Abstract
Objective
This study was designed to investigate the transcriptional response of OmpF and OmpC along with an antisense RNA, MicF under concentration gradient carbapenem exposure.
Result
An elevation in the expression of OmpF gene under concentration gradient imipenem stress from a particular concentration was observed. For OmpC gene a significant decrease in the expression was noticed under concentration gradient imipenem and meropenem stress. The study showed reduction in the expression of OmpC gene against imipenem and meropenem possibly preventing the entry of carbapenem antibiotic inside the cell indicating a possible role in carbapenem resistance.
Electronic supplementary material
The online version of this article (10.1186/s13104-019-4177-4) contains supplementary material, which is available to authorized users.
Keywords: OmpF, OmpC, micF, Carbapenem, Escherichia coli, Real time PCR
Introduction
OmpC and OmpF are two major outer membrane proteins in E. coli which serve as a barrier for antibiotics and other toxic agents entering inside the cell [1]. It is reported that sub lethal dose of antibiotics has protective role in bacterial cell against wide range of antimicrobials [2] and down regulation of porin genes are responsible in nonspecific resistance. OmpC and OmpF are known to be involved in non-specific solute transport and also it was reported that multidrug resistant E. coli had lower level of OmpC expression [3]. Also, previous reports suggest that OmpC and OmpF share reciprocal relationship [4].
Main text
Methodology
Bacterial strains
A total of 96 consecutive, non-duplicates, Escherichia coli clinical isolates resistant to at least one of the carbapenem antibiotic were collected from patients attended the clinic or admitted between August 2016 and July 2017 in Silchar Medical College and Hospital, Silchar, India.
Antibiotic susceptibility testing
A preliminary antibiotic susceptibility testing was performed to select the study isolates. Susceptibility was performed by Kirby Bauer method against different antibiotics viz, ciprofloxacin (5 μg),amikacin (30 μg),cefepime (30 μg), aztreonam (30 μg), ceftriaxone (30 μg), co-trimoxazole (25 μg), ceftazidime (30 μg), levofloxacin (5 μg) gentamicin (10 μg), carbenicillin (10 μg), ceftazidime (30 μg) and piperacillin-tazobactam (100/10 μg) (Hi-media, Mumbai, India) and results were interpreted as per Clinical and laboratory standards institute guidelines [5]. E. coli ATCC 25922 was used as control.
Minimal inhibitory concentration (MIC) assay
As the study intends to investigate the OmpC and OmpF transcriptional expression under carbapenem stress, the minimum inhibitory concentration of the test isolates was determined against imipenem, meropenem and ertapenem by agar broth dilution method at varied concentration ranging from 0.125 to 512 µg/ml and the results were interpreted as per CLSI guidelines [5].
Detection of efflux pump mediated carbapenem resistance using an inhibitor
Porin loss/mutation and increased efflux pump activity are major contributor of innate resistance mechanism. Therefore, the efflux pump mediated carbapenem resistance was assessed using an inhibitor. This test was carried out for all the test isolates with meropenem (10 µg, Himedia, Mumbai) with and without an efflux pump inhibitor carbonyl cyanide m-chlorophenylhyrazone (12.5 µM), [Himedia, Mumbai, India]. A difference between zone of inhibition of ≥ 5 mm with the inhibitor and the carbapenems alone confirms to be having efflux pump activity. Ethidium bromide was taken as control substrate for the efflux pump activity [6]. Disc with only CCCP was used to rule out any activity of the agent alone.
Detection of carbapenemases
To investigate the presence of carbapenemase activity among the selected isolates Modified Hodge test was performed. Further, polymerase chain reaction (PCR) assay was carried out in a 96 well thermal cycler (Applied Biosystems) in order to detect various carbapenemase-encoding genes which included blaKPC, blaIMP, blaVIM, blaNDM, blaOXA-23, blaOXA-48 and blaOXA-58 [7–10]. This experiment was performed to rule out the role of any acquired mechanism in carbapenem resistance among study isolates.
