Abstract
Background
A majority of women with polycystic ovary syndrome (PCOS) have metabolic dysfunction that results in an increased risk of type 2 diabetes. We previously developed a pubertal mouse model using the aromatase inhibitor, letrozole, which recapitulates many of the reproductive and metabolic features of PCOS. To further our understanding of the effects of androgen excess, we compared the effects of letrozole treatment initiated in puberty versus adulthood on reproductive and metabolic phenotypes as well as on the gut microbiome.
Results
Letrozole treatment of both pubertal and adult female mice resulted in reproductive hallmarks of PCOS, including hyperandrogenemia, anovulation and polycystic ovaries. However, unlike pubertal mice, treatment of adult female mice resulted in modest weight gain and abdominal adiposity, minimal elevation in fasting blood glucose and insulin levels, and no detectable insulin resistance. In addition, letrozole treatment of adult mice was associated with a distinct shift in gut microbial diversity compared to letrozole treatment of pubertal mice.
Conclusions
Our results indicate that dysregulation of metabolism and the gut microbiome in PCOS may be influenced by the timing of androgen exposure. In addition, the minimal weight gain and lack of insulin resistance in adult female mice after letrozole treatment indicates that this model may be useful for investigating the effects of hyperandrogenemia on the hypothalamic-pituitary-gonadal axis and the periphery without the influence of substantial metabolic dysregulation.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1425-7) contains supplementary material, which is available to authorized users.
Keywords: Gut microbiome, Polycystic ovary syndrome, Hyperandrogenism, Puberty
Background
Polycystic ovary syndrome (PCOS) is the most common endocrine disorder in reproductive-aged women with an estimated world-wide prevalence of 6–15%, but the etiology of PCOS is not well understood [1]. Heritability and twin studies have identified a strong genetic component that is likely polygenic [2–4]. Recent genome-wide association studies have reported multiple susceptibility loci associated with an increased risk of developing PCOS [5]. Environmental factors, such as prenatal exposure to androgens may also play a role in the etiology of PCOS [6]. Currently, diagnosis is made using the Rotterdam Consensus criteria (2003), which require at least two of the following: hyperandrogenism, oligo- or amenorrhea and polycystic ovaries [1].
Studies have shown that women with PCOS often suffer from profound, long-term health issues [7]. PCOS is the leading cause of anovulatory infertility in women and increases the likelihood of miscarriage and pregnancy complications [8, 9]. In addition, a majority of women with PCOS have abnormalities that increase their risk of developing metabolic disease [1, 10–15]. A large, retrospective study demonstrated that PCOS was associated with an increased risk of obesity (16 vs. 3.7%) and type 2 diabetes (12.5 vs. 3.8%) over a 15-year period [16]. Studies show that hyperandrogenism is strongly correlated with development of a metabolic phenotype. Metabolic dysfunction occurs predominantly in women diagnosed with hyperandrogenism and ovulatory dysfunction, independent of body mass index [17, 18].
A complex community of microorganisms (the microbiome) resides within the large intestine and is important for human health [19, 20]. Correlative studies have demonstrated that the gut microbiome of individuals with metabolic disorders, such as obesity and diabetes, differ significantly from healthy individuals [21–25]. In addition, mouse models of obesity are associated with gut microbiome dysregulation [26–31]. Studies have also shown that fecal transplantation of the gut microbiome from obese individuals into germ-free mice results in an obese phenotype [22, 32, 33], indicating a potential role of the gut microbiome in the development of metabolic disorders [34]. Recent studies indicate that changes in the gut microbiome are associated with PCOS. Women diagnosed with PCOS using the Rotterdam criteria were reported to have a significant reduction in the overall bacterial species richness (alpha diversity) of the gut microbial community and changes in the abundance of several bacterial taxa compared to healthy women [35–37]. Interestingly, a study from our lab also showed a significant correlation between hyperandrogenism and diversity of the gut microbiome, suggesting that androgens may influence the composition of the gut microbiome in women [37].
Since hyperandrogenism is associated with PCOS, researchers have created animal models to study the role of androgens in the development and pathology of PCOS [reviewed in [38–42]]. Several mouse models were developed using treatment with exogenous dihydrotestosterone but these models did not exhibit the elevated LH levels associated with PCOS [43–47]. We developed a PCOS mouse model in pubertal female mice using treatment with the aromatase inhibitor, letrozole, to limit the conversion of testosterone to estrogen which results in increased testosterone and decreased estrogen levels. This model is based on the findings that genetic variants of the aromatase gene are associated with the development of PCOS in women and that a higher androgen/estrogen ratio is found in the ovaries of women with PCOS [48–52]. We demonstrated that this mouse model has many hallmarks of PCOS including hyperandrogenemia, elevated LH levels, acyclicity, and polycystic ovaries [53, 54]. This model also exhibited a metabolic phenotype including weight gain, abdominal adiposity, dysglycemia, hyperinsulinemia, and insulin resistance after 5 weeks of letrozole treatment [55]. Similar to women with PCOS, we also showed that there was a significant decrease in the alpha diversity of the gut microbiome in the letrozole-induced PCOS mouse model that correlated with hyperandrogenism [54]. To gain more insight into the effects of androgen excess, we investigated whether the timing of testosterone exposure was important for the pathophysiology of PCOS by evaluating the effects of letrozole treatment on reproductive and metabolic phenotypes in pubertal versus adult female mice.
Results
Letrozole treatment of adult female mice resulted in reproductive hallmarks of PCOS
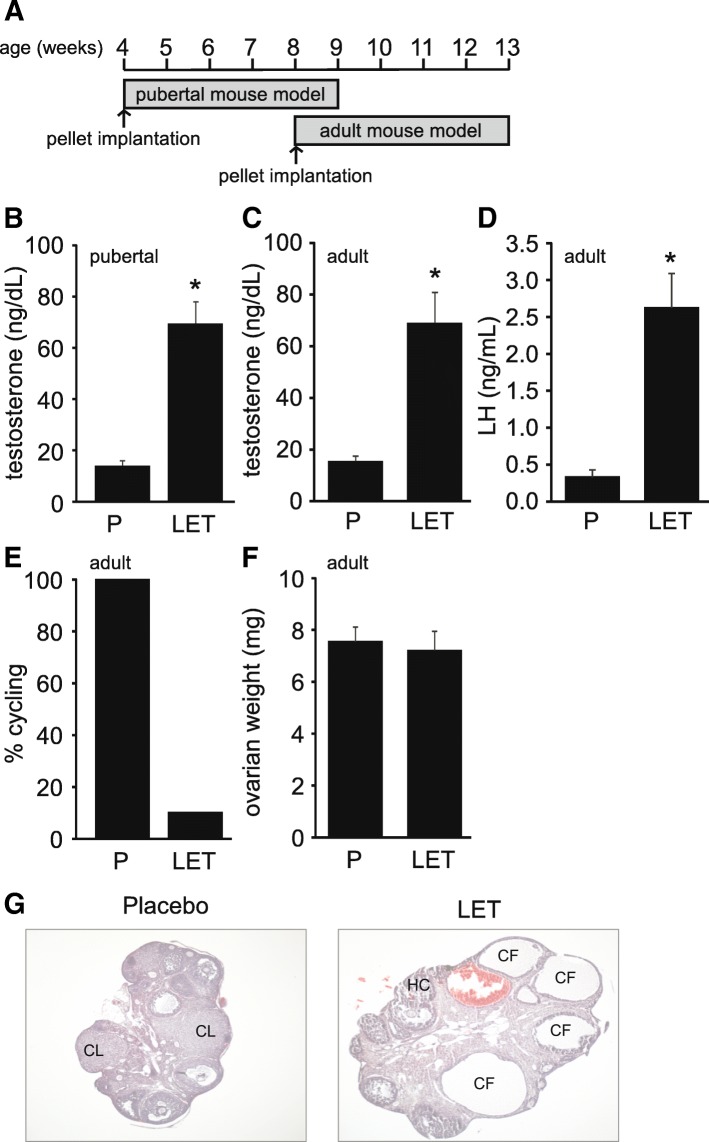
In this study, we investigated whether the age at which letrozole treatment was initiated affected development of the PCOS phenotype in female mice (Fig. 1a). Five weeks of letrozole treatment in pubertal and adult female mice resulted in elevated serum testosterone levels (Fig. 1b-c). Letrozole treatment in adult female mice also resulted in increased LH levels (Fig. 1d) and acyclicity (Fig. 1e). Interestingly, the ovarian weight was similar in placebo and letrozole-treated adult mice (Fig. 1f). This is in contrast to the increase in ovarian weight previously observed in letrozole-treated pubertal mice [53, 54]. Similar to pubertal mice, letrozole treatment of adult female mice resulted in ovaries with cystic follicles and hemorrhagic cysts (Fig. 1g). Ovaries in the letrozole-treated mice also lacked corpora lutea, indicating a lack of ovulation compared to placebo-treated mice.
Fig. 1.
