SUMMARY
Nutrient deprivation inhibits mRNA translation through mTOR and eIF2α signaling, but it is unclear how the translational program is controlled to reflect the degree of a metabolic stress. In a model of breast cellular transformation, various forms of nutrient deprivation differentially affect the rate of protein synthesis and its recovery over time. Genome-wide translational profiling of glutamine-deprived cells reveals a rapid upregulation of mRNAs containing uORFs and downregulation of ribosomal protein mRNAs, which are followed by selective translation of cytokine and inflammatory mRNAs. Transcription and translation of inflammatory and cytokine genes are stimulated in response to diverse metabolic stresses and depend on eIF2α phosphorylation, with the extent of stimulation correlating with the decrease in global protein synthesis. In accord with the inflammatory stimulus, glutamine deprivation stimulates the migration of transformed cells. Thus, pro-inflammatory gene expression is coupled to metabolic stress, and this can affect cancer cell behavior upon nutrient limitation.
Graphical Abstract
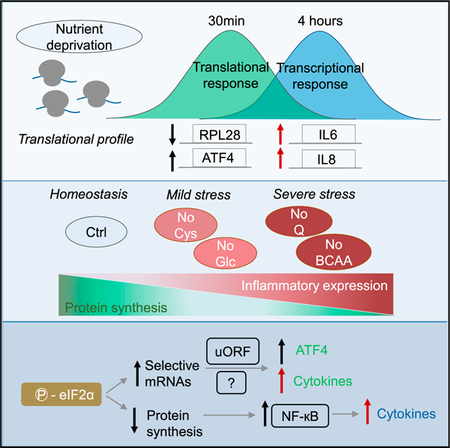
In Brief
Deprivation of some nutrients may impose more constraints on mRNA translation than others. Gameiro and Struhl describe a relationship between translational repression and pro-inflammatory gene expression in response to various metabolic stresses. The pro-inflammatory transcriptional and translational response is not triggered by mTOR inhibition, per se, and requires eIF2α phosphorylation.
INTRODUCTION
An adequate nutrient supply is essential for optimal mRNA translation, and eukaryotic cells have evolved nutrient-sensing pathways that coordinate protein synthesis with nutritional status. Nutrient deprivation inhibits global protein synthesis through modulation of the mechanistic target of rapamycin (mTOR) (Wullschleger et al., 2006) and integrated stress response (ISR) pathways (Holcik and Sonenberg, 2005).
The mTOR signaling pathway integrates nutritional and energy signals to regulate global translational rates via phosphorylation of 4E-BP1 and S6K1, both of which are important for cap-dependent translation (Wullschleger et al., 2006). In addition, mTOR preferentially regulates mRNAs containing TOP motifs in their 5′ UTR, which often encode ribosomal proteins that are translationally repressed in an mTOR-dependent manner (Tang et al., 2001; Thoreen et al., 2012).
In the ISR pathway, diverse stresses lead to the phosphorylation of the translation initiation factor eIF2α by four kinases, each of which is activated by specific stresses (Holcik and Sonenberg, 2005). Phosphorylation of eIF2α generally inhibits translation, but it enhances the translation of specific mRNAs, such as that encoding the transcription factor ATF4 or its yeast counterpart Gcn4 (Harding et al., 2000; Holcik and Sonenberg, 2005). Translational control of ATF4 is mediated by short upstream open reading frames (uORFs) that block translation of the downstream canonical ORF during normal growth but permit the scanning ribosome to initiate translation at the canonical ORF under starved conditions (Vattem and Wek, 2004). Selective translation of ATF4 is responsible for the transcriptional activation of adaptation genes under amino acid deprivation (Harding et al., 2003).
As dividing cells rely differently on glucose (Bauer et al., 2004; Warburg et al., 1927), glutamine (DeBerardinis et al., 2007; Newsholme et al., 1985), glycine (Jain et al., 2012), serine (Maddocks et al., 2013), and leucine (Sheen et al., 2011) to support specific metabolic functions, deprivation of some nutrients may impose more severe constraints on mRNA translation than others. In addition, oncogenes can drive translation of specific mRNAs involved in cell growth, metabolism, invasion, and metastasis (Truitt and Ruggero, 2016). However, there is limited knowledge about how different metabolic stresses control mRNA translation and how the translational program is linked to the degree of metabolic stress. Here, we show that a variety of nutrient stresses lead to an inflammatory response, at both the transcriptional and translational levels, that is linked to inhibition of global protein synthesis.
