Abstract
Thermus thermophilus is an extreme-thermophilic bacterium that can grow at a wide range of temperatures (50–83°C). To enable T. thermophilus to grow at high temperatures, several biomolecules including tRNA and tRNA modification enzymes show extreme heat-resistance. Therefore, the modified nucleosides in tRNA from T. thermophilus have been studied mainly from the view point of tRNA stabilization at high temperatures. Such studies have shown that several modifications stabilize the structure of tRNA and are essential for survival of the organism at high temperatures. Together with tRNA modification enzymes, the modified nucleosides form a network that regulates the extent of different tRNA modifications at various temperatures. In this review, I describe this network, as well as the tRNA recognition mechanism of individual tRNA modification enzymes. Furthermore, I summarize the roles of other tRNA stabilization factors such as polyamines and metal ions.
Keywords: methylation, RNA modification, thermophile, Thermus thermophilus, tRNA
Introduction
Thermus thermophilus is an extreme-thermophilic bacterium isolated from Mine Hot Spring in Japan that can grow at a wide range of temperatures (50–83°C) (Oshima and Imahori, 1974). This bacterium can grow under aerobic conditions and possesses only about 2200 genes. Therefore, T. thermophilus strain HB8 was selected as a model organism in the Structural-Biological Whole Cell Project in Japan (Yokoyama et al., 2000). A method for preparing gene disruptant strains of T. thermophilus has been established (Hoseki et al., 1999; Hashimoto et al., 2001). Furthermore, both expression vectors for T. thermophilus proteins in Escherichia coli cells and gene disruption vectors are available from RIKEN Bio Resource Center1. Today, T. thermophilus is one of the most studied thermophiles.
Transfer RNA is an adaptor molecule required for the conversion of genetic information encoded by nucleic acids into amino acid sequences in proteins. To date, more than 100 modified nucleosides have been found in tRNA from the three domains of life bacteria, archaea, and eukaryotes (Boccaletto et al., 2018). Modified nucleosides in tRNA primarily function in various steps of protein synthesis such as stabilization of tRNA structure (Motorin and Helm, 2010; Lorenz et al., 2017), codon-anticodon interaction (Takai and Yokoyama, 2003; Suzuki and Numata, 2014; Agris et al., 2018), prevention of frame-shift errors (Björk et al., 1989; Farabaugh and Björk, 1999; Urbonavicius et al., 2001), recognition by aminoacyl-tRNA synthetases (Muramatsu et al., 1988; Perret et al., 1990; Ikeuchi et al., 2010; Mandal et al., 2010), and recognition by translation factors (Aström and Byström, 1994). In other words, living organisms cannot synthesize proteins efficiently or correctly without tRNA modifications.
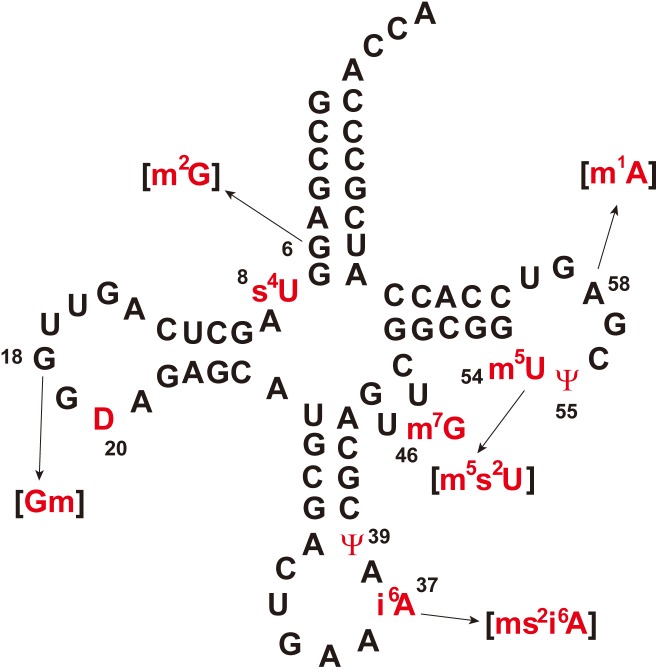
Figure 1 shows the sequence of tRNAPhe from T. thermophilus (Grawunder et al., 1992; Tomikawa et al., 2010). Eleven kinds of modified nucleosides indicated in red in Figure 1 have been found at ten positions in this tRNA (Figure 2); the abbreviations of modified nucleosides, related tRNA modification enzymes, and references (Watanabe et al., 1974; Caillet and Droogmans, 1988; Kammen et al., 1988; Nurse et al., 1995; Persson et al., 1997; Mueller et al., 1998; Esberg et al., 1999; Kambampati and Lauhon, 1999; Bishop et al., 2002; Hori et al., 2002; De Bie et al., 2003; Droogmans et al., 2003; Pierrel et al., 2004; Urbonavicius et al., 2005; Shigi et al., 2006a, 2016; Tomikawa et al., 2010; Ishida et al., 2011; Roovers et al., 2012; Shigi, 2012; Kusuba et al., 2015; Takuma et al., 2015; Yamagami et al., 2016; Bou-Nader et al., 2018) are summarized in Table 1. These modifications are post-transcriptionally conferred by tRNA modification enzymes, which generally act only at one position in tRNA. Thus, specific tRNA modification enzymes exist for the specific positions in specific tRNA species, even though they may synthesize the same type of modified nucleoside; for example, 2’-O-methylcytidine at position 32 (Cm32) in E. coli tRNAMet is conferred by TrmJ (Purta et al., 2006), whereas Cm34 in tRNALeu is synthesized by TrmL (Benítez-Páez et al., 2010). Moreover, in several cases, multiple tRNA modification enzymes and related proteins are required for synthesis of one modified nucleoside; for example, the m5s2U54 modification in T. thermophilus tRNAPhe requires methylation by TrmFO (Urbonavicius et al., 2005) and thiolation by TtuA, TtuB, TtuC, TtuD, and IscS (Shigi et al., 2006a, 2008, 2016; Shigi, 2012) (Table 1). In addition, the modifications in the anticodon-loop often require multiple enzymes and many substrates. For example, the synthesis of 5-methylaminomethyl-2-thiouridine at position 34 (mnm5s2U34) in E. coli tRNAs requires ten proteins (mnmA, mnmC, mnmE, mnmG, TusA, TusB, TusC, TusD, TusE, and IscS) and eight substrates (S-adenosyl-L-methionine, NH4+, ATP, GTP, 5, 10-methylenetetrahyrdofolate, NADH, glycine, and cysteine) (Armengod et al., 2014). Therefore, living organisms need numerous tRNA modification enzymes and related proteins beyond the multitude of modified nucleosides in tRNA.
FIGURE 1.
Cloverleaf representation of Thermus thermophilus tRNAPhe. Modified nucleosides are colored in red and numbers indicate the modification positions. Parentheses indicate that the modification is partial. The abbreviations of modified nucleosides are given in Table 1.
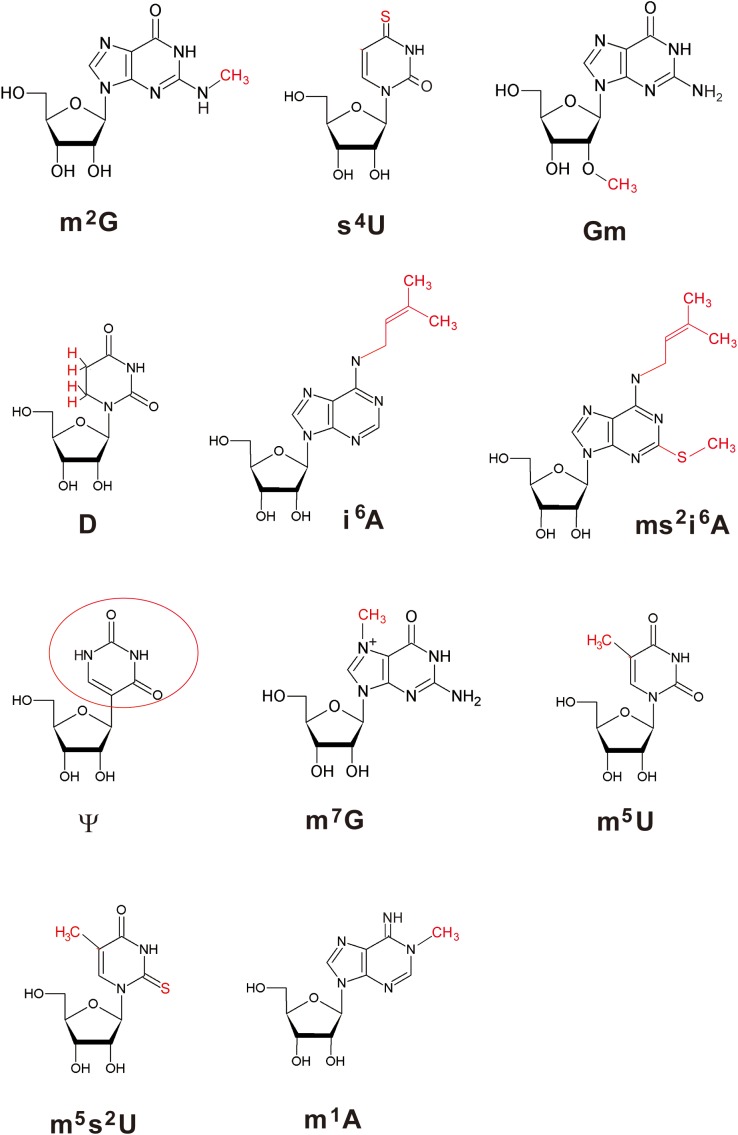
FIGURE 2.
Structures of modified nucleosides in T. thermophilus tRNAPhe. The modifications are highlighted in red. Because ψ is synthesized by isomerization of uridine, the base is enclosed by a red circle.
Table 1.
Modified nucleosides in tRNAPhe from T. thermophiles.
| Abbreviation and position | Modified nucleoside | Modification enzymes | Reference |
|---|---|---|---|
| m2G6 | N2-methylguanosine | TrmN | Roovers et al., 2012 |
| s4U8 | 4-thiouridine | ThiI | Mueller et al., 1998; Kambampati and Lauhon, 1999 |
| Gm18 | 2′-O-methylguanosine | TrmH | Persson et al., 1997; Hori et al., 2002 |
| D20 | dihydrouridine | DusA | Bishop et al., 2002; Kusuba et al., 2015; Bou-Nader et al., 2018 |
| i6A37 | N6-isopentenyladenosine | MiaA | Caillet and Droogmans, 1988 |
| ms2i6A37 | 2-methylthio- N6-isopentenyladenosine | MiaA and MiaB | Esberg et al., 1999; Pierrel et al., 2004 |
| ψ39 | pseudouridine | TruA | Kammen et al., 1988 |
| m7G46 | 7-methylguanosine | TrmB | De Bie et al., 2003; Tomikawa et al., 2010 |
| m5U54 | 5-methyluridine | TrmFO | Urbonavicius et al., 2005; Yamagami et al., 2016 |
| m5s2U54 | 5-methyl-2-thiouridine | TrmFO, TtuA, TtuB, TtuC, TtuD and IscS | Watanabe et al., 1974; Shigi et al., 2006a, 2016; Shigi, 2012 |
| ψ55 | pseudouridine | TruB | Nurse et al., 1995; Ishida et al., 2011 |
| m1A58 | N1-methyladenosine | TrmI | Droogmans et al., 2003; Takuma et al., 2015 |
Given that T. thermophilus grows at high temperatures, its biomolecules including tRNA and tRNA modification enzymes show extreme heat-resistance. As a result, modified nucleosides in tRNA from T. thermophilus have been studied from the view-point of stabilization of tRNA structure at high temperatures. In this review, I focus on the thermal adaptation system of tRNA modifications in T. thermophilus, which is regulated by many factors.