Letrozole treatment of adult female mice resulted in reproductive hallmarks of PCOS. Letrozole (LET) treatment was initiated at 8 weeks of age in the adult PCOS mouse model compared to 4 weeks of age in the pubertal PCOS mouse model (a). LET treatment of pubertal and adult female mice for 5 weeks resulted in elevated serum testosterone (b-c). LET treatment of adult female mice resulted in elevated LH levels (d), and decreased cyclicity as measured by percentage (%) of mice that had an estrous cycle between 4 and 5 weeks of treatment (e). In contrast to pubertal mice, LET treatment of adult female mice did not result in an increase in ovarian weight (f). Pubertal PCOS model (n = 24 placebo (P), n = 22 LET; adult PCOS model (n = 16 P, n = 14 LET). Student t-test; * p < 0.05. LET treatment of adult female mice resulted in ovaries lacking corpora lutea (CL) and containing cystic follicles (CF) and hemorrhagic cysts (HC) compared to placebo-treated mice (g)
Letrozole treatment of adult female mice resulted in minimal weight gain and abdominal adiposity after 5 weeks of treatment
Similar to previous reports [53, 54], letrozole treatment of pubertal female mice for 2 weeks resulted in substantial weight gain compared with placebo treatment, and weight was still increased at the end of the study (Fig. 2a). In contrast, letrozole treatment of adult female mice resulted in a more modest weight gain after 2 weeks of treatment and weight was not statistically different compared to placebo-treated mice after 5 weeks of treatment (Fig. 2a). Letrozole treatment of pubertal female mice resulted in a significant change in abdominal adiposity compared with placebo as reflected in an increase in the weight of the parametrial fat pad relative to total body weight (Fig. 2b). However, letrozole treatment of adult mice did not result in increased abdominal adiposity compared with placebo-treated mice (Fig. 2b).
Fig. 2.
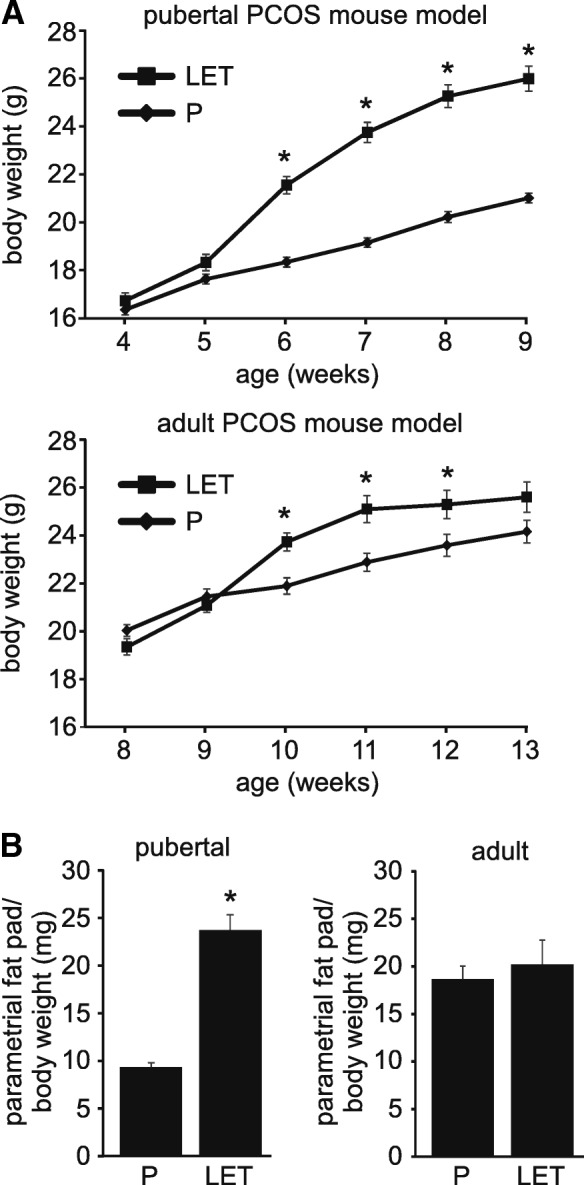
Five weeks of letrozole treatment of adult female mice did not result in substantial weight gain or abdominal adiposity. The phenotype of pubertal (4 week-old) versus adult (8 week-old) female mice treated with placebo (P) or letrozole (LET) for 5 weeks was compared. In contrast to pubertal mice, 5 weeks of LET treatment of adult female mice did not result in a significant increase in weight (a) or abdominal adiposity as measured by parametrial fat relative to total body weight (b). Pubertal PCOS model (n = 24 P, n = 22 LET; adult PCOS model (n = 16 P, n = 14 LET). Student t-test, * p < 0.05
Letrozole treatment of adult female mice resulted in less elevation of fasting blood glucose and insulin levels and did not result in insulin resistance
Both the pubertal and adult PCOS mouse models displayed dysglycemia and hyperinsulinemia but the phenotype was more modest in the adult model. Letrozole treatment of pubertal female mice resulted in elevated fasting blood glucose (FBG) levels and a 3-fold increase in fasting blood insulin levels (Fig. 3a-b). In contrast, letrozole treatment of adult female mice resulted in a slight but statistically significant increase in FBG and a 2-fold increase in insulin levels. There was no significant difference in the response to exogenous glucose in a glucose tolerance test in mice treated with letrozole compared to placebo in either the pubertal or adult PCOS mouse models (data not shown). Finally, the pubertal PCOS mouse model displayed signs of insulin resistance compared to placebo-treated mice while the adult PCOS mouse model remained insulin sensitive (Fig. 3c).
Fig. 3.
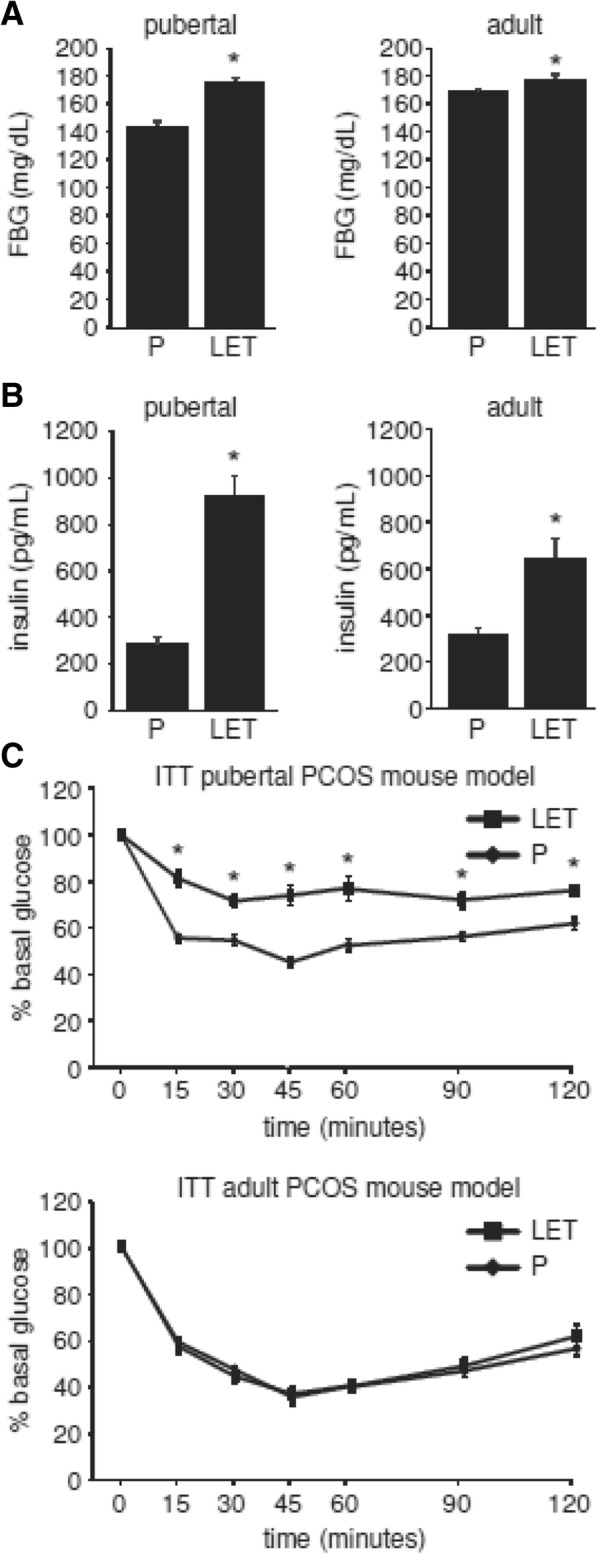
Five weeks of letrozole treatment of adult female mice resulted in a minimal increase in fasting blood glucose and insulin levels and did not result in insulin resistance. The metabolic phenotype of pubertal (4 week-old) versus adult (8 week-old) female mice treated with placebo (P) or letrozole (LET) for 5 weeks was compared. LET treatment of adult female mice resulted in reduced fasting blood glucose (FBG) or insulin levels compared to pubertal mice (a-b). Unlike pubertal female mice, LET treatment of adult female mice for five weeks did not result in insulin resistance (c). Pubertal PCOS model (n = 24 P, n = 22 LET; adult PCOS model (n = 8 P, n = 8 LET). Student t-test or two-way repeated-measures ANOVA with post-hoc Student t-tests to directly compare P versus LET at specific time points were performed; * p < 0.05
Letrozole treatment of adult female mice was not associated with a strong correlation between alpha diversity and time
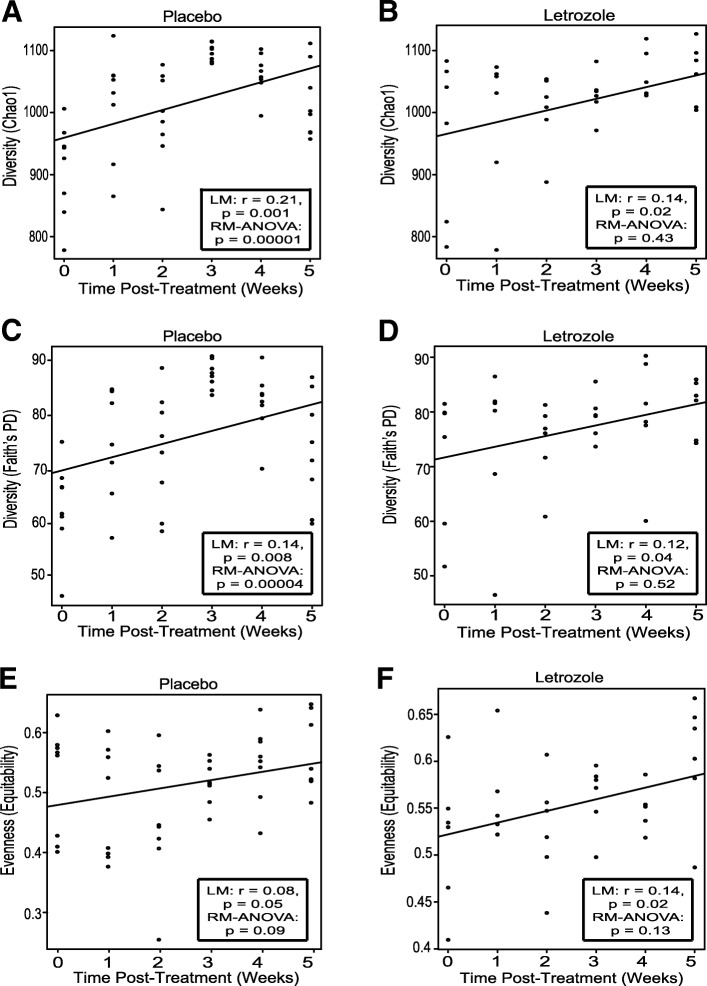
Gut microbial diversity profiles were generated from 84 fecal samples taken prior to and during 5 weeks of placebo or letrozole treatment (weeks 0–5). Sequences collected before placebo and letrozole treatment were compared for both the pubertal and adult mouse models. No significant difference in alpha and beta diversity was observed between the two treatment groups at time 0, indicating that the gut microbiomes of the groups were similar prior to treatment for both the pubertal and adult cohort (Additional file 1: Figure S1). Similar to a previous study in pubertal mice [54], placebo-treated adult mice showed a strong positive correlation between alpha diversity and time as measured by species richness and phylogenetic diversity but not evenness of their gut communities (Fig. 4a, c, e). In contrast, letrozole treatment of adult mice was associated with a relatively weak positive correlation between alpha diversity and time (Fig. 3b, d, f). To examine this further, we evaluated whether there was a significant difference amongst the time points using a repeated measures (RM) ANOVA. RM-ANOVA found a highly significant effect of time on species richness and phylogenetic diversity in placebo-treated mice but no difference in letrozole-treated mice.