RESULTS
Nutrient Limitation Differentially Affects Nascent Protein Synthesis
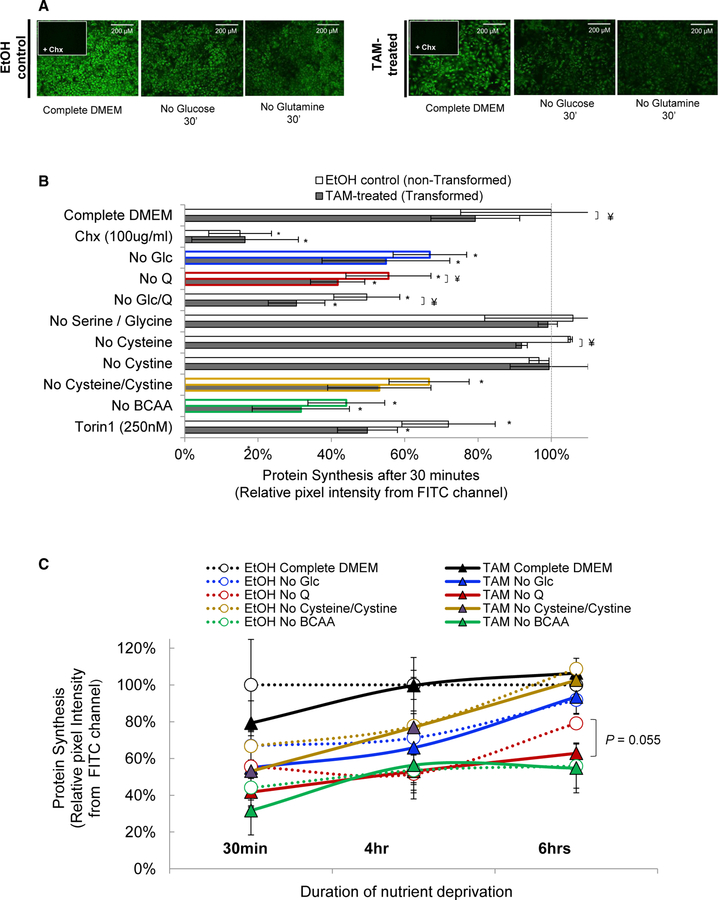
We analyzed the response to nutrient limitation in a model of cellular transformation that involves an immortalized breast epithelial cell line (MCF10A) containing an endoplasmic reticulum (ER)-Src fusion gene consisting of the v-Src oncogene and the ligand-binding domain of the estrogen receptor (Hirsch et al., 2010; Iliopoulos et al., 2009). Treatment of these cells with tamoxifen activates Src, which generates a rapid inflammatory stimulus causing an epigenetic switch from a non-transformed state to a stable transformed state (Iliopoulos et al., 2009). Using a fluorescent methionine analog, we quantified de novo protein synthesis in cells subjected to short-term (30 min) deprivation of glucose or amino acids with different bioenergetic functions. With the exception of glycine and serine deprivation, protein synthesis is inhibited to various extents depending on the limiting nutrient (Figures 1A and 1B), with deprivation of branched-chain amino acids (BCAAs) having the strongest effect. Protein synthesis is reduced in transformed cells, though the relative effects for the various nutrient stresses are similar in both cell types.
Figure 1. Quantification of Protein Synthesis in Response to Metabolic Stresses.
(A) Nascent proteins containing the “clickable” methionine analog (AHA) visualized by fluorescence microscopy.
(B) Quantification of nascent protein synthesis in tamoxifen (TAM)-treated and EtOH control cells under different metabolic conditions. CHX, cycloheximide.
(C) Recovery of nascent protein synthesis over time.
Error bars represent SD of three or more biological replicates, except for the 6-hr samples, and no cysteine or cystine at 30 min (two biological replicates). *p < 0.05, comparing nutrient-deprived to complete medium; ¥p < 0.05, comparing TAM-treated to EtOH control cells, as determined by t test. FITC, fluorescein isothiocyanate.
To analyze how cells recover from nutrient deprivation, we measured protein synthesis after 4 and 6 hr of nutrient deprivation (Figure 1C). Protein synthesis returns to normal or near-normal levels upon deprivation of cysteine + cysteine or of glucose, presumably due to metabolic adaptations. In contrast, cells deprived of BCAAs and glutamine recover poorly, presumably because they cannot synthesize these amino acids to sufficient levels. In all these cases, non-transformed and transformed cells behave similarly, though transformed cells seem to recover less well under glutamine deprivation (Figure 1C). The importance of glutamine in cancer cell metabolism (Altman et al., 2016) prompted us to investigate the translational program of transformed and non-transformed cells subjected to glutamine deprivation.
