, techniques for directing ADARs to specific points of intervention have been developed. This system, named RESTORE, uses a plasmid-borne guide RNA coupled to an ADAR recruiting domain to deliver ADAR2 directly to the region of interest. This approach has been used to successfully edit phosphotyrosine residues in STAT1 with resultant changes to the activity of this signaling protein (Merkle et al., 2019). There are also newly emerging techniques based on modified Cas technologies utilizing catalytically inactive Cas13-ADAR2 fusion proteins to bring about RNA editing (Cox et al., 2017). These early observations suggest that in the future, targeting ADARs or other regulators of RNA editing may prove promising points of traction for neurodevelopmental disorders and for cancer.
Modification of RNA Based Epitranscriptomics
Epitranscriptomic modification of DNA is well known, but it is now becoming increasingly evident that RNA is also epigenetically modified. RNA is subject to decoration with over 130 different modifications. Most of these map to very abundant RNAs such as rRNAs and tRNAs, but a subset are seen in mRNA, circRNA and lncRNA (Schaefer et al., 2017). The most common marks are N(6)-methyl-adenosine (m6A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C) and N1-methyladenosine (m1A), which have been shown to be widely present throughout the transcriptome by high throughput sequencing (Jung and Goldman, 2018). M6A is enriched in the last exon of genes and also occurs preferentially at 5′ untranslated regions (UTRs) (Ke et al., 2015; Meyer et al., 2015), whereas m1A is enriched in promoters and 5′ UTRs (Dominissini et al., 2016). m5C marks are often located at both 5′ and 3′ UTRs (Squires et al., 2012). RNA modifications can influence gene expression by a number of mechanisms, including influencing RNA structure, recruiting other regulatory proteins (e.g., splicing factors, RNA binding proteins involved in control of stability) or moderation of translation (Nachtergaele, 2017). RNA epitranscriptomic marks are added and removed by a series of writers (METTL3, METTL14, WTAP, KIAA1429, RBM15/15B, and METTL16) and erasers (FTO and ALKBH5) (Tong et al., 2018). Disruption of m6A disrupts RNA metabolism; m6A depleted transcripts have been reported to be unstable (Tang et al., 2018). Accordingly, mutations in the writer or eraser machinery have been associated with cancers such as hepatocellular carcinoma and acute myeloid leukemia (AML) (Vu et al., 2017; Chen et al., 2018), and with memory, fertility and metabolic phenotypes (Fischer et al., 2009; Zheng et al., 2013; Nainar et al., 2016). The RNA epigenomic writers and erasers are therefore promising future therapeutic targets. At present, the work in this area is mainly in cell and animal models. Silencing the METTL14 ‘writer’ led to restoration of differentiation of myeloid cells in AML and inhibited AML cell survival and proliferation (Weng et al., 2018). Similar strategies targeting ALKBH5 have showed promise as anti-tumor agents in glioblastoma stem cells (Schonberg et al., 2015). Studies have suggested that small molecule inhibitors of FTO may have potential utility as anticonvulsants in mouse models of epilepsy in vivo, by suppression of 2-oxoglutarate (2OG) through altering m6A levels (Zheng et al., 2014).
Modulation of RNA Export
The activity of genes is also dependent on the correct positioning of mRNAs within the cell. Once processed, RNAs are usually exported through the nuclear membrane into the cytoplasm ready to be translated. This is not a passive process; it is orchestrated by a portfolio of RNA export proteins which escort the RNA molecule through the nuclear pore. Messenger RNAs are primarily transported by Nxf1 and Xpo1, whereas miRNAs are exported by Xpot and Xpo5. The transcription Export complex 1 (Trex1) facilitates binding of Nxf1 to the processed mRNA, and together with a collection of other proteins such as karyopherins or importins causes the processed mRNA to associate with and transit through the nuclear pore (Viphakone et al., 2012). The nuclear pore itself is composed of a collection of nucleoporins, and comprises a multi-subunit structure consisting of a nuclear ring, a central transport channel and a basket-like structure (Kabachinski and Schwartz, 2015). Small molecules can diffuse across this barrier, but larger ones such an mRNA cannot. Some of the specificity of transport is achieved by the interaction of the nuclear transport machinery with specific signal sequences in the mRNA itself (Lee et al., 2006; Hutten and Kehlenbach, 2007), whereas other mRNAs rely upon adaptor proteins (Huang et al., 2017). The expression and localization of nuclear transporters is altered in certain cancers (Zhou et al., 2013; Talati and Sweet, 2018), and have been linked with some neurodegenerative disorders (Grima et al., 2017) and comprises important components of inflammatory and apoptotic response (Aggarwal and Agrawal, 2014; Kopeina et al., 2018). Individual components of the nuclear export machinery are currently under investigation as therapeutics. One of the most promising, Selinexor, targets exportin 1 (Xpo1) and is currently in pre-clinical trials and has shown efficacy against acute myeloid leukemia and multiple myeloma (Kashyap et al., 2016; Mahipal and Malafa, 2016).
Therapeutic Modulation of Non-coding RNA Regulators of Gene Expression
The repertoire of genes expressed by any given cell in any given circumstances is influenced by non-coding RNA (ncRNA) regulators of gene expression. These ncRNA genes do not encode proteins, but rather encode RNAs that contribute to the regulation of other RNAs. They are classified into 2 broad classes, short ncRNAs such as microRNAs (miRNAs) and longer ncRNAs such as long non-coding RNAS (lncRNAs) and circular RNAs (circRNAs).