Fig. 4.
Letrozole treatment of adult female mice did not result in a strong correlation between time and alpha diversity of the gut microbial community. Chao 1 species richness estimate per sample at each collection time for placebo (a) and letrozole-treated adult female mice (b), (n = 8 placebo, n = 6 letrozole). Faith’s phylogenetic diversity (PD) estimate per sample at each collection time for placebo (c) and letrozole-treated adult female mice (d). Equitability (evenness) estimate per sample at each collection time point for placebo (e) and letrozole-treated mice (f). Line of best fit along with results of linear regression (LM) and repeated measures (RM) ANOVA are shown in box inset
Letrozole treatment of adult female mice resulted in changes in gut microbiome beta diversity
UniFrac analyses were used to compare the similarity amongst gut microbial communities (beta diversity) in fecal samples from placebo versus letrozole–treated adult female mice. When all post-treatment data points were combined together, clustering of the data based on treatment was observed with unweighted UniFrac (Fig. 5a). When the samples were separated by the individual time points (Fig. 5b-f), Analysis of Similarity (ANOSIM) tests found a difference in the overall bacterial community composition of the gut microbiome between placebo and letrozole–treated adult female mice at weeks 4 and 5 post-treatment (p = 0.01 and p = 0.03 respectively). We also observed similar results using weighted UniFrac (data not shown).
Fig. 5.
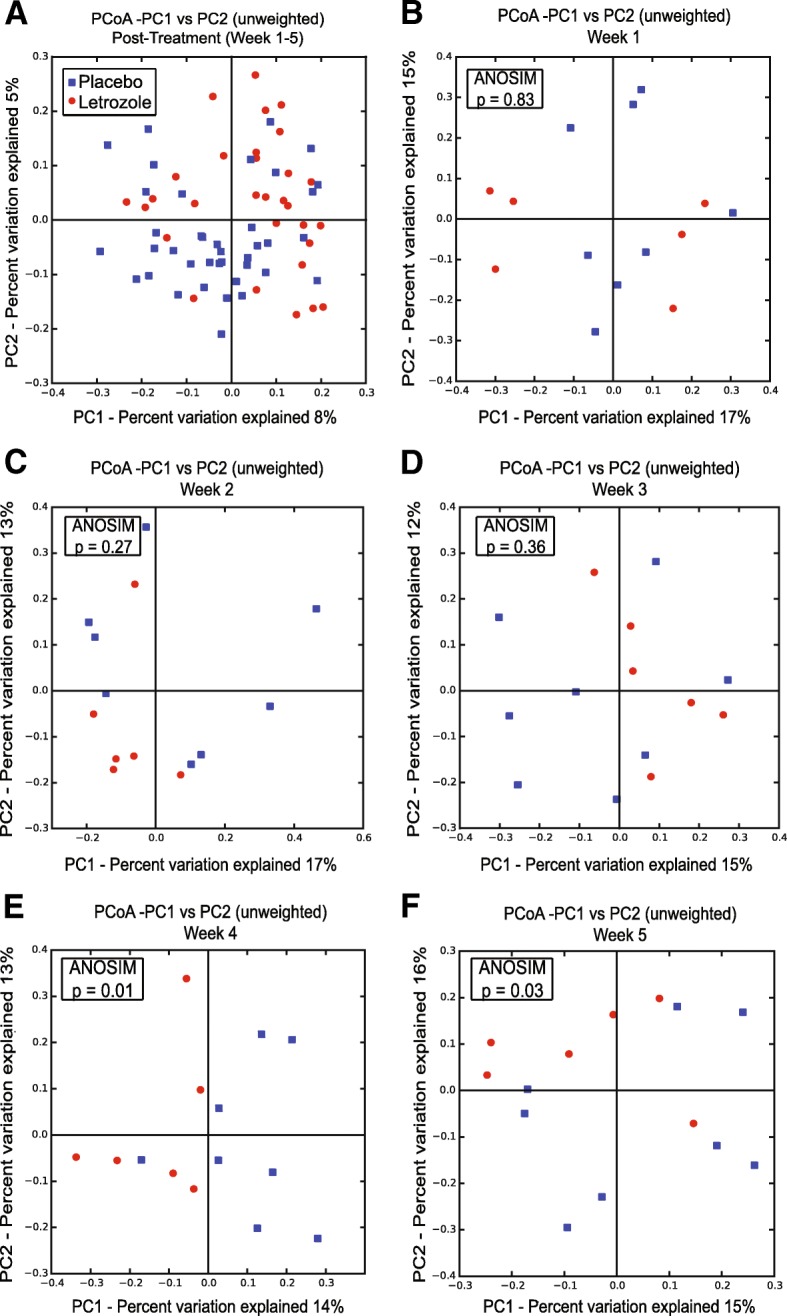
Letrozole treatment of adult female mice resulted in a significant shift in the beta diversity of the gut microbiome. Principal Coordinates Analysis (PCoA) of unweighted UniFrac for samples collected post-treatment (weeks 1–5) were compared between placebo (n = 8) and letrozole-treated (n = 6) mice (a). Proportion of variance explained by each principal coordinate axis is denoted in the corresponding axis label. Samples from placebo- and letrozole-treated mice were then compared for each time point (b-f). Results of Analysis of Similarity (ANOSIM) tests are shown in the box inset
Distinct bacterial genera discriminated between placebo and letrozole treatment in the pubertal and adult PCOS mouse models
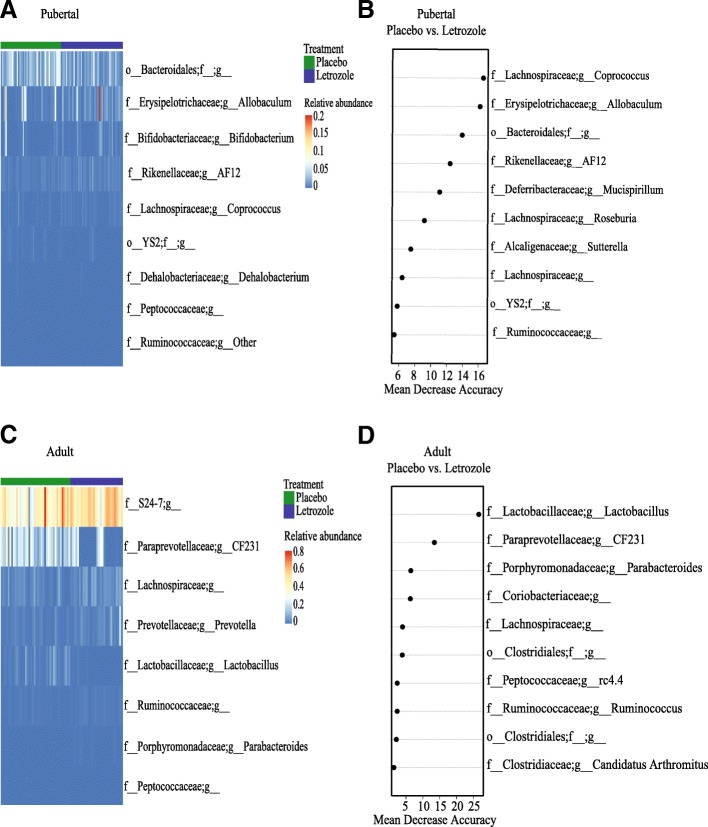
In addition to studying changes in alpha and beta diversity, we also investigated whether the age at which letrozole treatment was initiated was important for changes in the taxonomic composition of the gut microbiome. We combined the post-treatment data (weeks 1–5) from placebo and letrozole-treated mice in the pubertal and adult mouse models. Based on the Greengenes taxonomic database, we identified a total of 10 bacterial phyla and 51 bacterial genera in the four different groups. Similar to our previous study [54], the majority of Operational Taxonomic Units (OTUs) in the adult mouse fecal samples were identified as Bacteriodetes or Firmicutes (~ 84–95%). We used RM-ANOVA to determine if the mean relative abundances of specific bacterial genera were different in the gut microbiome of placebo versus letrozole-treated mice in the pubertal and adult mouse models. A heatmap was generated to represent the relative abundance of 9 different bacterial genera that changed significantly with letrozole treatment (FDR-corrected p < 0.05) in the pubertal mouse model (Fig. 6a). Letrozole treatment of pubertal female mice resulted in higher relative abundances of Coprococcus, Allobaculum, Bifidobacterium, and an undescribed genus belonging to the Ruminococcaceae, as well as a lower abundance of AF12, Dehalobacterium, taxa belonging to the uncultured order YS2, and undescribed genera of Peptococcaceae and Bacteroidales (Fig. 6a).
Fig. 6.
Repeated measures analysis of variance and Random Forest classification identified distinct bacteria associated with letrozole treatment in the pubertal versus adult PCOS mouse model. Repeated measures analysis of variance (corrected for multiple comparisons via FDR) was used to determine whether the abundance of specific bacterial genus differed in placebo versus letrozole-treated pubertal or adult mice. A heatmap was generated for the bacterial genera that had a FDR adjusted p-value of < 0.05 and mean taxa abundance above 0.001 (a, c). The Random Forest classifier was used to identify bacterial genera that distinguished between placebo and letrozole treatment in pubertal or adult female mice (b, d). The top ten most discriminant bacterial genera in the models were displayed in the variable importance plots using a total of forty-five genera in the analysis. An increase in mean decrease accuracy reflects the prediction strength of the variable in classifying the different treatment groups
