Abstract
NTRK gene fusions involving either NTRK1, NTRK2, or NTRK3 (encoding the neurotrophin receptors TRKA, TRKB, and TRKC, respectively) are oncogenic drivers of various adult and paediatric tumour types. These fusions can be detected in the clinic using a variety of methods, including tumour DNA and RNA sequencing and plasma cell-free DNA profiling. The treatment of patients with NTRK fusion-positive cancers with a first-generation TRK inhibitor, such as larotrectinib or entrectinib, is associated with high response rates (>75%), regardless of tumour histology. First-generation TRK inhibitors are well tolerated by most patients, with toxicity profiles characterized by occasional off-tumour, on-target adverse events (attributable to TRK inhibition in non-malignant tissues). Despite durable disease control in many patients, advanced-stage NTRK fusion-positive cancers eventually become refractory to TRK inhibition; resistance can be mediated by the acquisition of NTRK kinase domain mutations. Fortunately, certain resistance mutations can be overcome by second-generation TRK inhibitors, including LOXO-195 and TPX-0005 that are being explored in clinical trials. In this Review, we discuss the biology of NTRK fusions, strategies to target these drivers in the treatment-naive and acquired-resistance disease settings, and the unique safety profile of TRK inhibitors.
Introduction
The approach to the development of targeted therapies for oncogenic driver-positive cancers has historically been histology-specific. This strategy has resulted in the regulatory approval of several small-molecule inhibitors or antagonistic monoclonal antibodies for the treatment of patients with a single cancer type or, more commonly, a subtype that harbours a specific sensitizing molecular alteration. Examples include anti-HER2 monoclonal antibodies or HER2 tyrosine-kinase inhibitors (TKIs) for ERBB2 (HER2)-amplified breast cancer, EGFR, ALK, and ROS1 TKIs for EGFR-mutant, ALK-rearranged, and ROS1-rearranged non-small-cell lung cancer (NSCLC), respectively, and BRAF and MEK inhibitors for BRAF-mutant melanoma or NSCLC1–6.
With advances in clinical sequencing technologies, both new and known drivers of oncogenesis continue to be discovered across a variety of cancers. Many of these drivers, such as recurrent gene fusions involving RET, FGFR1, FGFR2, FGFR3, and NRG1, and mutations involving MET, ERBB2, PIK3CA, and AKT, are present across multiple cancer types and are potentially amenable to pharmacological inhibition, thus inspiring the basket trial design paradigm. In a basket trial, patients with tumours harbouring a particular genomic alteration are treated with a matched therapeutic regardless of tumour histology, with the goal of evaluating the activity of the agent against a range of cancers harbouring the qualifying genomic aberration5,7,8. The results of several of these studies have revealed that, depending on the molecular driver in question, responsiveness of the disease to therapy can either be histology-dependent or histology-independent. As an example of the former, considerably lower objective response rates (ORRs) are achieved with BRAF inhibition in patients with BRAFV600E-mutant colorectal cancers (0% ORR with vemurafenib)7 compared with the high rates achieved in those with BRAFV600E-mutant melanoma (48% ORR with vemurafenib)9 or NSCLC (42% ORR with vemurafenib)7. By contrast, the response of diverse cancers harbouring oncogenic fusions involving the neurotrophin receptor tyrosine kinase genes NTRK1, NTRK2, or NTRK3 (encoding TRKA, TRKB, and TRKC, respectively; collectively referred to hereafter as TRK) to TRK inhibition provides a prime example of the histology-independent activity of targeted therapy in a molecularly defined subset of cancers10,11.
In this Review, we describe the function of TRKA, TRKB, and TRKC as well as the biology of fusions involving NTRK1, NTRK2, and NTRK3. We also discuss current strategies to target these oncogenic drivers in patients (including those who have acquired resistance to a TRK inhibitor), and the mechanisms underlying the observed toxicities and resistance to TRK inhibition.
TRK biology and oncogenesis
NTRK proto-oncogenes
NTRK1 was first identified as an oncogene in 1982 by Mariano Barbacid and colleagues12 during gene transfer assays aimed at identifying genes with transforming capacities present in human tumour specimens (in this case, of a colon cancer) (FIG. 1). Specifically, the cDNA of the oncogene identified contained sequences of a non-muscle tropomyosin fused to sequences of a putative receptor tyrosine kinase13. In 1989, the same group isolated the cDNA of the NTRK1 proto-oncogene and described the gene product, TRKA, as a protein of 790 amino acids with features characteristic of cell surface receptor tyrosine kinases14 (FIG. 2). In 1991, two independent groups provided compelling evidence that TRKA was expressed in the nervous system and became phosphorylated in response to stimulation with the neurotrophin nerve growth factor (NGF), thus demonstrating the role of TRKA as a receptor for NGF15,16. This discovery paved the way for the identification of TRKB and TRKC as members of the same family of receptors17–20. These receptors are capable of binding with high affinity to the following ligands: NGF for TRKA, brain-derived neurotrophic factor (BDNF) or neurotrophin 4 (NT-4) for TRKB, and neurotrophin 3 (NT-3) for TRKC19,20. Of note, although NT-3 can bind with and activate all three TRK proteins, it has higher affinity for TRKC than for TRKA and TRKB (FIG. 2a, inset)21.
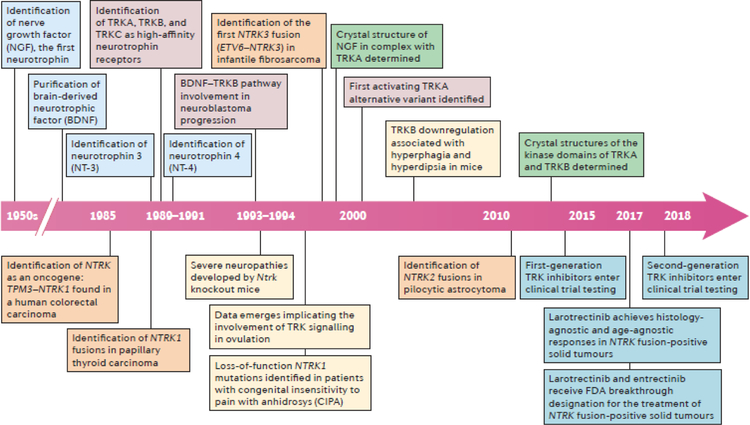
Fig. 1 |. Timeline of key advances relating to the biology and therapeutic targeting of TRK signalling.
Milestone discoveries that are relevant to normal TRK pathway biology (boxes above the timeline arrow) and NTRK fusions in cancer (boxes below the timeline arrow) are depicted. Key events relating to the following fields of study are colour coded as follows: neurotrophin identification (light blue), TRK function (red), TRK loss or NTRK-mutant phenotypes (yellow), TRK protein structure (green), TRK overexpression and splicing (grey), identification of NTRK fusions in clinical samples (orange), and clinical trials of TRK inhibitors (dark blue).