Analysis of efflux pump component acrA gene expression
To detect the presence of any isolates with overexpressed acrAB-tolC system, the non carbapenemase producers were further selected and subjected to quantitative Real Time PCR to examine the expressional level of the efflux pump gene acrA under normal condition. The total RNA was isolated using QIAGEN Rneasy Mini Kit (QIAGEN, Germany) following manufacturer’s instructions. Estimation of the isolated RNA was performed using Picodrop (Pico200, Cambridge, UK) and further cDNA was synthesized using prepared mRNA as template via Qiagen Reverse Transcription Kit (QIAGEN, Germany). Quantitative Real-time PCR of the cDNA prepared was done using power Sybrgreen PCR master mix reagents kit (Applied Biosystems, Austin, USA) in StepOnePlus quantitative Real Time-PCR (Applied Biosystems, USA) in triplicates using specific primers-acrA (F): 5′CTCTCAGGCAGCTTAGCCCTAA3′, acrA (R): 5′TGCAGAGGTTCAGTTTTGACTGTT3′ [11]. E. coli ATCC 25922 and RpslE was used as quality control and endogenous control.
Transcriptional expression of OmpF and OmpC and an antisense RNA gene micF gene
The overnight cultures of E. coli isolates on Luria Broth (Hi-media, Mumbai, India) were centrifuged and subjected to total RNA isolation using QIAGEN Rneasy Mini Kit (QIAGEN, Germany) according to manufacturer’s guidelines. The isolated RNA was then estimated with the use of Picodrop (Pico200, Cambridge, UK) and further synthesis of cDNA was done using Qiagen Reverse Transcription Kit (QIAGEN, Germany). Quantitative Real-time PCR of the cDNA prepared was performed by using power Sybrgreen PCR master mix reagents kit (Applied Biosystems, Austin, USA). Analysis of the synthesized cDNA levels was carried out in triplicate in StepOnePlus quantitative Real Time-PCR (Applied Biosystems, USA) using specific primers (Additional file 1: Table S1). E. coli ATCC 25922 was used as a reference for the analysis of relative fold changes in gene expression.
Transcriptional expression of OmpF and OmpC porin genes and MicF gene under concentration gradient carbapenem stress
To investigate the role of porin genes OmpF and OmpC during carbapenem stress the isolates were grown on LB broth containing 0.25 µg/ml, 0.5 µg/ml, 1 µg/ml, 2 µg/ml and 4 µg/ml of imipenem, meropenem and ertapenem individually. The culture was incubated for 16 h till late log phase. The mRNA was isolated immediately and reverse transcribed into cDNA. Quantitative Real Time PCR was performed to assess the transcriptional response of OmpF, OmpC and MicF against concentration gradient carbapenem stress. All the protocols were followed as described earlier in the text.
Statistical analysis
The relative expression differences of porin genes OmpF and OmpC along with an antisense RNA gene, MicF was compared with their respective wild type strain under concentration gradient carbapenem stress between samples were determined with the help of one-way ANOVA followed by Tukey–Kramer (Tukey’s W) multiple comparison test. Differences were considered statistically significant at both 5% and 1% level when p < 0.05. SPSS version 17.0 was used for statistical analysis.