The m5s2U54 Modification in T. thermophilus tRNA Is Essential for Protein Synthesis at High Temperatures
Discovery of m5s2U54 in tRNA From T. thermophilus
The m5s2U modification is a typical thermophile-specific modified nucleoside in tRNA (Hori et al., 2018). It was originally identified in RNase T1-digested RNA fragments derived from a tRNA mixture from T. thermophilus and sequence of the corresponding fragment strongly suggested that a portion of m5U at position 54 is replaced by m5s2U (Watanabe et al., 1974). Subsequently, it was found that this modified nucleoside was increased according to increasing in temperature of the cultures and melting temperature of tRNA was risen with increasing in the extent of m5s2U in tRNA (Watanabe et al., 1976). The presence of m5s2U54 was first confirmed in tRNAMetf1 and tRNAMetf2 (Watanabe et al., 1979), and then tRNA containing m5s2U54 was separated from tRNA containing m5U54 (Watanabe et al., 1983). The m5s2U54 modification has been identified in all T. thermophilus tRNA species sequenced so far [tRNAIle1 (Horie et al., 1985), tRNAAsp (Keith et al., 1993) and tRNAPhe (Grawunder et al., 1992; Tomikawa et al., 2010)].
Structural Effect of m5s2U54 on tRNA
The m5s2U54 modification forms a reverse-Hoogsteen base pair with A58 (or m1A58), and this base pair stacks with the G53-C61 base pair in the T-stem (Yokoyama et al., 1987). The hydrophobic interaction between the m5s2U54-A58 (or m1A58) and G53-C61 base pairs stabilizes the tertiary G18-ψ55 and G19-C56 base pairs between the T- and D-arms. Therefore, m5s2U54 contributes to stabilization of the L-shaped tRNA structure, and the melting temperature of tRNA is increased more than 3°C by the presence of m5s2U54 (Watanabe et al., 1976; Davanloo et al., 1979). Furthermore, the melting temperature of a tRNA mixture is maintained above 85°C by the 2-thiomodification in m5s2U54 (Shigi et al., 2006b), enabling T. thermophilus to grow at temperatures of 50–83°C.
Temperature-Dependent Regulation of m5s2U54 Content in tRNAPhe and Protein Synthesis
Figure 3, which is based on a combination of previous experimental results, shows the percentage of different modified nucleosides (m5s2U54, Gm18, m7G46, and m1A58) in tRNAPhe, purified from T. thermophilus cells cultured at 50, 60, 70, and 80°C (Tomikawa et al., 2010; Ishida et al., 2011; Yamagami et al., 2016). As shown in Figure 3, the extent of m5s2U54 in tRNAPhe increases with increasing culture temperatures. The balance between m5s2U54 and m5U54 regulates the rigidity (flexibility) of tRNA at a wide range of temperatures (50–83°C).
FIGURE 3.
Extents of m7G46, m1A58, Gm18, and m5s2U54 modifications in tRNAPhe from T. thermophilus. The extent of m7G46, m1A58, and Gm18 modifications was measured by a methylation assay with TrmB, TrmI, and TrmH, respectively. The extent of m5s2U54 modification was estimated from the peak areas of m5U54 and m5s2U54 on HPLC analysis. This figure is prepared from Figure 3 in a chapter “Regulation of Protein Synthesis via the Network Between Modified Nucleotides in tRNA and tRNA Modification Enzymes in T. thermophilus, a Thermophilic Eubacterium” of a book “Modified Nucleic Acids in Biology and Medicine,” Springer Nature 2016 with permission (4517441319562) from the publisher.
The presence of m5s2U54 in tRNA is required for efficient protein synthesis at high temperatures. A previous study measured the activity of Poly (U)-dependent poly-phenylalanine synthesis at several temperatures using various fractions (tRNAPhe, ribosome, and supernatant of 100,000 × g centrifugation fraction) prepared from T. thermophilus cells cultured at 50 and 80°C (Yokoyama et al., 1987). Poly-phenylalanine was effectively synthesized at 80°C only when tRNAPhe from cells cultured at 80°C was used. As shown in Figure 3, the proportion of m5s2U54 in tRNAPhe from cells cultured at 80°C is higher than that in tRNAPhe from cells cultured at 50°C. The melting temperature of a tRNA mixture from cells cultured at 70°C is 79.7°C in the presence of 50 mM Tris–HCl (pH7.5), 5 mM MgCl2 and 100 mM NaCl (Tomikawa et al., 2010); thus, the structure of tRNAPhe from cells cultured at 50°C seems to be looser than that from cells cultured at 80°C. Indeed, the gene disruption strain of ttuA encoding a component of sulfur-transfer complex for the m5s2U54 formation, results in a growth defect of the T. thermophilus strain at 80°C (Shigi et al., 2006a). Thus, the m5s2U54 modification is essential for effective protein synthesis in T. thermophilus at high temperatures.
m5s2U54 in tRNA From Other Thermophiles
In addition to T. thermophilus, m5s2U has been identified among the modified nucleosides in unfractionated tRNA from Thermotoga maritima (Edmonds et al., 1991) and at position 54 in tRNACys from Aquifex aeolicus (Awai et al., 2009). Furthermore, the m5s2U nucleoside has been identified among the modified nucleosides in unfractionated tRNA from some hyper-thermophilic archaea such as Thermococcus species (Edmonds et al., 1991) and Pyrococcus furiosus (Kowalak et al., 1994). Therefore, it is considered that these hyper-thermophiles also possess the m5s2U54 modification in tRNA.
Biosynthetic Pathway of m5s2U54 in Eubacteria and Archaea
Biosynthesis of m5s2U54 is accomplished in two steps, methylation of the C5 atom and the 2-thiolation. Although these steps occur independently at U54 in tRNA (Shigi et al., 2006a; Yamagami et al., 2016), the m5U54 modification in tRNA is almost fully formed in living cells even when T. thermophilus is cultured under nutrient-poor conditions (Yamagami et al., 2018). To date, therefore, an s2U54 modification has not been observed in tRNA from the T. thermophilus wild-type strain.
Formation of m5U54 is catalyzed by different tRNA methyltransferases in bacteria and archaea. A folate/FAD-dependent tRNA methyltransferase (TrmFO) catalyzes the methylation using 5, 10-methylenetetrafolate as a methyl donor in bacteria (Urbonavicius et al., 2005; Nishimasu et al., 2009; Yamagami et al., 2012; Hamdane et al., 2016), whereas an S-adenosyl-L-methionine (AdoMet)-dependent tRNA methyltransferase [RumA (or TrmA)-like enzyme] works in archaea (Urbonavicius et al., 2008). Notably, E. coli TrmA is an AdoMet-dependent tRNA (m5U54) methyltransferase (Ny and Björk, 1980), whereas E. coli RumA is an AdoMet-dependent 23S rRNA (m5U1939) methyltransferase (Agarwalla et al., 2002; Madsen et al., 2003). Although these RNA m5U methyltransferases belong to the same cluster of orthologous proteins (group COG2265) (Urbonavicius et al., 2008), archaeal tRNA (m5U54) methyltransferases structurally resemble RumA rather than TrmA (Walbott et al., 2008). Therefore, archaeal tRNA (m5U54) methyltransferases for m5s2U54 formation might have evolved from a RumA-type rRNA methyltransferase.
The 2-thiolation in m5s2U54 of T. thermophilus is conferred by multiple proteins, namely TtuA, TtuB, TtuC, TtuD and IscS (or SufS) (Shigi et al., 2006a, 2008, 2016; Shigi, 2012; Chen et al., 2017). In addition, the mechanism of the sulfur-transfer reaction carried out by TtuA from T. maritima has been recently proposed based on crystal structures of the enzyme (Arragain et al., 2017). The protein factors involved in 2-thiolation of m5s2U54 in archaea have not been confirmed experimentally.
tRNA Modification Enzymes Recognizes the Local Structure(s) in tRNA
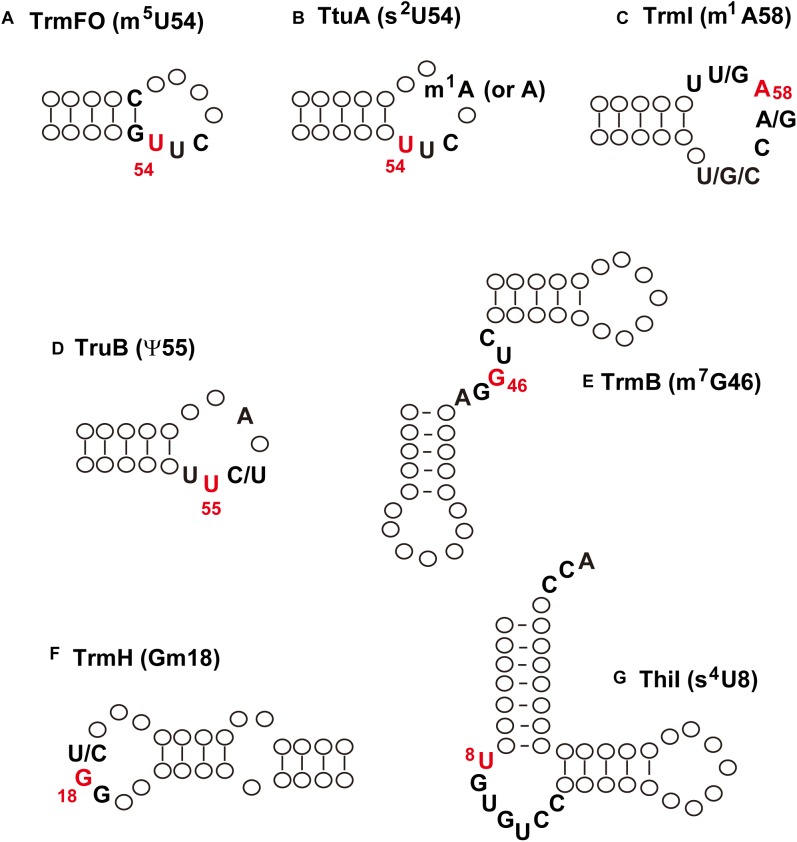
At the beginning of this century, the mechanisms of regulating the extent of modified nucleosides in tRNA from T. thermophilus were unknown. The transcriptional and/or translational regulations of amounts of tRNA modification enzyme(s) were assumed at the start of our studies, however, we noticed that this regulation might be explainable by the substrate tRNA recognition mechanisms of the tRNA modification enzymes. In general, tRNA modification enzymes recognize the local structure in tRNA. Figure 4 shows the minimum substrate or positive determinants for different tRNA modification enzymes, which I describe in more detail below.