Modulation of Small Non-coding RNAs
MicroRNAs (miRNAs) and siRNAs are short non-coding RNAs 20–25 bp in size. They interact with components of the RNA-induced silencing complex (RISC) to bring about translational blocking or RNA degradation. Each miRNA interacts with specific binding sites in the 3′ UTR of its target genes, which are 6–8 nt in length and are commonly found in the genome; each miRNA is thus capable of targeting hundreds of mRNA target genes simultaneously (Carthew and Sontheimer, 2009). Several classes of miRNAs have been associated with disease; these include the 17/92 cluster, the miR-24 cluster or miR-3676, all of which are associated with chronic lymphocytic leukemia (CLL) (Van Roosbroeck and Calin, 2016). Other examples include miR-21, miR-10b, miR-155, and Let-7a, which are associated with breast cancer (Khalighfard et al., 2018), and miR-192, miR200c and miR-17 which are associated with colon cancer (Ast et al., 2018). Similarly miR-33a and miR-33 have been associated with metabolic disease and atherosclerosis (Marquart et al., 2010; Rayner et al., 2011) and miR-155 has links with inflammatory diseases (Dorsett et al., 2008). The use of miRNAs as anti-tumor therapeutics is currently receiving much interest. Specific miRNAs can target the tumor suppressor machinery, and are commonly referred to as onco-miRs, or they may target the controls of cell cycle and act as tumor suppressors in their own right. The small size and relative stability of miRNAs and siRNAs, together with the observation that they are readily taken up in endosomes and microvesicles (Rani et al., 2017) renders them excellent candidates for therapeutic modulation or use as biomarkers of disease. This can take the form of antagomiRs that can target and silence endogenous miRNAs or chemically modified miRNA mimics that can increase regulation of their specific targets (Khvorova and Watts, 2017). To date, 20 clinical trials have been undertaken that exploit miRNA biology (Chakraborty et al., 2017), the first of which miravirsen, which is targeted to miR-122, is in phase II clinical trials for Hepatitis C (Lindow and Kauppinen, 2012). In August 2018, the first siRNA-related therapy, patisiran, was approved by the FDA for the treatment of peripheral nerve disease by targeting an abnormal form of the transthyretin (TTR) gene.
Modulation of LncRNAs
Long non-coding RNAs comprise a heterogeneous class of non-coding RNAs, which are longer than 200bp in length. They do not encode proteins, and originate from all most genomic regions. They can originate from the locus that they regulate, usually from the antisense strand, and regulate their target in cis (Natural antisense Transcripts (NATs), or they can map to entirely different genomic regions form their targets (introns, pseudogenes, and non-coding DNA) and cause regulation in trans. LncRNAs can also be associated with promoters, enhancers or other regulatory regions and do not have a homogeneous mode of action. They can activate or repress their targets and can work by a number of mechanisms. They are commonly involved in genomic imprinting; one of the first lncRNAs discovered, XIST, coordinates X chromosome inactivation (Brown et al., 1991). Other lncRNAs can act as guides. This class of lncRNA includes ANRIL, which directs the polycomb repressive complex to the site of action in the case of the CDKN2A and CDKN2B genes (Kotake et al., 2011) and the lncRNA HOTAIR, which has roles in colorectal cancer (Kogo et al., 2011). They can also act as scaffolds, directing the assembly of specific protein or RNA complexes to their sites of action. For example, one function of the lncRNA NEAT1, a multifunctional lncRNA with several roles in tumorigenesis (Ghaforui-Fard and Taheri, 2018) is to bring together the microRNA biogenesis machinery to enhance pri-miRNA processing (Jiang et al., 2017), and the lncRNA LINP1, which regulates the repair of DNA double strand breaks in breast cancer by acting as a scaffold for the ku80 and DNA-dependent protein kinase proteins (Zhang Y. et al., 2016). They can also repress expression by acting as decoys, co-regulators and inhibitors of RNA polymerase II. For example, the lncRNA PANDA acts by sequestering its transcription factor target NF-YA away from its site of action (Hung et al., 2011). They have roles in regulators of subcellular compartmentalization; the lncRNA MALAT is responsible for localizing splicing factors to the nuclear splicing speckles where they can be stored and regulated by phosphorylation (Bernard et al., 2010).
In accordance with their pivotal role in regulating gene expression, lncRNAs have been reported to be associated with several diseases such as cancer (Huarte, 2015; Parasramka et al., 2016; Peng et al., 2016), diabetes (Akerman et al., 2017; He et al., 2017; Leti and DiStefano, 2017), neurodegenerative disease (Riva et al., 2016) and cardiovascular disease (Hou et al., 2016; Haemmig et al., 2017; Gangwar et al., 2018). LncRNAs may represent promising therapeutic targets; they are responsive to small molecule therapeutics; a recent study documented 5916 lncRNAs that responded to 1262 small molecule drugs (Yang et al., 2017). Although progress toward the clinic has been slow, perhaps because of the diverse modes of actions of lncRNAs, there are some promising candidates. Several lncRNAs have been reported to be dysregulated in osteoarthritis (OA), including HOTAIR, RP11-445H22.4, GAS5, PMS2L2, H19, and CTD-2574D22.4 (Xing et al., 2014). At the present time, the majority of studies have not progressed beyond cell or animal models, several potential future therapeutic candidates have emerged; the lncRNA PCGEM1 was demonstrated to inhibit synoviocyte apoptosis on OA by moderation of its target miR-770 (Kang et al., 2016). Similarly, many lncRNAs have been identified as potential therapeutic targets in cardiovascular disease or cancer, including GAS5, LIPCAR, SENCR, ANRIL, SMILR, and MALAT (Gomes et al., 2017). ASP and siRNA approaches to therapeutically manipulate MALAT levels are in development in human cancer cells and in animal models (Arun et al., 2016). Targeting lncRNAs is subject to more difficulty than miRNAs, because of their larger size and the heterogeneity of their mode of action, which may explain why their evaluation is not as advanced as that of miRNAs. Nevertheless, they have significant potential as future therapeutic targets.
