Abstract
MicroRNAs are epigenetic regulators of gene expression at the posttranscriptional level. They are involved in intercellular communication and crosstalk between different organs. As key regulators of homeostasis, their dysregulation underlies several morbidities including kidney disease. Moreover, their remarkable stability in plasma and urine makes them attractive biomarkers. Beyond biomarker studies, clinical microRNA research in nephrology in recent decades has focused on the discovery of specific microRNA signatures and the identification of novel targets for therapy and/or disease prevention. However, much of this research has produced equivocal results and there is a need for standardization and confirmation in prospective trials. This review aims to provide an overview of general concepts and available clinical evidence in both the pathophysiology and biomarker fields for the role of microRNA in AKI and kidney transplantation.
Keywords: kidney transplantation, MicroRNAs, Prospective Studies, Acute Kidney Injury, Biomarkers, Body Fluids, Homeostasis
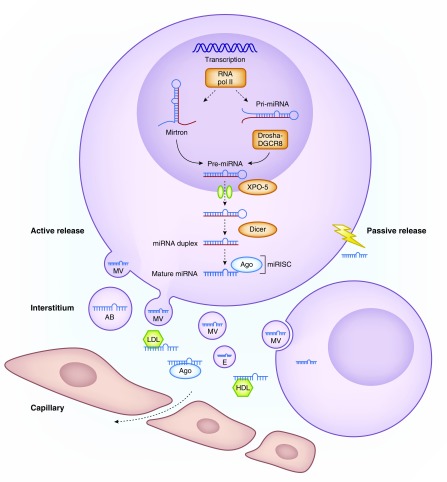
MicroRNAs (miRs), an evolutionary conserved class of noncoding RNAs, are negative regulators of post-transcriptional gene expression. Sequence-specific binding to the target mRNA results in translational inhibition or mRNA degradation. MicroRNA dysregulation is involved in the development and progression of numerous diseases, including cancer, cardiovascular and kidney disease. MicroRNA synthesis takes place through a canonical pathway involving four key enzymes or, alternatively, via the mirtron pathway (Figure 1). Genomic events or inhibition of regulatory enzymes all can lead to microRNA dysregulation in disease.
Figure 1.
MicroRNA biogenesis and function. microRNA coding regions in the human genome are found either intergenic or in the introns of annotated genes. microRNA synthesis starts in the nucleus where most of the miRs are transcribed by RNA polymerase II into primary miR transcripts (pri-miR) of several kilobases that contain local stem-loop structures. The first step of miR maturation is cleavage at the stem of the hairpin structure by a microprocessor complex consisting of Drosha (an RNase III protein) together with its cofactor DiGeorge Syndrome Critical Region 8 (DGCR8), which releases a small hairpin structure of 70 nucleotides that is termed a precursor miR (pre-miR). After nuclear processing, pre-miRs are exported to the cytoplasm by exportin 5 (XPO-5), where they are cleaved near the terminal loop by another RNase enzyme called Dicer, thereby releasing an approximately 22 nucleotide miR duplex. This duplex is loaded onto an AGO protein to generate the microRNA-induced silencing complex (miRISC). One strand (guide strand) remains in the AGO protein as a biologically active miR whereas the other stand (passenger strand, known as miR*) is degraded. The mature miR as part of the effector RISC binds to the 3′UTR region of the mRNA and mediates mRNA degradation, destabilization, or translational inhibition. Apart from this canonical pathway, there is an alternative “mirtron” pathway, independent from Drosha and DGCR8. Mirtrons are miRs that originate from spliced-out introns and are created when small RNAs bind to the termini of small intronic hairpins. Pre-microRNA hairpins with 3′ overhangs are so formed and can mature into 22 nucleotides structures, which look and function as normal miRs. microRNAs exert their repressive function intracellularly, but are also released into the extracellular compartment, with this initiating their role as important intercellular communicators as they are taken up by recipient cells. microRNA can be released passively after cell death or injury, or can be actively secreted in different types of extracellular vesicles, including exosomes, microvesicles and apoptotic bodies. Circulating microRNAs form complexes with RNA binding proteins including AGO2 proteins and lipoproteins (HDL and LDL), which protects them from RNAse-dependent degradation. AB, apoptotic body; E, exosome; MV, microvesicle; RNA pol II, RNA polymerase 2.
MicroRNAs execute not only their repressive function intracellularly, but are also released into the extracellular compartment where they act as hormones and/or biomarkers (Figure 1). Apart from being released passively as a result of cell death or injury, microRNAs are actively secreted in different types of extracellular vesicles, including exosomes, microvesicles, and apoptotic bodies. Circulating microRNAs form complexes with RNA binding proteins including Argonaute (AGO) 2 proteins and lipoproteins, which protects them from RNAse-dependent degradation (1). Interestingly, microRNAs are both important paracrine and endocrine intercellular communicators (2,3) (e.g., muscle–kidney crosstalk (4)). After uptake, specific microRNA can exert their silencing function in the recipient cells. In vivo modulation (merely inhibition) of microRNA expression as a therapeutic strategy is widely explored and in CKD, several microRNA-targeting drugs have entered clinical testing (5). In autosomal dominant polycystic kidney disease, insight in the pathogenetic role of miR-17 (6,7) recently lead to the start of a phase 1 trial of an anti–miR-17 compound (RGLS4326). Likewise, miR-21 inhibition in Alport syndrome shows promising results, both in an animal model and in vitro studies (8), and a phase 1 clinical trial is currently ongoing with an anti-miR compound (RG-012). In contrast, trials targeting microRNAs in the field of AKI or kidney transplantation have not entered the clinical phase yet.
This review provides an overview of the clinical evidence of both the pathogenic role and the diagnostic potential of miRs in AKI and kidney transplantation. A comprehensive review on the role of microRNA in CKD has recently been published (9). Tables 1 and 2 show the current evidence for AKI and transplantation-related disorders, respectively. Several caveats apply when interpreting the study results. First, the considerable heterogeneity in the techniques used (quantitative RT-PCR, microarrays, next generation sequencing) (10) and patient groups studied makes intergroup comparisons and independent validation difficult. Second, several of these studies investigated microRNA target prediction and mRNA interactions through biostatistical modeling. However, experimental validation remains important. Third, natural interindividual variation in microRNA expression levels is not well defined (11,12). Technological developments, principally the use of single-cell sequencing technologies, could add in-depth analysis to this area and enable a more comprehensive picture.
Table 1.