In contrast to the pubertal mice, letrozole treatment of adult female mice resulted in changes in the mean relative abundance of a distinct set of 8 bacterial genera (FDR-corrected p < 0.05). With the exception of uncultured members of the genus-level CF231 group, the rest of the genera from the Bacteroidetes phylum increased with letrozole treatment in adult female mice, including Prevotella, an uncultured genus within Parabacteroides and a genus-level group within the S24–7 family (Fig. 6c). Letrozole treatment of adult mice also resulted in a higher relative abundance of genera from Lachnospiraceae, Ruminococcaceae, and Peptococcaceae, as well as a lower abundance of Lactobacillus (Fig. 6c).
Random Forest classifier identified bacterial genera predictive of placebo and letrozole treatment in the pubertal and adult PCOS mouse models
The Random Forest (RF) classifier was trained to determine how well placebo or letrozole treatment could be predicted based on bacterial relative abundances in the two models (pubertal and adult). Forty-five of the 51 total bacterial genera identified in the four different groups were used for RF classification (six were excluded due to low relative abundances). Our results showed that RF predicted treatment category in the pubertal group with 78.5% accuracy while it predicted treatment category in the adult group with 84% accuracy (Table 1). Variable importance by mean decrease in accuracy was calculated for the RF models. Figure 6b and d illustrate 10 bacterial genera whose removal caused the greatest decrease in model accuracy (i.e. the most important for classification) in the pubertal and adult mouse models respectively. In the pubertal model, the removal of Coprococcus, Allobaculum, AF12, Mucispirillum, Roseburia, Sutterella, and an unknown genus from Bacteroidales had the greatest impact on classification (mean decrease accuracy > 8; Fig. 6b). In the adult mice, the removal of Lactobacillus, CF231 and Parabacteriodes caused the greatest decrease in prediction accuracy (mean decrease accuracy > 8; Fig. 6d).
Table 1.
Classification error rates carried out using Random Forest classifiers composed of 500 trees
| Predicted classes | Classification error rates | OOB estimate of error rate | Accuracy | ||
|---|---|---|---|---|---|
| Placebo | Letrozole | ||||
| Pubertal | 21.5% | 78.5% | |||
| Placebo | 32 | 7 | 0.18 | ||
| Letrozole | 10 | 30 | 0.25 | ||
| Adult | 16.0% | 84.0% | |||
| Placebo | 30 | 1 | 0.03 | ||
| Letrozole | 7 | 13 | 0.35 | ||
Discussion
Our study demonstrated that initiation of letrozole treatment during puberty or adulthood in female mice resulted in reproductive hallmarks of PCOS, including elevated testosterone levels, anovulation and ovaries with cystic follicles. This suggests that the timing of androgen exposure (puberty versus adulthood) may not be important for development of the PCOS reproductive phenotype. On the other hand, our study did find a clear divergence between the metabolic phenotypes of the pubertal and adult mouse models. Similar to previously published studies [53, 54], letrozole treatment in pubertal female mice resulted in multiple metabolic features of PCOS, including obesity, abdominal adiposity, hyperinsulinemia, and insulin resistance. On the other hand, letrozole treatment in adult female mice did not result in substantial weight gain, abdominal adiposity or insulin resistance, indicating that androgen exposure is not sufficient to induce the full PCOS-like metabolic phenotype in adult female mice. Interestingly, studies using post-natal treatment with DHT to create a hyperandrogenic mouse model observed a similar pattern: the metabolic phenotype depended on when DHT treatment was initiated. Compared with placebo-treated mice, female mice treated with DHT starting at 3 weeks of age gained significantly more weight, had greater levels of abdominal adiposity and were glucose intolerant [43, 47]. In contrast, while female mice treated with DHT in adulthood had impaired glucose tolerance, they did not become obese or display increased abdominal adiposity [56].
Our results also suggest that the timing of excess androgen exposure may be an important component in the development of the PCOS metabolic phenotype. Since PCOS often manifests in the early reproductive years, puberty has been suggested to be a critical developmental time period for the development and pathology of PCOS [6, 57]. Indeed, PCOS has been hypothesized to originate from abnormal pubertal development due to a lack of transition from an androgen-dominated state in early puberty to an estrogenic state in late puberty [58, 59]. Puberty is a time of considerable hormonal and metabolic change, including an increase in insulin resistance [60]. Although physiological insulin resistance is common in healthy adolescents, it usually resolves to prepubertal levels in adulthood [61]. Pubertal insulin resistance has been reported to increase the risk of developing type 2 diabetes along with accelerating the complications of diabetes [62–65]. Thus, it is possible that insulin resistance and the hyperinsulinemia that occurs during puberty may also contribute to the risk of developing obesity and metabolic dysfunction in PCOS.