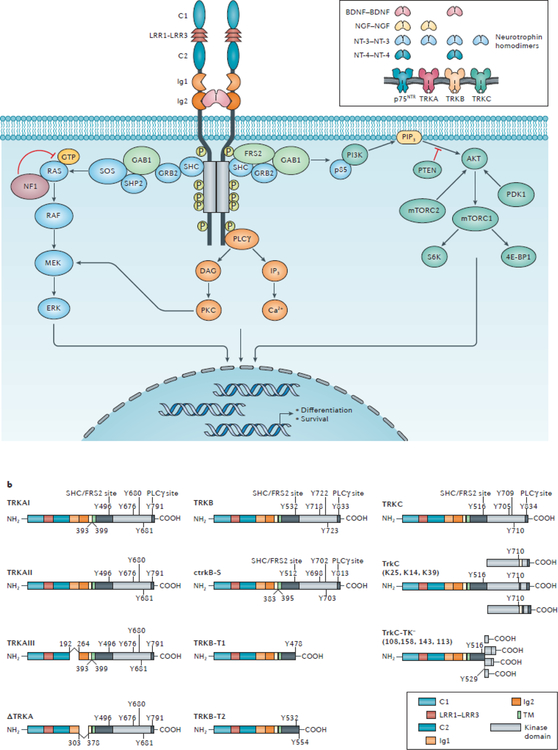
Fig. 2 |. TRK biology and signalling in the nervous system.
a | The inset image indicates the ligand specificity of the TRK proteins for the neurotrophins, brain-derived neurotrophic factor (BDNF), nerve-growth factor (NGF), neutrotrophin 3 (NT-3), and/or neurotrophin 4 (NT-4), which each bind to their cognate receptors as a homodimer. TRKA is the high affinity receptor for NGF, whereas TRKB has high affinity for both BDNF and NT-4. NT-3 can bind to all TRK receptors but has highest affinity for TRKC and is the sole ligand of this receptor. Additionally, the TNF receptor superfamily member p75NTR can bind to all neurotrophins with low affinity, resulting in enhanced TRK signalling and/or the activation of distinct signalling pathways. The main image depicts the structure of the TRK–neurotrophin complex and the signalling pathways activated by TRK upon neurotrophin stimulation. The cysteine clusters C1 and C2, leucine-rich regions (LRR) 1–3, the Ig1 and Ig2 immunoglobulin-like motifs, and the kinase domain (KD) are indicated. The binding of neurotrophins to the extracellular region of TRK proteins, predominantly at the Ig2 domain, results in ligand-dependent receptor homodimerization followed by transactivation of the intracellular tyrosine kinase domains and the recruitment of various cytoplasmic adaptors. The phosphorylation events that mediate activation of the kinase domain or binding of SHC-transforming protein (SHC), fibroblast growth factor receptor substrate 2 (FRS2), and phosphoinositide phospholipase Cɣ (PLCɣ) to TRK proteins are depicted. The recruited adaptor proteins activate downstream signalling pathways, including the MAPK, PI3K, and PKC pathways. Each of these signalling pathways also activates the transcription of genes involved in the differentiation and survival of neurons21. b | The schematic domain structures of known splice variants of TRKA, TRKB, and TRKC are shown. The docking residues for SHC–FRS2, and PLCɣ, and the three phosphorylated tyrosines within the activation loop of the kinase domain are also displayed. The ctrkB-S, TrkC, and TrkC-TK− schematics were generated using the amino acid sequences of chicken TrkB and porcine TrkC, respectively — the organisms in which these variants were initially discovered26. Of note, an alternative ATG transcription-initiation site in the NTRK2 gene has been reported; transcripts starting from this alternative site give rise to multiple additional TRKB-derived transcripts34 (not shown). 4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; DAG, diacylgycerol; GAB1, GRB2-associated-binding protein 1; GRB2, growth factor receptor-bound protein 2; IP3, inositol triphosphate; mTORC, mechanistic target of rapamycin complex; NF1, neurofibromin; p85, PI3K 85 kDa regulatory subunit α; PIP3, phosphatidylinositol 3,4,5-trisphosphate; SHP2, SH-PTP2 (also known as tyrosine-protein phosphatase non-receptor type 11); SOS, son of sevenless homologue.
The TRK family receptors are initially synthesized as precursor proteins; the post-translational glycosylation of the extracellular domains of these precursors yields the mature protein products TRKA (140 kDa), TRKB (145 kDa), and TRKC (145 kDa)22. All TRK proteins share similar structural domains in the extracellular region, including two immunoglobulin-like (Ig1 and Ig2) and three leucine-rich 24-residue motifs (LRR1–3; FIG. 2a). The LRR1–3 motifs, flanked by two cysteine clusters (C1 and C2), are specific to TRK proteins and are not found in other subfamilies of receptor tyrosine kinases22,23. TRK proteins interact with their cognate ligands predominantly via the Ig2 domain that is proximal to the transmembrane region, although other extracellular domains of TRK are known to be involved in neurotrophin binding. For example, the substitution of a conserved cysteine residue in the Ig1 domain of TRKA with a serine (C266S) abolishes NGF binding24. Similarly, the Ig1 domain of TRKB has been shown to be required, together with the Ig2 domain, for activation of this receptor by BDNF or NT-3 (REF.25). In addition, the LRR1 domain of TRKA has been found to be sufficient for NGF-dependent activation of an engineered TRKA–TRKB chimeric receptor in the presence of the low affinity receptor for neurotrophins p75NTR (REF.25) — an alternative pan-neurotrophin receptor that, unlike the TRK proteins, belongs to the TNF receptor superfamily member (and thus is also known as TNFRSF16). Intracellularly, TRK proteins possess a tyrosine kinase domain that becomes active following ligand binding and subsequent receptor homodimerization21.
A number of TRK splice variants with differential affinities for neurotrophins have been identified26,27 (FIG. 2b).The most abundant isoform of TRKA (TRKAII) is expressed in most non-neuronal tissues, consists of 796 amino acids, and is efficiently activated by both NGF and NT-328. The neuronal isoform, TRKAI, lacks six amino acids (residues 393–398) of the juxtamembrane domain, owing to omission of exon 9 through alternative splicing, and is efficiently activated only by NGF28,29. A third variant, TRKAIII, results from splicing out of exons 6, 7, and 9 and is constitutively active in a ligand-independent manner. This isoform is expressed by normal pluripotent neural stem and neural crest progenitor cells but was originally identified as an oncogenic driver in neuroblastoma28–31. One kinase-intact isoform of TRKB that lacks the residues encoded by exon 9 has been identified in chickens (ctrkB-S) (FIG. 2b); this variant can only be activated by BDNF and is predominantly expresses in a subpopulation of dorsal root ganglion neurons that is non-overlapping with the population expressing full-length TRKB (ctrkB-L)32, 33. In addition, a novel human NTRK2 transcript that encodes a truncated isoform of the TRKB protein (TRKB-T-TK) has been reported34. The TRKB-T-TK transcript includes an extended version of exon 22, which contains a translational stop-codon, and is predominantly expressed in the nervous system34. The resulting protein lacks the 114 C-terminal amino acids of the full-length receptor, including a binding site for phosphoinositide phospholipase Cɣ (PLCɣ)34 (FIG. 2b). Multiple isoforms of TRKC containing variable-sized amino acids insertions in the kinase domain have been described35 (FIG. 2b). These variants retain their ability to be activated by NT-3 but are not capable of mediating fibroblast proliferation or neuronal differentiation of rat pheochromocytoma PC12 cells35. Additionally, isoforms of TRKB and TRKC that lack the kinase domains have been reported28, 29 (FIG. 2b). These variants are expressed at high levels in mature neurons and were initially described to act as dominant negative isoforms by competing with the full-length receptors for the binding to neurotrophins36,37. More recent studies, however, have revealed that the truncated isoforms of TRK proteins can function as active signalling molecules by recruiting scaffolding proteins, including GRP1-associated scaffold protein and Rho GDP-dissociation inhibitor 1 (REFS.21,38,39). Together, these findings imply that TRK proteins can activate a wide variety of signalling pathways.