FIGURE 4.
Minimum substrate RNA or positive determinants for tRNA modification enzymes. (A) TrmFO; (B), TtuA; (C), TrmI; (D), TruB; (E), TrmB; (F) TrmH; (G) ThiI. The modification is indicated in parentheses; the modification site is colored in red. See the main text for details.
m5U54 Formation by TrmFO
TrmFO can methylate a micro-helix RNA, which mimics the T-arm structure (Yamagami et al., 2012; Figure 4A). The positive determinants for TrmFO are the stem-loop structure, G53-C61 base pair, and U54U55C56 sequence. Therefore, the substrate RNA recognition mechanism of TrmFO is very simple. This simple structure is also observed in the anticodon-loop of tRNAPro from T. thermophilus; however, A38 in the anticodon-loop prevents incorrect methylation by TrmFO (Yamagami et al., 2012). In some cases, therefore, there are negative determinants for tRNA modification enzymes.
s2U54 Formation by TtuA
In the case of TtuA (Figure 4B), only the modification patterns of tRNAAsp mutants expressed in T. thermophilus cells have been analyzed (Shigi et al., 2002); therefore, it is unknown whether TtuA can act on a micro-helix RNA. Nevertheless, it is clear that the positive determinants for the sulfur-transfer reaction of TtuA are also very simple. More recently, it was shown that the presence of m1A58 accelerates the velocity of sulfur-transfer (Shigi et al., 2006b). Thus, the sulfur-transfer reaction carried out by TtuA is regulated by another modification (m1A58).
m1A58 Formation by TrmI
The tRNA m1A58 methyltransferase TrmI can methylate a micro-helix RNA very slowly (Takuma et al., 2015; Figure 4C). The presence of an aminoacyl-stem or variable region accelerates the rate of methylation of truncated tRNA by TrmI. Furthermore, the presence of an m7G46 modification also accelerates the rate of methylation by TrmI (Tomikawa et al., 2010). Thus, the extent of m1A58 modification in tRNA is also controlled by the presence of another modification (m7G46).
Among T. thermophilus tRNAs, tRNAThrGGU exceptionally possesses C60 instead of U60, which is one of the positive determinants for TrmI. The proportion of m1A58 and m5s2U54 in tRNAThrGGU (Kazayama et al., 2015) is lower than that in tRNAPhe (Takuma et al., 2015), indicating that the extent of m1A58 modification has an effect on the extent of m5s2U54 modification, which is consistent with the observation of Shigi et al. (2006b). Thus, the degree of m1A58 modification in tRNAs differs according to the sequence of each tRNA. The sequence of tRNAThrGGU seems to be disadvantageous for survival of T. thermophilus at high temperatures, however, the physiological reason for the presence of C60 in tRNAThrGGU is unknown.
ψ55 Formation by TruB
Escherichia coli TruB can modify a micro-helix RNA, which mimics the T-arm structure (Gu et al., 1998). U54, U55, pyrimidine56 and A58 in the T-loop are important for the full activity of E. coli TruB (Figure 4D; Gu et al., 1998). The crystal structure of a complex of TruB and micro-helix RNA showed that other nucleotides in the T-loop contact with TruB (Hoang and Ferré-D’Amaré, 2001; Pan et al., 2003). The ribose-phosphate backbone in the T-arm structure, which is formed by the U54-A58 reverse Hoogsteen base pair, is important for the reaction by E. coli TruB. Although the substrate tRNA recognition mechanism of T. thermophilus TruB has not been confirmed experimentally, the high conservation of amino acid sequences between E. coli and T. thermophilus TruB proteins (Ishida et al., 2011) strongly suggests that these enzymes possess a common mechanism of tRNA recognition. Formation of ψ55 in T. thermophilus tRNAPhe transcript by TruB is very rapid at 55°C (Ishida et al., 2011), suggesting that T. thermophilus TruB does not require other modifications in tRNA in order to function.
m7G46 Formation by TrmB
Aquifex aeolicus TrmB can methylate a truncated tRNA (Figure 4E): its methylation speed for truncated RNA is comparable to that for the full-length tRNA transcript (Okamoto et al., 2004). In the truncated tRNA, the T-arm-like structure and five nucleosides corresponding to variable region are essential for methylation by TrmB. Thermophilic TrmB (A. aeolicus and T. thermophilus TrmB) share considerable amino acid sequence homology and exceptionally possess a long C-terminal region (Okamoto et al., 2004), which is involved in binding to AdoMet (Tomikawa et al., 2018). Therefore, the substrate tRNA recognition mechanism of thermophilic TrmB may be different from that of mesophilic TrmB (De Bie et al., 2003; Purta et al., 2005; Zegers et al., 2006; Zhou et al., 2009; Tomikawa, 2018).
Gm18 Formation by TrmH
Figure 4F shows the minimum substrate RNA of A. aeolicus TrmH (Hori et al., 2003). There are some differences in substrate tRNA recognition mechanism between A. aeolicus TrmH and T. thermophilus TrmH. For example, A. aeolicus TrmH cannot methylate tRNAs with A17 (Hori et al., 2003), but T. thermophilus TrmH methylates tRNASer, which contains A17 (Hori et al., 1998). Furthermore, the size and sequence of the D-loop have effects on methylation by A. aeolicus TrmH (Hori et al., 2003), however, T. thermophilus TrmH methylates mutant tRNAs irrespective of these D-loop features (Ochi et al., 2010). In short, T. thermophilus TrmH has been found to methylate all tRNAs tested so far (Hori et al., 1998). T. thermophilus TrmH can methylate a 5’-half fragment of tRNA (Matsumoto et al., 1990) but not a micro-helix that mimics the D-arm (Hirao et al., 1989). Therefore, T. thermophilus TrmH may recognize the bulge structure like A. aeolicus TrmH. This idea is consistent with observations that the cross-linking or chemical modification of s4U8 in substrate tRNA causes a decrease in methylation speed by TrmH (Hori et al., 1989). Full activity of TrmH requires the L-shaped structure formed by conserved nucleosides in tRNA (Hori et al., 1998). Furthermore, the speed of methylation by T. thermophilus TrmH for yeast tRNAPhe transcript is slower than that for native yeast tRNAPhe at 65°C, showing that other modified nucleosides have a positive effect on the methylation by TrmH (Hori et al., 1998). Indeed, the presence of m7G46 in tRNAPhe transcript accelerates methylation speed by TrmH (Tomikawa et al., 2010).
s4U8 Formation by ThiI
Escherichia coli and T. maritima ThiI (tRNA 4-thiouridine synthetase) can modify several truncated tRNAs that mimic the aminoacyl-stem and T-arm (Figure 4G; Lauhon et al., 2004; Neumann et al., 2014). Although the substrate tRNA recognition mechanism of T. thermophilus ThiI has not been investigated, it is likely that T. thermophilus ThiI also recognizes the local structure in tRNA. The THUMP domain in ThiI recognizes the CCA terminus of substrate RNA (Neumann et al., 2014). This feature has been identified in another tRNA modification enzyme, archaeal Trm11, which also possesses a THUMP domain (Hirata et al., 2016). Furthermore, given that TrmN and archaeal Trm14 have a THUMP domain (Menezes et al., 2011; Fislage et al., 2012; Roovers et al., 2012), these enzymes may recognize the CCA terminus in tRNA.
D20 and D20a Formations by DusA
In T. thermophilus, single Dus family protein, DusA synthesizes all D modifications (D20 and D20a) in tRNA (Kusuba et al., 2015); in E. coli, by contrast, three Dus family proteins share the modification sites in tRNA (Bishop et al., 2002; Bou-Nader et al., 2018). Among the T. thermophilus tRNA modification enzymes that act on the three-dimensional core in tRNA, DusA exceptionally recognizes the interaction between the T-arm and D-arm (Yu et al., 2011). For the reaction of DusA at high temperatures, therefore, stabilization of the L-shaped tRNA structure by other modified nucleosides is essential (Kusuba et al., 2015). Thus, D20 and D20a seem to be relatively late modifications in T. thermophilus tRNA.
Initial Binding and Induced-Fit Steps in Complex Formation Between tRNA Modification Enzymes and Substrate tRNA
As shown in Figure 4, many tRNA modification enzymes recognize the local structure in tRNA, while the interaction between the T-arm and D-arm in tRNA is not required for their activity. Indeed, in the case of TrmI, the methylation speed is faster for a mutant tRNA transcript, in which the interaction between the T-arm and D-arm is disrupted, than for the wild-type tRNA transcript (Takuma et al., 2015).
In many cases, the target modification site is embedded in the L-shaped tRNA structure. For example, G18 and U55 form a tertiary base pair and the uracil base in U55 is not localized at the surface of tRNA. Therefore, disruption of L-shaped tRNA structure is necessary for the reaction of TruB. Furthermore, to fit into the catalytic pocket of TruB, the uracil base must be flipped. These observations suggest that the reaction of tRNA modification enzyme comprises at least two steps, initial binding to the L-shaped tRNA, followed by a structural change (induced-fit) process in which the L-shaped tRNA structure is disrupted.