Another factor that changes during the transition from childhood to adulthood is the gut microbiome. Studies have shown that children or adolescents have a distinct gut microbial community compared to adults [66, 67]. Moreover, prepubertal mice were reported to have a different gut microbiome than adult mice [68, 69]. Studies in humans and mice have shown a strong positive association between gut bacterial alpha diversity and age, indicating that the complexity of the gut microbiome increases as the host ages [70–72]. In contrast to placebo, there was no significant effect of time on alpha diversity in letrozole-treated mice when the data was adjusted for within subject error using RM-ANOVA (Fig. 4) [54]. With regards to beta diversity, letrozole treatment of both pubertal and adult female mice resulted in a distinct shift in the gut microbial composition (Fig. 5). However, closer examination of the types of bacteria that changed after letrozole treatment showed that the taxa driving the shift in beta diversity were quite distinct in the two mouse models (Fig. 6).
Letrozole treatment initiated during puberty resulted in changes in the abundances of bacterial genera previously reported to be altered in diet-induced obesity mouse models. In the pubertal model, RF and statistical analysis of relative bacterial abundances determined that Coprococcus, Allobaculum and an unknown genus from Bacteroidales differentiated the gut microbiomes of placebo and letrozole-treated mice (Fig. 6a and b). Significant differences were also observed in the relative abundance of Bifidobacterium, reported to have strain-specific effects on weight gain in rodents [73], as well as Dehalobacterium and unknown genera belonging to the Rikenellaceae and Ruminococcaceae families, all of which have been associated with obesity [28, 74–77]. The genus with the strongest effect on RF classification, namely Coprococcus, was previously reported to be more abundant in obese individuals [78, 79], in agreement with the higher levels observed after letrozole treatment in pubertal mice. The second most important genus in terms of classification, Allobaculum, was reported to be lower in the gut of obese mice fed a high-fat diet [80, 81], in contrast to the increase in Allobaculum observed after letrozole treatment.
In comparison to pubertal mice, letrozole treatment of adult female mice had a distinct impact on the composition of the gut microbial community. With the exception of a genus within the Peptococcaceae, the bacterial genera most affected by letrozole treatment in adult females were not altered in pubertal mice and vice versa (Fig. 6a, c). It should be noted that the genera that changed in the pubertal and adult female mice after letrozole treatment were present at both ages, indicating that the differential effects of letrozole treatment in the two models was not due to the absence of specific bacteria. The most striking difference in letrozole treatment of adult female mice was the importance that Lactobacillus, Parabacteroides and the uncultured Paraprevotellaceae group CF231 played in classifying the treatment groups (Fig. 6d). The mean relative abundance of these bacteria changed significantly after letrozole treatment in adult female mice (Fig. 6c). This is in contrast to the increased abundance of some Lactobacillus species observed in obese humans [82–84], though direct comparisons are difficult since there may be strain-specific effects of Lactobacillus on weight gain [85]. While CF231 has not been described in much detail, members of the Paraprevotellaceae are found in the gut of many mammals [86, 87] and have been suggested to be involved in the degradation of plant polysaccharides into short chain fatty acids [88]. Parabacteroides are also known to metabolize non-digestible carbohydrates, but the increase in Parabacteroides relative abundance after 5 weeks of letrozole treatment contrasts with the decrease observed in mice fed a high-fat diet [27, 89].
Conclusions
In summary, our study demonstrated that the timing of androgen exposure may be important for development of the PCOS metabolic phenotype and associated changes in the gut microbiome. While letrozole treatment of female mice during puberty and in adulthood both resulted in reproductive hallmarks of PCOS, including hyperandrogenemia, anovulation and polycystic ovaries, letrozole treatment in adulthood did not result in the weight gain, abdominal adiposity or insulin resistance observed in the pubertal PCOS mouse model. In addition, letrozole treatment in adulthood resulted in distinct changes in the gut microbiome, particularly in Lactobacillus. Although evidence is accumulating that changes in steroid hormones are associated with an altered gut microbiome [90], the mechanisms involved in steroid hormone/gut microbe interactions are currently unknown. Future studies investigating whether steroid hormones regulate the gut microbiome through actions in the gastrointestinal tract, immune system or other tissues will begin to address the mechanisms involved.
Given that many of the previous studies that report an association of specific bacterial genera with obesity in humans and high fat diet-induced mouse models are contradictory, it is possible that these results are due to modulation of specific bacterial species and strains within genera. Future studies should employ higher resolution methods such as metagenomic sequencing or quantitative PCR to fully understand the effects of hyperandrogenism on the gut microbiome. Moreover, since many studies of the role of the gut microbiome in obesity are confounded by the effect of diet on the microbiome, the letrozole-induced PCOS mouse model provides an opportunity to study the effects of androgen excess on the gut microbiome and metabolism in a diet-independent setting, since food intake is not altered by letrozole treatment [55]. Moreover, the adult PCOS mouse model can be used to study the effects of hyperandrogenism in female mice without the confounding variable of insulin resistance. Further studies addressing whether the gut microbiome plays a causal role in the development of PCOS or if manipulation of the gut microbiome can improve the PCOS phenotype will be informative. In addition, prospective studies with adolescent girls may be crucial to understand the etiology and development of PCOS, particularly the metabolic dysregulation and changes in the gut microbiome associated with this disease.
Methods
PCOS mouse model
C57BL/6NHsd female mice purchased from Envigo were housed in a vivarium for one week under specific pathogen-free conditions with an automatic 12 h:12 h light/dark cycle (light period: 06.00–18.00) and ad libitum access to water and food (Teklad Global 18% Protein Extruded Diet, Envigo). Prior to the beginning of the study, the mice were sorted by weight to ensure that the starting weight was similar between the two treatment groups. To establish the pubertal or adult PCOS models, 4 or 8 week-old female mice, respectively were implanted subcutaneously with a placebo or 3 mg letrozole pellet (3 mm diameter; Innovative Research of America) that provided a slow, constant release of letrozole (50 μg/day) over 5 weeks. For the duration of the experiment, the mice were housed 2 per cage: 2 placebo or 2 letrozole-treated mice. Placebo and letrozole-treated mice were not housed together to avoid the influence of coprophagy on the PCOS mouse model. At the end of the study, the mice were sacrificed using 2.5% isoflurane delivered with a precision vaporizer followed by a physical method of euthanasia.
Analysis of reproductive and metabolic phenotype
The mice were weighed weekly. The stage of the estrous cycle for placebo and letrozole-treated mice was determined from the predominant cell type in vaginal epithelium smears obtained during weeks 4–5 of treatment. After 5 weeks of placebo or letrozole treatment, the mice were fasted for 5 h and blood from the tail vein was collected to measure fasting insulin levels. Blood glucose was measured using a handheld glucometer (One Touch UltraMini, LifeScan, Inc) and an intraperitoneal (IP) insulin tolerance test (ITT) was performed. Tail vein blood glucose was measured just before (time 0) an IP injection of insulin (0.75 U/kg in sterile saline; Humulin R U-100, Eli Lilly) was given and at 15, 30, 45, 60, 90, and 120 min post injection.
At the end of the experiment, blood was collected from the posterior vena cava, parametrial fat pads were dissected and weighed, and the ovaries were dissected, weighed, fixed in 4% paraformaldehyde at 4 °C overnight, and stored in 70% ethanol before processing for histology. Paraffin-embedded ovaries were sectioned at 10 μm and stained with hematoxylin and eosin (Zyagen). Serum testosterone were measured using a mouse ELISA (range 10–800 ng/dL) while LH levels were measured using a radioimmunoassay (range 0.04–75 ng/mL) by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core Facility. Serum insulin was measured using a mouse ELISA (ALPO) by the University of California, Davis Mouse Metabolic Phenotyping Center. The data from four mice in the pubertal and adult letrozole-treated groups were removed from the analyses because these mice did not have a significant elevation in serum testosterone when compared to the average of the placebo-treated mice. The analysis of the reproductive and metabolic phenotypes was performed with data from 2 unpublished cohorts of the adult PCOS model (total n = 16 placebo, n = 14 letrozole) and 3 cohorts of the pubertal PCOS model (total n = 24 placebo, n = 22 letrozole) including 2 unpublished and 1 previously published cohort [54]. Differences between placebo and letrozole treatment were analyzed by Student t-test or two-way repeated measures ANOVA followed by post-hoc comparisons of individual time points.
Fecal sample collection and DNA isolation
Fecal samples were collected from one cohort of 8-week-old female mice (n = 8/group) prior to treatment with placebo or letrozole and once per week thereafter for 5 weeks. Fecal samples were frozen immediately after collection and stored at − 80 °C. Bacterial DNA was extracted from the fecal samples using the MoBio PowerSoil DNA Extraction Kit following the manufacturer’s protocol, and the DNA was stored at − 80 °C.