The Immediate Translational Response to Glutamine Deprivation: Downregulation of mRNAs Involved in Translation and Upregulation of mRNAs Containing uORFs
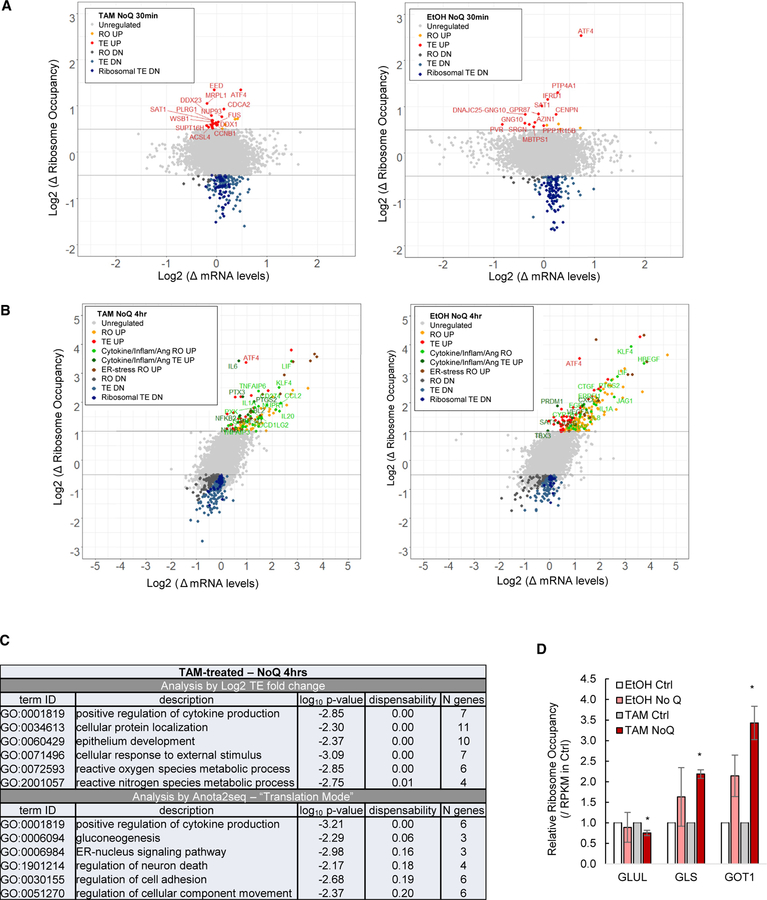
We performed ribosome profiling (Ingolia et al., 2012), sequencing of ribosome-protected RNA, on transformed and non-transformed cells cultured with either complete or glutamine-depleted medium for 30 min. Using RibORF (Ji et al., 2015), we identified ~5 million unique exon-mapped, ribosome footprints corresponding to ~5,700 translated ORFs (Table S1). For each mRNA, we log2 divided the reads per kilobase of transcript per million mapped reads (RPKM) measured by ribosome profiling over the RPKM measured by RNA sequencing (RNA-seq) to obtain translation efficiencies (TEs), and then determined differential TE induced by glutamine deprivation. We also analyzed the ribosome occupancy (RO) as determined by the RPKM of ribosome-protected mRNA fragments. Approximately 140 mRNA transcripts have 1.5-fold or lower TE values, and these translationally inhibited mRNAs are enriched for ribosomal and translation proteins (Figures 2A and S1A), and most of them contain TOP motifs in their 5′ UTRs (Figure S1B). This regulatory response resembles that observed upon inhibiting TOR activity (Thoreen et al., 2012).
Figure 2. Regulation of mRNAs upon Glutamine Deprivation.
(A) Changes in mRNA and ribosome occupancy levels upon 30-min deprivation of glutamine relative to complete medium in transformed (left) and non-transformed (right) cells.
(B) Similar analysis in cells grown for 4 hr in glutamine-deprived versus complete medium.
(C) GO analysis of translationally regulated mRNAs as determined by log2 TE (top) and APV by anota2seq (bottom).
(D) Relative RO of glutamine-catabolizing enzymes.
Error bars represent SD of two biological replicates. *p < 0.05, comparing glutamine-deprived to complete medium, as determined by t test.
See also Figure S1.
Conversely, only 13 (non-transformed cells) or 22 (transformed cells) mRNAs are translationally stimulated after 30 min of glutamine deprivation (Figure 2A), the most dramatic of which encodes ATF4. Like ATF4, some upregulated transcripts are linked to translational control and have translated uORFs. PPP1R15B (2 uORFs) is a phosphatase that dephosphorylates eIF2α, AZIN1 (6 uORFs) is an antizyme inhibitor that stimulates polyamine levels necessary for mRNA translation, and SAT1 (2 uORFs) inhibits translation through polyamine depletion. ATF4 and SAT1 are the only uORF-containing mRNAs upregulated in both transformed and non-transformed cells.
Selective Translation of Cytokine and Inflammatory mRNAs upon Glutamine Deprivation
To investigate the gene expression program during the recovery from glutamine deprivation, we performed ribosome and mRNA profiling of cells cultured for 4 hr with and without glutamine (Table S1). In contrast to the 30-min response (Figure 2A), the fold changes in mRNA and ribosome occupancy are strongly correlated (Figure 2B), indicating a strong transcriptional contribution to the response following 4 hr of glutamine deprivation. In addition, the cells are still metabolically stressed, as evidenced by decreased translational efficiency of ribosomal proteins and increased translational efficiency of ATF4 (Figure 2B).