Result
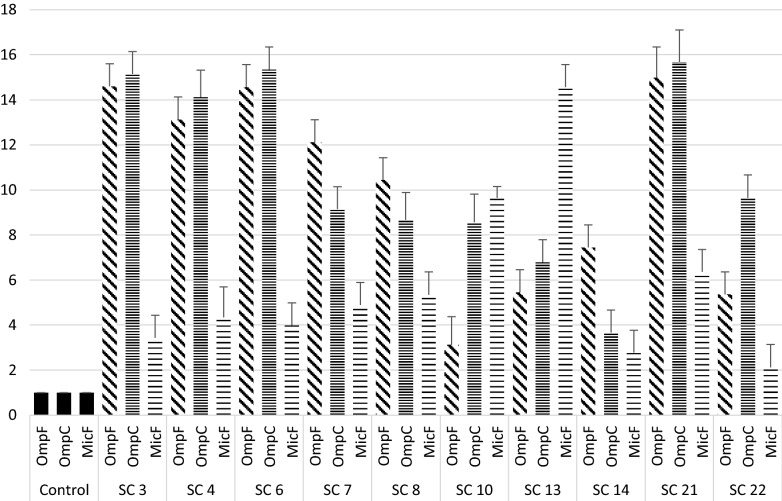
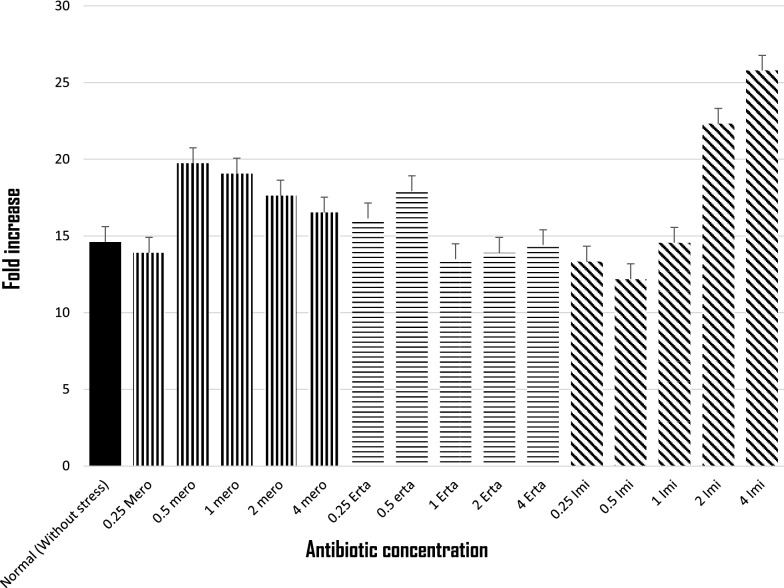
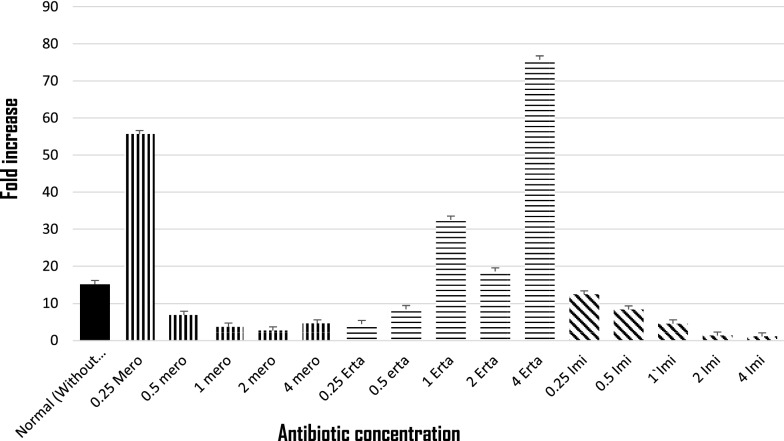
Among 96 carbapenem non-susceptible and carbapenemase non-producer E. coli, 39.5% (38/96) were selected based on their resistance towards at least one of the carbapenems tested. Of which 57.89% (22/38) isolates showed efflux pump positive activity phenotypically when tested with meropenem along with an efflux pump inhibitor CCCP. Antimicrobial susceptibility of the isolates showed highest susceptibility towards amikacin (86.36%) followed by cefepime (59.09%) and co-trimoxazole (40.90%). MIC result revealed that more than half of the isolates were found to be above the break point against ertapenem (90.9%), followed by imipenem (77.2%) and meropenem (59.0%) (Additional file 2: Table S2). Further, the transcriptional expression of acrA exhibited over expression of acrA gene in 12 isolates and 10 isolates showed low level of expression (Additional file 3: Figure S1) when compared with E. coli ATCC 25922. Isolates with reduced expression of acrA gene were selected and further studied for mRNA transcript of the porin genes OmpF and OmpC along with MicF (Fig. 1), of which all the three genes exhibited higher expression. However, when the expressional level was compared with the stressed condition, expression of OmpF gene was elevated against concentration gradient imipenem stress from 0.5 µg/ml, whereas; under meropenem stress a steady decrease in the expression was recorded (Fig. 2). Ertapenem exposures showed random transcriptional level (Fig. 2). For OmpC gene a significant decrease in the expression was noticed under concentration gradient imipenem and meropenem stress (Fig. 3). While against ertapenem stress an asymmetric pattern of expression was observed (Fig. 3). In case of MicF a steady decrease in the expression after meropenem exposure was observed and ertapenem and imipenem stress showed an arbitrary pattern in the expression (Additional file 4: Figure S2).