TRK activation results in the autophosphorylation of intracellular tyrosine residues. In particular, Y676, Y680, and Y681 of human TRKA, and their corresponding residues in TRKB and TRKC, which are located within the activation loop of the kinase domain; phosphorylation of these tyrosines is required for the full activation of the kinase40. Phosphorylation of Y496 and Y791 of TRKA, and the corresponding tyrosine residues in TRKB and TRKC, drives downstream signalling. Specifically, phosphorylated Y496 directly binds and activates the SHC-transforming protein (SHC) and fibroblast growth factor receptor substrate 2 (FRS2), whereas phosphorylated Y791 interacts directly with PLCɣ41. Other intracellular downstream effectors activated by these adaptors include members of the MAPK, PI3K, and protein kinase C (PKC) pathways (FIG. 2a). Each of these signalling pathways ultimately regulates the transcription of genes involved in the differentiation and survival of neurons (such as those encoding the cyclic AMP-responsive element-binding (CREB) transcriptional co-activator proteins)21.
Almost all studies describing TRK signalling refer to the MAPK, PI3K, and PKC pathways as the main downstream effectors responsible for the activation of neuronal survival and differentiation pathways; however, mice homozygous for a Y-to-F mutation at the SHC binding site of TRKB (Y532) or TRKC (Y516) have limited loss of sensory neurons, whereas mice homozygous for kinase-dead TRKB or TRKC mutants have severe neuronal loss42,43, suggesting that Y496/Y532/Y516-independent interactions and the activation of SHC-independent signalling pathways promote neuronal survival. Thus, additional studies are required to fully delineate the signalling network activated by TRK receptors in the nervous system. Importantly, Y516 of TRKC is absent from the oncoprotein generated by the ETV6–NTRK3 fusion owing to the genomic location of the breakpoint in NTRK344, supporting the hypothesis that this specific tyrosine residue and, thus, SHC binding are not required for efficient activation of the kinase domain or TRK signalling.
TRK in development and physiology
As alluded to above, TRK pathway signalling has crucial roles in neuronal development and differentiation. Gene-expression studies have shown that NTRK genes are predominantly transcribed in the nervous system in adult tissues, as well as during embryonic development45. The types and amounts of neurotrophins and neurotrophin receptors available in different compartments of the nervous system are crucial to maintaining normal neuronal homeostasis46. Indeed, neurotrophins were initially identified as survival molecules for sensory and sympathetic neurons; however, their functions are now known to be more diverse and complex47. For example, whereas neurotrophin-mediated activation of TRK mainly promotes neuronal growth, differentiation, and survival, binding of neurotrophins to their alternative receptor, p75NTR, primarily results in the activation of the JNK signalling cascades as well as the p75NTR-interacting protein (NRAGE, also known as melanoma-associated antigen D1) and p75NTR-associated cell death executor (NADE) adaptors that directly promote cell cycle arrest and apoptosis48,49,50. Importantly, p75NTR can directly bind to TRKA, TRKB, or TRKC, and this interaction can modulate the function of TRK receptors and their responsiveness to neurotrophins25. For example, the presence of p75NTR results in increase rate of NGF association with TRKA51 and is required for high-affinity interactions between NGF and TRKA52. In addition, p75NTR expression enhances the specificity of TRKB for its primary ligand BDNF and partially suppresses the ubiquitination of TRKA and TRKB receptors, thereby delaying their degradation54,55. Furthermore, the apoptotic signalling activated by p75NTR in the presence of NGF can be inhibited by overexpressing TRKA56,57. The findings of these studies indicate that the effects of p75NTR on the cellular response to neurotrophins might be dependent on the concentration of neurotrophins, the type and the abundance of neurotrophin receptors expressed in the cells, and the stage of cell differentiation. Thus, the signalling pathways activated by TRK proteins and p75NTR upon neurotrophin stimulation can affect many diverse neuronal functions, including the survival and differentiation of neurons, synapse formation and plasticity, membrane trafficking, and axon and dendrite formation27.
TRK activation in cancer
NTRK mutations, splice variants, and TRK overexpression.
TRK proteins can potentially be activated by a variety of mechanisms. First, somatic NTRK mutations have been identified in various tumour types, including colorectal58, lung cancers (large-cell neuroendocrine carcinoma and NSCLC)59,60, as well as melanoma61, and acute myeloid leukaemia58–62. For example, NTRK2 mutations affecting TRKB have been reported at two different kinase domain sites (T695I and D751N) in patients with colorectal cancer, within the extracellular region (L138F) in the NCI-H2009 lung adenocarcinoma cell line, and at a site proximal to the kinase domain (P507L) in the MDA-MB-435 melanoma cell line58. The effects of these TRKB mutations have been characterized using in vitro assays and the T695I and D751N variants were found to display reduced kinase activity and, consequentially, were associated with a reduced capability for tumour formation in vivo, as compared with wild-type TRKB, when transduced into rat immortalized epithelial cells58; the activity of the L138F and P507L variants was similar to wild-type TRKB. Additional mutations affecting the activation loop residues of TRKB (M713I, R715G, and R734C) have been described in NSCLC specimens59. These mutants also had less activity than full-length TRKB (indicated by lower levels of autophosphorylation) and were less efficient in promoting cell migration and transformation when transduced into mouse NIH-3T3 and BaF3 cells59. Moreover, pharmacological TRK inhibition was effective in abrogating the migration and growth of wild-type TRKB overexpressing cells but not those expressing the mutant proteins59. More recently, mutations in TRKA (M379I and R577G), affecting the Ig2 and the kinase domain of TRKA, respectively have been identified in melanomas61. Characterization studies revealed, once again, that these mutants were functionally indistinguishable from the wild-type receptor when transiently or stably transfected into HeLa or NIH-3T3 cells, respectively61. Hence, the potential role of NTRK mutations in promoting tumourigenesis and cancer progression has not yet been established.
Second, activating splice variants of the NTRK1 gene have been identified and characterized. In particular, the NTRK1 splice variant TRKAIII and a genomic in-frame deletion mutant (ΔTRKA) detected in human neuroblastoma31 and acute myeloid leukaemia63 specimens, respectively, have been reported to be oncogenic. Interestingly, both these oncogenic variants of TRKA lack of regions of the extracellular domain that are known to be highly glycosylated and are involved in ligand binding (FIG. 2b). ΔTRKA, for example, lacks 75 amino acids encompassing the ligand-binding Ig2 domain (owing to partial deletion of exon 8); this mutant protein has a constitutively active kinase domain, independent of ligand binding, and is capable of transforming fibroblastic, epithelial, and myeloid cells in vitro63. The TRKAIII splice variant results in the omission of exons 6, 7, and 9 and thus the functional Ig1 domain24. Similar to ΔTRKA, this protein is constitutively active, can transform mouse NIH-3T3 fibroblasts in vitro, and enables tumour formation by these cells in nude mice; TRKAIII expression also increased the growth of a SY5Y-derived human neuroblastoma cell line in a mouse tumour model30,31. This evidence suggests that regulatory auto-inhibitory domains lie within the regions excluded from each of these TRKA variants. This hypothesis is supported by the results of mutagenesis assays demonstrating that the substitution of a single residue of TRKA (P203A) within the Ig1 domain is sufficient to generate a TRKA variant that is capable of spontaneous dimerization and harbours transforming properties24.