Modulation of CircRNAs
Circular RNAs (circRNAs) are a relatively newly discovered class of non-coding RNA regulators found in multiple species (Haque and Harries, 2017). They are formed from ‘backsplicing’ events of linear genes, and comprise circular molecules, which are therefore relatively immune to exonucleases (Cocquerelle et al., 1993; Schwanhausser et al., 2011; Jeck et al., 2013; Lan et al., 2016; Lasda and Parker, 2016). Like lncRNAs, circRNAs have been reported to influence gene expression by a variety of mechanisms including action as miRNA sponges or mRNA traps, as well as comprising modifiers of transcription. translation, or splicing (Haque and Harries, 2017). Circular RNAs have been suggested to have roles in many cellular processes, including embryonic development (Xia et al., 2016), metabolism (Xu et al., 2015), regulation of cell cycle (Zheng et al., 2016) and regulation of cellular stress (Burd et al., 2010). In accordance with this observation, dysregulated circRNA expression has been associated with multiple human diseases such as cancer (Yao et al., 2017), neurological disease (Khoutorsky et al., 2013), osteoarthritis (Liu et al., 2016), cardiovascular disease (Taibi et al., 2014; Wang et al., 2016), type 2 diabetes (Gu et al., 2017), pre-eclampsia (Zhang Y.G. et al., 2016) and impaired immune responses (Ng et al., 2016). Although the study of circRNAs is in its infancy compared with other ncRNAs, they too have potential as future therapeutic targets.
Remaining Barriers and Future Prospects
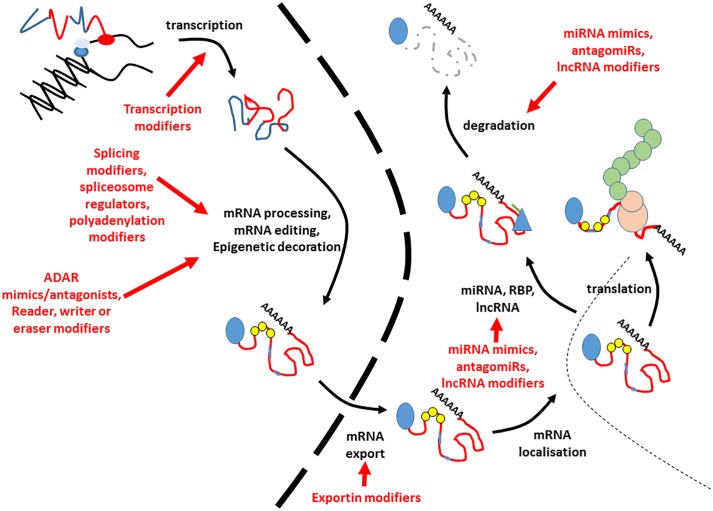
This is an exciting time for RNA-based therapeutics, with several notable examples making it as far as license for clinical usage. Over the next decade, it is likely that there will be a large expansion in the breadth and scope of human disorders that can be treated using these, and similar approaches. Most developed at the present time, are interventions targeted at specific splice events and those involving small RNAs, but future work may harness the potential of targeting other parts of the RNA regulatory milieu (Figure 2).
FIGURE 2.
Potential points of intervention for RNA based therapies. This figure indicates the potential points at which interventions could be made to alter the amount or nature of expressed RNA. Blue lines in the transcript refer to introns and untranslated regions, whilst exons are indicted by red lines. The 5′ cap is indicated by a blue circle. Small yellow circles indicate epitranscriptomic decoration, whilst pale blue lines within the exons refer to RNA editing events. The nuclear envelope is indicated by a large dashed line. RNA binding proteins modifying stability are given by blue triangles, and miRNAs by green lines. The translating ribosome is indicated by beige circles. Nascent polypeptide is given by green interlocked circles. Each potential point of intervention is given by a red arrow. Degraded RNA is indicated by a gray dashed line.