MicroRNAs in AKI
| Phenotype | Study | Study Population | Sample | miR | Internal Validation (Independent Cohort) | Overlap with Other Studies | |
|---|---|---|---|---|---|---|---|
| Upregulation | Downregulation | ||||||
| MicroRNA and pathophysiology | |||||||
| AKI | Lan et al. (14) | Critical patients with AKI (n=16) | Serum and urine samples | ↑ Urinary miR-494 in patients with AKI | — | — | — |
| Critical patients without AKI (n=10) | |||||||
| Healthy controls (n=14) | |||||||
| AKI | Wang et al. (16) | Septic AKI (n=15) | Circulating endothelial cells | ↑ miR-107 in septic patients with AKI | — | — | — |
| Non-septic AKI (n=15) | |||||||
| Septic non-AKI (n=15) | |||||||
| Healthy volunteers (n=15) | |||||||
| AKI | Chen et al. (15) | Critical patients with AKI (n=11) | Serum and urine samples | ↑ Urinary let-7d, life-26–3p, miR-16, miR-451, miR-486–5p, miR-518*, miR-720 | miR-16 was further validated in animal studies | — | — |
| Critical patients without AKI (n=7) | ↓ 21 miRs | ||||||
| Healthy volunteers (n=4) | |||||||
| AKI | Kang et al. (43) | Children after cardiac surgery | Plasma and urine | ↑ miR-21 after RIPC | — | — | — |
| Control group (n=249): | |||||||
| - AKI (n=115) | |||||||
| - Non-AKI (n=134) | |||||||
| RIPC group (n=200): | |||||||
| - AKI (n=38) | |||||||
| - Non-AKI (n=162) | |||||||
| AKI | Guo et al. (18) | Patients with AKI with cisplatin (n=21) | Kidney biopsy | ↑ miR-709 | — | — | — |
| AKI | Liu et al. (13) | Patients with AKI (n=4) | PBMC | Overexpression of miR-101 led to reduced c-Rel and IL-2 expression | |||
| AKI | Ge et al. (17) | Discovery cohort: Septic AKI (n=6) | Serum | 40 miRs were differentially expressed between patients with and without AKI | ↑ miR-4270, miR-4321, miR-3165 | — | — |
| Septic non-AKI (n=6) | ↓ miR-142–5p, miR-22–3p, miR-191–5p, miR-23a-3p, miR-4456 | ||||||
| Controls (n=3) | |||||||
| Validation cohort: Septic AKI (n=35) | |||||||
| Septic non-AKI (n=30) | |||||||
| MicroRNA as biomarkers | |||||||
| AKI | Lorenzen et al. (38) | Discovery cohort: Critically ill patients with AKI (n=5) | Plasma | 13 miRs were different between patients with AKI and healthy controls | ↓miR-16 and miR-320 in AKI | — | — |
| Healthy controls (n=5) | ↑ miR-210 in AKI | ||||||
| Validation cohort: AKI (n=77) | |||||||
| Healthy controls (n=30) | |||||||
| AMI (n=18) | |||||||
| Ischemic or septic AKI | Saikumar et al. (36) | Critically ill patients with elevated Scr and elevated levels of urinary KIM-1 (n=22) | Urine | ↑ miR-21 in AKI | — | ↑ miR-21 (39,40) | — |
| Healthy volunteers (n=25) | ↓ miR-155 in AKI | ||||||
| Severe AKI | Du et al. (40) | Stage 1 or 2 AKI defined by AKIN after cardiac surgery (n=80) | Urine and plasma | ↑ miR-21 in AKI in both urine and plasma samples | — | ↑ miR-21 (36,39) | — |
| Non-AKI group (n=40) | |||||||
| AKI | Ramachandran et al. (39) | Discovery cohort: ICU patients with AKI (n=6) | Urine | 378 miRs were selected for validation with qPCR in the validation cohort | ↑ miR-21, miR-200c, miR-423 in patients with AKI | ↑ miR-21 (36,40) | — |
| Healthy volunteers (n=6) | ↓ miR-4640 in patients with AKI | ||||||
| Validation cohort: | |||||||
| Healthy volunteers (n=74) | |||||||
| ICU patients without kidney disease (n=23) | |||||||
| ICU patients with AKI (n=71) | |||||||
| Kidney transplant patients with tubular injury (n=27) | |||||||
| AKI | Aguado-Fraile et al. (44) | Discovery cohort: ICU patients (n=4) | Serum | 10 miRs were selected (more than two-fold change) | ↓ miR-101–3p, miR-127–3p, miR-210–3p, miR126–3p, miR-26b-5p, miR-29a-3p, miR-146a-5p, miR-27a-3p, miR-93–3p, miR-10a-5p in AKI in ICU patients | — | — |
| Healthy volunteers (n=10) | ↓ miR-127–3p, miR-26b-5p, miR-146a-5p, miR-93–3p in patients after CS | ||||||
| Validation cohort: ICU patients (n=35) | |||||||
| Cardiac surgery patients (n=41) | |||||||
| Healthy volunteers (n=20) | |||||||
| AKI | Zou et al. (46) | AKI after cardiac surgery (n=27) | Urine | ↑ miR-30c-5p, miR-192–5p, miR-378a-3p | — | ↑ miR-30c (47) | — |
| Non-AKI after cardiac surgery (n=44) | ↑ miR-192 (45) | ||||||
| Contrast-induced AKI | Gutiérrez-Escolano et al. (47) | Patients with contrast-induced nephropathy (n=92) | Plasma | ↑ miR-30a, -c, and -e | — | ↑ miR-30a (48) | — |
| Patients without contrast-induced nephropathy (n=92) | ↑ miR-30c (46) | ||||||
| ↑ miR-30e (48) | |||||||
| Contrast-induced AKI | Sun et al. (48) | Patients with AKI after elective coronary angiography or percutaneous coronary intervention (n=71) | Plasma | ↑ miR-188, miR-30a and -e | — | ↑ miR-30a (47) | — |
| ↑ miR-30e (47) | |||||||
| AKI | Arvin et al. (42) | Stage 2–3 AKI after cardiac surgery (n=18) | Serum and urine | ↓ Urinary and serum miR-21 | — | — | ↓serum miR-21 (41) |
| Stage 0–1 AKI after cardiac surgery (n=97) | |||||||
| AKI | Gaede et al. (41) | AKI after cardiac surgery (n=14) | Serum | ↓ Serum miR-21 | — | — | ↓serum miR-21 (42) |
| Non-AKI after cardiac surgery (n=14) | |||||||
| AKI | Zhang et al. (45) | AKI after cardiac surgery (n=35) | Plasma | ↑ miR-192 | — | ↑ miR-192 (46) | |
| Non-AKI after cardiac surgery (n=35) | |||||||
miR, microRNA; RIPC, remote ischemic preconditioning; AMI, acute myocardial infarction; Scr, serum creatinine; KIM-1, kidney injury molecule-1; AKIN, acute kidney injury network; ICU, intensive care unit; qPCR, quantitative PCR; CS, cardiac surgery.
Table 2.