Finally, TRK overexpression has been reported in a variety of cancers, including breast, cutaneous (for example, basal cell carcinoma), and lung cancers, neuroblastoma, cylindroma and others64–67. In patients with neuroblastoma, TRKA and TRKC overexpression is strongly predictive of favourable outcomes, while TRKB is mainly expressed in higher-grade tumours that also harbour MYCN amplification68. Multiple studies have reported promising activity of TRK inhibitors against TRKB-expressing neuroblastoma cell lines in vitro and in vivo. For example, Evans and colleagues69,70 demonstrated that the potent TRK inhibitor CEP-751 (KT-6587) was highly effective in inhibiting the growth of TRKB-overexpressing SY5Y-derived neuroblastoma xenografts in nude mice. The related TRK inhibitor lestaurtinib (previously known as CEP-701 and KT-5555) has been tested in various clinical trials, including a phase I study in patients with neuroblastoma; however, a response to therapy was only observed in a minority of these patients, and whether clinical responses occurred secondary to TRK inhibition, recognizing that lestaurtinib is a multikinase inhibitor, remains unclear71. In breast cancer models, ectopic overexpression of TRKA promoted tumour cell proliferation, migration, and invasion through MAPK and PI3K pathway activation65, thus suggesting that TRKA overexpression correlates with tumour aggressiveness in this malignancy. TRKB and/or TRKC overexpression has been reported in patients with cylindroma, a tumour that can develop in patients with germline mutations in the tumour-suppressor gene CYLD, and in sporadic basal cell carcinomas67. Interestingly, lestaurtinib treatment reduced colony formation and proliferation in 3D primary cell cultures established from CYLD-mutant tumours67.
NTRK fusions.
Fusions involving NTRK1, NTRK2, or NTRK3 are the most common mechanisms of oncogenic TRK activation72. Typically, intrachromosomal or interchromosomal rearrangements form hybrid genes in which 3′ sequences of NTRK1, NTRK2, or NTRK3 that include the kinase domain are juxtaposed to 5′ sequences of a different gene (FIG. 3). The product of the fusion is a chimeric oncoprotein characterized by ligand-independent constitutive activation of the TRK kinase.
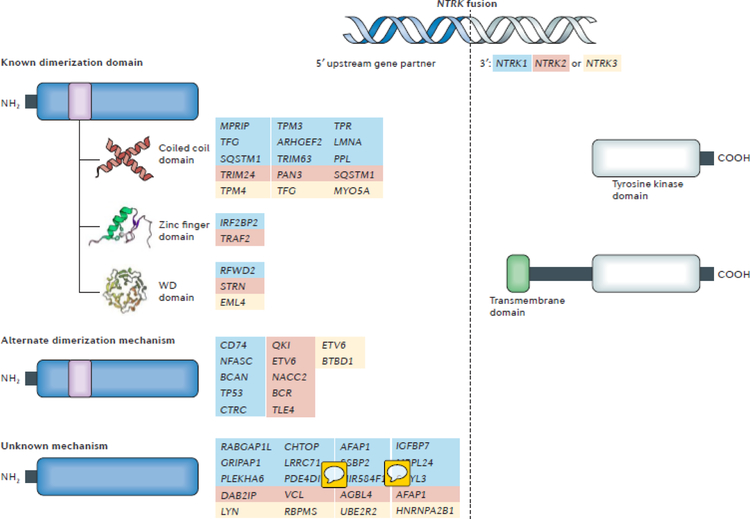
Fig. 3 |. Activating mechanisms of NTRK fusions.
The structure of a representative NTRK fusion gene with sequences of the NTRK gene (grey) and the upstream partner gene (blue) is shown. Most NTRK fusion partners are hypothesized to activate the downstream TRK kinase domain and thus aberrant TRK signalling via dimerization. A list of the known upstream partners, stratified according to the type of oligomerization domain that they contain (coiled coil, zinc finger, or WD domains), is provided. Of note, not all fusion partners harbour typical dimerization domains and the upstream genes for which an alternate mechanism of dimerization is potentially responsible for fusion activation, or the mechanism by which fusion activation is unknown are also listed. Partners of NTRK1, NTRK2, and NTRK3 are indicated by blue, red, and yellow text, respectively. The TRK kinase domain is always included in the oncogenic fusion protein. By contrast, the transmembrane domain of the TRK protein is only present in select fusions, suggesting that this domain is not required for activation of the TRK kinase; however, incorporation of the transmembrane domain might have an effect on cellular localization of the fusion protein (for example, to the plasma membrane).
In fusion biology, upstream gene partners often contain oligomerization domains, such as coiled-coil domains, zinc finger domains, or WD repeats, that in some cases are required for full activation of the downstream kinase14,73–75. Consistent with this paradigm, the majority of NTRK fusion partners harbour oligomerization domains (FIG. 3). Nevertheless, examples of fusion partners that lack known dimerization domains do exist. In these instances, the contribution of the upstream partner in promoting activation of the downstream TRK kinase is unclear. One possibility is that the sequences derived from the gene partner simply replace the auto-inhibitory domains present in the extracellular domain of the TRK proteins. Alternatively, the partner can be actively involved in the transformation process: immunohistochemical analyses of TRK expression in tumours harbouring NTRK fusions have revealed that the kinase partner might determine the subcellular localization of the fusion products76, similar to the finding that transforming acidic coiled-coil-containing protein 3 (TACC3) is responsible for the localization of oncogenic FGFR–TACC3 fusion proteins to the mitotic spindle poles77. These data underscore the importance of the upstream partners in the oncogenic activation of different TRK fusion proteins, by various mechanisms.
TRK fusion proteins can signal through the same downstream pathways activated by full-length TRK proteins upon neurotrophin binding. In studies using transduced models focused on the identification of substrates directly activated by TPR–TRKA (encoded by TPR–NTRK1) and TPM3–TRKA (encoded by TPM3–NTRK1)78–80, similar to full-length TRKA, both fusion oncoproteins were able to bind to SHC, insulin receptor substrate 1 (IRS1), IRS2, FRS2, and FRS3. These activated adaptors promoted the recruitment of the PI3K 85 kDa regulatory subunit α (p85), SH-PTP2 (SHP2, also known as tyrosine-protein phosphatase non-receptor type 11), and growth factor receptor-bound protein 2 (GRB2), resulting in subsequent activation of the PI3K and the MAPK pathways78–80. Additionally, the direct binding of IRS1 and IRS2 to the TPR–TRKA oncoprotein was shown to activate serum responsive elements associated with, and thus transcription of, the Fos gene78–80.