Fig. 1.
Expression of OmpF, OmpC and micF gene under normal condition (without stress) relative to E. coli ATCC 25922
Fig. 2.
Expression of OmpF gene under concentration gradient carbapenem stress relative to the normal one without stress
Fig. 3.
Expression of OmpC gene under concentration gradient carbapenem stress relative to the normal one without stress
Discussion
This study deals with the outer-membrane porin genes OmpF, OmpC and MicF gene which perhaps has a role in developing resistance towards carbapenems. As reported by Nikaido and Vaara, the expression of OmpF gene is reduced in high osmolarity condition whereas, for OmpC the same environment was found to be favorable [12]. However, efflux pump gene AcrB has also been reported to repress the expression of the porin OmpF gene [13, 14]. In this study carbapenem resistant E. coli isolates were studied of which decrease in the expression of OmpF and OmpC genes were observed in AcrAB overexpressed isolates which is in agreement with previous studies [15–17]. In a study done by Yigit et al., in Enterobacter strain 810, alteration in the porin genes OmpF and OmpC resulted in imipenem resistance along with reduced susceptibility towards meropenem and cefepime [18]. After, exposure to sub inhibitory concentration of imipenem down regulation of OmpC gene was also observed which is in support of the present study. However, it was observed under imipenem stress an increase in the expression of OmpF gene was observed which has not been reported earlier. The expression level of OmpF is also affected by a small antisense RNA, MicF which inhibits the expression of OmpF gene [19]. However, in the present study meropenem stress reduced the expression of OmpF and micF as well, indicating that there may be other factors responsible for the down regulation of OmpF against carbapenem exposure. Therefore, it is probably the major porin that prevents entry of carbapenem inside the cell which calls for further investigation.
Limitation
This study warrants a proteomic approach to analyze the expression of porin genes at translational level.
Additional files
Additional file 1: Table S1. Oligonucleotides used in the study.
Additional file 2: Table S2. Minimum inhibitory concentration range of carbapenems tested.
Additional file 3: Figure S1. Expression of acrA gene under normal condition (without stress) relative to Escherichia coli ATCC 25922.
Additional file 4: Figure S2. Expression of MicF gene under concentration gradient carbapenem stress relative to normal one without stress.
Authors’ contributions
SC performed the experimental work, data collection and analysis and prepared the manuscript. MS, DB and KN analyzed the data. DDC and AC have designed work plan and corrected manuscript. AB has conceived the plan and supervised the whole study. All authors ensured that this is the case. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to acknowledge Assam University Biotech Hub for providing infrastructure facility.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the data generated in this research work are presented in this research article. In case of any additional information requirement corresponding author will be providing the necessary information as per ethical guidelines.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The work was approved by Institutional Ethical committee of Assam University, Silchar. The authors confirm that participants provided their written informed consent to participate in this study.
Funding
The study was supported by Indian Council of Medical Research (Letter No: 2018-3430/CMB-BMS), dated: 12.07.2018 by awarding Senior Research Fellowship to Shiela Chetri. The study design and protocol was approved by the funding agency. However, the funding body has no role in analysis and interpretation of data and in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- OmpF
outer membrane protein F
- OmpC
outer membrane protein C
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
Contributor Information
Shiela Chetri, Email: sony240689@gmail.com.
Melson Singha, Email: melsonsingha012@gmail.com.
Deepshikha Bhowmik, Email: krm.deepshikha@gmail.com.
Kathakali Nath, Email: kathakalinath@gmail.com.
Debadatta Dhar Chanda, Email: drdebadattadhar@rediffmail.com.
Atanu Chakravarty, Email: atanu_chakravarty@rediffmail.com.
Amitabha Bhattacharjee, Email: ab0404@gmail.com.