MicroRNA in kidney transplantation
| Phenotype | Study | Study Population | Sample | miR | Internal Validation | Overlap with Other Studies | |
|---|---|---|---|---|---|---|---|
| Upregulation | Downregulation | ||||||
| MicroRNA and pathophysiology | |||||||
| ATN/delayed graft function | Wilflingseder et al. (19) | ATN (n=14) normal PBX (n=10) | Biopsy | ↑ 7 miRs | — | ↑ miR-21–3p (20) | — |
| ↑ miR-182–5p (20) | |||||||
| ATN/delayed graft function | Wilflingseder et al. (20) | ATN+T0BX (n=8) normal PBX+T0BX (n=10) | Biopsy | ↑ 29 miRs | — | ↑ miR-21–3p (19) | — |
| ↑ miR-182–5p (19) | |||||||
| ATN/delayed graft function | Amrouche et al. (21) | ATN (n=19) Normal PBX (n=15) | Biopsy | ↑ miR-146a | — | — | — |
| ATN/delayed graft function | McGuinness et al. (22) | Discovery cohort: T0BX good performing allografts within 2 yr post-Tx (n=5) | Biopsy | 11 Differentially expressed miRs (fold changes not reported) | ↓ miR-125b | — | — |
| T0BX poor performing allografts within 2 yr post-Tx (n=5) | ↓ miR-217 | ||||||
| Validation cohort: T0BX delayed graft function (n=27) | |||||||
| T0BX no delayed graft function (n=67) | |||||||
| Model validation cohort: T0BX delayed graft function (n=10) | |||||||
| T0BX no delayed graft function (n=14) | |||||||
| Acute T cell–mediated rejection (Banff I) | Sui et al. (28) | T cell–mediated rejection (n=3) | Biopsy | ↑ 8 miRs | — | ↑ miR-125a (26) | ↓ miR-17–3p (26) |
| resected tissue RCC (n=3) | ↓ 12 miRs | ↑ miR-602 (26) | ↓ miR-330 (26) | ||||
| ↑ miR-628 (26) | ↓ miR-483 (26) | ||||||
| ↑ miR-658 (26) | ↓ miR-611 (26) | ||||||
| ↓ miR-663 (26) | |||||||
| Acute T cell–mediated rejection (Banff I) | Anglicheau et al. (24) | Discovery cohort: T cell–mediated rejection (n=3) | Biopsy | ↑ 10 miRs | ↑ miR-142–5p | ↑ miR-142–3p (25,27,29) | ↓ miR-30c (26,27) |
| normal PBX (n=4) | ↓ 43 miRs | ↑ miR-155 | ↑ miR-155 (19,25,26) | ↓ miR-125a (19,27) | |||
| Validation cohort: T cell–mediated rejection (n=9) | ↑ miR-223 | ↑ miR-223 (25–27) | ↓ miR-204 (27,29) | ||||
| normal PBX (n=17) | ↓ miR-10b | ↑ miR-342–3p (27,29) | ↓ miR-30a-5p (27) | ||||
| ↓ miR-30a-3p | ↑ miR-142–5p (25) | ↓ miR-30d-5p (27) | |||||
| ↑ miR-21 (26) | ↓ miR-32 (26) | ||||||
| ↑ miR-146a (26) | ↓ miR-125b-5p (27) | ||||||
| ↑ miR-650 (26) | ↓ miR-193b (19) | ||||||
| ↓ miR-99a-5p (27) | |||||||
| ↓ miR-100–5p (27) | |||||||
| ↓ miR-126–3p (27) | |||||||
| ↓ miR-130a-3p (27) | |||||||
| ↓ miR-10b (26) | |||||||
| ↓ miR-30a-3p (26) | |||||||
| ↓ miR-27b (19) | |||||||
| Acute T cell–mediated rejection (Banff I–II) | Wilflingseder et al. (19) | T-cell mediated rejection (n=30) | Biopsy | ↑ 4 miRs | — | ↑ miR-155 (24–26) | ↓ miR-125a (24,27) |
| normal PBX (n=10) | ↓ 18 miRs | ↑ miR-150–5p (27) | ↓ miR-27b (24) | ||||
| ↓ miR-193b (24) | |||||||
| ↓ miR-181a (29) | |||||||
| ↓ miR-23b-3p (27) | |||||||
| ↓ miR-99b-5p (27) | |||||||
| Acute rejection | Oghumu et al. (27) | AR (heterogeneous) (n=5) | Biopsy | ↑ 13 miRs | — | ↑ miR-142–3p (24,25,29) | ↓ miR-30c-5p (24,26) |
| normal T0BX (n=4) | ↓ 16 miRs | ↑ miR-223–3p (24–26) | ↓ miR-125a-5p (19,24) | ||||
| ↑ miR-342–3p (24,29) | ↓ miR-204–5p (24,29) ↓ miR-23b-3p (19) | ||||||
| ↑ miR-150–5p (19) | ↓ miR-30a-5p (24) | ||||||
| ↓ miR-30d-5p (24) | |||||||
| ↓ miR-99b-5p (19) | |||||||
| ↓ miR-99a-5p (24) | |||||||
| ↓ miR-100–5p (24) | |||||||
| ↓ miR-125b-5p (24) | |||||||
| ↓ miR-126–3p (24) | |||||||
| ↓ miR-130a-3p (24) | |||||||
| Acute rejection | Liu et al. (26) | AR (n.o.s) (n=15) | Biopsy | 75 Differentially expressed miRs (fold changes not reported) | — | ↑ miR-155 (19,24,25) | ↓ miR-30c (24,27) |
| normal TxBX (n=15) | ↑ miR-223 (24,25,27) | ↓ miR-10b (24) | |||||
| ↑ miR-21 (24) | ↓ miR-17–3p (28) | ||||||
| ↑ miR-125a (28) | ↓ miR-30a-3p (24) | ||||||
| ↑ miR-146a (24) | ↓ miR-32 (24) | ||||||
| ↑ miR-602 (28) | ↓ miR-330 (28) | ||||||
| ↑ miR-628 (28) | ↓ miR-483 (28) | ||||||
| ↑ miR-629 (28) | ↓ miR-611 (28) | ||||||
| ↑ miR-650 (24) | ↓ miR-663 (28) | ||||||
| Acute T cell–mediated rejection | Bijkerk et al. (54) | Discovery cohort: stable Tx (clinical) (n=4) | Plasma | Not all differentially expressed miRs reported | ↑ miR-17 | ||
| Validation cohort: stable Tx (clinical) (n=13) | ↑ miR-140–3p | ||||||
| T cell–mediated rejection (n=13) | ↑ miR-130b | ||||||
| ↑ miR-122 | |||||||
| ↑ miR-192 | |||||||
| ↓ miR-135a | |||||||
| ↓ miR-199a-3p | |||||||
| ↓ miR-15a | |||||||
| Acute T cell–mediated rejection | Vitalone et al. (29) | T cell–mediated rejection (n=29) | Biopsy | ↑ 3 miRs | — | ↑ miR-142–3p (24,25,27) | ↓ miR-204 (24,27) |
| normal TxBX (n=68) | miR-142–3p | ↑ miR-342–3p (24,27) | ↓ miR 181a (19) | ||||
| miR-342–3p | |||||||
| miR-25 | |||||||
| ↓ 6 miRs | |||||||
| miR-181a | |||||||
| miR-192 | |||||||
| miR-204 | |||||||
| miR-215 | |||||||
| miR-10b-3p | |||||||
| miR-615–3p | |||||||
| Acute T cell–mediated rejection (Banff I) | Soltaninejad et al. (25) | T cell–mediated rejection (n=17) | Biopsy | ↑ 4 miR | — | ↑ miR-155 (19,24,26) | — |
| normal TxBX (n=18) | miR-142–5p | ↑ miR-142–3p (24,27,29) | |||||
| miR-155 | ↑ miR-223 (24,26,27) | ||||||
| miR-142–3p | ↑ miR-142–5p (24) | ||||||
| miR-223 | |||||||
| Acute antibody-mediated rejection | Wilflingseder et al. (19) | Morphologic antibody-mediated rejection (n=11) | Biopsy | ↑ 6 miRs | — | — | — |
| normal PBX (n=10) | |||||||
| Chronic antibody-mediated rejection | Danger et al. (30) | Chronic antibody-mediated rejection (n=18) | PBMC biopsy | Not all differentially expressed miRs reported | ↑ miR-142–5p | — | — |
| Stable Tx (clinical) (n=30) | ↑ miR-142–5p | — | — | ||||
| AR (heterogeneous) (n=9) | |||||||
| Chronic antibody-mediated rejection (n=21) | |||||||
| normal TxBx (n=18) | |||||||
| Chronic antibody-mediated rejection | Rascio et al. (31) | Discovery cohort: chronic ABRM (n=5) | PBMC | ↓ 16 miRs | ↓ miR-148b-3p | — | — |