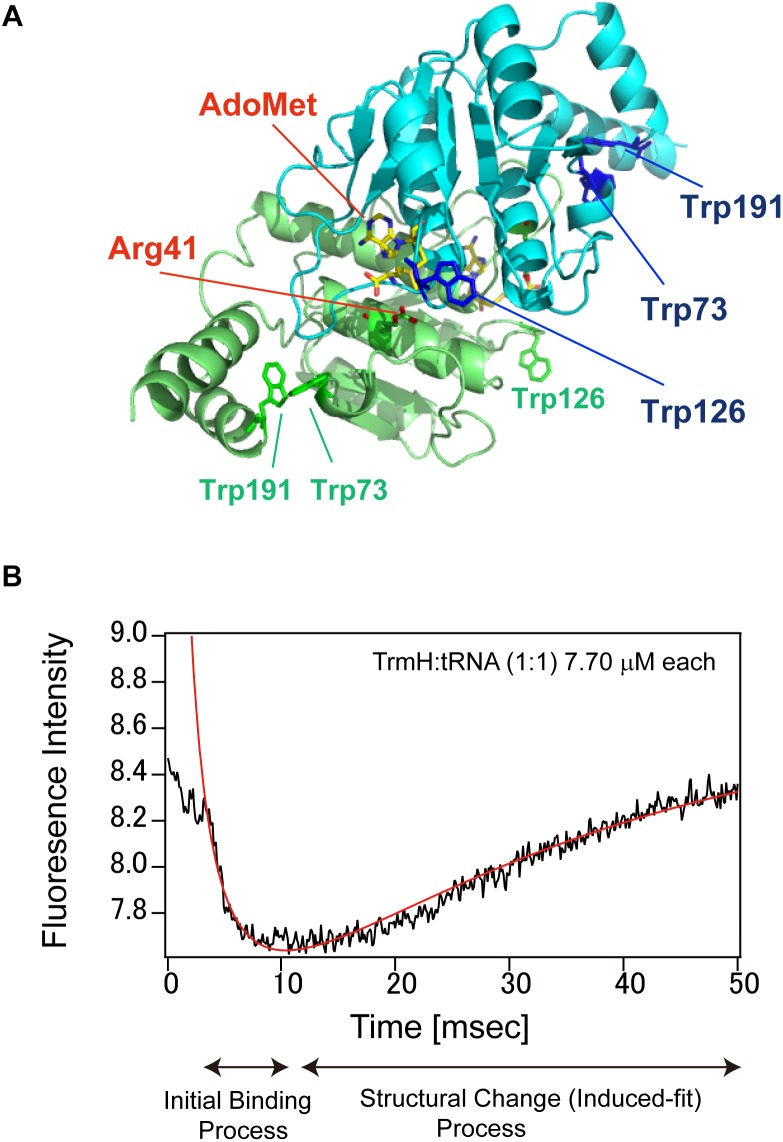
These steps have been monitored for complex formation between T. thermophilus TrmH and tRNAPhe transcript by using a stopped-flow fluorescence measurement system (Figure 5; Ochi et al., 2010, 2013). T. thermophilus TrmH is a member of the SpoU-TrmD (so-called SPOUT) methyltransferase superfamily (Anantharaman et al., 2002; Nureki et al., 2004; Hori, 2017), and site-directed mutagenesis studies (Nureki et al., 2004; Watanabe et al., 2005) suggest that arginine at position 41 (Arg41) in TrmH acts as the catalytic center. As shown in Figure 5A, TrmH is a dimeric enzyme with three tryptophan residues (Trp73, Trp126, and Trp191) in each subunit. The fluorescence intensity at 320 nm derived from these tryptophan residues was measured during complex formation between TrmH and tRNA. Figure 5B shows the one result obtained when 7.70 μM TrmH–AdoMet complex and 7.70 μmM tRNAPhe transcript were mixed by the stopped-flow system at 25°C (Ochi et al., 2010). A very fast decrease in fluorescence was observed in the initial 10 ms, followed by relatively slow increase in fluorescence from 10 to 50 ms. In general, a decrease of tryptophan fluorescence intensity suggests an increase in accessibility of the residue to solvent water. The obtained data could be fitted to an equation, which showed that the reaction was bi-molecular binding reaction. In the case of TrmH, therefore, the initial decrease of fluorescence suggests that a tryptophan residue(s) is moved to the solvent during the initial binding process. Subsequently, this residue was confirmed as Trp126 by measurements on TrmH mutant proteins (Ochi et al., 2013). The slow increase in fluorescence from 10 to 50 ms reflects the structural change (induced-fit) process; detailed measurements on several concentrations of TrmH and tRNA indicate that the slow increase in fluorescence is fitted to a combination of unimolecular (first-order) reactions. The methylation was not completed within 50 ms, showing that the slow increase in fluorescence was not caused by dissociation of tRNA from the enzyme after the reaction. In the structural change process, Trp126 is moved to a hydrophilic environment. During the induced fit process, disruption of L-shaped tRNA structure, recognition of 6-oxygen in G18, and introduction of ribose into the catalytic pocket are occurred (Ochi et al., 2010).
FIGURE 5.
TrmH and the pre-steady state analysis of complex formation between TrmH and tRNA. (A) Dimer structure of TrmH. AdoMet, Arg41 and tryptophan residues are highlighted by stick-models. The two subunits of TrmH are distinguished by green and blue coloring. Trp126 residue is located near AdoMet in the blue subunit and Arg41 in the green subunit. (B) Stopped-flow fluorescence measurement of TrmH–tRNA complex formation. See main text for details.
The folding of tRNA and rigidity (flexibility) of the local structure in tRNA affect the speed of the initial binding and induced-fit processes. Thus, other modifications in tRNA, temperature, and RNA stabilization factors such as Mg2+ ions and polyamines are all likely to influence on the initial binding and induced-fit processes.
A Network Between Modified Nucleosides in tRNA and tRNA Modification Enzymes Controls the Flexibility (Rigidity) of tRNA in T. thermophilus at a Wide Range of Temperatures
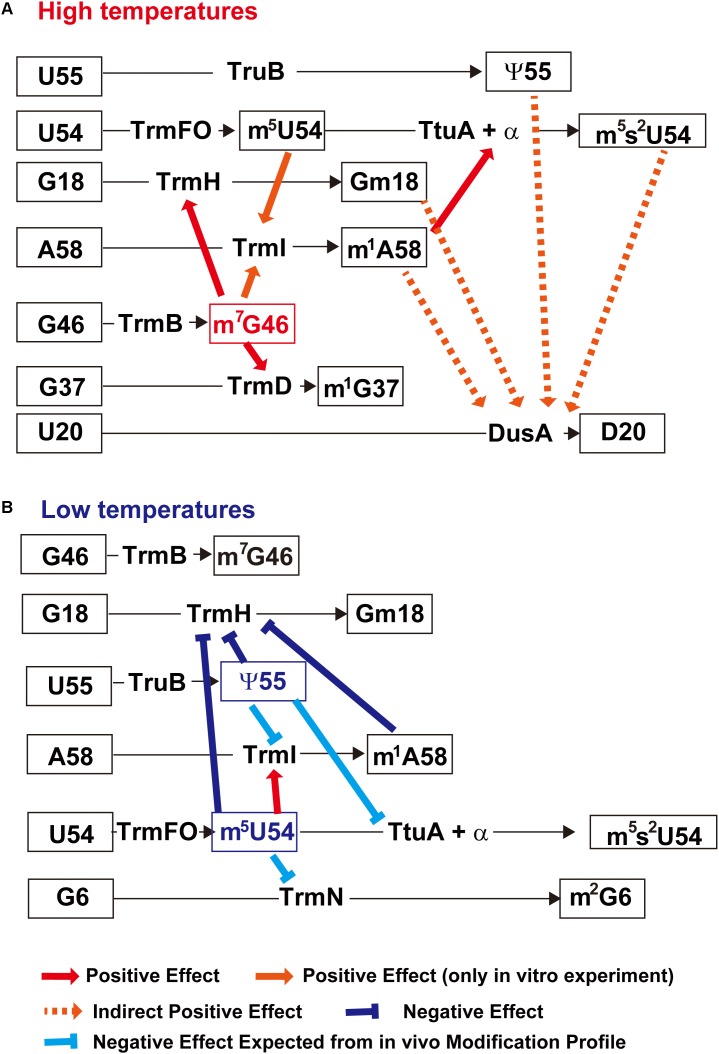
A method for preparing gene disruptant strain of T. thermophilus was developed at the beginning of this century (Hoseki et al., 1999; Hashimoto et al., 2001). This gene-disruption system, coupled with biochemical studies, has been used to elucidate the regulatory network between modified nucleosides in tRNA and tRNA modification enzymes in T. thermophilus cells. Figure 6 summarizes the network between modified nucleosides in tRNA and tRNA modification enzymes. Although each tRNA modification enzymes can act on unmodified tRNA transcript (or truncated tRNA transcript as shown in Figure 4), the presence of modified nucleosides often accelerates (or slows down) the speed of modification by other tRNA modification enzymes depending on the environmental temperatures.
FIGURE 6.
Network between modified nucleosides in tRNA and tRNA modification enzymes in T. thermophilus. (A) Network at high temperatures (>75°C). The m7G46 modification (highlighted in red) is a key factor in this network. Its presence accelerates the speed of other tRNA modification enzymes such as TrmH, TrmD, and TrmI. In addition, the presence of m5U54 also increases the methylation speed of TrmI. The increase in m1A58 due to accelerated TrmI activity further increases the speed of sulfur-transfer by TtuA and that of related proteins, and results in an increased percentage of m5s2U54. The introduced modifications coordinately stabilize the L-shaped tRNA structure. (B) Network at low temperatures (<55°C). In this network, the ψ55 modification stabilizes the local structure in tRNA and slows down the speed of tRNA modification enzymes. The m5U54 modification plays a role in maintaining the balance of modifications at the elbow region in tRNA. This figure is prepared from Figure 4 in a chapter “Regulation of Protein Synthesis via the Network Between Modified Nucleotides in tRNA and tRNA Modification Enzymes in T. thermophilus, a Thermophilic Eubacterium” of a book “Modified Nucleic Acids in Biology and Medicine” Springer Nature 2016 with permission (4517441319562) from the publisher.
T. thermophilus lives in hot springs. The temperature of hot spring water can change for several reasons including an influx of river water, snowfall, and eruption of hot water. Therefore, the ability of protein synthesis to adapt to temperature changes via the flexibility (rigidity) of tRNA is very important for survival of T. thermophilus. One of advantages of the network is that it does not require protein synthesis. As a result, it can respond rapidly. Furthermore, the network may be a survival strategy of eubacteria, which have a limited genome size.
Network at High Temperatures (>75°C)
At high temperatures (>75°C), m7G46 modification by TrmB is one of the key modifications in the network (Figure 6A). It has been shown that the trmB gene deletion strain does not grow at 80°C and has several hypo-modifications in tRNA (Tomikawa et al., 2010). When the culture temperature is shifted from 70 to 80°C, tRNAPhe and tRNALys are degraded and protein synthesis is impaired in this strain. Particular, heat shock proteins are not synthesized efficiently in the trmB gene deletion strain. Thus, the m7G46 modification is essential for survival of T. thermophilus at high temperatures.
The positive effects of m7G46 on TrmH, TrmD, and TrmI activity have been confirmed by in vitro experiments. Because the m1G37 modification conferred by TrmD is not present in T. thermophilus tRNAPhe, yeast tRNAPhe transcript was used in these experiments (Tomikawa et al., 2010). TrmH can methylate a 5’-half fragment of tRNA (Matsumoto et al., 1990), but its full activity requires the three-dimensional core structure of tRNA (Hori et al., 1998). m7G46 forms a tertiary base pair with the C13-G22 base pair in the D-arm; thus, this tertiary base pair seems to have a positive effect on TrmH activity. TrmD can act on a truncated tRNA (Redlak et al., 1997) and micro-helix RNA (Takeda et al., 2006), but foot-printing analyses have shown that the D-arm and variable region are protected in addition to the anticodon-arm (Gabryszuk and Holmes, 1997). Furthermore, the crystal structure of the TrmD–tRNA complex revealed that the C-terminal domain of TrmD makes contacts with the D-arm in tRNA (Ito et al., 2015). Therefore, the positive effect of m7G46 on TrmD activity can be explained by stabilization of the D-arm via the formation of an m7G46-C13-G22 tertiary base pair.
In the case of TrmI, the presence of aminoacyl-stem or variable region increases the methyl-group acceptance activity of a truncated tRNA transcript (Takuma et al., 2015). Although there is no docking model of TrmI with tRNA, positively-charged grooves, which are present on the surface of TrmI (Barraud et al., 2008), may capture the aminoacyl-stem and variable region in tRNA.
The positive effect of m5U54 on TrmI activity was also confirmed in vitro (Yamagami et al., 2012). However, the tRNA fraction from a trmFO gene deletion strain cultured at 70°C contained the same amount of m1A nucleoside as that from the wild-type strain (Yamagami et al., 2016). Therefore, there seems to be a sufficient amount of TrmI to maintain the extent of m1A58 in tRNA in the T. thermophilus trmFO gene deletion strain. The positive effect of m5U54 on the TrmI activity can be explained by stabilizing effect of the m5U54-A58 reverse Hoogsteen base pair.