References
- 1.Igwe JC, Olayinka BO, Ehnimidu JO, Onaolapo JA. Impact of outer membrane protein Ompc and Ompf on antibiotics resistance of E. coli isolated from UTI and diarrhoeic patients in Zaria, Nigeria. Clin Microbiol. 2016;5(6):1000267. doi: 10.4172/2327-5073.1000267. [DOI] [Google Scholar]
- 2.Agafitei O, Kim EJ, Maguire T, Sheridan J. The role of Escherichia coli porins OmpC and OmpF in antibiotic cross resistance induced by sub-inhibitory concentrations of kanamycin. J Exp Microbiol Immunol. 2010;14:34–39. [Google Scholar]
- 3.Cohen SP, McMurry LM, Hooper DC, Wolfson JS, Levy SB. Cross resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/AAC.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norioka S, Ramakrishnan G, Ikenaka K, Inouye M. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. J Biol Chem. 1986;261:17113–17119. [PubMed] [Google Scholar]
- 5.CLSI. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute.
- 6.Quale J, Bratu S, Landman D, Heddurshetti R. Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin Infect Dis. 2003;37:214–220. doi: 10.1086/375821. [DOI] [PubMed] [Google Scholar]
- 7.Yong D, Toleman M, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yum JH, Yi K, Lee H, Yong D, Lee K, Kim JM, Rossolini GM, Chong Y. Molecular characterization of metallo-b-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla VIM-2 gene cassettes. J Antimicrob Chemother. 2002;49(5):837–840. doi: 10.1093/jac/dkf043. [DOI] [PubMed] [Google Scholar]
- 9.Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A. The emergence of OXA-48 and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17:1130–1133. doi: 10.1016/j.ijid.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Mendes R, Bell JM, Turnidge JD, Castahneira M, Jones RN. Emergence and widespread dissemination of OXA-23. OXA-24/40 and OXA-58 carbapenemases among Acinetobacter spp. in Asia-pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63:55–59. doi: 10.1093/jac/dkn434. [DOI] [PubMed] [Google Scholar]
- 11.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother. 2011;55:921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 14.Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pagès JM. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets. 2008;9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 15.Koyano S, Saito R, Nagai R, Tatsuno K, Okugawa S, Okamura N, Moriya K. Molecular characterization of carbapenemase-producing clinical isolates of Enterobacteriaceae in a teaching hospital, Japan. J Med Microbiol. 2013;62(3):446–450. doi: 10.1099/jmm.0.050708-0. [DOI] [PubMed] [Google Scholar]
- 16.Philippe N, Maigre L, Santini S, Pinet E, Claverie JM, Davin-Régli AV, Pagès JM, Masi M. In vivo evolution of bacterial resistance in two cases of Enterobacter aerogenes infections during treatment with imipenem. PLoS ONE. 2015;10(9):e0138828. doi: 10.1371/journal.pone.0138828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaskulski MR, Medeiros BC, Borges JV, Zalewsky R, Fonseca MEC, Marinowic DR, Rocha MP, Nodari P, Machado DC. Assessment of extended-spectrum β-lactamase, KPC carbapenemase and porin resistance mechanisms in clinical samples of Klebsiella pneumoniae and Enterobacter spp. Int J Antimicrob Agents. 2013;42(1):76–79. doi: 10.1016/j.ijantimicag.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Yigit H, Anderson GJ, Biddle JW, Steward CD, Rasheed JK, Valera LL, McGowan JE, Tenover FC. Carbapenem resistance in a clinical isolate of Enterobacter aerogenes is associated with decreased expression of OmpF and OmpC porin analogs. Antimicrob Agents Chemother. 2002;46(12):3817–3822. doi: 10.1128/AAC.46.12.3817-3822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pagès JM, Amaral L. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE. 2007;2(4):e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Oligonucleotides used in the study.
Additional file 2: Table S2. Minimum inhibitory concentration range of carbapenems tested.
Additional file 3: Figure S1. Expression of acrA gene under normal condition (without stress) relative to Escherichia coli ATCC 25922.
Additional file 4: Figure S2. Expression of MicF gene under concentration gradient carbapenem stress relative to normal one without stress.
Data Availability Statement
All the data generated in this research work are presented in this research article. In case of any additional information requirement corresponding author will be providing the necessary information as per ethical guidelines.