| normal PBX (n=5) | ↓ miR-769–5p | ||||||
| Validation cohort: chronic antibody-mediated rejection (n=5) | ↓ miR-29b-3p | ||||||
| normal PBX (n=5) | |||||||
| Acute pyelonephritis | Oghumu et al. (27) | APN (n=11) | Biopsy | ↑ 24 miRs | — | — | — |
| AR (heterogeneous) (n=5) | ↓ 1 miR | ||||||
| IF/TA | Scian et al. (34) | Discovery cohort: IF/TA (n=13) | Biopsy | 56 Differentially expressed miRs (fold changes not reported) | ↑ miR-142–3p | ↑ miR-142–3p (32,33) | ↓ miR-211 (33) |
| normal PBX (n=5) | ↑ miR-32 | ||||||
| Validation cohort: IF/TA (n=19) | ↓ miR-204 | ||||||
| normal PBX (n=8) | ↓ miR-107 | ||||||
| ↓ miR-211 | |||||||
| IF/TA | Ben-Dov et al. (32) | Discovery cohort: n=4 IF/TA | Biopsy | ↑ 28 miRs | ↑ miR-142–3p | ↑ miR-142–3p (33,34) | — |
| n=4 normal PBX | ↓ 7 miRs | ↑ miR-142–5p | ↑ miR-21–5p (35) | ||||
| Validation cohort: n=10 IF/TA | ↑ miR-21–5p | ↑ miR-142–5p (33) | |||||
| n=8 normal PBX | ↑ miR-21–3p | ||||||
| ↑ miR-223 | |||||||
| ↓ miR-30b | |||||||
| ↓miR-30c | |||||||
| ↓ miR-338–3p | |||||||
| IF/TA | Glowacki et al. (35) | Severe graft fibrosis (explant) (n=11) | Biopsy | ↑ miR-21 | — | ↑ miR-21 (32) | — |
| nonpathologic parenchyma of urologic cancer (kidney/urinary tract) (n=12) | |||||||
| IF/TA | Soltaninejad et al. (33) | IF/TA (n=16) | Biopsy | ↑ miR-142–3p | — | ↑ miR-142–3p (32,34) | — |
| Normal TxBX (n=17) | ↑ miR-142–5p | ↑ miR-142–5p (32) | |||||
| ↓ miR-211 | |||||||
| MicroRNAs as biomarkers | |||||||
| Ischemia/reperfusion injury | Amrouche et al. (21) | LD (n=16) | Urine pellet | ↑ miR-146a | — | — | — |
| DD (n=35) | |||||||
| Acute T cell–mediated rejection (borderline, Banff I–II) | Lorenzen et al. (56) | Discovery cohort: T cell–mediated rejection (n=5) | Total urine | ↑ 5 miRs | ↑ miR-10a | — | — |
| normal PBX (n=5) | ↓ 16 miRs | ↓ miR-210 | |||||
| Validation cohort: T cell–mediated rejection (n=68) | ↓ miR-10b | ||||||
| normal PBX (n=20) | |||||||
| UTI (n=13) | |||||||
| Acute T cell–mediated rejection (Banff I–II) | Betts et al. (49) | T cell–mediated rejection (n=8) | Serum | ↑ miR-223 | — | — | — |
| pre-T cell–mediated rejection (n=3) | ↑ miR-10a | ||||||
| post-T cell–mediated rejection (n=6) | |||||||
| 1 yr T cell–mediated rejection (n=6) | |||||||
| healthy controls (n=4) | |||||||
| Acute rejection | Tao et al. (55) | Discovery cohort: AR (n.o.s) (n=4) | Serum | ↑ 6 miRs | ↑ miR-99a | — | — |
| stable Tx (clinical) (n=4) | ↑ miR-100 | ||||||
| Validation cohort: AR (n.o.s) (n=12) | |||||||
| delayed graft function (n=15) | |||||||
| stable Tx (clinical) (n=11) | |||||||
| T cell–mediated rejection | Millán et al. (50) | T cell–mediated rejection (n=8) | Urine pellet | ↑ miR-155 | — | — | — |
| no T cell–mediated rejection (n=71) | ↑ miR-142–3p | ||||||
| ↓ miR-210–3p | |||||||
| Acute T cell–mediated vascular rejection (Banff II–III) | Matz et al. (53) | Discovery cohort: stable Tx (clinical) (n=4) | Blood cells | 29 miRs differentially expressed miRs (fold changes not reported) | ↓ miR-15b | — | — |
| T cell–mediated vascular rejection (n=4) | ↓ miR-16 | ||||||
| Validation cohort: T cell–mediated vascular rejection (Banff II–III) (n=24) | ↓ miR-106a | ||||||
| stable Tx (clinical) (n=40) | ↓ miR-103a | ||||||
| UTI (n=11) | ↓ miR-107 | ||||||
| Borderline (n=17) | ↓ miR-15a | ||||||
| Banff IA–IB (n=15) | |||||||
| antibody-mediated rejection (n=15) | |||||||
| Mixed T cell–mediated rejection antibody-mediated rejection (n=6) | |||||||
| IF/TA (n=33) | |||||||
| Chronic antibody-mediated rejection | Danger et al. (30) | Discovery cohort: chronic antibody-mediated rejection (n=9) | PBMC | Not all differentially expressed miRs reported | ↑ miR-142–5p | — | — |
| stable Tx (clinical) (n=10) | |||||||
| Validation cohort: chronic antibody-mediated rejection (n=18) | |||||||
| stable Tx (clinical) (n=30) | |||||||
| AR (heterogeneous) (n=9) | |||||||
| IF/TA | Scian et al. (34) | IF/TA (n=7) | Urine pellet | ↑ miR-142–3p | — | ↑ miR-142–3p (51) | ↓ miR-211 (51) |
| normal PBX (n=7) | ↓ miR-211 | ↓ miR-204 (51) | |||||
| Prospective validation cohort: n=36 kidney Tx recipients (108 samples) | ↓ miR-204 | ||||||
| IF/TA | Maluf et al. (51) | First discovery cohort: IF/TA (n=10) | Urine pellet | First discovery cohort: ↑ 10 miRs | ↑ miR-142–3p | ↑ miR-142–3p (34) | ↓ miR-204 (34) |
| normal PBX (n=12) | ↓ 12 miRs | ↓ miR-125b | ↓ miR-211 (34) | ||||
| Validation cohort: IF/TA (n=7) | Second discovery cohort | ↓ miR-203 | |||||
| normal PBX (n=10) | 48 miRs differentially expressed miRs (fold changes not reported) | ↓ miR-204 | |||||
| Second discovery cohort (3 mo post Tx with IF/TA 24 mo) (n=10) | ↓ miR-211 | ||||||
| (3 mo post Tx without IF/TA 24 mo) (n=10) | Prospective validation cohort | ||||||
| Prospective validation cohort: n=66 kidney Tx recipients | ↑miR-200 | ||||||
| (132 samples) | ↑miR-140–3p | ||||||
| 3–6 mo | ↓ miR-99a | ||||||
| 18–24 mo | ↓miR-200b | ||||||
| IF/TA | Glowacki et al. (35) | IF/TA grade I (n=12) | Serum | ↑ miR-21 | — | — | — |
| IF/TA grade II (n=7) | |||||||
| IF/TA grade III (n=10) | |||||||
| no IF/TA (n=13) | |||||||
| IF/TA | Zununi et al. (52) | stable Tx (clinical) (n=27) | Plasma | ↑ miR-150 | — | — | — |
| IF/TA (n=26) | ↑ miR 423–3p | ||||||
| (grade I: n=16, grade III: n=10) | ↓ miR-192 (only IF/TA grade III) | ||||||
| ↓ miR-200b | |||||||
ATN, acute tubular necrosis; PBX, protocol biopsy; Scr, serum creatinine; miR, microRNA; T0BX, time-zero biopsy; RCC, renal cell carcinoma; AR, acute rejection; n.o.s., not otherwise specified; TxBX, transplant biopsy; Tx, transplantation; ABMR, antibody-mediated rejection; APN, acute pyelonephritis; IF/TA, interstitial fibrosis/tubular atrophy; LD, living donor; DD, deceased donor; UTI, urinary tract infection.