In 1998, the ETV6–NTRK3 gene fusion was discovered in congenital fibrosarcoma tumours by Sorensen and colleagues75. This chromosomal rearrangement results in a hybrid gene encoding the helix-loop-helix (HLH) protein dimerization domain of the transcription factor ETV6 fused with the kinase domain of TRKC. As mentioned previously, this chimeric protein lacks the SHC binding site of TRKC and, therefore, cannot bind to this adaptor protein. Nonetheless, ETV6–NTRK3 expression leads to the constitutive activation of the same major signalling cascades activated by the full-length TRKC: the MAPK and PI3K pathways81. Functional studies have elucidated that, among a series of TRK signalling adaptors investigated including p85, GRB2, and PLCɣ, only the latter associates with ETV6–TRKC (REF.44). Moreover, an ETV6–TRKC kinase-active mutant that is unable to bind PLCɣ did not have defective transformation capabilities when transduced in NIH-3T3 cells, suggesting that PLCɣ does not play a crucial part in the oncogenic activity of ETV6–NTRK3 (REF.44). Further work by the same team has revealed that ETV6–TRKC binds to the IGF1R–IRS1 complex at the plasma membrane; this interaction promotes the recruitment of Src, with consequent activation of the PI3K–AKT signalling cascade and upregulation of MAPK activity and cyclin D1 expression82. Similarly, results of a more recent study by our group83 demonstrate that the product of the ETV6–NTRK3 oncogene that is detected in almost all patients with mammary analogue secretory carcinoma (MASC) mainly signals through the MAPK, signal transducer and activator of transcription 3 (STAT3), and PLCɣ pathways when expressed in BaF3 cells.
These findings indicate that TRK fusions can, in part, signal via the same pathways activated by full-length TRK proteins; however, tissue histology might be a critical determinant of the principal pathway activated. In studies conducted in NTRK1 fusion-positive primary colorectal and lung cancer cell lines, activation of PI3K and STAT3 signalling was detected, although signalling through the SHC–RAS–MAPK pathway was predominant in these cell lines, as indicated by a greater degree of dephosphorylation of MAPKs versus other canonical targets of TRK proteins following TRK inhibition84. The same group also elegantly demonstrated that activation of EGFR signalling provides an adaptive survival mechanism in NTRK1 fusion-positive lung and colorectal cancer cell lines upon TRK inhibition, suggesting potential crosstalk between TRKA fusion proteins and receptor tyrosine kinases in these malignancies85. The patient-derived acute promyelocytic leukaemia (APML) cell line AP-1060 has been shown to harbour the ETV6–NTRK3 fusion and to be sensitive to TRK inhibition86; this is the only haematological cancer model driven by an endogenous NTRK translocation reported in the literature to date. Thus, only limited data on the potentially histology-dependent signalling downstream of TRK fusion proteins are currently available from primary cancer cell lines. Importantly, however, genetically engineered mouse models of NTRK fusions-positive malignancies have been generated in the past 2 years; specifically, a conditional knock-in model of acute lymphoblastic leukaemia (ALL) carrying the ETV6–NTRK3 fusion and a glioma model expressing the BCAN–NTRK1 fusion87,88. In both of these models, the presence of the fusion gene triggers the formation of highly aggressive tumours that could be effectively controlled using TRK inhibitors.
Finally, the fusion partner-mediated subcellular localization of different TRK fusions can also influence signalling. For example, the N-terminal domain of the TRK-fused gene (TFG) has been shown to localize the TFG–TRKA fusion to endoplasmic reticulum exit sites where constitutive TRKA kinase activity promotes the hyperactivation of ERK1 and ERK2 (REF.89). The use of primary cell lines and patient-derived xenograft (PDX) models could provide the possibility to study histology-specific and/or fusion-specific TRK-mediated signalling.
NTRK fusion frequency and diagnosis
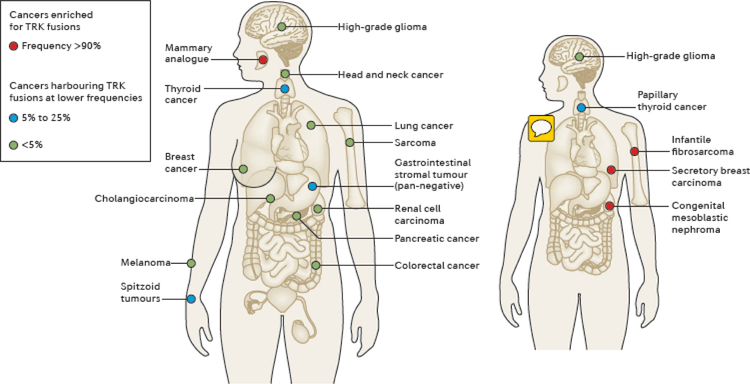
Initially identified in colorectal and papillary thyroid carcinomas (FIG. 1), NTRK fusions have since been found in multiple tumour types from both adult and paediatric patients10,72,90. These cancers can be grouped into two general categories according to the frequency at which these fusions are detected. First, rare cancer types highly enriched for NTRK fusions (FIG. 4). For example, the ETV6–NTRK3 fusion is considered practically pathognomonic in secretory breast carcinoma, MASC, congenital mesoblastic nephroma (cellular or mixed subtypes), and infantile fibrosarcomas, with a prevalence of >90% in select series of patients83,91–94. Second, other cancer types in which NTRK fusions are found at much lower frequencies (5–25% or <5%), including rare subsets of some common tumours (such as breast, lung, and colorectal cancers, and melanoma). Papillary thyroid cancers, Spitzoid neoplasms, gastrointestinal stromal tumours (GIST) lacking canonical KIT, PDGFRA, or RAS alterations, and certain paediatric gliomas are among the tumours types that have been shown to harbour NTRK fusions with frequencies of 5–25% (FIG. 4)90. NTRK fusions can be detected in <5% (predominantly <1%) of lung or pancreatic adenocarcinomas, head and neck squamous cell, bile duct, breast, colorectal, and renal cell carcinomas, melanomas, primary brain tumours of adulthood (such as astrocytomas or glioblastomas), and non-GIST soft-tissue sarcomas (FIG. 4)10,90. Interestingly, some haematological malignancies, such as acute lymphoblastic and acute myeloid leukaemias (ALL and AML, respectively), have also been shown to harbour NTRK fusions at low frequencies95 (FIG. 4). Importantly, a partial response to the TRK inhibitor larotrectinib has been observed in a patient with AML harbouring an ETV6–NTRK2 fusion, thus suggesting that TRK inhibition is a valid alternative to be considered in the treatment of NTRK-rearranged haematological tumours95.
Fig. 4 |. Distribution and frequency of NTRK fusions in adult and paediatric tumours.
NTRK fusions are identified across multiple paediatric and adult cancer histologies. The frequency of these fusions varies from <1% in cancer types including lung, colorectal, pancreatic, breast cancers, melanoma, and other solid or haematological cancers (green circles), up to 25% in tumours including thyroid, spitzoid, and gastrointestinal stromal tumours (blue circles), to >90% in rare tumours types, specifically secretory breast carcinoma, mammary analogue secretory carcinoma (MASC), congenital infantile fibrosarcoma, and cellular or mixed congenital mesoblastic nephroma (red circles) for which the NTRK fusions are considered practically pathognomonic.