Several barriers do, however, remain to the wide implementation of these opportunities which are focused mainly on delivery, specificity and duration of treatment. Firstly, delivery of specific molecules to their site of action may be challenging. For some applications, such as skin, which may be treated topically or lung, which may be treated via inhalation, therapeutic delivery of interventions may be easier. Delivery to internal organs such as brain, liver or pancreas will require different and systemic approaches. One reason why AONs, readthrough agents and small RNAs have been at the forefront of this emerging field is that their small size and relative stability means that they can be more easily introduced into cells. This may not be true of entities such as lncRNAs or large circRNAs, which may be large molecules with potentially challenging secondary or tertiary structure. Small molecules can readily be introduced into cells using lipid-mediated transfer agents, or endogenous structures such as endosomes or microvesicles, which could be harnessed to deliver cargoes. Secondly, there are questions of specificity. One feature of the therapies that are in clinic currently is their specificity to their sites of action. Gene expression and the regulation thereof is highly tissue specific, and genes may often be required to be expressed only at a specified time, or in response to specific circumstances. It may not be advantageous to produce changes in all tissues or at all times, and effects must of course be limited to their intended targets. Specificity of effect can be achieved by choosing targets that are only present at their sites of action, or by modifying delivery so that cargoes are only delivered to their intended place of action. For example, strategies are emerging now which allow selective delivery of senolytic cargoes to senescent cells only using galactosaccharide nanoparticles, which harness the observation that senescent cells harbor large quantities of lysosomal β-galactosidase (Munoz-Espin et al., 2018). Similarly, strategies could be developed that introduce therapeutic oligonucleotides under the control of gene regulatory elements expressed only in the intended target tissues. Lastly, one needs to consider the potential need for repeated treatments. The approaches discussed here differ from emerging “gene editing” technologies such as CRISPR, in that they are not transmitted to future generations, and may require repeated treatments. This can be considered both a caveat and an advantage. The need for repeated treatments may be burdensome for patients, but in reality, the vast majority of currently available treatments for human disorders fall into this category. Conversely, the need to deliver repeated doses introduces a degree of flexibility, and allows treatments to be quickly discontinued or changed if adverse effects occur. We are at a time of huge advances in our understanding of how our genome is curated and regulated and how our genes are expressed.
The multifactorial control of gene expression, and the complexity of this progress offers multiple points of potential intervention for therapeutic benefit. Over the coming decades, there is likely to be a huge increase in the number of therapies for human diseases that target not the genes themselves, but the expression and regulation of those genes. We are at the dawning of the era of genomic medicine, and the future looks bright.
Author Contributions
LH planned and wrote the manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The Harries lab is funded by the Dunhill Medical Trust (grant number R386/1114).
References
- Aartsma-Rus A. (2010). Antisense-mediated modulation of splicing: therapeutic implications for Duchenne muscular dystrophy. RNA Biol. 7 453–461. [DOI] [PubMed] [Google Scholar]
- Aggarwal A., Agrawal D. K. (2014). Importins, and exportins regulating allergic immune responses. Med. Inflamm. 2014:476357. 10.1155/2014/476357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman I., Tu Z., Beucher A., Rolando D. M. Y., Sauty-Colace C., Benazra M., et al. (2017). Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab. 25 400–411. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S., Dairiki R., Nakayama Y., Murai A., Miyashita R., Iwatani M., et al. (2015). Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PLoS One 10:e0116929. 10.1371/journal.pone.0116929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G., Diermeier S., Akerman M., Chang K. C., Wilkinson J. E., Hearn S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30 34–51. 10.1101/gad.270959.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast V., Kordass T., Oswald M., Kolte A., Eisel D., Osen W., et al. (2018). MiR-192, miR-200c and miR-17 are fibroblast-mediated inhibitors of colorectal cancer invasion. Oncotarget 9 35559–35580. 10.18632/oncotarget.26263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. O., Morris J. C., Oltean S., Donaldson L. F. (2017). Pharmacology of modulators of alternative splicing. Pharmacol. Rev. 69 63–79. 10.1124/pr.115.011239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bava F. A., Eliscovich C., Ferreira P. G., Minana B., Ben-Dov C., Guigo R., et al. (2013). CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495 121–125. 10.1038/nature11901 [DOI] [PubMed] [Google Scholar]
- Beaulieu C. L., Majewski J., Schwartzentruber J., Samuels M. E., Fernandez B. A., Bernier F. P., et al. (2014). FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 94 809–817. 10.1016/j.ajhg.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. (2002). The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14 336–342. [DOI] [PubMed] [Google Scholar]
- Bernard D., Prasanth K. V., Tripathi V., Colasse S., Nakamura T., Xuan Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29 3082–3093. 10.1038/emboj.2010.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson A., Barbash S., Shaltiel G., Goll Y., Hanin G., Greenberg D. S., et al. (2012). Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 4 730–742. 10.1002/emmm.201100995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. (1988). Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2 754–765. [DOI] [PubMed] [Google Scholar]
- Blencowe B. J. (2000). Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25 106–110. [DOI] [PubMed] [Google Scholar]
- Borbolis F., Syntichaki P. (2015). Cytoplasmic mRNA turnover and ageing. Mech. Ageing Dev. 152 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Lafreniere R. G., Powers V. E., Sebastio G., Ballabio A., Pettigrew A. L., et al. (1991). Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349 82–84. [DOI] [PubMed] [Google Scholar]