MicroRNA in the Pathophysiology of AKI and Transplant-Related Disorders
AKI
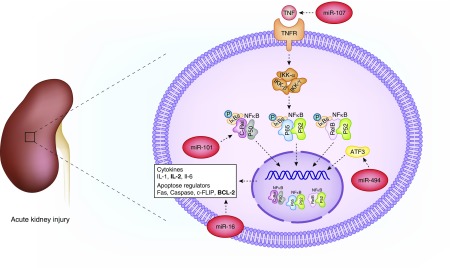
From limited human experiments, microRNAs appear to act via different mechanisms. Some microRNAs repress pathways that play a protective role in kidney physiology, whereas a proinflammatory effect by inhibition of anti-inflammatory pathways or mitochondrial function have also been described. Several microRNAs appear to play a role in AKI, where the release of multiple IL precedes structural kidney damage (Figure 2). These include miR-101 (IL2, NFκB pathway) (13), miR-494 (activating transcription factor 3 in NFκB pathway) (14), miR-16 (BCL-2) (15), and miR-107 (TNF) (16). Interestingly, urinary miR-494 levels, as opposed to serum levels, increase early in critically ill patients with AKI. In turn, miR-494 inhibits the expression of the kidney protective gene ATF3, resulting in aggravated kidney injury (14). C/EBP-β (C/enhancer binding protein-β) upregulated miR-16, which in turn blocked one of the antiapoptotic genes, BCL-2, after ischemia/reperfusion-induced injury (15). In septic patients with AKI, increased miR-107 induced TNF secretion by targeting DUSP7 (dual specificity protein phosphatase 7) in endothelial cells, which may directly cause tubular injury (16). In vitro inhibition of this microRNA resulted in attenuated TNF secretion and prevented subsequent tubular cell injury (16). In a study by Ge et al. (17), 37 microRNAs were differentially expressed in the serum of patients with sepsis-induced AKI versus those with non-sepsis AKI. Eight of them were associated with 13 genes involved in mitochondrial oxidative stress and dysfunction response, including peroxisome proliferator-activated receptor γ coactivator 1-α, sirtuin 1, mammalian target of rapamycin, oxidative stress responsive 1, and NADPH oxidase 5 (17). Congruent with these observations, upregulation of kidney tubular miR-709 after cisplatin-induced AKI hampers mitochondrial function and induces cell apoptosis (18).
Figure 2.
Several microRNAs are involved in the NFκB pathway in AKI. Several microRNA contribute to AKI by targeting the NFκB pathway. ATF3, activating transcription factor 3; BCL-2, B cell lymphoma-2; c-FLIP, cellular FLICE-like inhibitory protein; c-Rel, P50, P52, P65 and RelB, NF-κB transcription factor family members; Fas, first apoptosis signal; IKK, IκB kinase; TNFR, TNF receptor.
Kidney Transplantation
Ischemia/Reperfusion Injury and Delayed Graft Function.
A schematic overview of the pathogenic role of different microRNAs in transplantation-related kidney disease is given in Figure 3. MicroRNAs are involved in the regulation of angiogenesis and apoptosis through TGF-β, endothelin, vascular endothelial growth factor, and PDGF signaling (19). Upregulation of miR-182–5p, miR-21–3p, and miR-146a have been reported (20,21). The overexpression of miR-146a probably represents a compensatory mechanism because in vitro experiments identified the role of miR-146a as a negative regulator of inflammation in tubular cells by downregulation of the NFκB/C-X-C motive chemokine ligand 8 pathway (21). In multivariable logistic regression analysis, the expression of miR-217 and miR-125b (both targeting cyclin-dependent kinase inhibitor 2 loci) in preimplantation biopsy samples together with donor age and type were independently associated with delayed graft function, which could be predicted in 84% of cases (22). Recently, delayed graft function was identified as a manifestation of allostatic overload at a transcriptional level (23). A composite indicator of accumulated biologic stress over the life course is defined as allostatic load, which predisposes to morbidity in case of chronic or repeated stress exposure. Organs undergoing delayed graft function exhibited a greater magnitude of change in transcriptional amplitude and elevated expression of noncoding RNAs and pseudogenes, consistent with increased allostatic load than in those showing immediate graft function. Notably, this study incorporated a validation biopsy set and individual validation of targets transcriptionally and post-transcriptionally. Additionally, it undertook a crosscomparison with publicly available data sets for kidney pathologies, used to identify significant transcriptional commonality for over 20 delayed graft function transcripts providing a clear molecular signature for the burden of “wear and tear” within the kidney and age-related physiologic capability and resilience. The expression of the CDKN2 locus transcripts in this cohort related to the delayed graft function outcome and perfusion status at the transcript level. These results indicate that CDKN2A/p16INK4, ARF/p14, and CDKN2B reflected the allostatic load (and biologic age) of these organs preperfusion. Regulation of these loci by miR-125b is a notable feature.
Figure 3.
Several specific microRNAs are up- or downregulated in the kidney graft in transplantation-related kidney disease. Functional processes are represented in the inner circle, and the outer circle represents molecular pathways of interest for the particular clinical entity. Only the biologically confirmed microRNA–target interactions are highlighted. The color code of the microRNAs indicates whether the tissue microRNA are mimicked in biofluids. Red represents parallel findings in plasma/serum for that particular microRNA. Orange represents parallel findings in urine for that particular microRNA and blue represents parallel findings in PBMC. Combination of the colors is possible. ABMR, antibody-mediated rejection; BCL-2, B cell lymphoma-2; CDKN2, cyclin-dependent kinase inhibitor 2; CXCL8, C-X-X motive chemokine ligand 8; ECM, extracellular matrix; EDN, endothelin; IRI/DGF, ischemia/reperfusion injury and delayed graft function; NFκB/CXCL-8, NFκB/C-X-C motive chemokine ligand 8; TCMR, T cell–mediated rejection; TGFβ/FOXP3, TGFβ/forkhead box P3; TGFβ/SMAD7, TGFβ/SMAD family member 7; VEGF, vascular endothelial growth factor.
T Cell–Mediated Rejection.
Global miR expression profiling of grafts with T cell–mediated rejection showed that miR-142–5p, miR-155, and miR-223 each predict T cell–mediated rejection with high sensitivity and specificity (area under the curve, 0.96–0.99) (24). Their correlation with intragraft CD3 and CD20 mRNA levels suggests that these miRs originate from immune cells infiltrated in the graft (24). Other groups have shown similar patterns for miR-142–5p (25), miR-155 (19,25,26), and miR-223 (25–27). In addition, miR-10b (antiapoptotic targeting BCL211 (26)) and miR-30a-3p appeared to be downregulated in rejecting graft tissue, as well as correlating with kidney tubule–specific mRNAs (Na+-K+-2Cl− cotransporter) (24). In a small set of biopsy specimens, eight miRs were upregulated and 12 miRs were downregulated in T cell–mediated rejection (28).
Vitalone et al. (29) identified 19 microRNAs that may target the differentially expressed mRNAs in T cell–mediated rejection. Validation of these microRNAs revealed significant upregulation of three miRs (Table 1) and downregulation of six miRs in rejecting versus nonrejecting grafts. All upregulated miRs were associated with tubulitis and interstitial inflammation, suggesting infiltrating lymphocytes as the origin of these miRs, whereas miR-204, miR-210, and miR-10b-3p negatively correlated with Banff scores. The TGF-β signaling pathway, and in particular forkhead box P3 regulated transcription, is common to the regulatory action of all these miRs (29).
Oghumu et al. (27) have identified a panel of 25 miRs significantly differently expressed in grafts from recipients with acute rejection compared with acute pyelonephritis. Interestingly, some previously reported downregulated miRs in T cell–mediated rejection grafts, including miR-23–3p (19), miR-30a-5p (24), miR-30d-5p (24), miR-30c-5p (24,26), and miR-99b-5p (19), were significantly upregulated in acute pyelonephritis compared with rejection biopsy specimens (27).
Antibody-Mediated Rejection.
Upregulation of miR-146–5p, miR-182, miR-21–3p, miR-1228, and let-7i, involved in inflammation, chemokine and cytokine signaling, apoptosis, and IL signaling, has been observed in grafts with antibody-mediated rejection (19). Increased miR-142–5p expression levels were observed in PBMC and grafts from recipients with chronic antibody-mediated rejection compared with normal allografts and PBMC from stable kidney transplant recipients. miR-142–5p overexpression was associated with downregulation of 41 genes related to a cell-mediated immune response (30). Of note, miR-146–5p as well as miR-142–5p were also significantly upregulated in grafts with T cell–mediated rejection (24,25).
To unravel the molecular mechanisms underlying chronic antibody-mediated rejection, Rascio et al. (31) performed a combined mRNA and miR expression analysis in PBMC from kidney transplant recipients with chronic antibody-mediated rejection and normal allografts. Four miRs were found to be modulators of six mRNAs involved in the type I IFN signaling network. miR validation in an independent set of PBMC revealed a significant downregulation of miR-148b-3p, miR-29b-3p, and miR-769–5p. No overlapping miR signature could be identified with the data from Danger et al. (30), possibly because of methodological differences and definition of the controls.