16S rRNA amplicon sequencing
The V4 hypervariable region of the 16S rRNA gene was PCR amplified with primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) [91]. The reverse primers contained unique 12 base pair Golay barcodes that were incorporated into the PCR amplicons [92]. The barcoded primers allowed for pooling of multiple PCR amplicons in a single sequencing run. Thermocycling parameters were as follows: denaturing at 98 °C for 2 min followed by amplification for 35 cycles at 98 °C for 30 s, 50 °C for 30 s and 72 °C for 60 s, and a final extension of 72 °C for 10 min. The resulting amplicons were submitted to The Scripps Research Institute Next Generation Sequencing Core Facility where they were cleaned using Zymo DNA Clean & Concentrator™-25 columns, quantified using a Qubit Flourometer (Life Technologies) and pooled. Pooled PCR products were size selected on a 2% agarose gel (290–350 bp), purified using a Zymoclean™ Gel DNA recovery kit and used to prepare sequencing libraries following the recommended Illumina protocol involving end-repair, A-tailing and adapter ligation. The DNA library was then size selected on a 2% agarose gel (410–470 bp), cleaned using the Agencourt SPRI system (Beckman Coulter, Inc.) and PCR amplified with HiFi Polymerase (Kapa Biosystems) for 12 cycles. The amplified DNA products were again size selected on a 2% agarose gel and purified using the Zymoclean™ Gel DNA recovery kit. The purified DNA library was quantitated, denatured in 0.1 N NaOH and diluted to a final concentration of 5 pM before being loaded onto the Illumina single read flow-cell for sequencing on the Illumina MiSeq system along with 4 pM PhiX control library.
16S rRNA gene sequence analysis
16S rRNA sequences for the adult mice were de-multiplexed using the Quantitative Insights Into Microbial Ecology (QIIME v.1.9.1, http://www.qiime.org) pipeline [93] using the default split_libraries.py script parameter [94]. This resulted in approximately 4.3 million Illumina sequences across all samples with an average of 50,000 sequences per sample. Sequences from two mice in the letrozole-treated group were removed from the analysis because these mice did not have a significant elevation in serum testosterone levels compared to the average of the placebo-treated mice. The 16S rRNA gene sequencing quality control and analysis for the samples from the adult mice followed the same pipeline as the samples in a previously published study with placebo or letrozole-treated pubertal female mice [54]. Sequences were clustered using the pick_de_novo_otus.py script with usearch [95]. Sequences were assigned to OTUs with an assumed 97% threshold of pairwise identity for bacterial species by comparison with the Greengenes reference database [96] using the RDP classifier [97]. Before performing downstream analysis, singletons and OTUs present in less than 25% of the samples were discarded from the database to minimize the effect of spurious, low abundance sequences using the filter_otus_from_otu_table.py script. Sequences were then aligned using PyNast [93] and a phylogenetic tree constructed using FastTree [98]. The alpha_diversity.py script was used to estimate several different attributes of alpha diversity. Species richness was estimated using Chao1 to define the total number of unique species in a community [99]. Faith’s Phylogenetic Diversity was used to measure the phylogenetic diversity of a community by calculating the total branch lengths on a phylogenetic tree of all members of the community [100]. Evenness was estimated using the Equitability index [101]. The beta_diversity_through_plots.py script was used to compute weighted and unweighted UniFrac distances [102]. The smaller the UniFrac distance between two microbial communities, the more similar the communities are in their overall diversity. The weighted UniFrac distance metric incorporates the abundance of specific taxa in each community into the UniFrac distance calculation while unweighted UniFrac ignores abundance information. Taxonomic distributions across sample categories were calculated (from phylum to genus) using the summarize_taxa_through_plots.py script. Several bacterial genera such as Anaeroplasma and an unknown Enterobacteriaceae were excluded from the analysis because of extremely low abundance, suggesting that they may have been artifacts. Sequences from placebo-treated samples collected during week 5 (9 weeks of age) of the pubertal cohort were compared to samples collected from week 1 (9 weeks of age) from the adult cohort. No significant difference in alpha and beta diversity was observed between the two cohorts, indicating that the gut microbiome at the end of placebo treatment in the pubertal cohort was similar to the gut microbiome at the beginning of placebo treatment in the adult cohort.
Statistical analysis
Pearson’s product-moment correlation was performed when analyzing alpha diversity over time using the RStudio statistical package (version 0.99.893). RM-ANOVA was used to model alpha diversity measures accounting for within subject error. Two-dimensional PCoA plots were constructed using the make_2d_plots.py script in QIIME. ANOSIM tests for weighted and unweighted UniFrac distances between treatments were performed using the compare_categories.py script. The biom table of post treatment samples (weeks 1–5) from the adult study was merged with the biom table from the pubertal study, resulting in approximately 6.2 million sequences from 170 samples (pubertal = 100 samples; adult = 70 samples). The merged dataset was used to compare differences among treatment group and developmental stage. RM-ANOVA adjusting for within subject error (corrected for multiple comparisons via FDR) was used to determine whether the abundance of specific bacterial genus differed between treatments. The RF supervised machine-learning classifier was used to determine how well a given set of factors (e.g. bacterial genera) classified discrete categories and which factors were most important for the classification [70, 116]. RF was implemented in R using the “randomForest” library, and was used to identify bacterial genera that differentiated placebo and letrozole treatment within pubertal or adult mice.
Additional file
Figure S1. No differences in gut microbial community diversity between placebo and letrozole-treated mice were observed prior to treatment. No significant differences in gut microbiome alpha diversity (Faith’s PD) between placebo- and letrozole-treated mice were observed prior to treatment (week 0) in the pubertal (placebo n = 10, letrozole n = 10) (A) or adult (placebo n = 8, letrozole n = 6) (B) model. Similarly, no differences in beta diversity (unweighted UniFrac) were observed between placebo- and letrozole-treated mice prior to treatment in the pubertal (C) or adult model (D). Student t-test was used to compare alpha diversity between groups and Analysis of Similarity (ANOSIM) test was used to compare beta diversity between groups. (PDF 1335 kb)
Acknowledgements
We thank members of the Kelley and Thackray labs for insightful comments and suggestions. Hormone levels were measured by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core Facility (P50 HD28934) and the University of California, Davis Mouse Metabolic Phenotyping Core (U24 DK092993).
Funding
This work was funded by the National Institute of Child Health and Human Development through a cooperative agreement as part of the National Centers for Translational Research in Reproduction and Infertility (P50 HD012303) as well as Grants T32 HD007203 and F32 HD074414 to D.V.S. D.V.S. was also funded by the National Institute of General Medical Sciences Grant K12 GM068524. L.S. was funded by a Doris A. Howell Foundation Research Scholarship for Women’s Health and A.R.A was funded by a Ledell Family Endowed Research Scholarship for Science and Engineering.
Availability of data and materials
The 16S rRNA sequence dataset generated and analyzed during the current study is available in the European Nucleotide Archive (Study Accession Number PRJEB20895).
Abbreviations
- ANOSIM
Analysis of similarity
- FBG
Fasting blood glucose
- IP
Intraperitoneal
- ITT
Insulin tolerance test
- OTU
Operational taxonomic unit
- PCOS
Polycystic ovary syndrome
- QIIME
Quantitative Insights Into Microbial Ecology
- RF
Random Forest
- RM-ANOVA
Repeated measures analysis of variance
Authors’ contributions
VT and SK conceived and designed the study; PJT, DVS, BSH, LS and ARA performed reproductive and metabolic assessments, and collected samples; PJT performed DNA extractions and PCR amplifications; PJT, VT and SK analyzed the data; PT, SK and VT wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal procedures in this study were approved by the University of California, San Diego Institutional Animal Care and Use Committee (protocol number S14011).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pedro J. Torres, Email: pjtorres88@gmail.com
Danalea V. Skarra, Email: dskarra@gmail.com
Bryan S. Ho, Email: bsho@ucsd.edu
Lillian Sau, Email: lsau@ucsd.edu.