NTRK fusions can be diagnosed using a variety of methods. Historically, the clinical detection of other recurrent gene rearrangements, such as those involving ALK and ROS1, has relied largely on fluorescence in situ hybridization (FISH) or reverse-transcriptase PCR (RT-PCR), although with a contemporary shift towards the use of comprehensive DNA-based next-generation sequencing (NGS) in select centres. By contrast, the clinical detection of NTRK fusions has predominantly been based on NGS10,11. This strategy has generally proved to be effective. Care must be exercised, however, in selecting NGS platforms that enable reliable detection of NTRK fusions because not all assays are optimized to identify these drivers: even the most advanced DNA-based NGS platforms might not enable the identification of all NTRK fusions, especially those involving NTRK2 and NTRK3, the detection of which is complicated by the presence of large intronic regions. Targeted RNA sequencing can serve as a complementary method to DNA-based NGS assays. Anchored multiplex PCR, a unidirectional targeted RNA sequencing approach that allows for the detection of both known and unknown upstream gene partners of varying lengths, is one example. In one patient series, such targeted RNA sequencing enabled the identification of a variety of gene fusions, including those involving NTRK in lung adenocarcinomas for which no known driver was previously identified on prior DNA-based NGS96.
FISH and RT-PCR have been successfully used in the clinic to detect NTRK fusions93 and are reasonable alternatives to NGS, especially for tumour histologies with a high prevalence of NTRK fusions involving recurrent partners (MASC, infantile fibrosarcomas, secretory breast carcinoma, and cellular or mixed congenital mesoblastic nephromas). FISH and RT-PCR can be performed relatively quickly and at lower costs; however, these tests are largely limited to the detection of a single driver alteration, whereas NGS offers the ability to detect multiple potential drivers in addition to NTRK fusions — a clear advantage in the analysis of many tumour types that can harbour any of a number of oncogenes (lung adenocarcinomas are a classic example). Beyond tumour-based testing of DNA or RNA, NTRK fusions can potentially be detected through plasma-based cell-free DNA (cfDNA) testing97,98.
Immunohistochemistry (IHC) is another complementary method that can enable the detection of TRK overexpression as a surrogate for the potential presence of an NTRK fusion. In a series of solid tumours harbouring NTRK fusions, IHC using a pan-TRK antibody revealed positivity for TRK expression in 20 of 21 cases76. Similar results were obtained in a panel of 79 paediatric patients with mesenchymal tumours, whereby positive staining with the pan-TRK antibody identified TRK fusions with a sensitivity of 97% and a specificity of 98%99. Interestingly, the IHC staining patterns (membranous, cytoplasmic, perinuclear, or nuclear) varied99, presumably secondary to differences in subcellular localization mediated by the type of upstream gene partner. In clinical settings with limited resources and/or access to NGS, considering IHC as a screening tool for NTRK fusions is not unreasonable, although preferably with confirmatory nucleic acid-based testing performed for those who test positive, when possible.
TRK inhibitor therapy
Several TKIs with varying degrees of activity against TRKA, TRKB, and/or TRKC are available (FIG. 5), which can broadly be grouped into multi-kinase inhibitors with activity against a range of targets including TRK or more-selective TRK inhibitors. The multi-kinase inhibitor group includes entrectinib, crizotinib, cabozantinib, lestaurtinib, altiratinib, foretinib, ponatinib, nintedanib, merestinib, MGCD516, PLX7486, DS-6051b, and TSR-011. Larotrectinib is currently the most specific TRK inhibitor being testing in patients with cancer10. Of these agents, larotrectinib and entrectinib are furthest along in clinical development. Entrectinib is an orally available pan-TRK inhibitor with additional activity against ROS1 and ALK100. Larotrectinib is a potent and selective inhibitor of all three TRK proteins101. On the basis of in vitro kinase assays, both compounds inhibit TRKA, TRKB, and TRKC with IC50 values in the low nanomolar range100,101 (FIG. 5). Correspondingly, both entrectinib and larotrectinib are highly effective in inhibiting the growth of BaF3 cells transduced with different NTRK fusions and of primary cancer cell lines harbouring NTRK rearrangements in vitro and in mice83,98,101. Treatment of TRK fusion-containing tumour models with entrectinib or larotrectinib results in the inhibition of the MAPK, PI3K–AKT, PKC, and STAT3 pathways83,98,101.
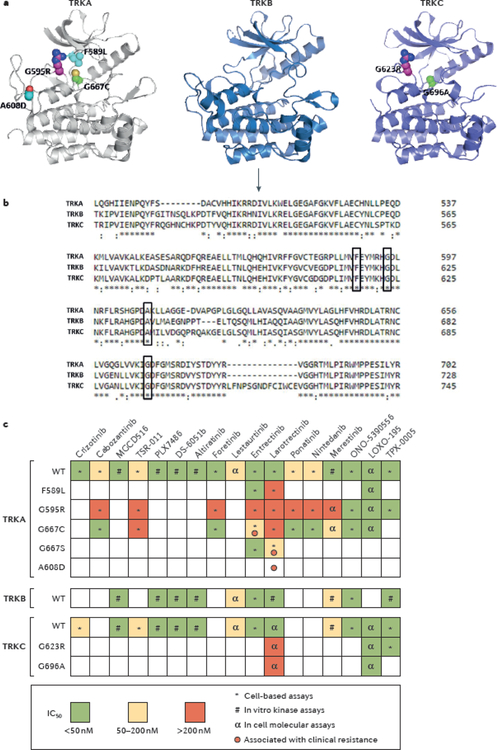
Fig. 5 |. Mechanisms of acquired resistance to TRK inhibitors and profiles of TRK inhibitor activity.
a | Structural modelling of the tyrosine kinase domains of TRKA, TRKB, and TRKC showing amino acid substitutions resulting from somatic mutations in NTRK1, NTRK2, and NTRK3 fusions, respectively, that have been associated with acquired resistance to first-generation TRK inhibitors in patients: the TRKA G595R and TRKC G623R solvent-front substitutions; the TRKA F589L gatekeeper mutation; G667C and G696A substitutions within the xDFG-motif of the kinase activation loops of TRKA and TRKC, respectively; and the TRKA A608D mutation. No substitutions involving TRKB have yet been identified in patient samples. Paralogous mutations in TRKA and TRKC are shown in the same colour. b | Homology alignment of the kinase domains of TRKA, TRKB, and TRKC indicates that these resistant mutations affect highly conserved amino acid residues across the TRK proteins (black boxes). The arrow indicates the beginning of the kinase domains. c | A heat map of the half maximal inhibitory concentration (IC50) values of selected multi-kinase or TRK-specific inhibitors is shown. These drugs have different levels of activity against wild-type (WT) or mutant TRK proteins (for example, those with solvent-front, gatekeeper, or xDFG substitutions). The symbols in each box indicate which type of assay has been used to determine the IC50. Red circles represent reports of clinical resistance that have been observed for each mutant. Of note, some mutants that have been described to be sensitive to specific inhibitors in preclinical analyses were instead found to confer resistance to the same drugs in patients. xDFG, X-aspartate-phenylalanine-glycine.