As described in the Section “tRNA Modification Enzymes Recognizes the Local Structure(s) in tRNA”, the m1A58 modification conferred by TrmI is a positive determinant for the sulfur-transfer system (TtuA and related proteins) (Shigi et al., 2006b). Therefore, TrmI is essential for survival of T. thermophilus at high temperatures (Droogmans et al., 2003). Furthermore, the m5s2U54 modification is essential for stabilizing the tRNA structure, as described in the Section “The m5s2U54 Modification in T. thermophilus tRNA is Essential for Protein Synthesis at High Temperatures.” As a result, the sulfur-transfer system for s2U54 formation is also essential for the survival of T. thermophilus at high temperatures (Shigi et al., 2006a). In contrast, TrmFO is not essential and the trmFO gene deletion strain can grow at 80°C (Yamagami et al., 2016). Collectively, these modified nucleosides in tRNA coordinately stabilize the tRNA structure at high temperatures.
Given that the formation of D20 by DusA requires the interaction between D-arm and T-arm (Yu et al., 2011), stabilization of the L-shaped tRNA structure is essential for D20 formation at high temperatures (Kusuba et al., 2015). Therefore, several modifications, including m5s2U54, m1A58, Gm18 and ψ55, seem to be required for sufficient activity of DusA at high temperatures.
Among the modified nucleosides in T. thermophilus tRNAPhe, m2G6 by TrmN, s4U8 by ThiI, i6A37 and ms2i6A37 by MiaA and MiaB, and ψ39 by TruA have not been investigated as yet. However, it is possible that some of them may affect the stability of tRNA in T. thermophilus at high temperatures. For example, it has been recently reported that the melting temperature of tRNA from E. coli thiI gene disruptant strain is lower than that from the wild-type strain (Nomura et al., 2016). Therefore, s4U8 may contribute to stabilization of the structure of tRNA. In the case of anticodon-loop modifications (ψ38, i6A37, and ms2i6A37), their deletion may severely impair protein synthesis because they are important for the structure of anticodon-loop and function directly in protein synthesis (Spenkuch et al., 2014; Grosjean and Westhof, 2016; Schweizer et al., 2017). Furthermore, i6A37 is required for the 2’-O-methylation conferred by TrmL at position 34 in E. coli tRNALeu (Benítez-Páez et al., 2010; Zhou et al., 2015). In the case of T. thermophilus tRNALeu, therefore, TrmL is probably included in the network.
Network at Low Temperatures (<55°C)
At low temperatures (<55°C), the ψ55 modification conferred by TruB works as a key factor in the network (Figure 6B). In the truB gene deletion strain, excess amounts of Gm18, m1A58 and m5s2U54 are introduced into tRNAs at low temperatures and the melting temperature of tRNA mixture is increased by more than 8°C (Ishida et al., 2011). This excess rigidity of tRNA results in a disorder of protein synthesis, and cold shock proteins are not synthesized efficiently in the truB gene deletion strain. Therefore, ψ55 in tRNA is required for survival of T. thermophilus at low temperatures. The m5U54 modification aids ψ55 in maintaining the balance of other modifications in tRNA (Yamagami et al., 2016). The ψ55 modification stabilizes the structure of elbow region in tRNA and slows down the formation speed of other modifications around ψ55 (Gm18, m1A58 and m5s2U54) (Ishida et al., 2011). The positive effect of m5U54 on m1A58 modification was confirmed both by the in vivo methylation profile and by in vitro experiments (Yamagami et al., 2016) and is probably due to the stabilization of the m5U54-A58 reverse Hoogsteen base pair.
What Stabilizes the Structure of Unmodified Precursor tRNA in T. thermophilus at 80°C?
Unmodified tRNA transcript cannot maintain its L-shaped tRNA structure at high temperatures. Primary transcript (precursor tRNA), which is synthesized by RNA polymerase, is unmodified. Therefore, even though several tRNA modification enzymes from T. thermophilus can act on unmodified tRNA transcript, their activities cannot be measured at 80°C due to the disrupted structure of substrate RNA. For example, T. thermophilus TrmH methylates tRNA effectively only at temperatures below the melting temperature (Matsumoto et al., 1987). These observations raise an important question, what stabilizes the structure of unmodified precursor tRNA in T. thermophilus at 80°C? If there were no stabilization factors in living cells, tRNA modification enzymes from T. thermophilus would not be able to act on precursor tRNA at 80°C.
Unique Polyamines in T. thermophilus and Their Interaction With tRNA
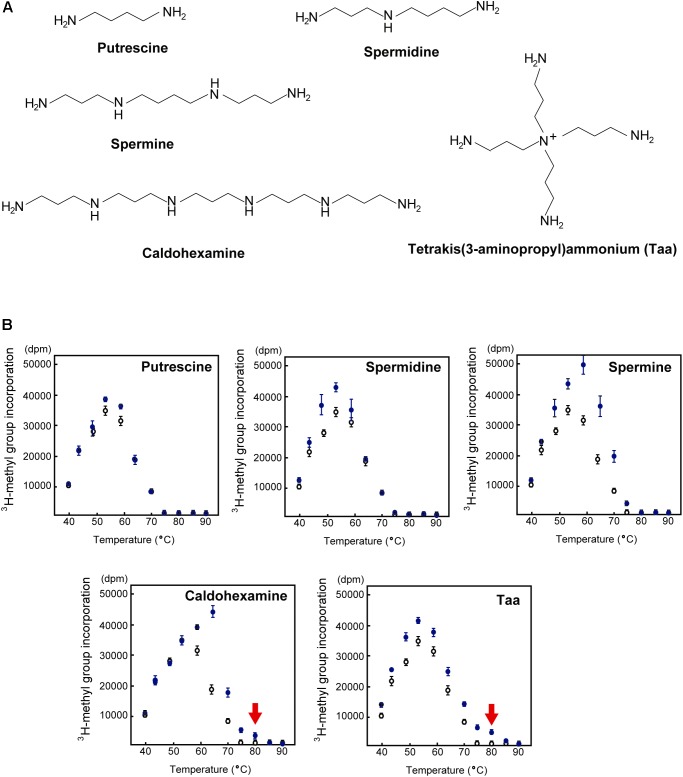
In general, living organisms produce three standard polyamines (putrescine, spermidine and spermine) (Bae et al., 2018; Igarashi and Kashiwagi, 2018). In addition to these standard polyamines, T. thermophilus produces at least 16 polyamine species, including long and branched polyamines (Figure 7A; Hamana et al., 1991; Oshima, 2007; Oshima et al., 2011).
FIGURE 7.
Effect of polyamines on TrmH activity. (A) Typical polyamines. Putrescine, spermidine, and spermine are the three standard polyamines that are found in all living organisms. Unique long and branched polyamines such as caldohexamine and tetrakis(3-aminopropyl)ammonium (Taa) exist in T. thermophilus. (B) TrmH activity was measured in the presence (blue filled circles) or absence (black open circles) of polyamines at 40–90°C. Only in the presence of appropriate concentrations of caldohexamine (1.5 mM) or Taa (0.25 mM), T. thermophilus TrmH can methylate yeast tRNAPhe transcript, of which the melting temperature is 69°C, at 80°C (red arrow).
Because polyamines have positive charges and hydrophilic regions, they have the potential to interact with nucleic acids and phospholipids. Indeed, there have been several studies on the interaction between polyamines and tRNA. For example, addition of polyamines shifted the melting temperature of native tRNAPhe from Saccharomyces cerevisiae to higher temperatures in accordance with polyamine length (Terui et al., 2005). The crystal structure of the complex of yeast tRNAPhe and spermine revealed that two spermine molecules bind to two sites in one tRNAPhe molecule (Quigley et al., 1978). Furthermore, FT-IR analysis of tRNA in the presence of putrescine, spermidine, and spermine showed that similar to spermine, putrescine and spermidine bind to the connection region between the D-arm and anticodon-stem (Ouameur et al., 2010). Moreover, a 13C-NMR study reported that 14 spermidine-binding sites were present in one tRNA molecule and that three spermidine molecules stably bound to tRNA in the presence of Mg2+ (Frydman et al., 1990). It has also been reported that a branched polyamine (Figure 7A), tetrakis(3-aminopropyl)ammonium (Taa), slightly stimulates the activity of archaeal Trm1 and TrmI (Hayrapetyan et al., 2009), which are archaeal tRNA methyltransferases for the formation of m22G26 (or m2G26) (Constantinesco et al., 1999) and m1A57 and m1A58 (Roovers et al., 2004), respectively.
TrmH Methylates Unmodified tRNA Transcript at 80°C Only in the Presence of Long or Branched Polyamine
The effect of polyamine on the methyl-transfer speed of TrmH for yeast tRNAPhe transcript has been measured at various temperatures (Figure 7B; Hori et al., 2016). All polyamines were found to increase the speed of methylation by TrmH at appropriate temperatures, although the positive effect of putrescine was relatively weak and observed only at low temperatures. As the length of polyamine increased, the optimum temperature for methyl-transfer shifted to higher temperatures, however, standard polyamines did not work at 80°C. By contrast, very weak but clear methyl-transfer activity of TrmH for unmodified tRNAPhe was observed at 80°C in the presence of optimum concentration (1.5 mM) of caldohexamine, a long polyamine, (red arrow in Figure 7B). Addition of branched polyamine Taa had a stronger positive effect on TrmH activity at 80°C (red arrow in Figure 7B). Thus, long and branched polyamines can support the methylation by TrmH at 80°C.
If initial modifications are introduced into unmodified precursor tRNA, they stabilize the local structure in tRNA enabling the tRNA modification enzymes to function on the precursor tRNA. Introduction of initial modifications into tRNA probably occurs in the presence of polyamines. Among polyamines, long and branched polyamines are effective at very high temperatures.
Because TrmB, which confers the m7G46 modification, is a key enzyme in the network at high temperatures, the effects of polyamines on TrmB activity should be clarified. At present, however, this is not possible due to a technical problem: T. thermophilus TrmB, which is expressed in E. coli cells, is partially degraded in these cells and purification of intact TrmB is difficult.
Long and Branched Polyamines Are Required for Maintenance of 70S Ribosome and Several tRNAs
The biosynthetic pathway from arginine to spermidine in T. thermophilus is different from that in eukaryotes, archaea, and other bacteria because the intermediate in T. thermophilus is N1-aminopropylagmatine (Ohnuma et al., 2005, 2011). S-Adenosyl-L-methionine decarboxylase-like protein 1 (SpeD1) is required for the biosynthesis of N1-aminopropylagmatine from arginine, and aminopropylagmatine ureohydrolase (SpeB) catalyzes the conversion of N1-aminopropylagmatine to spermidine. Because long and branched polyamines are synthesized from spermidine, the T. thermophilus speD1 or speB gene deletion strain cannot produce long and branched polyamines (Nakashima et al., 2017) and cannot grow at high temperatures (>75°C) unless polyamines are added to the medium. When the speD1 and speB deletion strains were cultured at 70°C in minimal medium until mid-log phase and then the culture temperature was shifted to 80°C, they could survive for 10 h. Although abnormal modifications in tRNA were expected, at least the m5U54, m7G46, Gm18 and m1A58 modifications were present as normal in the tRNA mixture from these strains. Given that the transcription of tRNA and the introduction of major modifications into tRNA are expected to occur mainly before the mid-log phase, it seems that these modification can be introduced into tRNA at 70°C without the presence of long and branched polyamines: the expression patterns of mRNAs in the wild-type strain can be obtained from the database (NCBI/GEO2) (Shinkai et al., 2007). After the temperature shift, tRNAHis, tRNATyr and 70S ribosome were gradually degraded in the speD1 and speB deletion strains and protein synthesis was severely impaired (Nakashima et al., 2017). Thus, long and branched polyamines are required to maintain several tRNAs at high temperatures in T. thermophilus in addition to regulating the extent of modified nucleosides in tRNA.