Arya R. Anvar, Email: aryaranvar@gmail.com
Scott T. Kelley, Email: skelley@sdsu.edu
Varykina G. Thackray, Email: vthackray@ucsd.edu
References
- 1.Fauser BC, Tarlatzis BC, Rebar RW, Legro RS, Balen AH, Lobo R, Carmina E, Chang J, Yildiz BO, Laven JS, et al. Consensus on women's health aspects of polycystic ovary syndrome (PCOS): the Amsterdam ESHRE/ASRM-sponsored 3rd PCOS consensus workshop group. Fertil Steril. 2012;97(1):28–38. doi: 10.1016/j.fertnstert.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 2.Barber TM, Franks S. Genetics of polycystic ovary syndrome. Front Horm Res. 2013;40:28–39. doi: 10.1159/000341682. [DOI] [PubMed] [Google Scholar]
- 3.Legro RS, Driscoll D, Strauss JF, 3rd, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci U S A. 1998;95(25):14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91(6):2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 5.Jones MR, Goodarzi MO. Genetic determinants of polycystic ovary syndrome: progress and future directions. Fertil Steril. 2016;106(1):25–32. doi: 10.1016/j.fertnstert.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 6.Abbott DH, Bacha F. Ontogeny of polycystic ovary syndrome and insulin resistance in utero and early childhood. Fertil Steril. 2013;100(1):2–11. doi: 10.1016/j.fertnstert.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teede H, Deeks A, Moran L. Polycystic ovary syndrome: a complex condition with psychological, reproductive and metabolic manifestations that impacts on health across the lifespan. BMC Med. 2010;8:41. doi: 10.1186/1741-7015-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balen AH, Tan SL, Jacobs HS. Hypersecretion of luteinising hormone: a significant cause of infertility and miscarriage. Br J Obstet Gynaecol. 1993;100(12):1082–1089. doi: 10.1111/j.1471-0528.1993.tb15170.x. [DOI] [PubMed] [Google Scholar]
- 9.Boomsma CM, Eijkemans MJ, Hughes EG, Visser GH, Fauser BC, Macklon NS. A meta-analysis of pregnancy outcomes in women with polycystic ovary syndrome. Hum Reprod Update. 2006;12(6):673–683. doi: 10.1093/humupd/dml036. [DOI] [PubMed] [Google Scholar]
- 10.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, et al. The androgen excess and PCOS society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril. 2009;91(2):456–488. doi: 10.1016/j.fertnstert.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 11.Carmina E, Campagna AM, Lobo RA. A 20-year follow-up of young women with polycystic ovary syndrome. Obstet Gynecol. 2012;119:263–269. doi: 10.1097/AOG.0b013e31823f7135. [DOI] [PubMed] [Google Scholar]
- 12.Diamanti-Kandarakis E, Spritzer PM, Sir-Petermann T, Motta AB. Insulin resistance and polycystic ovary syndrome through life. Curr Pharm Des. 2012;18(34):5569–5576. doi: 10.2174/138161212803307590. [DOI] [PubMed] [Google Scholar]
- 13.Wild RA, Carmina E, Diamanti-Kandarakis E, Dokras A, Escobar-Morreale HF, Futterweit W, Lobo R, Norman RJ, Talbott E, Dumesic DA. Assessment of cardiovascular risk and prevention of cardiovascular disease in women with the polycystic ovary syndrome: a consensus statement by the androgen excess and polycystic ovary syndrome (AE-PCOS) society. J Clin Endocrinol Metab. 2010;95(5):2038–2049. doi: 10.1210/jc.2009-2724. [DOI] [PubMed] [Google Scholar]
- 14.Wild S, Pierpoint T, McKeigue P, Jacobs H. Cardiovascular disease in women with polycystic ovary syndrome at long-term follow-up: a retrospective cohort study. Clin Endocrinol. 2000;52(5):595–600. doi: 10.1046/j.1365-2265.2000.01000.x. [DOI] [PubMed] [Google Scholar]
- 15.Moran LJ, Misso ML, Wild RA, Norman RJ. Impaired glucose tolerance, type 2 diabetes and metabolic syndrome in polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod Update. 2010;16(4):347–363. doi: 10.1093/humupd/dmq001. [DOI] [PubMed] [Google Scholar]
- 16.Hart R, Doherty DA. The potential implications of a PCOS diagnosis on a Woman's long-term health using data linkage. J Clin Endocr Metab. 2015;100(3):911–919. doi: 10.1210/jc.2014-3886. [DOI] [PubMed] [Google Scholar]
- 17.Barber TM, Wass JA, McCarthy MI, Franks S. Metabolic characteristics of women with polycystic ovaries and oligo-amenorrhoea but normal androgen levels: implications for the management of polycystic ovary syndrome. Clin Endocrinol. 2007;66(4):513–517. doi: 10.1111/j.1365-2265.2007.02764.x. [DOI] [PubMed] [Google Scholar]
- 18.Moghetti P, Tosi F, Bonin C, Di Sarra D, Fiers T, Kaufman JM, Giagulli VA, Signori C, Zambotti F, Dall'Alda M, et al. Divergences in insulin resistance between the different phenotypes of the polycystic ovary syndrome. J Clin Endocrinol Metab. 2013;98(4):E628–E637. doi: 10.1210/jc.2012-3908. [DOI] [PubMed] [Google Scholar]
- 19.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res. 2013;69(1):75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 26.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chassaing B, Aitken JD, Gewirtz AT, Vijay-Kumar M. Gut microbiota drives metabolic disease in immunologically altered mice. Adv Immunol. 2012;116:93–112. doi: 10.1016/B978-0-12-394300-2.00003-X. [DOI] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 33.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindheim L, Bashir M, Munzker J, Trummer C, Zachhuber V, Leber B, Horvath A, Pieber TR, Gorkiewicz G, Stadlbauer V, et al. Alterations in gut microbiome composition and barrier function are associated with reproductive and metabolic defects in women with polycystic ovary syndrome (PCOS): a pilot study. PLoS One. 2017;12(1):e0168390. doi: 10.1371/journal.pone.0168390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Zhang CH, Shi Y, Zhang F, Li LX, Wang XJ, Ling YX, Fu HQ, Dong WP, Shen J, et al. Dysbiosis of gut microbiota associated with clinical parameters in polycystic ovary syndrome. Front Microbiol. 2017;8:324. doi: 10.3389/fmicb.2017.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres PJ, Siakowska M, Banaszewska B, Pawelczyk L, Duleba AJ, Kelley ST, Thackray VG. Gut Microbial Diversity in Women with Polycystic Ovary Syndrome Correlates with Hyperandrogenism. J Clin Endocrinol Metab. 2018;103(4):1502–1511. doi: 10.1210/jc.2017-02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmanabhan V, Veiga-Lopez A. Animal models of the polycystic ovary syndrome phenotype. Steroids. 2013;78(8):734–740. doi: 10.1016/j.steroids.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi D, Vine DF. Animal models of polycystic ovary syndrome: a focused review of rodent models in relationship to clinical phenotypes and cardiometabolic risk. Fertil Steril. 2012;98(1):185–193. doi: 10.1016/j.fertnstert.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 40.van Houten EL, Visser JA. Mouse models to study polycystic ovary syndrome: a possible link between metabolism and ovarian function? Reprod Biol. 2014;14(1):32–43. doi: 10.1016/j.repbio.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Walters KA, Allan CM, Handelsman DJ. Rodent models for human polycystic ovary syndrome. Biol Reprod. 2012;86(5):149. doi: 10.1095/biolreprod.111.097808. [DOI] [PubMed] [Google Scholar]
- 42.Walters KA. Androgens in polycystic ovary syndrome: lessons from experimental models. Current opinion in endocrinology, diabetes, and obesity. 2016;23(3):257–263. doi: 10.1097/MED.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 43.Caldwell ASL, Middleton LJ, Jimenez M, Desai R, McMahon AC, Allan CM, Handelsman DJ, Walters KA. Characterization of reproductive, metabolic, and endocrine features of polycystic ovary syndrome in female Hyperandrogenic mouse models. Endocrinology. 2014;155(8):3146–3159. doi: 10.1210/en.2014-1196. [DOI] [PubMed] [Google Scholar]
- 44.Moore AM, Prescott M, Campbell RE. Estradiol negative and positive feedback in a prenatal androgen-induced mouse model of polycystic ovarian syndrome. Endocrinology. 2013;154(2):796–806. doi: 10.1210/en.2012-1954. [DOI] [PubMed] [Google Scholar]
- 45.Roland AV, Nunemaker CS, Keller SR, Moenter SM. Prenatal androgen exposure programs metabolic dysfunction in female mice. J Endocrinol. 2010;207(2):213–223. doi: 10.1677/JOE-10-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witham EA, Meadows JD, Shojaei S, Kauffman AS, Mellon PL. Prenatal exposure to low levels of androgen accelerates female puberty onset and reproductive senescence in mice. Endocrinology. 2012;153(9):4522–4532. doi: 10.1210/en.2012-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Houten EL, Kramer P, McLuskey A, Karels B, Themmen AP, Visser JA. Reproductive and metabolic phenotype of a mouse model of PCOS. Endocrinology. 2012;153(6):2861–2869. doi: 10.1210/en.2011-1754. [DOI] [PubMed] [Google Scholar]
- 48.Barontini M, Garcia-Rudaz MC, Veldhuis JD. Mechanisms of hypothalamic-pituitary-gonadal disruption in polycystic ovarian syndrome. Arch Med Res. 2001;32(6):544–552. doi: 10.1016/s0188-4409(01)00325-3. [DOI] [PubMed] [Google Scholar]
- 49.Nelson VL, Legro RS, Strauss JF, 3rd, McAllister JM. Augmented androgen production is a stable steroidogenic phenotype of propagated theca cells from polycystic ovaries. Mol Endocrinol. 1999;13(6):946–957. doi: 10.1210/mend.13.6.0311. [DOI] [PubMed] [Google Scholar]
- 50.Naessen T, Kushnir MM, Chaika A, Nosenko J, Mogilevkina I, Rockwood AL, Carlstrom K, Bergquist J, Kirilovas D. Steroid profiles in ovarian follicular fluid in women with and without polycystic ovary syndrome, analyzed by liquid chromatography-tandem mass spectrometry. Fertil Steril. 2010;94(6):2228–2233. doi: 10.1016/j.fertnstert.2009.12.081. [DOI] [PubMed] [Google Scholar]