- Burd C. E., Jeck W. R., Liu Y., Sanoff H. K., Wang Z., Sharpless N. E. (2010). Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6:e1001233. 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Chew S. L., Krainer A. R. (2002). Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3 285–298. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A. R., Sharma G., Doss C. G. P., Lee S. S. (2017). Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 8 132–143. 10.1016/j.omtn.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. (2013). Characterization and comparison of human nuclear and cytosolic editomes. Proc. Natl. Acad. Sci. U.S.A. 110 E2741–E2747. 10.1073/pnas.1218884110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wei L., Law C. T., Tsang F. H., Shen J., Cheng C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67 2254–2270. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- Chen W., Jia Q., Song Y., Fu H., Wei G., Ni T. (2017). Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics 15 287–300. 10.1016/j.gpb.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnop M., Abdulkarim B., Bottu G., Cunha D. A., Igoillo-Esteve M., Masini M., et al. (2014). RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 63 1978–1993. 10.2337/db13-1383 [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. (1993). Mis-splicing yields circular RNA molecules. FASEB J. 7 155–160. [DOI] [PubMed] [Google Scholar]
- Cox D. B. T., Gootenberg J. S., Abudayyeh O. O., Franklin B., Kellner M. J., Joung J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358 1019–1027. 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M. S., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530 441–446. 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., McBride K. M., Jankovic M., Gazumyan A., Thai T. H., Robbiani D. F., et al. (2008). MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 28 630–638. 10.1016/j.immuni.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. Y., Elgart A., Farrell C., Barnett-Griness O., Rabinovich-Guilatt L., Spiegelstein O. (2017). A population pharmacokinetic meta-analysis of custirsen, an antisense oligonucleotide, in oncology patients and healthy subjects. Br. J. Clin. Pharmacol. 83 1932–1943. 10.1111/bcp.13287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Ugalde A. P., Agami R. (2013). Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14 496–506. 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- Eran A., Li J. B., Vatalaro K., McCarthy J., Rahimov F., Collins C., et al. (2013). Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry 18 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajollahi S., Maas S. (2010). Molecular diversity through RNA editing: a balancing act. Trends Genet. 26 221–230. 10.1016/j.tig.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlin R. A. (2015). Personalized immunotherapy ( AGS-003 ) when combined with sunitinib for the treatment of metastatic renal cell carcinoma. Expert Opin. Biol. Ther. 15 1241–1248. 10.1517/14712598.2015.1063610 [DOI] [PubMed] [Google Scholar]
- Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Bruning J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458 894–898. 10.1038/nature07848 [DOI] [PubMed] [Google Scholar]
- Fukao A., Aoyama T., Fujiwara T. (2015). The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 12 922–926. 10.1080/15476286.2015.1073436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao A., Fujiwara T. (2017). The coupled and uncoupled mechanisms by which trans-acting factors regulate mRNA stability and translation. J. Biochem. 161 309–314. 10.1093/jb/mvw086 [DOI] [PubMed] [Google Scholar]
- Gallo A., Vukic D., Michalik D., O’Connell M. A., Keegan L. P. (2017). ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 136 1265–1278. 10.1007/s00439-017-1837-0 [DOI] [PubMed] [Google Scholar]
- Ganem N. S., Ben-Asher N., Lamm A. T. (2017). In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic. Drug Resist. Updat. 32 16–22. 10.1016/j.drup.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Ganem N. S., Lamm A. T. (2017). A-to-I RNA editing - thinking beyond the single nucleotide. RNA Biol. 14 1690–1694. 10.1080/15476286.2017.1364830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar R. S., Rajagopalan S., Natarajan R., Deiuliis J. A. (2018). Noncoding RNAs in cardiovascular disease: pathological relevance and emerging role as biomarkers and therapeutics. Am. J. Hypertens. 31 150–165. 10.1093/ajh/hpx197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaforui-Fard S., Taheri M. (2018). Nuclear Enriched Abundant Transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis. Biomed. Pharmacother. 111 51–59. 10.1016/j.biopha.2018.12.070 [DOI] [PubMed] [Google Scholar]
- Goemans N. M., Tulinius M., van den Akker J. T., Burm B. E., Ekhart P. F., Heuvelmans N., et al. (2011). Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 364 1513–1522. 10.1056/NEJMoa1011367 [DOI] [PubMed] [Google Scholar]
- Gomes C. P. C., Spencer H., Ford K. L., Michel L. Y. M., Baker A. H., Emanueli C., et al. (2017). The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol. Ther. Nucleic Acids 8 494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B., Marshall E., McFarland M. R., Romano M. C., Stansfield I. (2014). Controlling translation elongation efficiency: tRNA regulation of ribosome flux on the mRNA. Biochem. Soc. Trans. 42 160–165. 10.1042/BST20130132 [DOI] [PubMed] [Google Scholar]
- Grima J. C., Daigle J. G., Arbez N., Cunningham K. C., Zhang K., Ochaba J., et al. (2017). Mutant huntingtin disrupts the nuclear pore complex. Neuron 94 93–107.e6. 10.1016/j.neuron.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Ke G., Wang L., Zhou E., Zhu K., Wei Y. (2017). Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalm. Res. 58 176–184. 10.1159/000479156 [DOI] [PubMed] [Google Scholar]
- Haemmig S., Simion V., Yang D., Deng Y., Feinberg M. W. (2017). Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr. Opin. Cardiol. 32 776–783. 10.1097/HCO.0000000000000454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque S., Harries L. W. (2017). Circular RNAs (circRNAs) in health and disease. Genes 8 E353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y., Parker R. (2010). No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA 1 132–141. 10.1002/wrna.17 [DOI] [PubMed] [Google Scholar]
- He X., Ou C., Xiao Y., Han Q., Li H., Zhou S. (2017). LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget 8 71325–71341. 10.18632/oncotarget.19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Motorin Y. (2017). Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18 275–291. 10.1038/nrg.2016.169 [DOI] [PubMed] [Google Scholar]