Interestingly, 34 and 56 mRNAs are translated more efficiently in transformed and non-transformed cells (Figure 2B), respectively, and these are enriched for genes involved in cytokine activity, inflammation, and angiogenesis (Figures 2B and 2C; Table S2). Some cytokines and inflammatory mRNAs are induced transcriptionally but do not show increased TE (Figure 2B; Table S2). As TE values tend to be correlated with cytosolic mRNA levels (Larsson et al., 2010), we also performed an analysis of the partial variance (APV) of ribosome occupancy and mRNA levels to identify differential translation with higher sensitivity (Larsson et al., 2010). The APV shows increased translation of cytokine mRNAs mainly in transformed cells (Figure 2C) and increased ribosome occupancy (combined transcription and translation) of inflammatory mRNAs in both cell types (Figure S1C; Table S2). As expected from the induction of the ISR pathway, the mRNAs with increased ribosome occupancy are also involved in the ER stress response, which includes the transcription factors DDIT3 and XBP1, the canonical targets CHAC1 and PPP1R15A, and aminoacyl-tRNA synthetases (Figure 2B and S1C). Non-transformed cells also exhibit increased expression of cytokine and angiogenic mRNAs, but this response is less pronounced and, primarily, transcriptionally mediated (Figures 2B, S1C, and S1D; Table S2). Thus, glutamine deprivation stimulates inflammatory mRNAs, and this response is both transcriptionally and translationally enhanced in transformed cells.
Although MCF10A cells express glutamine synthetase (GLUL) and can proliferate without glutamine supplementation in the medium, GLUL is not regulated by glutamine deprivation (Figure 2D). In contrast, genes involved in glutamine catabolism, such as glutaminase (GLS) and glutamate-oxaloacetic transaminase 1 (GOT1), are induced (Figure 2D). In general, genes encoding enzymes in the tricarboxylic acid (TCA) cycle are mostly unaffected by glutamine deprivation.
Cytokine and Inflammatory Gene Expression Is Coupled to Translational Inhibition upon Nutrient Deprivation
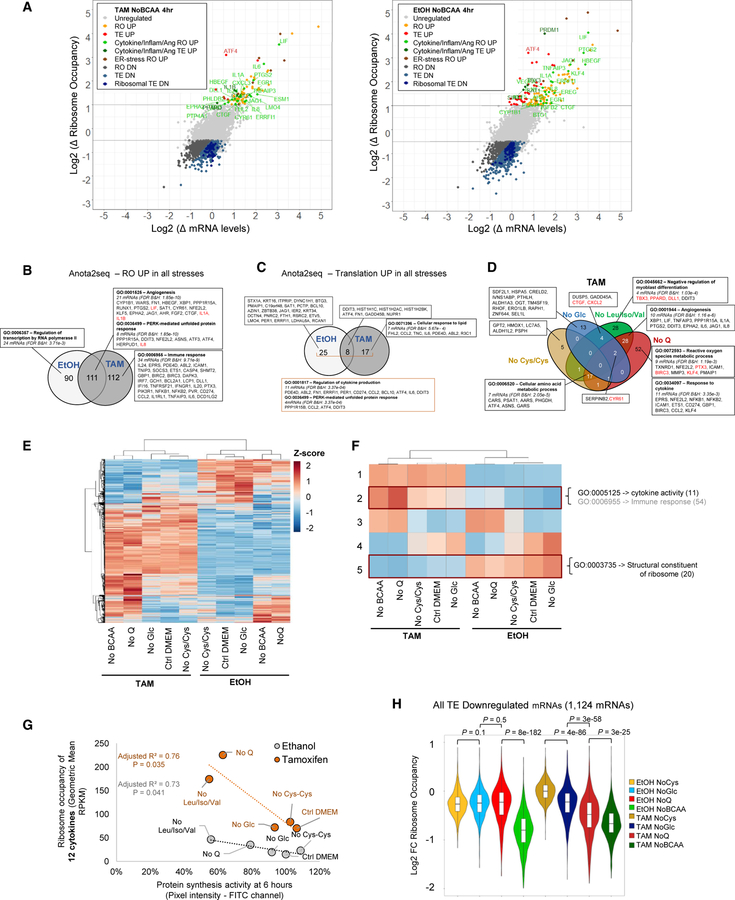
We performed similar experiments in cells subjected to 4 hr of deprivation of glucose, cysteine/cystine, and BCAA. As observed upon glutamine deprivation, cytokine and inflammatory mRNAs are enhanced at both the transcriptional and TE levels upon deprivation of BCAA (Figures 3A and S2A; Table S2). Cells deprived of glucose or cysteine/cystine-deprived cells behave similarly, although to a lesser extent (Figures S2B and S2C). In contrast to the 30-min dataset, the 22 inflammatory mRNAs with increased TE in all stresses (Figure S2D) do not contain many uORFs (Figure S2E; Table S2). As also determined by the APV model, the overall inflammatory response by nutrient depletion is induced both at the transcriptional level (Figure 3B) and translational level (Figure 3C), and it is more pronounced in the transformed state. Intriguingly, stresses causing the strongest inhibition of protein synthesis after 30 min and weakest recovery at 4 and 6 hr (Figure 1, depletion of glutamine or BCAA) also cause the most pronounced increase in cytokine and inflammatory expression (Figures 3D and S2F). In addition, the number of TE-downregulated mRNAs is higher in more “severe” stresses (Figures 2B, 3A, S2B, and S2C).