Larotrectinib in clinical trials
The activity of the TRK-selective inhibitor larotrectinib in patients with tumours harbouring NTRK fusions has been explored in three clinical trials: a phase I trial in adults (NCT02122913), a phase I/II trial in paediatric patients (SCOUT, NCT02637687), and a phase II trial involving adults and adolescents (NAVIGATE, NCT02576431). Data on the first 55 consecutively evaluable patients treated with larotrectinib on these trials have been analysed10. The ORR according to independent radiology review, the primary end point of this combined analysis, was 75% (95% CI 61–85%) and the investigator assessed ORR was 80% (95% CI 67–90%); the complete response rates were 13% and 16%, respectively10 (TABLE 1). The median time to response was 1.8 months, consistent with the time at which the first protocol-mandated follow-up imaging assessment was performed10. Patients with a total of 17 unique cancer types harbouring NTRK fusions, including MASC, infantile fibrosarcoma, melanoma, GIST, and thyroid, colorectal, or lung adenocarcinoma, were included in this analysis. Importantly, responses were observed in a histology-agnostic fashion, regardless of fusion type (NTRK1, NTRK2, or NTRK3) or upstream partner, and independent of age (patients aged from 4 months to 76 years were treated)10. While the median progression-free survival duration had not been reached at the time of the last data cut-off, 55% of the patients remained progression-free after 1 year of treatment, and 71% of the responses were ongoing10.
Table 1 |.
Clinical activity and safety profile of first-generation TRK inhibitors
| Drug | NTRK fusion-positive cancers treated (n; % of total) | ORR (95% CI; n) |
PR rate (n) | CR rate (n) | Most common drug-related AEs of any gradea |
|---|---|---|---|---|---|
| Larotrectinib10 | MASC or other salivary gland cancer (12; 22%) | 75% (61–85%; 41/55) | 62% (34/55) | 13% (7/55) | Increased serum AST and/or ALT levels (38%) |
| Non-GIST STS (11; 20%) | Dizziness (25%) | ||||
| Infantile fibrosarcoma (7; 13%) | Fatigue (16%) | ||||
| Thyroid cancer (5; 9%) | Nausea (16%) | ||||
| CRC (4; 7%) | Constipation (16%) | ||||
| NSCLC (4; 7%) | Vomiting (11%) | ||||
| Melanoma (4; 7%) | Weight gain (11%) | ||||
| GIST (3; 5%) | Decreased neutrophil counts (9%) | ||||
| Cholangiocarcinoma (2; 4%) | |||||
| Appendix cancer (1; 2%) | |||||
| Breast cancer (1; 2%) | |||||
| PDAC (1; 2%) | |||||
| Entrectinib11 | CRC (1; 25%) | 100% (44–100%; 3/3b) | 100% (3/3b) | 0% (0/3) | Fatigue (46%) |
| Glioneuronal tumour (1; 25%) | Dysgeusia (42%) | ||||
| MASC (1; 25%) | Paresthesia (29%) | ||||
| NSCLC (1; 25%) | Nausea (28%) | ||||
| Myalgia (23%) | |||||
| Diarrhoea (19%) | |||||
| Vomiting (17%) | |||||
| Arthralgia (16%) | |||||
| Dizziness (16%) | |||||
| Constipation (12%) | |||||
| Weight gain (10%) |
Data on the activity of larotrectinib in NTRK fusion-positive tumours were derived with regulatory input from three trials: a phase I trial in adult patients (NCT02122913), a phase I/II trial in paediatric patients (SCOUT; NCT02637687), and an phase II basket trial in adult or adolescent patients (NAVIGATE; NCT02576431). Data on the activity of entrectinib in NTRK fusion-positive tumours were derived from two phase I trials in adult patients (ALKA-372-001 and STARTRK-1; EudraCT 2012-000148-88 and NCT02097810, respectively). AEs, adverse events; ALT, alanine transaminase; AST, aspartate transaminase; CI, confidence interval; CR, complete response; CRC, colorectal carcinoma; GIST, gastrointestinal stromal tumour; MASC, mammary analogue secretory carcinoma; NSCLC, non-small-cell lung cancer; ORR, objective response rate; PDAC, pancreatic ductal adenocarcinoma; PR, partial response; STS, soft-tissue sarcoma.
The frequencies indicated for entrectinib refer to those observed in all 119 adult patients enrolled in the ALKA-372–001 and STARTRK-1 phase I trials, only 4 of whom had tumours harbouring NTRK fusions).
The patient with glioneuronal tumour with stable disease by RECIST was excluded from the analysis but had unconfirmed disease regression according to an exploratory volumetric assessment and a clinical response (improvement in symptoms).
Of note, while most patients included in these three trials received larotrectinib for metastatic disease, select patients had locally advanced disease. For example, two children with locally advanced infantile fibrosarcoma of the knee had substantial tumour shrinkage that enabled limb-sparing surgery with curative intent instead of a possible amputation10. These cases highlight the potential utility of TRK inhibition as neoadjuvant therapy for patients with non-metastatic cancers harbouring NTRK fusions.
Entrectinib in clinical trials
Entrectinib has been tested in four clinical trials: a phase I study in adults with NTRK, ROS1, or ALK rearrangements (ALKA 372–001, EudraCT 2012-000148-88); a phase I trial in adults with various NTRK, ROS1, or ALK aberrations (STARTRK-1, NCT02097810); a phase I/Ib study in children or young adults (aged 2–22 years) with or without NTRK, ROS1, or ALK fusions (STARTRK-NG, NCT02650401); and a phase II basket trial in adults with NTRK, ROS1, or ALK fusions (STARTRK-2, NCT02568267). Data on the clinical activity of entrectinib observed in the ALKA 372–001 and STARTRK-1 trials have been published11. In these two studies, four patients with tumours harbouring NTRK fusions were treated with entrectinib, one with colorectal carcinoma, one with MASC, one with lung adenocarcinoma, and one with glioneural tumour; the first three patients had confirmed partial responses, and the patient with glioneuronal tumour had disease regression by an exploratory volumetric assessment11 (TABLE 1). Overall, the ORR in tyrosine kinase inhibitor treatment-naive patients with cancers harbouring an NTRK, ROS1, or ALK fusion were 100% (3 of 3 patients), 86% (12 of 14 patients), and 57% (4 of 7 patients), respectively. In 53 additional patients with tumours harbouring point mutations, amplifications, copy-number variants, or insertions or deletions involving NTRK, ROS1, or ALK, no objective responses to entrectinib treatment were observed, except in patients with ALK-mutant neuroblastoma, highlighting the utility of recurrent gene rearrangements as better clinical predictors of potential benefit.
Other agents
The following multi-target TKIs with varying degrees of inhibitory activity against TRK have been approved for indications outside of the treatment of patients with NTRK fusions: crizotinib (ALK-rearranged and ROS1-rearranged NSCLC)102,103, cabozantinib (renal cell carcinoma and medullary thyroid carcinoma)104, ponatinib (chronic myelogenous leukaemia)105, and nintedanib (idiopathic pulmonary fibrosis)106. Crizotinib was originally developed as a MET inhibitor107 and thereafter also identified as an ALK107–109, ROS1110, and TRK109 inhibitor, but has a substantially lower affinity for TRK than for MET, ALK, and ROS1. Cabozantinib targets several receptor tyrosine kinases including MET, RET, AXL, TRKA, and TRKB111. Ponatinib, initially developed as a BCR–ABL1 inhibitor112 (with activity against the majority of BCR–ABL1 resistance mutations), was also found to suppress the growth of NTRK fusion-positive tumours in preclinical models113. Nintedanib is an anti-angiogenic drug (VEGFR TKI) that also inhibits PDGFR, FGFR, and TRK kinases113,114.