- Holly A. C., Melzer D., Pilling L. C., Fellows A. C., Tanaka T., Ferrucci L., et al. (2013). Changes in splicing factor expression are associated with advancing age in man. Mech. Ageing Dev. 134 356–366. 10.1016/j.mad.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Zhou C., Long H., Zheng S., Guo T., Wu Q., et al. (2016). Long noncoding RNAs: novel molecules in cardiovascular biology, disease and regeneration. Exp. Mol. Pathol. 100 493–501. 10.1016/j.yexmp.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Huang J. H., Ku W. C., Chen Y. C., Chang Y. L., Chu C. Y. (2017). Dual mechanisms regulate the nucleocytoplasmic localization of human DDX6. Sci. Rep. 7:42853. 10.1038/srep42853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21 1253–1261. [DOI] [PubMed] [Google Scholar]
- Huisman C., van der Wijst M. G., Falahi F., Overkamp J., Karsten G., Terpstra M. M., et al. (2015). Prolonged re-expression of the hypermethylated gene EPB41L3 using artificial transcription factors and epigenetic drugs. Epigenetics 10 384–396. 10.1080/15592294.2015.1034415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43 621–629. 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Kehlenbach R. H. (2007). CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17 193–201. [DOI] [PubMed] [Google Scholar]
- Iadevaia V., Gerber A. P. (2015). Combinatorial control of mRNA Fates by RNA-binding proteins and non-coding RNAs. Biomolecules 5 2207–2222. 10.3390/biom5042207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson J. E., Reenan R. A. (2008). RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta 1779 459–470. [DOI] [PubMed] [Google Scholar]
- Jiang L., Shao C., Wu Q. J., Chen G., Zhou J., Yang B., et al. (2017). NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 24 816–824. 10.1038/nsmb.3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Goldman D. (2018). Role of RNA modifications in brain and behavior. Genes Brain Behav. 17:e12444. 10.1111/gbb.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabachinski G., Schwartz T. U. (2015). The nuclear pore complex–structure and function at a glance. J. Cell Sci. 128 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Song J., Kim D., Ahn C., Park S., Chun C. H., et al. (2016). PCGEM1 stimulates proliferation of osteoarthritic synoviocytes by acting as a sponge for miR-770. J. Orthop. Res. 34 412–418. 10.1002/jor.23046 [DOI] [PubMed] [Google Scholar]
- Kashyap T., Argueta C., Aboukameel A., Unger T. J., Klebanov B., Mohammad R. M., et al. (2016). Selinexor, a selective inhibitor of nuclear export (SINE) compound, acts through NF-kappaB deactivation and combines with proteasome inhibitors to synergistically induce tumor cell death. Oncotarget 7 78883–78895. 10.18632/oncotarget.12428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E. T., Airoldi E. M., Burge C. B. (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Alemu E. A., Mertens C., Gantman E. C., Fak J. J., Mele A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′. UTR regulation. Genes Dev. 29 2037–2053. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalighfard S., Alizadeh A. M., Irani S., Omranipour R. (2018). Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci. Rep. 8:17981. 10.1038/s41598-018-36321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A., Yanagiya A., Gkogkas C. G., Fabian M. R., Prager-Khoutorsky M., Cao R., et al. (2013). Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron 78 298–311. 10.1016/j.neuron.2013.02.025 [DOI] [PubMed] [Google Scholar]
- Khvorova A., Watts J. K. (2017). The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35 238–248. 10.1038/nbt.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauer A. A., van Hoof A. (2012). Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA 3 649–660. 10.1002/wrna.1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71 6320–6326. 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- Kopeina G. S., Prokhorova E. A., Lavrik I. N., Zhivotovsky B. (2018). Alterations in the nucleocytoplasmic transport in apoptosis: caspases lead the way. Cell Prolif. 51:e12467. 10.1111/cpr.12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., et al. (2011). Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30 1956–1962. 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P. H., Liu Z. H., Pei Y. J., Wu Z. G., Yu Y., Yang Y. F., et al. (2016). Landscape of RNAs in human lumbar disc degeneration. Oncotarget 7 63166–63176. 10.18632/oncotarget.11334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E., Parker R. (2016). Circular RNAs Co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One 11:e0148407. 10.1371/journal.pone.0148407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E., Birar V. C., Sheerin A. N., Jeynes J. C. C., Hooper A., Dawe H. R., et al. (2017). Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence. BMC Cell Biol. 18:31. 10.1186/s12860-017-0147-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E., Ostler E. O., Faragher R. G. A., Harries L. W. (2018). FOXO1 and ETV6 genes may represent novel regulators of splicing factor expression in cellular senescence. FASEB J. 33 1086–1097. 10.1096/fj.201801154R [DOI] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Suel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80 155–165. [DOI] [PubMed] [Google Scholar]
- Lejeune F. (2017). Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep. 50 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leti F., DiStefano J. K. (2017). Long Noncoding RNAs as Diagnostic and Therapeutic Targets in Type 2 Diabetes and Related Complications. Genes 8 E207. 10.3390/genes8080207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow M., Kauppinen S. (2012). Discovering the first microRNA-targeted drug. J. Cell Biol. 199 407–412. 10.1083/jcb.201208082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J., et al. (2016). Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci. Rep. 6:22572. 10.1038/srep22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson C. L., Hahnen E., Androphy E. J., Wirth B. (1999). A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 96 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J. E., Hayden E. J., Rohn T. T. (2015). Alternative splicing in Alzheimer’s disease. J. Parkinsons Dis. Alzheimers Dis. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. X., Huang Q., Park J. W., Shen S., Lin L., Tokheim C. J., et al. (2015). Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res. 13 305–318. 10.1158/1541-7786.MCR-14-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahipal A., Malafa M. (2016). Importins, and exportins as therapeutic targets in cancer. Pharmacol. Ther. 164 135–143. [DOI] [PubMed] [Google Scholar]
- Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., et al. (2002). Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298 2205–2208. [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Brooks L. D., Collins F. S. (2008). A HapMap harvest of insights into the genetics of common disease. J. Clin. Investig. 118 1590–1605. 10.1172/JCI34772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart T. J., Allen R. M., Ory D. S., Baldan A. (2010). miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. U.S.A. 107 12228–12232. 10.1073/pnas.1005191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrou A., Brakspear K., Hamdollah-Zadeh M., Damodaran G., Babaei-Jadidi R., Oxley J., et al. (2015). Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene 34 4311–4319. 10.1038/onc.2014.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. R., Shilling C., Leslie N. D., Flanigan K. M., al-Dahhak R., Gastier-Foster J., et al. (2012). Evidence-based path to newborn screening for duchenne muscular dystrophy. Ann. Neurol. 71 304–313. 10.1002/ana.23528 [DOI] [PubMed] [Google Scholar]
- Merkle T., Merz S., Reautschnig P., Blaha A., Li Q., Vogel P., et al. (2019). Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37 133–138. 10.1038/s41587-019-0013-6 [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′ UTR m(6)A promotes cap-independent translation. Cell 163 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A. P., Bertelson C. J., Liechti-Gallati S., Moser H., Kunkel L. M. (1988). An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2 90–95. [DOI] [PubMed] [Google Scholar]
- Munoz-Espin D., Rovira M., Galiana I., Gimenez C., Lozano-Torres B., Paez-Ribes M., et al. (2018). A versatile drug delivery system targeting senescent cells. EMBO Mol. Med. 10:e9355. 10.15252/emmm.201809355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S. (2017). He C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 14 156–163. 10.1080/15476286.2016.1267096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainar S., Marshall P. R., Tyler C. R., Spitale R. C., Bredy T. W. (2016). Evolving insights into RNA modifications and their functional diversity in the brain. Nat. Neurosci. 19 1292–1298. 10.1038/nn.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Hori Y., Terano H., Okuhara M., Manda T., Matsumoto S., et al. (1996). New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. 49 1204–1211. [DOI] [PubMed] [Google Scholar]
- Namgoong J. H., Bertoni C. (2016). Clinical potential of ataluren in the treatment of Duchenne muscular dystrophy. Degener. Neurol. Neuromuscul. Dis. 6 37–48. 10.2147/DNND.S71808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L., Marinov G. K., Liau E. S., Lam Y. L., Lim Y. Y., Ea C. K. (2016). Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 13 861–871. 10.1080/15476286.2016.1207036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niks E. H., Aartsma-Rus A. (2017). Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 17 225–236. 10.1080/14712598.2017.1271872 [DOI] [PubMed] [Google Scholar]
- Nishikura K. (2010). Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79 321–349. 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17 83–96. 10.1038/nrm.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S., Paz Yaacov N., Safran M., Moshkovitz S., Shtrichman R., Sherf O., et al. (2010). Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One 5:e11173. 10.1371/journal.pone.0011173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40 1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Parasramka M. A., Maji S., Matsuda A., Yan I. K., Patel T. (2016). Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 161 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente V., Corti S. (2018). Advances in spinal muscular atrophy therapeutics. Ther. Adv. Neurol. Disord. 11:1756285618754501. 10.1177/1756285618754501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yuan X., Jiang B., Tang Z. (2016). Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 37 2779–2788. 10.1007/s13277-015-4663-9 [DOI] [PubMed] [Google Scholar]
- Peng P. L., Zhong X., Tu W., Soundarapandian M. M., Molner P., Zhu D., et al. (2006). ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 49 719–733. [DOI] [PubMed] [Google Scholar]
- Peng Z., Cheng Y., Tan B. C., Kang L., Tian Z., Zhu Y., et al. (2012). Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30 253–260. 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- Rani A., O’Shea A., Ianov L., Cohen R. A., Woods A. J., Foster T. C. (2017). miRNA in circulating microvesicles as biomarkers for age-related cognitive decline. Front. Aging Neurosci. 9:323. 10.3389/fnagi.2017.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner K. J., Sheedy F. J., Esau C. C., Hussain F. N., Temel R. E., Parathath S., et al. (2011). Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 121 2921–2931. 10.1172/JCI57275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G. I., Kasher P. R., Forte G. M., Mannion N. M., Greenwood S. M., Szynkiewicz M., et al. (2012). Mutations in ADAR1 cause aicardi-goutieres syndrome associated with a type I interferon signature. Nat. Genet. 441243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva P., Ratti A., Venturin M. (2016). The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 13 1219–1231. [DOI] [PubMed] [Google Scholar]
- Rizk M., Tuzmen S. (2017). Update on the clinical utility of an RNA interference-based treatment: focus on Patisiran. Pharmgenomics Pers. Med. 10 267–278. 10.2147/PGPM.S87945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S., Slack F. J. (2008). The let-7 family of microRNAs. Trends Cell Biol. 18 505–516. [DOI] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008). Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science 320 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Kapoor U., Jantsch M. F. (2017). Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 7:170077. 10.1098/rsob.170077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg D. L., Miller T. E., Wu Q., Flavahan W. A., Das N. K., Hale J. S., et al. (2015). Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell 28 441–455. 10.1016/j.ccell.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Shi Y., Di Giammartino D. C., Taylor D., Sarkeshik A., Rice W. J., Yates J. R., III, et al. (2009). Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33 365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Valcarcel J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25 381–388. [DOI] [PubMed] [Google Scholar]