Figure 3. Regulation of mRNAs upon 4 hr of Metabolic Stress Conditions.
(A) Changes in mRNA and ribosome occupancy levels upon 4-hr deprivation of BCAAs relative to complete medium in transformed (left) and non-transformed (right) cells. Ang, angiogenesis; DN, down-regulated; RO, ribosome occupancy; UP, up-regulated.
(B and C) Venn diagram showing the anota2seq analysis of mRNAs with increased (B) transcriptional and (C) translational activity in all metabolic stresses in transformed versus non-transformed cells.
(D) Venn diagram showing the mRNAs with increased ribosome occupancy in each metabolic stress condition in transformed cells; cytokines are highlighted in red in non-inflammatory GO terms.
(E and F) Shown here: (E) hierarchical and (F) k-means clustering of ribosome occupancy across metabolic conditions.
(G) Correlation between protein synthesis and ribosome occupancy of regulated cytokines among nutrient-deprived conditions.
(H) Ribosome occupancy of all TE-downregulated mRNAs in each metabolic condition; p values are derived from the Wilcoxon test.
See also Figures S2 and S3.
To examine more rigorously whether increased cytokine expression is linked to translational stress, we performed hierarchical clustering of all conditions based on ribosome occupancy (Figure 3E). Aggregation of all mRNAs into five k-means clusters shows that cytokine and inflammatory mRNAs are highly coregulated among the metabolic conditions with the highest upregulation in BCAA and glutamine-deprived transformed cells and the lowest expression in non-transformed cells grown with complete, cysteine/cystine- or glucose-free DMEM (Figure 3F; Table S2, cluster 2). Conversely, the cluster with the strongest downregulation in BCAA and glutamine-deprived transformed cells is strongly enriched for ribosomal proteins (Figure 3F; Table S2, cluster 5). Cluster 1 consists of immune-related mRNAs and is, overall, upregulated in transformed cells (Iliopoulos et al., 2009). In addition, recovery of protein synthesis 6 hr after the stress is inversely correlated with the geometric mean in ribosome occupancy of 12 cytokines (Figure 3G; Table S2), but not with aminoacyl-tRNA synthetases, whose transcription is induced by amino-acid deprivation (Figure S3A), or ATF4, the canonical example of selective mRNA translation (Figure S3B). Likewise, the ribosome occupancy of all downregulated mRNAs (Figure 3H) and 176 ribosomal factor mRNAs in Gene Ontology (GO): 0003735 (Figure S3C) is lower in the more severe stresses, though the pattern is less evident for the latter, as ribosomal factor mRNAs are proportionally more represented in the glucose stress (Figure S3D).
Enhanced Cytokine Expression Requires Translational Inhibition by eIF2α Phosphorylation
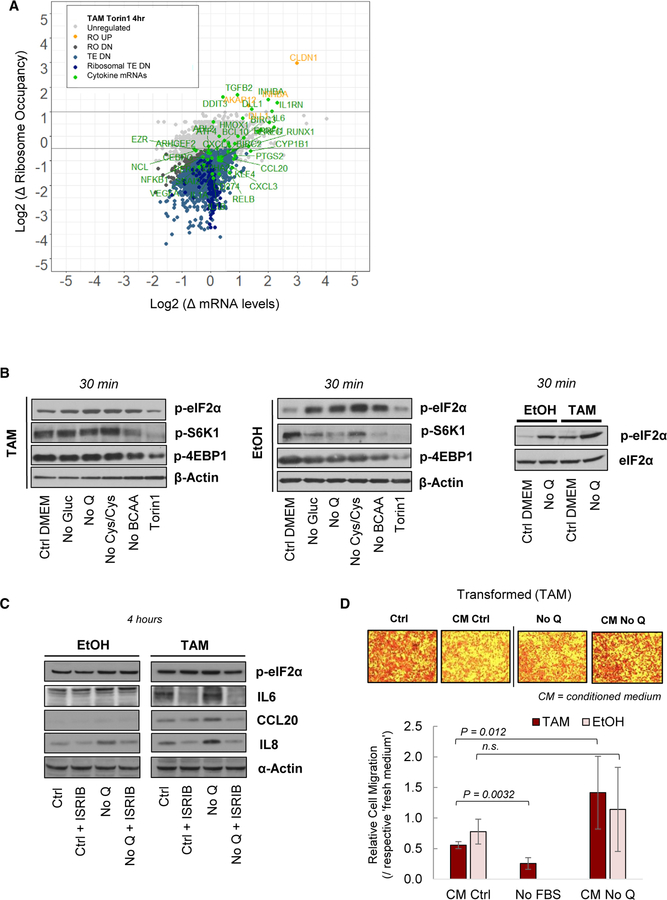
To examine whether inhibition of protein synthesis, per se, leads to an inflammatory response, we performed ribosome profiling of transformed cells treated with torin1, an inhibitor of mTOR. As expected (Thoreen et al., 2012), and in accord with what occurs upon nutrient depletion, torin1-treated cells show decreased levels of protein synthesis (Figure 1B), with translation of ribosomal proteins being particularly affected (Figure 4A). Nutrient deprivation mitigates the phosphorylation of 4EBP1 and S6K1 at 30 min and 4 hr to different extents (Figures 4B and S4A). However, in contrast to conditions of nutrient depletion, torin1 markedly inhibits the phosphorylation of mTOR targets at 30 min and 4 hr (Figures 4B and S4A) but does not lead to increased translation of inflammatory genes (Figure 4A). Indeed, the TE of some cytokine mRNAs actually decreases upon torin1 treatment, and the level of transcriptional induction is considerably below what occurs upon nutrient stresses.