Other Regulatory Factors for tRNA Stability
RNA binding proteins, Mg2+ ions and K+ ions can stabilize tRNA structure.
In A. aeolicus, a hyper-thermophilic bacterium, tRNA-binding protein 111 (Trbp111) stabilizes the three-dimensional core of tRNA (Morales et al., 1999; Swairjo et al., 2000). However, Trbp111 is specific to A. aeolicus and not found in T. thermophilus. Archease is also an RNA-binding protein that changes the specificity of archaeal Trm4 (Auxilien et al., 2007) and is required for tRNA splicing (Desai et al., 2014; Popow et al., 2014). Although neither trm4 nor a tRNA gene with an intron-coding region is not encoded in the T. thermophilus genome, an archease-like protein gene (TTHA1745) exists. Therefore, it is possible that an archease-like protein stabilizes tRNA structure at high temperatures in T. thermophilus cells.
Mg2+ ions are required for folding of tRNA (Lorenz et al., 2017) and are important in considering the structural effects of several modifications in tRNA (Yue et al., 1994; Agris, 1996; Nobles et al., 2002). However, the precise concentrations of Mg2+ ions in T. thermophilus cells are unknown. K+ ions are also important for correct folding of tRNA. The concentrations of K+ ions in the cells of several thermophilic archaea are reported to be extremely high (>700 mM) (Hensel and Konig, 1988). However, the intracellular concentrations of K+ ions of T. thermophilus have not been reported. The concentrations of Mg2+ and K+ ions may change depending on the growth environment.
Perspective
2018 marked the 50th anniversary year of the first isolation of T. thermophilus. In these past 50 years, T. thermophilus has been studied as a model organism that can adapt to extremely high temperatures. In this regard, the modifications in tRNA have been studied mainly from the viewpoint of the stabilization of tRNA structure at high temperatures. In particular, the role of m5s2U54 in tRNA has been clarified, the genes responsible for almost all tRNA modifications in T. thermophilus have been annotated and the regulatory network between modified nucleosides and tRNA modification enzymes has been identified. Furthermore, numerous tRNA modification enzymes from T. thermophilus have been used in structural studies (reviewed in Hori et al., 2018).
Nevertheless, several enigmas remain, even today. For example, the functions of tRNA modifications in the anticodon-loop at high temperatures have not been studied in detail. Studies on anticodon-loop modifications are difficult because in vitro protein synthesis of T. thermophilus does not work effectively at high temperatures (>75°C). Although there have been many attempts to synthesize proteins at high temperatures using a T. thermophilus cell-free translation system (Ohno-Iwashita et al., 1975; Uzawa et al., 1993a,b; Zhou et al., 2012), it remains difficult to monitor protein synthesis at 80°C. As a result, our knowledge about the effects of tRNA modifications on codon-anticodon interactions, on maintenance of reading frame and on dynamics of tRNA on ribosome at high temperatures is limited. Furthermore, recent studies on mesophiles reported that tRNA modifications occur in response to environmental stresses or function as stress resistance factors (Nawrot et al., 2011; Preston et al., 2013; Endres et al., 2015; Jaroensuk et al., 2016; Campos Guillen et al., 2017). As yet, however, there are no studies from this viewpoint for T. thermophilus. To understand the roles of tRNA modifications in totality, further studies will be required.
Author Contributions
HH determined the concept of this review and prepared the manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by a Grant-in-Aid for Scientific Research (16H04763 to HH) from the Japan Society for the Promotion of Science (JSPS).
References
- Agarwalla S., Kealey J. T., Santi D. V., Stroud R. M. (2002). Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 277 8835–8840. 10.1074/jbc.M111825200 [DOI] [PubMed] [Google Scholar]
- Agris P. F. (1996). The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53 79–129. 10.1016/S0079-6603(08)60143-9 [DOI] [PubMed] [Google Scholar]
- Agris P. F., Eruysal E. R., Narendran A., Väre V. Y. P., Vangaveti S., Ranganathan S. V. (2018). Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15 537–553. 10.1080/15476286.2017.1356562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Koonin E. V., Aravind L. (2002). SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4 71–75. [PubMed] [Google Scholar]
- Armengod M. E., Meseguer S., Villarroya M., Prado S., Moukadiri I., Ruiz-Partida R., et al. (2014). Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes. RNA Biol. 11 1495–1507. 10.4161/15476286.2014.992269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arragain S., Bimai O., Legrand P., Caillat S., Ravanat J. L., Touati N., et al. (2017). Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 114 7355–7360. 10.1073/pnas.1700902114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aström S. U., Byström A. S. (1994). Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell 79 535–546. 10.1016/0092-8674(94)90262-3 [DOI] [PubMed] [Google Scholar]
- Auxilien S., El Khadali F., Rasmussen A., Douthwaite S., Grosjean H. (2007). Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J. Biol. Chem. 282 18711–18721. 10.1074/jbc.M607459200 [DOI] [PubMed] [Google Scholar]
- Awai T., Kimura S., Tomikawa C., Ochi A., Ihsanawati I. S., Bessho Y., et al. (2009). Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 284 20467–20478. 10.1074/jbc.M109.020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae D. H., Lane D. J. R., Jansson P. J., Richardson D. R. (2018). The old and new biochemistry of polyamines. Biochem. Biophys. Acta Gen. Subj. 1862 2053–2068. 10.1016/j.bbagen.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Barraud P., Golinelli-Pimpaneau B., Atmanene C., Sanglier S., Van Dorsselaer A., Droogmans L., et al. (2008). Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 377 535–550. 10.1016/j.jmb.2008.01.041 [DOI] [PubMed] [Google Scholar]
- Benítez-Páez A., Villarroya M., Douthwaite S., Gabaldón T., Armengod M. E. (2010). YibK is the 2’-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 16 2131–2143. 10.1261/rna.2245910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Xu J., Johnson R. C., Schimmel P., de Crécy-Lagard V. (2002). Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 277 25090–25095. 10.1074/jbc.M203208200 [DOI] [PubMed] [Google Scholar]
- Björk G. R., Wikstrom P. M., Byström A. S. (1989). Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244 986–989. 10.1126/science.2471265 [DOI] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M. A., Purta E., Piatkowski P., Baginski B., Wirecki T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46 D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Nader C., Montémont H., Guérineau V., Jean-Jean O., Brégeon D., Hamdane D. (2018). Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario. Nucleic Acids Res. 46 1386–1394. 10.1093/nar/gkx1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet J., Droogmans L. (1988). Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentenyl adenosine in tRNA. J. Bacteriol. 170 4147–4152. 10.1128/jb.170.9.4147-4152.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos Guillen J., Jones G. H., Saldaña Gutiérrez C., Hernández-Flores J. L., Cruz Medina J. A., Valenzuela Soto J. H., et al. (2017). Critical minireview: the fate of tRNACys during oxidative stress in bacillus subtilis. Biomolecules 7:E6. 10.3390/biom7010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Asai S.-I., Narai S., Nambu S., Omura N., Sakaguchi Y., et al. (2017). Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA. Proc. Natl. Acad. Sci. U.S.A. 114 4954–4959. 10.1073/pnas.1615585114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco F., Motorin Y., Grosjean H. (1999). Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 291 375–392. 10.1006/jmbi.1999.2976 [DOI] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Watanabe K., Albani M., Kersten H. (1979). Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 6 1571–1581. 10.1093/nar/6.4.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie L. G., Roovers M., Oudjama Y., Wattiez R., Tricot C., Stalon V., et al. (2003). The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J. Bacteriol. 185 3238–3243. 10.1128/JB.185.10.3238-3243.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K. K., Cheng C. L., Bingman C. A., Phillips G. N., Jr., Raines R. T. (2014). A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 42 3931–3942. 10.1093/nar/gkt1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L., Roovers M., Bujnicki J. M., Tricot C., Hartsch T., Stalon V., et al. (2003). Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 31 2148–2156. 10.1093/nar/gkg314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds C. G., Crain P. F., Gupta R., Hashizume T., Hocart C. H., Kowalak J. A., et al. (1991). Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 173 3138–3148. 10.1128/jb.173.10.3138-3148.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres L., Dedon P. C., Begley T. J. (2015). Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 12 603–614. 10.1080/15476286.2015.1031947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg B., Leung H. C., Tsui H. C., Björk G. R., Winkler M. E. (1999). Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181 7256–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Björk G. R. (1999). How translational accuracy influences reading frame maintenance. EMBO J. 18 1427–1434. 10.1093/emboj/18.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fislage M., Roovers M., Tuszynska I., Bujnicki J. M., Droogmans L., Versées W. (2012). Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res. 40 5149–5161. 10.1093/nar/gks163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman B., de los Santos C., Frydman R. B. (1990). A 13C NMR study of [5,8-13C2]spermidine binding to tRNA and to Escherichia coli macromolecules. J. Biol. Chem. 265 20874–20878. [PubMed] [Google Scholar]
- Gabryszuk J., Holmes W. M. (1997). tRNA recognition for modification: solution probing of tRNA complexed with Escherichia coli tRNA (guanosine-1) methyltransferase. RNA 3 1327–1336. [PMC free article] [PubMed] [Google Scholar]
- Grawunder U., Schön A., Sprintzl M. (1992). Sequence and base modifications of two phenylalanine-tRNAs from Thermus thermophilus HB8. Nucleic Acids Res. 20:137. 10.1093/nar/20.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Westhof E. (2016). An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44 8020–8040. 10.1093/nar/gkw608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Yu M., Ivanetich K. M., Santi D. V. (1998). Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37 339–343. 10.1021/bi971590p [DOI] [PubMed] [Google Scholar]
- Hamana K., Niitsu M., Samejima K., Matsuzaki S. (1991). Polyamine distributions in thermophilic eubacteria belonging to Thermus and Acidothermus. J. Biochem. 109 444–449. 10.1093/oxfordjournals.jbchem.a123401 [DOI] [PubMed] [Google Scholar]
- Hamdane D., Grosjean H., Fontecave M. (2016). Flavin-dependent methylation of RNAs: complex chemistry for a simple modification. J. Mol. Biol. 428 4867–4881. 10.1016/j.jmb.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Yano T., Kuramitsu S., Kagamiyama H. (2001). Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 506 231–234. 10.1016/S0014-5793(01)02926-X [DOI] [PubMed] [Google Scholar]
- Hayrapetyan A., Grosjean H., Helm M. (2009). Effect of a quaternary pentamine on RNA stabilization and enzymatic methylation. Biol. Chem. 390 851–861. 10.1515/BC.2009.096 [DOI] [PubMed] [Google Scholar]