- 51.Xita N, Lazaros L, Georgiou I, Tsatsoulis A. CYP19 gene: a genetic modifier of polycystic ovary syndrome phenotype. Fertil Steril. 2010;94(1):250–254. doi: 10.1016/j.fertnstert.2009.01.147. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Li Q, Wang T, Yang G, Wang Y, Zhang X, Sang Q, Wang H, Zhao X, Xing Q, et al. A common polymorphism in the human aromatase gene alters the risk for polycystic ovary syndrome and modifies aromatase activity in vitro. Mol Hum Reprod. 2011;17(6):386–391. doi: 10.1093/molehr/gar007. [DOI] [PubMed] [Google Scholar]
- 53.Kauffman AS, Thackray VG, Ryan GE, Tolson KP, Glidewell-Kenney CA, Semaan SJ, Poling MC, Iwata N, Breen KM, Duleba AJ, et al. A novel letrozole model recapitulates both the reproductive and metabolic phenotypes of polycystic ovary syndrome in female mice. Biol Reprod. 2015;93(3):69. doi: 10.1095/biolreprod.115.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelley ST, Skarra DV, Rivera AJ, Thackray VG. The gut microbiome is altered in a Letrozole-induced mouse model of polycystic ovary syndrome. PLoS One. 2016;11(1):e0146509. doi: 10.1371/journal.pone.0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skarra DV, Hernandez-Carretero A, Rivera AJ, Anvar AR, Thackray VG. Hyperandrogenemia induced by Letrozole treatment of pubertal female mice results in hyperinsulinemia prior to weight gain and insulin resistance. Endocrinology. 2017;158(9):2988–3003. doi: 10.1210/en.2016-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrisse S, Childress S, Ma YP, Billings K, Chen Y, Xue P, Stewart A, Sonko ML, Wolfe A, Wu S. Low-dose dihydrotestosterone drives metabolic dysfunction via cytosolic and nuclear hepatic androgen receptor mechanisms. Endocrinology. 2017;158(3):531–544. doi: 10.1210/en.2016-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson AD, Solorzano CM, McCartney CR. Childhood obesity and its impact on the development of adolescent PCOS. Semin Reprod Med. 2014;32(3):202–213. doi: 10.1055/s-0034-1371092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ankarberg C, Norjavaara E. Diurnal rhythm of testosterone secretion before and throughout puberty in healthy girls: correlation with 17 beta-estradiol and dehydroepiandrosterone sulfate. J Clin Endocr Metab. 1999;84(3):975–984. doi: 10.1210/jcem.84.3.5524. [DOI] [PubMed] [Google Scholar]
- 59.Nader S. Hyperandrogenism during puberty in the development of polycystic ovary syndrome. Fertil Steril. 2013;100(1):39–42. doi: 10.1016/j.fertnstert.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Moran A, Jacobs DR, Jr, Steinberger J, Hong CP, Prineas R, Luepker R, Sinaiko AR. Insulin resistance during puberty: results from clamp studies in 357 children. Diabetes. 1999;48(10):2039–2044. doi: 10.2337/diabetes.48.10.2039. [DOI] [PubMed] [Google Scholar]
- 61.Hannon TS, Janosky J, Arslanian SA. Longitudinal study of physiologic insulin resistance and metabolic changes of puberty. Pediatr Res. 2006;60(6):759–763. doi: 10.1203/01.pdr.0000246097.73031.27. [DOI] [PubMed] [Google Scholar]
- 62.Cho YH, Craig ME, Donaghue KC. Puberty as an accelerator for diabetes complications. Pediatr Diabetes. 2014;15(1):18–26. doi: 10.1111/pedi.12112. [DOI] [PubMed] [Google Scholar]
- 63.Goran MI, Ball GDC, Cruz ML. Obesity and risk of type 2 diabetes and cardiovascular disease in children and adolescents. The Journal of Clinical Endocrinology & Metabolism. 2003;88(4):1417–1427. doi: 10.1210/jc.2002-021442. [DOI] [PubMed] [Google Scholar]
- 64.Kelly LA, Lane CJ, Weigensberg MJ, Toledo-Corral CM, Goran MI. Pubertal changes of insulin sensitivity, acute insulin response, and β-cell function in overweight Latino youth. J Pediatr. 2011;158(3):442–446. doi: 10.1016/j.jpeds.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelsey MM, Zeitler PS. Insulin Resistance of Puberty. Current Diabetes Reports. 2016;16:7. doi: 10.1007/s11892-016-0751-5. [DOI] [PubMed] [Google Scholar]
- 66.Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, Raza S, Doddapaneni HV, Metcalf GA, Muzny DM, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77(2):404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 69.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saraswati S, Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Front Microbiol. 2014;5:764. doi: 10.3389/fmicb.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Toyomasu Y, Hayashi Y, Linden DR, Szurszewski JH, Nelson H, Farrugia G, Kashyap PC, Chia N, Ordog T. Altered gut microbiota in female mice with persistent low body weights following removal of post-weaning chronic dietary restriction. Genome Med. 2016;8(1):103. doi: 10.1186/s13073-016-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin YN, Yu QF, Fu NA, Liu XW, Lu FG. Effects of four Bifidobacteria on obesity in high-fat diet induced rats. World J Gastroentero. 2010;16(27):3394–3401. doi: 10.3748/wjg.v16.i27.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang HS, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu YS, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY, Mao YJ, Zhang XJ, Pang XY, Wei CC, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice (vol 4, pg 232, 2010) ISME J. 2010;4(2):312–313. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 76.Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104(6):919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 77.Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS One. 2012;7:10. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong ML, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13(6):514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- 79.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20(4):738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Everard A, Lazarevic V, Gaia N, Johansson M, Stahlman M, Backhed F, Delzenne NM, Schrenzel J, Francois P, Cani PD. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8(10):2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in lactobacillus in obese patients and methanogens in anorexic patients. PLoS One. 2009;4(9):e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stsepetova J, Sepp E, Kolk H, Loivukene K, Songisepp E, Mikelsaar M. Diversity and metabolic impact of intestinal lactobacillus species in healthy adults and the elderly. Br J Nutr. 2011;105(8):1235–1244. doi: 10.1017/S0007114510004770. [DOI] [PubMed] [Google Scholar]
- 84.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obesity. 2012;36(6):817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53(2):100–108. doi: 10.1016/j.micpath.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 86.Kubasova T, Davidova-Gerzova L, Merlot E, Medvecky M, Polansky O, Gardan-Salmon D, Quesnel H, Rychlik I. Housing Systems Influence Gut Microbiota Composition of Sows but Not of Their Piglets. PLoS One. 2017;12:1. doi: 10.1371/journal.pone.0170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Fan H, Han Y, Zhao JZ, Zhou ZJ. Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyrosequencing analysis. Asian Austral J Anim. 2017;30(1):100–110. doi: 10.5713/ajas.16.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morotomi M, Nagai F, Sakon H, Tanaka R. Paraprevotella clara gen. Nov., sp nov and Paraprevotella xylaniphila sp nov., members of the family 'Prevotellaceae' isolated from human faeces. Int J Syst Evol Micr. 2009;59:1895–1900. doi: 10.1099/ijs.0.008169-0. [DOI] [PubMed] [Google Scholar]
- 89.Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. Nov., comb. nov., Parabacteroides goldsteinii comb. nov and Parabacteroides merdae comb. nov. Int J Syst Evol Micr. 2006;56:1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 90.Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015;39(4):509–521. doi: 10.1093/femsre/fuu010. [DOI] [PubMed] [Google Scholar]
- 91.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navas-Molina JA, Peralta-Sanchez JM, Gonzalez A, McMurdie PJ, Vazquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 96.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 100.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 101.Sheldon AL. Equitability Indices - Dependence on Species Count. Ecology. 1969;50(3):466-&. [Google Scholar]
- 102.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. No differences in gut microbial community diversity between placebo and letrozole-treated mice were observed prior to treatment. No significant differences in gut microbiome alpha diversity (Faith’s PD) between placebo- and letrozole-treated mice were observed prior to treatment (week 0) in the pubertal (placebo n = 10, letrozole n = 10) (A) or adult (placebo n = 8, letrozole n = 6) (B) model. Similarly, no differences in beta diversity (unweighted UniFrac) were observed between placebo- and letrozole-treated mice prior to treatment in the pubertal (C) or adult model (D). Student t-test was used to compare alpha diversity between groups and Analysis of Similarity (ANOSIM) test was used to compare beta diversity between groups. (PDF 1335 kb)
Data Availability Statement
The 16S rRNA sequence dataset generated and analyzed during the current study is available in the European Nucleotide Archive (Study Accession Number PRJEB20895).