Figure 4. Cytokine Expression under Inhibition of mTOR and eIF2α Signaling and Migration of Glutamine-Deprived MCF10A-ER-Src Cells.
(A) Changes in mRNA and ribosome occupancy levels in transformed cells subjected to 4-hr treatment with torin1 (500 nM) relative to complete medium.
(B) Immunoblot analysis of mTOR-phosphorylated proteins and eIF2α phosphorylation after 30 min of nutrient deprivation or Torin1 treatment (500 nM).
(C) Immunoblot analysis of cytokine protein levels after 4 hr of glutamine deprivation and/or ISRIB treatment (250 nM).
(D) Microscopy images and quantification of cell migration through a basement toward a chamber with different test media. Error bars represent SD of three or more biological replicates. The p values are derived from the t test. n.s., not significant.
See also Figure S4.
In accord with these observations, eIF2α phosphorylation increases upon all forms of nutrient deprivation tested here, but it is not affected by torin1 treatment (Figure 4B). In addition, non-stressed transformed cells exhibit higher levels of eIF2α phosphorylation and lower levels of protein synthesis than non-transformed cells (Figure 4B). To test whether the inflammatory response is linked to eIF2α phosphorylation, we measured cytokine levels in nutrient-deprived cells treated with ISRIB, a potent inhibitor of the integrated stress response pathway (Sidrauski et al., 2013). Cytokine protein levels are induced after glutamine depletion and in response to the other amino-acid stresses (Figure S4B). Treatment with the ISRIB inhibitor decreases the induced expression of IL8, IL6, and CCL20 in transformed cells, and of IL8 in non-transformed cells, after 4 hr of glutamine deprivation (Figure 4C). In accord with the literature, ISRIB does not affect the levels of phosphorylated eIF2α (Sidrauski et al., 2013). Thus, translational repression by eIF2α phosphorylation is necessary for the enhanced cytokine expression under metabolic stress.
Short-Term Nutrient Deprivation Increases Cell Migration
As secreted chemokines can induce the directed migration of leukocytes and neighbor cells, we examined whether glutamine deprivation affects cell migration. Migration of transformed cells through a basement membrane is enhanced toward conditioned medium (CM) from glutamine-deprived cells relative to fresh glutamine-depleted medium, whereas complete CM inhibits cell migration when compared to fresh complete medium (Figure 4D). Migration of non-transformed cells is not affected through the basement membrane (Figure 4D), but their motility is enhanced in a wound healing assay in CM from glutamine- or BCAA-deprived cells (Figure S4C). Transformed cells do not exhibit increased motility in the wound-healing assay (Figure S4C). Thus, short-term glutamine deprivation can drive cancer cell behavior independently of proliferation.
DISCUSSION
As defined by the resulting gene expression patterns, depletion of nutrients with different metabolic functions cause similar, but not identical, inflammatory responses in transformed and non-transformed cells at the transcriptional and translational levels. It is likely that the transcriptional response involves the transcription factor NF-κB (nuclear factor κB), as glutamine deprivation leads to increased NF-κB activity (Hou et al., 2012; Kim et al. 2014). Conversely, glutamine supplementation prevents NF-κB activity and cytokine expression (Chen et al., 2008; Singleton et al., 2005). Glutamine deprivation also triggers co-localization of autophagosomes, lysosomes, and the Golgi apparatus into a subcellular structure whose integrity is essential for IL-8 secretion, a process regulated by mTOR and JNK kinases (Shanware et al., 2014).