- Hensel R., Konig H. (1988). Thermoadaptation of metanogenic bacteria by interacellular ion concentration. FEMS Microbiol. Lett. 49 75–79. 10.1111/j.1574-6968.1988.tb02685.x 2721925 [DOI] [Google Scholar]
- Hirao I., Ishikawa M., Hori H., Watanabe K., Miura K. (1989). Synthesis of nonadeca- and octadecaribonucleotides using the solid-phase phosphotriester with tetrahydropyranyl groups as the 2’-Hydroxyl-protecting group. Bull. Chem. Soc. Jpn. 62 1995–2001. 10.1246/bcsj.62.1995 [DOI] [Google Scholar]
- Hirata A., Nishiyama S., Tamura T., Yamauchi A., Hori H. (2016). Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 44 6377–6390. 10.1093/nar/gkw561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C., Ferré-D’Amaré A. R. (2001). Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107 929–939. 10.1016/S0092-8674(01)00618-3 [DOI] [PubMed] [Google Scholar]
- Hori H. (2017). Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules 7:E23. 10.3390/biom7010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Kawamura T., Awai T., Ochi A., Yamagami R., Tomikawa C., et al. (2018). Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganism 6:E110. 10.3390/microorganisms6040110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Kubota S., Watanabe K., Kim J. M., Ogasawara T., Sawasaki T., et al. (2003). Aquifex aeolicus tRNA (Gm18) methyltransferase has unique substrate specificity: tRNA recognition mechanism of the enzyme. J. Biol. Chem. 278 25081–25090. 10.1074/jbc.M212577200 [DOI] [PubMed] [Google Scholar]
- Hori H., Saneyoshi M., Kumagai I., Miura K., Watanabe K. (1989). Effects of modification of 4-thiouridine in E. coli tRNAMetf on its methyl acceptor activity by thermostable Gm-methylases. J. Biochem. 106 798–802. 10.1093/oxfordjournals.jbchem.a122933 [DOI] [PubMed] [Google Scholar]
- Hori H., Suzuki T., Sugawara K., Inoue Y., Shibata T., Kuramitsu S., et al. (2002). Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells 7 259–272. 10.1046/j.1365-2443.2002.00520.x [DOI] [PubMed] [Google Scholar]
- Hori H., Terui Y., Nakamoto C., Iwashita C., Ochi A., Watanabe K., et al. (2016). Effects of polyamines from Thermus thermophilus, an extreme-thermophilic eubacterium, on tRNA methylation by tRNA (Gm18) methyltransferase (TrmH). J. Biochem. 159 509–517. 10.1093/jb/mvv130 [DOI] [PubMed] [Google Scholar]
- Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., et al. (1998). Substrate recognition of tRNA (Guanosine-2’-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 273 25721–25727. 10.1074/jbc.273.40.25721 [DOI] [PubMed] [Google Scholar]
- Horie N., Hara-Yokoyama M., Yokoyama S., Watanabe K., Kuchino Y., Nishimura S., et al. (1985). Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24 5711–5715. 10.1021/bi00342a004 [DOI] [PubMed] [Google Scholar]
- Hoseki J., Yano T., Koyama Y., Kuramitsu S., Kagamiyama H. (1999). Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 126 951–956. 10.1093/oxfordjournals.jbchem.a022539 [DOI] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (2018). Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 293:jbc.TM118.003465. 10.1074/jbc.TM118.003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y., Kimura S., Numata T., Nakamura D., Yokogawa T., Ogata T., et al. (2010). Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6 277–282. 10.1038/nchembio.323 [DOI] [PubMed] [Google Scholar]
- Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., et al. (2011). Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39 2304–2318. 10.1093/nar/gkq1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Masuda I., Yoshida K., Goto-Ito S., Sekine S., Suh S. W., et al. (2015). Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc. Natl. Acad. Sci. U.S.A. 112 E4197–E4205. 10.1073/pnas.1422981112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroensuk J., Atichartpongkul S., Chionh Y. H., Wong Y. H., Liew C. W., McBee M. E., et al. (2016). Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa. Nucleic Acids Res. 44 10834–10848. 10.1093/nar/gkw870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambampati R., Lauhon C. T. (1999). IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry 38 16561–16568. 10.1021/bi991119r [DOI] [PubMed] [Google Scholar]
- Kammen H. O., Marvel C. C., Hardy L., Penhoet E. E. (1988). Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J. Biol. Chem. 263 2255–2263. [PubMed] [Google Scholar]
- Kazayama A., Yamagami R., Yokogawa T., Hori H. (2015). Improved solid-phase DNA probe method for tRNA purification: large-scale preparation and alteration of DNA fixation. J. Biochem. 157 411–418. 10.1093/jb/mvu089 [DOI] [PubMed] [Google Scholar]
- Keith G., Yusupov M., Briand C., Moras D., Kern D. (1993). Sequence of tRNAAsp from Thermus thermophilus HB8. Nucleic Acids Res. 21:4399 10.1093/nar/21.18.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak J. A., Dalluge J. J., McCloskey J. A., Stetter K. O. (1994). The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33 7869–7876. 10.1021/bi00191a014 [DOI] [PubMed] [Google Scholar]
- Kusuba H., Yoshida T., Iwasaki E., Awai T., Kazayama A., Hirata A., et al. (2015). In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium. J. Biochem. 158 513–521. 10.1093/jb/mvv066 [DOI] [PubMed] [Google Scholar]
- Lauhon C. T., Erwin W. M., Ton G. N. (2004). Substrate specificity for 4-thiouridine modification in Escherichia coli. J. Biol. Chem. 279 23022–23029. 10.1074/jbc.M401757200 [DOI] [PubMed] [Google Scholar]
- Lorenz C., Lünse C. E., Mörl M. (2017). tRNA Modifications: impact on structure and thermal adaptation. Biomolecules 7:E35. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C. T., Mengel-Jørgensen J., Kirpekar F., Douthwaite S. (2003). Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 31 4738–4746. 10.1093/nar/gkg657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D., Köhrer C., Su D., Russell S. P., Krivos K., Castleberry C. M., et al. (2010). Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl. Acad. Sci. U.S.A. 107 2872–2877. 10.1073/pnas.0914869107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Nishikawa K., Hori H., Ohta T., Miura K., Watanabe K. (1990). Recognition sites of tRNA by a thermostable tRNA(guanosine-2’-)-methyltransferase from Thermus thermophilus HB27. J. Biochem. 107 331–338. 10.1093/oxfordjournals.jbchem.a123047 [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Ohta T., Kumagai I., Oshima T., Murao K., Hasegawa T., et al. (1987). A thermostable Gm-methylase recognizes the tertiary structure of tRNA. J. Biochem. 101 1191–1198. 10.1093/oxfordjournals.jbchem.a121983 [DOI] [PubMed] [Google Scholar]
- Menezes S., Gaston K. W., Krivos K. L., Apolinario E. E., Reich N. O., Sowers K. R., et al. (2011). Formation of m2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res. 39 7641–7655. 10.1093/nar/gkr475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales A. J., Swairjo M. A., Schimmel P. (1999). Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J. 18 3475–3483. 10.1093/emboj/18.12.3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Helm M. (2010). tRNA stabilization by modified nucleotides. Biochemistry 49 4934–4944. 10.1021/bi100408z [DOI] [PubMed] [Google Scholar]
- Mueller E. G., Buck C. J., Palenchar P. M., Barnhart L. E., Paulson J. L. (1998). Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 26 2606–2610. 10.1093/nar/26.11.2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., et al. (1988). Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336 179–181. 10.1038/336179a0 [DOI] [PubMed] [Google Scholar]
- Nakashima M., Yamagami R., Tomikawa C., Ochi Y., Moriya T., Asahara H., et al. (2017). Long and branched polyamines are required for maintenance of the ribosome, tRNAHis and tRNATyr in Thermus thermophilus cells at high temperatures. Genes Cells 22 628–645. 10.1111/gtc.12502 [DOI] [PubMed] [Google Scholar]
- Nawrot B., Sochacka E., Duchler M. (2011). tRNA structural and functional changes induced by oxidative stress. Cell Mol. Life Sci. 68 4023–4032. 10.1007/s00018-011-0773-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Lakomek K., Naumann P. T., Erwin W. M., Lauhon C. T., Ficner R. (2014). Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 42 6673–6685. 10.1093/nar/gku249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Ishitani R., Yamashita K., Iwashita C., Hirata A., Hori H., et al. (2009). Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 106 8180–8185. 10.1073/pnas.0901330106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles K. N., Yarian C. S., Liu G., Guenther R. H., Agris P. F. (2002). Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 30 4751–4760. 10.1093/nar/gkf595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Ohno S., Nishikawa K., Yokogawa T. (2016). Correlation between the stability of tRNA tertiary structure and the catalytic efficiency of a tRNA-modifying enzyme, archaeal tRNA-guanine transglycosylase. Genes Cells 21 41–52. 10.1111/gtc.12317 [DOI] [PubMed] [Google Scholar]
- Nureki O., Watanabe K., Fukai S., Ishii R., Endo Y., Hori H., et al. (2004). Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure 12 593–604. 10.1016/j.str.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Nurse K., Wrzesinski J., Bakin A., Lane B. G., Ofengand J. (1995). Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. RNA 1 102–112. [PMC free article] [PubMed] [Google Scholar]
- Ny T., Björk G. R. (1980). Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J. Bacteriol. 142 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi A., Makabe K., Kuwajima K., Hori H. (2010). Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes. J. Biol. Chem. 285 9018–9029. 10.1074/jbc.M109.065698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi A., Makabe K., Yamagami R., Hirata A., Sakaguchi R., Hou Y. M., et al. (2013). The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process. J. Biol. Chem. 288 25562–25574. 10.1074/jbc.M113.485128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Iwashita Y., Oshima T., Imahori K. (1975). In vitro protein synthesis at elevated temperature by an extract of an extreme thermophile. Arch. Biochem. Biophys. 171 490–499. 10.1016/0003-9861(75)90058-2 [DOI] [PubMed] [Google Scholar]
- Ohnuma M., Ganbe T., Terui Y., Niitsu M., Sato T., Tanaka N., et al. (2011). Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus. J. Mol. Biol. 408 971–986. 10.1016/j.jmb.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Ohnuma M., Terui Y., Tamakoshi M., Mitome H., Niitsu M., Samejima K., et al. (2005). N1-aminopropylagmatine, a new polyamine produced as a key intermediate in polyamine biosynthesis of an extreme thermophile, Thermus thermophilus. J. Biol. Chem. 280 30073–30082. 10.1074/jbc.M413332200 [DOI] [PubMed] [Google Scholar]
- Okamoto H., Watanabe K., Ikeuchi Y., Suzuki T., Endo Y., Hori H. (2004). Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 279 49151–49159. 10.1074/jbc.M408209200 [DOI] [PubMed] [Google Scholar]
- Oshima T. (2007). Unique polyamines produced by an extreme thermophiles, Thermus thermophilus. Amino Acids 33 367–372. 10.1007/s00726-007-0526-z [DOI] [PubMed] [Google Scholar]
- Oshima T., Imahori K. (1974). Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium form a japanese thermal Spa. Int. J. Syst. Bacteriol. 24 102–112. 10.1099/00207713-24-1-102 [DOI] [Google Scholar]
- Oshima T., Moriya T., Terui Y. (2011). Identification, chemical synthesis, and biological functions of unusual polyamines produced by extreme thermophiles. Methods Mol. Biol. 720 81–111. 10.1007/978-1-61779-034-8_5 [DOI] [PubMed] [Google Scholar]