Interstitial Fibrosis and Tubular Atrophy.
Fifteen miRs have been identified as being of interest in interstitial fibrosis and tubular atrophy (IF/TA), relating to regulation of lymphocyte proliferation and B, T, and natural killer cell activation/differentiation. The expression of five of these miRs has been independently validated, with miR-142–3p (32,33) and miR-32 being upregulated and miR-204, miR-107, and miR-211 (33) being downregulated in grafts with IF/TA (34). The upregulation of miR-142–5p, miR-21 (35) (target: SMAD7, an inhibitor of TGFβ-mediated fibrosis), and miR-223, and downregulation of miR-30b, miR-30c, and miR-338–3p were confirmed in an independent but small set of IF/TA biopsy specimens (36). Similar miR expression data for miR-142–3p (24,25,27,29), miR-142–5p (24,25), miR-223 (24–26), miR-204 (24,27,29), miR-30c (24,26,27), and miR-30b (24) was found in intragraft miR profiling studies on rejecting allografts, particularly T cell–mediated rejection.
MicroRNA as Biomarkers of Kidney Disease
Because microRNAs are highly stable in plasma and urine, they are attractive biomarkers.
AKI.
AKI coincides with reduced expression levels of most, but not all, circulating miRs (37). In the plasma of patients with AKI, miR-16 and miR-320 were found to be down-regulated, whereas miR-210 was upregulated (38). This upregulation is a strong independent prognostic factor for 28-day survival of critically ill patients with AKI (38). Likewise, urinary levels of miR-21 and miR-155 could successfully distinguish patients with and without AKI (36). This is in keeping with other findings that urinary miR-21 appeared to be more associated with AKI prognosis than plasma miR-21 levels (39,40). Serum miR-21 levels were associated with the development of AKI when sampled before cardiac surgery (41) and 6 hours after cardiac surgery (42). In the latter study, urinary miR-21 levels were also associated with AKI (42). Interestingly, ischemic preconditioning could increase endogenous miR-21 expression and further protect kidney function (43).
Urinary miR-200c and miR-423 were upregulated in AKI on the basis of microRNA array analyses (39). A panel of 10 miRs could be used for AKI diagnosis in patients in intensive care with nearly 100% sensitivity and specificity and four of them were associated with AKI severity (44). Another set of four miRs was associated with AKI development before S-creatinine rose after cardiac surgery (44). miR-192 was assessed for diagnosis of AKI when sampled 2 hours after cardiac surgery; however, it had a rather poor sensitivity and specificity (45). Likewise, urinary miR-30c-5p performed well as a biomarker of AKI after cardiac surgery, and even better compared with protein-based markers such as neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 (46). In contrast-induced nephropathy, miR-30a, c, and e appeared to be significantly higher in comparison with patients who received contrast but without nephropathy (47). Although all three miRs correlated positively with serum creatinine, they only increased in 55.5% of patients with contrast-induced nephropathy. The positive predictive value of these three miRs varied between 91.3% and 94.9%, whereas the negative predictive value varied between 61.3% and 78.2% (47). Sun et al. (48) confirmed the increase of miR-30a and e and, in addition, they found miR-188 to be increased in a similar population.
Transplantation.
The value of microRNA as biomarkers of different graft-associated pathologies were investigated either by quantification of a set of miRs known to be dysregulated in the graft (21,25,33–35,49–51) or involved in pathways of interest (52), or by performing a global miR profiling on blood cells (30,53), serum/plasma (54,55), and urine (51,56).
Ischemic-Reperfusion Injury.
A significant upregulation of miR-146a was observed in urine samples of recipients transplanted with a deceased donor compared with a living donor, and was thus suggested as a diagnostic marker for ischemia/reperfusion injury (21).
T Cell–Mediated Rejection.
Both senescence associated miR-223 and miR-142–3p are upregulated in the graft and PBMC of patients with acute T cell–mediated rejection (25). Increased miR-223 levels, along with increased levels of miR-10a, were also observed in the serum of a small number of transplant recipients during T cell–mediated rejection (49). The upregulation of serum miR-99a and miR-100 was also reported in kidney transplant recipients with T cell–mediated rejection (55). However, previous miR profiling studies reported decreased levels of miR-99a expression in T cell–mediated rejection kidney allografts (24,27). Paired tissue and blood analysis should therefore be performed to determine the significance of these conflicting results.
In multivariable logistic regression analysis, a panel of five miRs isolated from blood cells (miR-15b, miR-16, miR103a, miR106a, and miR-107) could accurately discriminate an acute vascular rejection (Banff II–III) from stable graft function. The difference between T cell–mediated vascular rejection and other phenotypes was less distinct (53).
Urinary levels of miR-10a were significantly upregulated, whereas miR-10b and miR-210 were downregulated in urine samples of recipients with acute T cell–mediated rejection. Furthermore, expression levels of urinary miR-210, involved in cellular aging, were related to biopsy-proven rejection severity with levels normalizing after rejection treatment. However, receiver operating characteristic (ROC) analysis revealed a weak specificity and sensitivity for the distinction between acute rejection and stable graft function (56). A decreased expression of miR-210–3p was confirmed in urine pellets from transplant recipients with T cell–mediated rejection (50). In this study, higher expression levels of urine miR-155–5p, also reported as highly expressed in the graft during acute T cell–mediated rejection (19,24–26), were more discriminative for the diagnosis of acute rejection (50).
Antibody-Mediated Rejection.
In PBMC from recipients with chronic, but not acute, antibody-mediated rejection, miR-142–5p was upregulated. In ROC analysis, the discriminative capacity for chronic antibody-mediated rejection versus stable controls was fair (30). Although the authors suggest the specificity of this miR in chronic antibody-mediated rejection, other groups also reported increased levels of this hematopoietic miR in PBMC and grafts of recipients with an acute T cell–mediated rejection (24,25).
IF/TA.
Lower miR-211 and miR-204 expression levels and an upregulation of miR-142–3p were found in urine pellets of recipients with biopsy-proven IF/TA compared with recipients with a normal histology and graft function (34). miR levels in the urine appeared to be correlated with miR expression levels in the graft (34). These findings were confirmed in a cohort of recipients with established IF/TA (51). A significant downregulation of miR-200b, miR-375, and miR-193b, and upregulation of miR-423–5p and miR-345 has been observed in these two miR discovery data sets. A larger prospective validation study revealed a significant downregulation of miR-200b in urine of recipients with established IF/TA 1 year after kidney transplantation. No correlation was found between the expression of miR-200b and proteinuria (51). A significant downregulation of miR-200b was also reported in plasma of recipients with IF/TA (52). Higher expression levels of miR-21 were measured from recipients with biopsy-proven IF/TA, thereby showing a gradually increase with IF/TA severity. Furthermore, no correlations were found between miR-21 levels and the presence of other acute or chronic Banff lesions in this study (35).
Discussion
Insight in the pathophysiological role of miRs in CKD is growing, and miR targeting therapies are being introduced in the clinic. In contrast, the role of microRNAs as therapeutic targets in the pathophysiology of AKI and kidney transplantation is still in the exploratory phase and needs confirmation and validation. Few articles show that in AKI, microRNAs are strong regulators of the NFκB pathway as a major target in proinflammatory diseases. However, the NFκB pathway also plays a role in anti-inflammatory processes via its antiapoptotic functions. This dual effect has hampered the development of anti-NFκB pathway drugs. In the kidney transplantation field, more insights in the pathophysiology of transplant-related processes, as well as biomarkers for diagnosis, are awaited. Given their remarkable stability, several microRNAs have been put forward as biomarkers for the diagnosis of kidney diseases and transplant related pathologies. Rather poor performance in ROC analyses currently hampers the clinical implementation of microRNAs as diagnostic or prognostic markers. However, after kidney transplantation, a combined panel of five miRs was able to discriminate T cell–mediated vascular rejection form stable graft function (53). Thus, it is likely that the use of microRNA combinations (panels) will increase their clinical utility. Because most studies remain in the exploratory phase, there is a need for larger clinical prospective trials to validate the results and thoroughly investigate their diagnostic and prospective potential. A standardized method for sampling and analysis is recommended to improve between-group comparison in external validation set-ups.