Translational stimulation of cytokine and inflammatory mRNAs upon nutrient or other stresses has not been described previously. However, based on published ribosome profiling data, we noticed that cells treated with thapsigargin (Reid et al., 2014), a drug that induces ER stress, show increased translation of inflammatory mRNAs. Our observation that nutrient (and, more generally, ER) stress affects inflammatory genes at both the transcriptional and translational levels underscores the biological importance of this response.
The relationship between the inflammatory response and metabolic stress is relevant to transformed cells. First, as NF-κB and STAT3 are critical for inflammation and tumor development (Grivennikov et al., 2010), increased inflammation in transformed cells is likely to have a greater impact in nutrient-deprived tumors than in normal tissues. As the concentrations of glucose and glutamine are lower in tumors than in normal tissues (Cairns et al., 2011; Warburg et al., 1927), pro-inflammatory translation may be enhanced in a tumor microenvironment. Second, increased expression of cytokines in a nutrient-limited environment would be an autocrine mechanism for tumor cells to invade and metastasize. As glutamine depletion, per se, negatively modulates cell migration, cytokine production under this condition might permit cancer cells to migrate toward a region with higher glutamine concentration. In addition, breast cancer cells selected to grow in the absence of glutamine become more metastatic and tumorigenic and overexpress cyclooxygenase-2 (COX-2 or PTGS2) (Singh et al., 2012), one of the more efficiently translated mRNAs in glutamine-deprived cells in our experiments.
The inverse relationship between the inflammatory response and general protein synthesis suggests a mechanistic connection between these two processes. Inhibition of mTOR activity induces eIF2α phosphorylation in a context-dependent manner (Cherkasova and Hinnebusch, 2003; Gandin et al., 2016; Thoreen et al., 2009; Wengrod et al., 2015). Inhibition of mTOR does not explain our findings, because torin1 decreases protein synthesis to an extent comparable to that of many conditions of nutrient depletion but does not lead to a similar inflammatory response. In fact, torin1-treated cells exhibit a lower translation of various cytokine and inflammatory mRNAs, so basal mTOR activity is probably required for the inflammatory response. In contrast, ISRIB reverses the effects of eIF2α phosphorylation (Sidrauski et al., 2013) and diminishes enhanced cytokine expression in glutamine-deprived cells, suggesting that inhibition of translation initiation by eIF2α phosphorylation triggers the inflammatory response. eIF2α phosphorylation leads to activation of NF-κB due to decreased levels of the IκB inhibitor arising from the combination of its short half-life and the reduced translation (Deng et al., 2004; Jiang et al., 2003). This mechanism may explain why the transcriptional induction of cytokines does not occur rapidly upon nutrient deprivation but, rather, takes several hours.
The inflammatory response arising upon nutrient depletion also occurs at the translation level. Though unknown, the mechanistic basis of this translational control likely involves eIF2α phosphorylation and may act in a uORF-independent manner, as 15 of the 22 TE upregulated inflammatory mRNAs do not contain uORFs. Cytokine mRNAs can be stabilized by the mitogen-activated protein (MAP) kinase/p38 pathway via aurich element (ARE)-binding proteins that interact with sequences within the 3′ UTR (Hoffmann et al., 2002), so ARE- or other RNA-binding proteins might interact with translation factors and/or ribosomes to affect the translation of cytokine mRNAs. Whatever the precise mechanisms involved, the opposing effects of eIF2α phosphorylation on general translation and inflammatory response provides a mechanism to coordinate these processes in response to a wide range of nutritional states.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF10A-ER-Src cells were grown at 37°C with 5% CO2 in phenol red-free DMEM-F12 medium containing 5% charcoal-stripped horse serum, penicillin-streptomycin (1×), epidermal growth factor (EGF) (200 ng/mL), hydrocortisone (0.5 µg/mL, final), cholera toxin (100 ng/mL), and insulin (10 µg/mL), as described previously (Iliopoulos et al., 2009). Cells were transformed by treatment with 1 µM 4-hydroxy-tamoxifen for 24 hr, and ethanol was used as vehicle control in non-transformed cells. For metabolic experiments, cells were grown in the same medium but lacking nutrients as indicated.
Measurement of Nascent Protein Synthesis
Protein synthesis was assessed by treating cells with the “clickable” methionine analog L-azidohomoalanine (AHA) for 30 min, 4 hr, or 6 hr and detecting nascent proteins by fluorescence microscopy using the 488-nm laser channel according to the manufacturer’s instructions (Thermo Fisher Scientific, catalog no. C10289). Details are given in the Supplemental Experimental Procedures.
Ribosome Profiling and RNA-Seq
Details pertaining high-throughput sequencing data acquisition and analyses are given in the Supplemental Experimental Procedures. Ribosome profiling and RNA-seq were performed as described previously (Ingolia et al., 2012). For determination of gene expression levels of protein-coding genes, we removed refSeq-defined coding regions overlapping with uORFs defined in Ji et al. (2015), 15 amino acids downstream of the start codons and 5 amino acids upstream of the stop codons. The gene expression levels were calculated as RPKMs in the remaining coding regions, and we considered only mRNAs expressed at RPKM > 10 in at least one of the metabolic conditions (Table S1). Differential translation was assessed by (1) TE, and (2) partial variance of RO and RNA levels (Larsson et al., 2010).