- Ouameur A. A., Bourassa P., Tajmir-Riahi H.-A. (2010). Probing tRNA interaction with biogenic polyamines. RNA 16 1968–1979. 10.1261/rna.1994310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Agarwalla S., Moustakas D. T., Finer-Moore J., Stroud R. M. (2003). Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. U.S.A. 100 12648–12653. 10.1073/pnas.2135585100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. (1990). Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature 344 787–789. 10.1038/344787a0 [DOI] [PubMed] [Google Scholar]
- Persson B. C., Jäger G., Gustafsson C. (1997). The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2’-O-methyltransferase activity. Nucleic Acid Res. 25 3969–3973. 10.1093/nar/25.20.4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F., Douki T., Fontecave M., Atta M. (2004). MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 279 47555–47563. 10.1074/jbc.M408562200 [DOI] [PubMed] [Google Scholar]
- Popow J., Jurkin J., Schleiffer A., Martinez J. (2014). Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511 104–107. 10.1038/nature13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston M. A., D’Silva S., Kon Y., Phizicky E. M. (2013). tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae. RNA 19 243–256. 10.1261/rna.035808.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purta E., van Vliet F., Tkaczuk K. L., Dunin-Horkawicz S., Mori H., Droogmans L., et al. (2006). The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol. Biol. 7:23. 10.1186/1471-2199-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purta E., van Vliet F., Tricot C., De Bie L. G., Feder M., Skowronek K., et al. (2005). Sequence-structure-function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins 59 482–488. 10.1002/prot.20454 [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. (1978). Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 75 64–68. 10.1073/pnas.75.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlak M., Andraos-Selim C., Giege R., Florentz C., Holmes W. M. (1997). Interaction of tRNA with tRNA (guanosine-1) methyltransferase: binding specificity determinants involve the dinucleotide G36pG37 and tertiary structure. Biochemistry 36 8699–8709. 10.1021/bi9701538 [DOI] [PubMed] [Google Scholar]
- Roovers M., Oudjama Y., Fislage M., Bujnicki J. M., Versees W., Droogmans L. (2012). The open reading frame TTC1157 of Thermus thermophilus HB27 encodes the methyltransferase forming N-methylguanosine at position 6 in tRNA. RNA 18 815–824. 10.1261/rna.030411.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M., Wouters J., Bujnicki J. M., Tricot C., Stalon V., Grosjean H., et al. (2004). A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32 465–476. 10.1093/nar/gkh191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U., Bohleber S., Fradejas-Villar N. (2017). The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 14 1197–1208. 10.1080/15476286.2017.1294309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N. (2012). Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 287 17568–17577. 10.1074/jbc.M112.359844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N., Asai S.-I., Watanabe K. (2016). Identification of a rhodanese-like protein involved in thiouridine biosynthesis in Thermus thermophilus tRNA. FEBS Lett. 590 4628–4637. 10.1002/1873-3468.12499 [DOI] [PubMed] [Google Scholar]
- Shigi N., Sakaguchi Y., Asai S., Suzuki T., Watanabe K. (2008). Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 27 3267–3278. 10.1038/emboj.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N., Sakaguchi Y., Suzuki T., Watanabe K. (2006a). Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 281 14296–14306. [DOI] [PubMed] [Google Scholar]
- Shigi N., Suzuki T., Terada T., Shirouzu M., Yokoyama S., Watanabe K. (2006b). Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281 2104–2113. [DOI] [PubMed] [Google Scholar]
- Shigi N., Suzuki T., Tamakoshi M., Oshima T., Watanabe K. (2002). Conserved bases in the TPsi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 277 39128–39135. 10.1074/jbc.M207323200 [DOI] [PubMed] [Google Scholar]
- Shinkai A., Ohbayashi N., Terada T., Shirouzu M., Kuramitsu S., Yokoyama S. (2007). Identification of promoters recognized by RNA polymerase-sigmaE holoenzyme from Thermus thermophilus HB8. J. Bacteriol. 189 8758–8764. 10.1128/JB.01076-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenkuch F., Motorin Y., Helm M. (2014). Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 11 1540–1554. 10.4161/15476286.2014.992278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Numata T. (2014). Convergent evolution of AUA decoding in bacteria and archaea. RNA Biol. 11 1586–1596. 10.4161/15476286.2014.992281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swairjo M. A., Morales A. J., Wang C. C., Ortiz A. R., Schimmel P. (2000). Crystal structure of trbp111: a structure-specific tRNA-binding protein. EMBO J. 19 6278–6298. 10.1093/emboj/19.23.6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Yokoyama S. (2003). Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 31 6383–6391. 10.1093/nar/gkg839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., Toyooka T., Ikeuchi Y., Yokobori S., Okadome K., Takano F., et al. (2006). The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells 11 1353–1365. 10.1111/j.1365-2443.2006.01022.x [DOI] [PubMed] [Google Scholar]
- Takuma H., Ushio N., Minoji M., Kazayama A., Shigi N., Hirata A., et al. (2015). Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI). J. Biol. Chem. 290 5912–5925. 10.1074/jbc.M114.606038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y., Ohnuma M., Hiraga K., Kawashima E., Oshima T. (2005). Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile, Thermus thermophilus. Biochem. J. 388 427–433. 10.1042/BJ20041778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C. (2018). 7-Methylguanosine modifications in transfer RNA (tRNA). Int. J. Mol. Sci. 19:E4080. 10.3390/ijms19124080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C., Takai K., Hori H. (2018). Kinetic characterization of substrate-binding sites of thermostable tRNA methyltransferase (TrmB). J. Biochem. 163 133–142. 10.1093/jb/mvx068 [DOI] [PubMed] [Google Scholar]
- Tomikawa C., Yokogawa T., Kanai T., Hori H. (2010). N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability through a tRNA modification network. Nucleic Acids Res. 38 942–957. 10.1093/nar/gkp1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Auxilien S., Walbott H., Trachana K., Golinelli-Pimpaneau B., Brochier-Armanet C., et al. (2008). Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol. Microbiol. 67 323–335. 10.1111/j.1365-2958.2007.06047.x [DOI] [PubMed] [Google Scholar]
- Urbonavicius J., Qian Q., Durand J. M., Hagervall T. G., Björk G. R. (2001). Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20 4863–4873. 10.1093/emboj/20.17.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Skouloubris S., Myllykallio H., Grosjean H. (2005). Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria–evolutionary implications. Nucleic Acids Res. 33 3955–3964. 10.1093/nar/gki703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa T., Hamasaki N., Oshima T. (1993a). Effects of novel polyamines on cell-free polypeptide synthesis catalyzed by Thermus thermophilus HB8 extract. J. Biochem. 114 478–486. [DOI] [PubMed] [Google Scholar]
- Uzawa T., Yamagishi A., Ueda T., Chikazumi N., Watanabe K., Oshima T. (1993b). Effects of polyamines on a continuous cell-free protein synthesis system of an extreme thermophile, Thermus thermophilus. J. Biochem. 114 732–734. [DOI] [PubMed] [Google Scholar]
- Walbott H., Leulliot N., Grosjean H., Golinelli-Pimpaneau B. (2008). The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res. 36 4929–4940. 10.1093/nar/gkn437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Kuchino Y., Yamaizumi Z., Kato M., Oshima T., Nishimura S. (1979). Nucleotide sequence of formylmethionine tRNA from an extreme thermophile, Thermus thermophilus HB8. J. Biochem. 86 893–905. 10.1093/oxfordjournals.jbchem.a132621 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Nureki O., Fukai S., Ishii R., Okamoto H., Yokoyama S., et al. (2005). Roles of conserved amino acid sequence motifs in the SpoU (TrmH) RNA methyltransferase family. J. Biol. Chem. 280 10368–10377. 10.1074/jbc.M411209200 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Hansske F., Ohta T. (1983). Separation and comparison of 2-thioribothymidine-containing transfer ribonucleic acid and the ribothymidine-containing counterpart from cells of Thermus thermophilus HB 8. Biochemistry 22 98–102. 10.1021/bi00270a014 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Saneyoshi M., Nishimura S. (1974). Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 43 59–63. 10.1016/0014-5793(74)81105-1 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Sinma M., Oshima T., Nishimura S. (1976). Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72 1137–1144. 10.1016/S0006-291X(76)80250-1 [DOI] [PubMed] [Google Scholar]
- Yamagami R., Miyake R., Fukumoto A., Nakashima M., Hori H. (2018). Consumption of N5, N10-methylenetetrahydrofolate in Thermus thermophilus under nutrient-poor condition. J. Biochem. 164 141–152. 10.1093/jb/mvy037 [DOI] [PubMed] [Google Scholar]
- Yamagami R., Tomikawa C., Shigi N., Kazayama A., Asai S., Takuma H., et al. (2016). The folate/FAD-dependent tRNA methyltransferase (TrmFO) from Thermus thermophilus regulates the other modifications in tRNA at low temperatures. Genes Cells 21 740–754. 10.1111/gtc.12376 [DOI] [PubMed] [Google Scholar]
- Yamagami R., Yamashita K., Nishimasu H., Tomikawa C., Ochi A., Iwashita C., et al. (2012). The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). J. Biol. Chem. 287 42480–42494. 10.1074/jbc.M112.390112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Hirota H., Kigawa T., Yabuki T., Shirouzu M., Terada T., et al. (2000). Structural genomics projects in Japan. Nat. Struct. Biol. 7 943–945. 10.1038/80712 [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe K., Miyazawa T. (1987). Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv. Biophys. 23 115–147. 10.1016/0065-227X(87)90006-2 [DOI] [PubMed] [Google Scholar]
- Yu F., Tanaka Y., Yamashita K., Suzuki T., Nakamura A., Hirano N., et al. (2011). Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci. U.S.A. 108 19593–19598. 10.1073/pnas.1112352108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue D., Kintanar A., Horowitz J. (1994). Nucleoside modifications stabilize Mg2+ binding in Escherichia coli tRNA(Val): an imino proton NMR investigation. Biochemistry 33 8905–8911. 10.1021/bi00196a007 [DOI] [PubMed] [Google Scholar]
- Zegers I., Gigot D., van Vliet F., Tricot C., Aymerich S., Bujnicki J. M., et al. (2006). Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 34 1925–1934. 10.1093/nar/gkl116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu Q., Yang W., Gao Y., Teng M., Niu L. (2009). Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion. Proteins 76 512–515. 10.1002/prot.22413 [DOI] [PubMed] [Google Scholar]
- Zhou M., Long T., Fang Z. P., Zhou X. L., Liu R. J., Wang E. D. (2015). Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2’-O-methyltransferase. RNA Biol. 12 900–911. 10.1080/15476286.2015.1050576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Asahara H., Gaucher E. A., Chong S. (2012). Reconstitution of translation from Thermus thermophilus reveals a minimal set of components sufficient for protein synthesis at high temperatures and functional conservation of modern and ancient translation components. Nucleic Acids Res. 40 7932–7945. 10.1093/nar/gks568 [DOI] [PMC free article] [PubMed] [Google Scholar]