Search Terms
The following databases were used: PubMed and Web of Science. No limits were applied on publication date and the last database search was performed on May 25, 2018. The following MeSH terms were used: “microRNA or microRNA or miR AND AKI”; microRNA or microRNA or miR AND renal function”; “microRNA or microRNA or miR AND acute renal impairment”; and “MicroRNAs AND Kidney Transplantation.” Only articles on human research were withheld for this review.
Disclosures
None.
Acknowledgments
We thank Erik Snelders for his help with the figures. Drawings are on the basis of material from the Servier Medical Art image bank (https://smart.servier.com/, creative commons license https://creativecommons.org/licenses/by/3.0/).
E.M.G. is supported by the Research Foundation Flanders (PhD fellowship 1136317N).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Michell DL, Vickers KC: Lipoprotein carriers of microRNAs. Biochim Biophys Acta 1861[12 Pt B]: 2069–2074, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayraktar R, Van Roosbroeck K, Calin GA: Cell-to-cell communication: MicroRNAs as hormones. Mol Oncol 11: 1673–1686, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Zhou X, Zhang H, Yao Q, Liu Y, Dong Z: Extracellular vesicles in diagnosis and therapy of kidney diseases. Am J Physiol Renal Physiol 311: F844–F851, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang A, Li M, Wang B, Klein JD, Price SR, Wang XH: miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia Muscle 9: 755–770, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupaimoole R, Slack FJ: MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16: 203–222, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Patel V, Williams D, Hajarnis S, Hunter R, Pontoglio M, Somlo S, Igarashi P: miR-17∼92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci U S A 110: 10765–10770, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajarnis S, Lakhia R, Yheskel M, Williams D, Sorourian M, Liu X, Aboudehen K, Zhang S, Kersjes K, Galasso R, Li J, Kaimal V, Lockton S, Davis S, Flaten A, Johnson JA, Holland WL, Kusminski CM, Scherer PE, Harris PC, Trudel M, Wallace DP, Igarashi P, Lee EC, Androsavich JR, Patel V: microRNA-17 family promotes polycystic kidney disease progression through modulation of mitochondrial metabolism. Nat Commun 8: 14395, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, Xia TH, Liu X, Borza DB, Grafals M, Shankland SJ, Himmelfarb J, Portilla D, Liu S, Chau BN, Duffield JS: Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest 125: 141–156, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv W, Fan F, Wang Y, Gonzalez-Fernandez E, Wang C, Yang L, Booz GW, Roman RJ: Therapeutic potential of microRNAs for the treatment of renal fibrosis and CKD. Physiol Genomics 50: 20–34, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevaert AB, Witvrouwen I, Vrints CJ, Heidbuchel H, Van Craenenbroeck EM, Van Laere SJ, Van Craenenbroeck AH: MicroRNA profiling in plasma samples using qPCR arrays: Recommendations for correct analysis and interpretation. PLoS One 13: e0193173, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClelland R, Christensen K, Mohammed S, McGuinness D, Cooney J, Bakshi A, Demou E, MacDonald E, Caslake M, Stenvinkel P, Shiels PG; work was done on behalf of the pSoBiD team: Accelerated ageing and renal dysfunction links lower socioeconomic status and dietary phosphate intake. Aging (Albany NY) 8: 1135–1149, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiels PG, McGuinness D, Eriksson M, Kooman JP, Stenvinkel P: The role of epigenetics in renal ageing. Nat Rev Nephrol 13: 471–482, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Hua R, Gong Z, Shang B, Huang Y, Guo L, Liu T, Xue J: Human amniotic epithelial cells inhibit CD4+ T cell activation in acute kidney injury patients by influencing the miR-101-c-Rel-IL-2 pathway. Mol Immunol 81: 76–84, 2017 [DOI] [PubMed] [Google Scholar]
- 14.Lan YF, Chen HH, Lai PF, Cheng CF, Huang YT, Lee YC, Chen TW, Lin H: MicroRNA-494 reduces ATF3 expression and promotes AKI. J Am Soc Nephrol 23: 2012–2023, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HH, Lan YF, Li HF, Cheng CF, Lai PF, Li WH, Lin H: Urinary miR-16 transactivated by C/EBPβ reduces kidney function after ischemia/reperfusion-induced injury. Sci Rep 6: 27945, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Zhang Z, Wang J, Miao H: MiR-107 induces TNF-α secretion in endothelial cells causing tubular cell injury in patients with septic acute kidney injury. Biochem Biophys Res Commun 483: 45–51, 2017 [DOI] [PubMed] [Google Scholar]
- 17.Ge QM, Huang CM, Zhu XY, Bian F, Pan SM: Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS One 12: e0173292, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Ni J, Chen S, Bai M, Lin J, Ding G, Zhang Y, Sun P, Jia Z, Huang S, Yang L, Zhang A: MicroRNA-709 mediates acute tubular injury through effects on mitochondrial function. J Am Soc Nephrol 29: 449–461, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilflingseder J, Regele H, Perco P, Kainz A, Soleiman A, Mühlbacher F, Mayer B, Oberbauer R: miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation 95: 835–841, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilflingseder J, Sunzenauer J, Toronyi E, Heinzel A, Kainz A, Mayer B, Perco P, Telkes G, Langer RM, Oberbauer R: Molecular pathogenesis of post-transplant acute kidney injury: Assessment of whole-genome mRNA and miRNA profiles. PLoS One 9: e104164, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amrouche L, Desbuissons G, Rabant M, Sauvaget V, Nguyen C, Benon A, Barre P, Rabaté C, Lebreton X, Gallazzini M, Legendre C, Terzi F, Anglicheau D: MicroRNA-146a in human and experimental ischemic AKI: CXCL8-dependent mechanism of action. J Am Soc Nephrol 28: 479–493, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGuinness D, Leierer J, Shapter O, Mohammed S, Gingell-Littlejohn M, Kingsmore DB, Little AM, Kerschbaum J, Schneeberger S, Maglione M, Nadalin S, Wagner S, Königsrainer A, Aitken E, Whalen H, Clancy M, McConnachie A, Koppelstaetter C, Stevenson KS, Shiels PG: Identification of molecular markers of delayed graft function based on the regulation of biological ageing. PLoS One 11: e0146378, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuinness D, Mohammed S, Monaghan L, Wilson PA, Kingsmore DB, Shapter O, Stevenson KS, Coley SM, Devey L, Kirkpatrick RB, Shiels PG: A molecular signature for delayed graft function. Aging Cell 17: e12825, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M: MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A 106: 5330–5335, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soltaninejad E, Nicknam MH, Nafar M, Ahmadpoor P, Pourrezagholi F, Sharbafi MH, Hosseinzadeh M, Foroughi F, Yekaninejad MS, Bahrami T, Sharif-Paghaleh E, Amirzargar A: Differential expression of microRNAs in renal transplant patients with acute T-cell mediated rejection. Transpl Immunol 33: 1–6, 2015 [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Dong C, Jiang Z, Wu WK, Chan MT, Zhang J, Li H, Qin K, Sun X: MicroRNA-10b downregulation mediates acute rejection of renal allografts by derepressing BCL2L11. Exp Cell Res 333: 155–163, 2015 [DOI] [PubMed] [Google Scholar]