- Sperling R. (2017). The nuts and bolts of the endogenous spliceosome. Wiley Interdiscip. Rev. RNA 8 e1377. 10.1002/wrna.1377 [DOI] [PubMed] [Google Scholar]
- Squires J. E., Patel H. R., Nousch M., Sibbritt T., Humphreys D. T., Parker B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B. (2018). RNA localization and transport. Biochim. Biophys. Acta Gene Regul. Mech. 1861 938–951. [DOI] [PubMed] [Google Scholar]
- Taibi F., Metzinger-Le Meuth V., Massy Z. A., Metzinger L. (2014). miR-223: an inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta 1842 1001–1009. 10.1016/j.bbadis.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Talati C., Sweet K. L. (2018). Nuclear transport inhibition in acute myeloid leukemia: recent advances and future perspectives. Int. J. Hematol. Oncol. 7:IJH04. 10.2217/ijh-2018-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 115 E325–E333. 10.1073/pnas.1717794115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Graber J. H. (2012). Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA 3 385–396. 10.1002/wrna.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J. R., Wang Z., Hortobagyi T., Witten J. T., Zarnack K., Kayikci M., et al. (2011). Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 21 1572–1582. 10.1101/gr.122226.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Flavell R. A., Li H. B. (2018). RNA m(6)A modification and its function in diseases. Front. Med. 12:481–489. 10.1007/s11684-018-0654-8 [DOI] [PubMed] [Google Scholar]
- Uil T. G., Haisma H. J., Rots M. G. (2003). Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities. Nucleic Acids Res. 31 6064–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roosbroeck K., Calin G. A. (2016). MicroRNAs in chronic lymphocytic leukemia: miRacle or miRage for prognosis and targeted therapies? Semin. Oncol. 43 209–214. 10.1053/j.seminoncol.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N., Hautbergue G. M., Walsh M., Chang C. T., Holland A., Folco E. G., et al. (2012). TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 3:1006. 10.1038/ncomms2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L. P., Pickering B. F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23 1369–1376. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Winkler G. S. (2013). RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 1829 561–570. 10.1016/j.bbagrm.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J. X., Liu C. Y., Zhao B., et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 37 2602–2611. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- Wein N., Vulin A., Findlay A. R., Gumienny F., Huang N., Wilton S. D., et al. (2017). Efficient skipping of single exon duplications in DMD Patient-derived cell lines using an antisense oligonucleotide approach. J. Neuromuscul. Dis. 4 199–207. 10.3233/JND-170233 [DOI] [PubMed] [Google Scholar]
- Weng H., Huang H., Wu H., Qin X., Zhao B. S., Dong L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22 191–205.e9. 10.1016/j.stem.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. L., Luhrmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Ngo L. H., Wickramasinghe V. O. (2018). Nuclear export of RNA: different sizes, shapes and functions. Semin. Cell Dev. Biol. 75 70–77. 10.1016/j.semcdb.2017.08.054 [DOI] [PubMed] [Google Scholar]
- Xia S., Feng J., Lei L., Hu J., Xia L., Wang J., et al. (2016). Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 18 984–992. 10.1093/bib/bbw081 [DOI] [PubMed] [Google Scholar]
- Xing D., Liang J. Q., Li Y., Lu J., Jia H. B., Xu L. Y., et al. (2014). Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop. Surg. 6 288–293. 10.1111/os.12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Guo S., Li W., Yu P. (2015). The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 5:12453. 10.1038/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonovitch A. L., Deng P., Jacobson D., Li J. B. (2017). The evolution and adaptation of A-to-I RNA editing. PLoS Genet. 13:e1007064. 10.1371/journal.pgen.1007064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Shang D., Xu Y., Zhang C., Feng L., Sun Z., et al. (2017). The LncRNA connectivity map: using LncRNA signatures to connect small molecules, LncRNAs, and diseases. Sci Rep. 7:6655. 10.1038/s41598-017-06897-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Huang P., Li F., Zhao L., Zhang Y., Li S., et al. (2012). c-Jun amino-terminal kinase-1 mediates glucose-responsive upregulation of the RNA editing enzyme ADAR2 in pancreatic beta-cells. PLoS One 7:e48611. 10.1371/journal.pone.0048611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Muzny D. M., Reid J. G., Bainbridge M. N., Willis A., Ward P. A., et al. (2013). Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369 1502–1511. 10.1056/NEJMoa1306555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Luo J., Hu K., Lin J., Huang H., Wang Q., et al. (2017). ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 11 422–437. 10.1002/1878-0261.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., He Q., Hu Z., Feng Y., Fan L., Tang Z., et al. (2016). Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 23 522–530. 10.1038/nsmb.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. G., Yang H. L., Long Y., Li W. L. (2016). Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG 123 2113–2118. 10.1111/1471-0528.13897 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tjian R. (2018). Measuring dynamics of eukaryotic transcription initiation: challenges, insights and opportunities. Transcription 9 159–165. 10.1080/21541264.2017.1363017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Tian B. (2014). RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv. Exp. Med. Biol. 825 97–127. 10.1007/978-1-4939-1221-6_3 [DOI] [PubMed] [Google Scholar]
- Zheng G., Cox T., Tribbey L., Wang G. Z., Iacoban P., Booher M. E., et al. (2014). Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem. Neurosci. 5 658–665. 10.1021/cn500042t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Qiu W., Yao R., Xiang J., Sun X., Liu S., et al. (2013). CRM1 is a novel independent prognostic factor for the poor prognosis of gastric carcinomas. Med. Oncol. 30:726. 10.1007/s12032-013-0726-1 [DOI] [PubMed] [Google Scholar]
- Zipeto M. A., Court A. C., Sadarangani A., Delos Santos N. P., Balaian L., Chun H. J., et al. (2016). ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell 19 177–191. 10.1016/j.stem.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]