Statistical Analyses
Statistical analyses and visualizations were performed in R or Excel. The hierarchical and k-means clustering was performed using the pheatmap package with rlog-transformed ribosome occupancy. The association between protein synthesis and ribosome occupancy of cytokine mRNAs was tested using the Pearson correlation coefficient. Other statistical significance was assessed using either the Wilcoxon rank-sum test or t test, as indicated.
Supplementary Material
Highlights.
A translational profile upon depletion of glucose, glutamine, cysteine/cystine, and BCAA
Metabolic stresses differentially inhibit protein synthesis and translation factors
Transcription and translation of cytokine mRNAs are enhanced by metabolic stresses
Cytokine gene expression correlates with the degree of translational inhibition
ACKNOWLEDGMENTS
We would like to thank Ji Zhe for help and expert advice with the RibORF pipeline and gene expression profile. This work was supported by grant CA 107486 to K.S. from the NIH.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE114794.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.07.021.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Altman BJ, Stine ZE, and Dang CV (2016). From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 773. [DOI] [PubMed] [Google Scholar]
- Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, Riley JL, and Thompson CB (2004). Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J 18, 1303–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, and Mak TW (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Chen G, Shi J, Qi M, Yin H, and Hang C (2008). Glutamine decreases intestinal nuclear factor kappa B activity and pro-inflammatory cytokine expression after traumatic brain injury in rats. Inflamm. Res 57, 57–64. [DOI] [PubMed] [Google Scholar]
- Cherkasova VA, and Hinnebusch AG (2003). Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev 17, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, and Thompson CB (2007). Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 104, 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, and Ron D (2004). Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol 24, 10161–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Cargnello M, Gyenis L, McLaughlan S, Cai Y, Tenkerian C, Morita M, Balanathan P, Jean-Jean O, et al. (2016). mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat. Commun 7, 11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Liu XS, and Struhl K (2010). A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell 17, 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Dittrich-Breiholz O, Holtmann H, and Kracht M (2002). Multiple control of interleukin-8 gene expression. J. Leukoc. Biol 72, 847–855. [PubMed] [Google Scholar]
- Holcik M, and Sonenberg N (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol 6, 318–327. [DOI] [PubMed] [Google Scholar]
- Hou Y-C, Chiu W-C, Yeh C-L, and Yeh S-L (2012). Glutamine modulates lipopolysaccharide-induced activation of NF-κB via the Akt/mTOR pathway in lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol 302, L174–L183. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, and Struhl K (2009). An epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, and Mootha VK (2012). Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, and Struhl K (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H-Y, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, and Wek RC (2003). Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol. Cell. Biol 23, 5651–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-H, Kim A, Yu JH, Lim JW, and Kim H (2014). Glutamine deprivation induces interleukin-8 expression in ataxia telangiectasia fibroblasts. Inflamm. Res 63, 347–356. [DOI] [PubMed] [Google Scholar]
- Larsson O, Sonenberg N, and Nadon R (2010). Identification of differential translation in genome wide studies. Proc. Natl. Acad. Sci. USA 107, 21487–21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, and Vousden KH (2013). Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, and Ardawi MS (1985). The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci. Rep 5, 393–400. [DOI] [PubMed] [Google Scholar]
- Reid DW, Chen Q, Tay AS-L, Shenolikar S, and Nicchitta CV (2014). The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158, 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanware NP, Bray K, Eng CH, Wang F, Follettie M, Myers J, Fantin VR, and Abraham RT (2014). Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat. Commun 5, 4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J-H, Zoncu R, Kim D, and Sabatini DM (2011). Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 19, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KKH, Wilson C, et al. (2013). Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Tai K, Madan S, Raythatha MR, Cady AM, Braunlin M, Irving LR, Bajaj A, and Lucci A (2012). Selection of metastatic breast cancer cells based on adaptability of their metabolic state. PLoS ONE 7, e36510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton KD, Beckey VE, and Wischmeyer PE (2005). Glutamine prevents activation of NF-κB and stress kinase pathways, attenuates inflammatory cytokine release, and prevents acute respiratory distress syndrome (ARDS) following sepsis. Shock 24, 583–589. [DOI] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, and Meyuhas O (2001). Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol 21, 8671–8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, and Gray NS (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, and Ruggero D (2016). New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O, Wind F, and Negelein E (1927). The metabolism of tumors in the body. J. Gen. Physiol 8, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengrod J, Wang D, Weiss S, Zhong H, Osman I, and Gardner LB (2015). Phosphorylation of eIF2α triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci. Signal 8, ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, and Hall MN (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.