- 27.Oghumu S, Bracewell A, Nori U, Maclean KH, Balada-Lasat JM, Brodsky S, Pelletier R, Henry M, Satoskar AR, Nadasdy T, Satoskar AA: Acute pyelonephritis in renal allografts: A new role for microRNAs? Transplantation 97: 559–568, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H: Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol 19: 81–85, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Vitalone MJ, Sigdel TK, Salomonis N, Sarwal RD, Hsieh SC, Sarwal MM: Transcriptional perturbations in graft rejection. Transplantation 99: 1882–1893, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danger R, Paul C, Giral M, Lavault A, Foucher Y, Degauque N, Pallier A, Durand M, Castagnet S, Duong Van Huyen JP, Delahousse M, Renaudin K, Soulillou JP, Brouard S: Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PLoS One 8: e60702, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rascio F, Pontrelli P, Accetturo M, Oranger A, Gigante M, Castellano G, Gigante M, Zito A, Zaza G, Lupo A, Ranieri E, Stallone G, Gesualdo L, Grandaliano G: A type I interferon signature characterizes chronic antibody-mediated rejection in kidney transplantation. J Pathol 237: 72–84, 2015 [DOI] [PubMed] [Google Scholar]
- 32.Ben-Dov IZ, Muthukumar T, Morozov P, Mueller FB, Tuschl T, Suthanthiran M: MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation 94: 1086–1094, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soltaninejad E, Nicknam MH, Nafar M, Sharbafi MH, Keshavarz Shahbaz S, Barabadi M, Yekaninejad MS, Bahrami T, Ahmadpoor P, Amirzargar A: Altered expression of microRNAs following chronic allograft dysfunction with interstitial fibrosis and tubular atrophy. Iran J Allergy Asthma Immunol 14: 615–623, 2015 [PubMed] [Google Scholar]
- 34.Scian MJ, Maluf DG, David KG, Archer KJ, Suh JL, Wolen AR, Mba MU, Massey HD, King AL, Gehr T, Cotterell A, Posner M, Mas V: MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant 11: 2110–2122, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glowacki F, Savary G, Gnemmi V, Buob D, Van der Hauwaert C, Lo-Guidice JM, Bouyé S, Hazzan M, Pottier N, Perrais M, Aubert S, Cauffiez C: Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One 8: e58014, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, Brown RP, Bijol V, Park PJ, Waikar SS, Vaidya VS: Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci 129: 256–267, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vliegenthart AD, Shaffer JM, Clarke JI, Peeters LE, Caporali A, Bateman DN, Wood DM, Dargan PI, Craig DG, Moore JK, Thompson AI, Henderson NC, Webb DJ, Sharkey J, Antoine DJ, Park BK, Bailey MA, Lader E, Simpson KJ, Dear JW: Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury. Sci Rep 5: 15501, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kümpers P, Faulhaber-Walter R, Haller H, Fliser D, Thum T: Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol 6: 1540–1546, 2011 [DOI] [PubMed] [Google Scholar]
- 39.Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, Waikar SS, Vaidya VS: Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem 59: 1742–1752, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, Cao X, Zou L, Chen Y, Guo J, Chen Z, Hu S, Zheng Z: MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS One 8: e63390, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaede L, Liebetrau C, Blumenstein J, Troidl C, Dörr O, Kim WK, Gottfried K, Voss S, Berkowitsch A, Walther T, Nef H, Hamm CW, Möllmann H: Plasma microRNA-21 for the early prediction of acute kidney injury in patients undergoing major cardiac surgery. Nephrol Dial Transplant 31: 760–766, 2016 [DOI] [PubMed] [Google Scholar]
- 42.Arvin P, Samimagham HR, Montazerghaem H, Khayatian M, Mahboobi H, Ghadiri Soufi F: Early detection of cardiac surgery-associated acute kidney injury by microRNA-21. Bratisl Lek Listy 118: 626–631, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Kang Z, Li Z, Huang P, Luo J, Liu P, Wang Y, Xia T, Zhou Y: Remote ischemic preconditioning upregulates microRNA-21 to protect the kidney in children with congenital heart disease undergoing cardiopulmonary bypass. Pediatr Nephrol 33: 911–919, 2018 [DOI] [PubMed] [Google Scholar]
- 44.Aguado-Fraile E, Ramos E, Conde E, Rodríguez M, Martín-Gómez L, Lietor A, Candela Á, Ponte B, Liaño F, García-Bermejo ML: A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS One 10: e0127175, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Xu Y, Xue S, Wang X, Dai H, Qian J, Ni Z, Yan Y: Implications of dynamic changes in miR-192 expression in ischemic acute kidney injury. Int Urol Nephrol 49: 541–550, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou YF, Wen D, Zhao Q, Shen PY, Shi H, Zhao Q, Chen YX, Zhang W: Urinary microRNA-30c-5p and microRNA-192-5p as potential biomarkers of ischemia-reperfusion-induced kidney injury. Exp Biol Med (Maywood) 242: 657–667, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutiérrez-Escolano A, Santacruz-Vázquez E, Gómez-Pérez F: Dysregulated microRNAs involved in contrast-induced acute kidney injury in rat and human. Ren Fail 37: 1498–1506, 2015 [DOI] [PubMed] [Google Scholar]
- 48.Sun SQ, Zhang T, Ding D, Zhang WF, Wang XL, Sun Z, Hu LH, Qin SY, Shen LH, He B: Circulating microRNA-188, -30a, and -30e as early biomarkers for contrast-induced acute kidney injury. J Am Heart Assoc 5: piie004138, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betts G, Shankar S, Sherston S, Friend P, Wood KJ: Examination of serum miRNA levels in kidney transplant recipients with acute rejection. Transplantation 97: e28–e30, 2014 [DOI] [PubMed] [Google Scholar]
- 50.Millán O, Budde K, Sommerer C, Aliart I, Rissling O, Bardaji B, Matz M, Zeier M, Silva I, Guirado L, Brunet M: Urinary miR-155-5p and CXCL10 as prognostic and predictive biomarkers of rejection, graft outcome and treatment response in kidney transplantation. Br J Clin Pharmacol 83: 2636–2650, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maluf DG, Dumur CI, Suh JL, Scian MJ, King AL, Cathro H, Lee JK, Gehrau RC, Brayman KL, Gallon L, Mas VR: The urine microRNA profile may help monitor post-transplant renal graft function. Kidney Int 85: 439–449, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zununi Vahed S, Poursadegh Zonouzi A, Mahmoodpoor F, Samadi N, Ardalan M, Omidi Y: Circulating miR-150, miR-192, miR-200b, and miR-423-3p as non-invasive biomarkers of chronic allograft dysfunction. Arch Med Res 48: 96–104, 2017 [DOI] [PubMed] [Google Scholar]
- 53.Matz M, Fabritius K, Lorkowski C, Dürr M, Gaedeke J, Durek P, Grün JR, Goestemeyer A, Bachmann F, Wu K, Rudolph B, Schmidt D, Weber U, Haftmann C, Unterwalder N, Lachmann N, Radbruch A, Neumayer HH, Mashreghi MF, Budde K: Identification of T cell-mediated vascular rejection after kidney transplantation by the combined measurement of 5 specific microRNAs in blood. Transplantation 100: 898–907, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Bijkerk R, Florijn BW, Khairoun M, Duijs JMGJ, Ocak G, de Vries APJ, Schaapherder AF, Mallat MJK, de Fijter JW, Rabelink TJ, van Zonneveld AJ, Reinders MEJ: Acute rejection after kidney transplantation associates with circulating microRNAs and vascular injury. Transplant Direct 3: e174, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao J, Yang X, Han Z, Lu P, Wang J, Liu X, Wu B, Wang Z, Huang Z, Lu Q, Tan R, Gu M: Serum microRNA-99a helps detect acute rejection in renal transplantation. Transplant Proc 47: 1683–1687, 2015 [DOI] [PubMed] [Google Scholar]
- 56.Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, Gwinner W, Thum T: Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant 11: 2221–2227, 2011 [DOI] [PubMed] [Google Scholar]