Other TKIs are in various stages of early phase clinical testing. As discussed, lestaurtinib, targets of which include TRK, JAK2, and FLT3, has been explored in patients with neuroblastoma, a cancer known to have overexpression of TRK proteins71. PLX7486 is a dual TRK and CSF-1R inhibitor115. MGCD516 is a multi-kinase inhibitor with activity against TRK proteins116. TSR-011 inhibits both ALK and TRK with nanomolar IC50 potency117. DS-6051b has shown potent antitumour activity in ROS1-driven or TRK-dependent preclinical models118. Altiratinib binds TRK proteins with low nanomolar affinity, in addition to MET, TIE2 and VEGFR2, and has preclinical activity against a number of TRK fusion-positive cancers119,120. Merestinib has been extensively used as an experimental MET inhibitor121,122, but suppresses other kinases including MST1R, FLT3, AXL, MERTK, ROS1, DDR1, DDR2, and TRK at nanomolar concentrations123,124.
The clinical activity of these other multi-target TRK inhibitors in patients with NTRK fusion-positive cancers is not well characterized. Whether the clinical development of these drugs will be pursued further in this disease setting is uncertain, given the already established data on larotrectinib and entrectinib, and the fact that both larotrectinib and entrectinib have received breakthrough designation status by the US FDA for the treatment of cancers harbouring NTRK fusions.
Acquired resistance to TRK inhibition
Resistance mechanisms
In general, acquired resistance to TKI therapy in patients with fusion-positive cancers can be mediated by on-target and off-target mechanisms. To date, the acquisition of on-target mutations in the NTRK kinase domain of the oncogenic fusion is the only mechanism of secondary resistance identified in patients following TRK inhibition73, although one can reasonably assume that off-target or bypass resistance mechanisms also exist. The resistance mutations can result in amino acid substitutions involving the solvent front, activation loop xDFG motif, or so-called gatekeeper residue of TRKA or TRKC (FIG. 5a). No mutations in TRKB have been identified in clinic to date, probably owing to the limited number of patients with NTRK2 fusions analysed.
The first case of acquired resistance to TRK inhibition was reported in 2016 by Russo and colleagues98, and occurred in a patient with LMNA–NTRK1 rearranged colorectal cancer after 4 months of treatment with entrectinib. Plasma cfDNA was collected longitudinally during treatment in order to monitor the patient’s response, and sequencing of cfDNA at the time of relapse revealed the emergence of two mutations resulting in substitutions in the kinase domain of TRKA — G595R and G667C. The G595R substitution is located in the solvent front region of the TRK kinase domain (FIG. 5a) and is the paralogue of the ALK G1202R and ROS1 G2032R substitutions that are known to cause acquired resistance to TKIs in patients with ALK-rearranged and ROS1-rearranged lung cancers, respectively125,126. The G667C substitution maps to the ‘x’ position of the xDFG motif in the highly conserved activation segment of TRKA kinase domain (FIG. 5a–b) and is the paralogue of ALK G1269A, which is likewise associated with acquired resistance to crizotinib in patients with ALK-rearranged lung cancers127.
In the same year, acquired resistance to TRK inhibition was reported in a patient with MASC who initially responded but then progressed on entrectinib therapy83. Sequencing of DNA from the resistant tumour identified an acquired TRKC G623R substitution (paralogous to the TRKA G595R substitution), confirming that solvent front substitutions are recurrent mechanisms of acquired resistance to TRK inhibition (FIG. 5a). These findings were further supported by data from the larotrectinib clinical trials10. In the aforementioned analysis of these trials10, post-progression tumour or plasma cfDNA samples of seven patients who initially responded to larotrectinib and then progressed on therapy were found to carry mutations resulting in the G595R substitution in TRKA or the G623R substitution in TRKC. In addition, a gatekeeper F589L substitution and a novel A608D mutation in TRKA as well as novel substitutions involving the xDFG site of TRKA (G667S) and TRKC (G696A) were identified after patients progressed on therapy10 (FIG. 5a). Data from structural modelling analyses of these mutants (except the A608D mutant, for which no modelling analyses are available to date) suggest that the resultant amino acid substitutions prevent the binding of entrectinib and larotrectinib to the kinase owing to steric hindrance10,98; however, in vitro kinase assays have revealed that the G595R-mutant TRKA has increased affinity for ATP compared with that of the wild-type protein, suggesting that multiple factors might contribute to the resistant phenotype97. Interestingly, although the majority of these TRK mutants are resistant to many multi-kinase inhibitors with activity against TRK, data from in vitro cell models demonstrate that some TKIs (cabozantinib, ponatinib, foretinib, and nintedanib) retain strong inhibitory capacity against the G667C or G667S mutant113 (FIG. 5c). These findings suggest that a careful biochemical characterization of the different mutants is required to better understand their mechanisms of activation and oncogenicity as well as their sensitivity to distinct TRK inhibitors.
Next-generation TRK inhibitors
Next-generation TRK inhibitors that overcome acquired resistance to first-generation TKIs are fortunately already in development. In particular, LOXO-195 (REF.97), TPX-0005 (REF.128), and ONO-5390556 (REF.129) have demonstrated in vitro activity against many of the aforementioned TRK mutants in the low nanomolar concentration range (FIG. 5c). The development of LOXO-195 paralleled the early phases of the clinical characterization of larotrectinib. In fact, targeted mutagenesis experiments and modelling studies of predicted mechanisms of acquired resistance enabled the preclinical validation of LOXO-195 as a second-generation TRK inhibitor before acquired resistance to larotrectinib was observed in the clinic97. Importantly, proof-of-concept data on the utility of sequential TKI use in patients with NTRK fusion-positive cancers has been published97. One adult and one paediatric patient, both with advanced-stage NTRK-rearranged cancers, developed TRK solvent front mutation-mediated acquired resistance to larotrectinib. Subsequently, treatment with LOXO-195 was administered on compassionate use, single-patient, first-in-human protocols that involved rapid dose titration guided by pharmacokinetic assessments, resulting in confirmed objective responses in both patients97. The safety and efficacy of LOXO-195 is currently being explored in a phase I/II trial involving patients aged ≥1 month with NTRK-rearranged cancers after prior treatment with a different TRK inhibitor (NCT03215511).
TPX-0005 is a next-generation TRK, ROS1, and ALK inhibitor that likewise has activity against TRK variants with kinase domain substitutions128 (FIG. 5c). In an ongoing phase I/II study of TPX-0005 (TRIDENT-1; NCT03093116), a patient with MASC that had acquired resistance to entrectinib mediated by solvent front mutation had a response to TPX-0005 therapy130. No data on the clinical development of ONO-5390556 are available at present.
Consequences of TRK inhibition
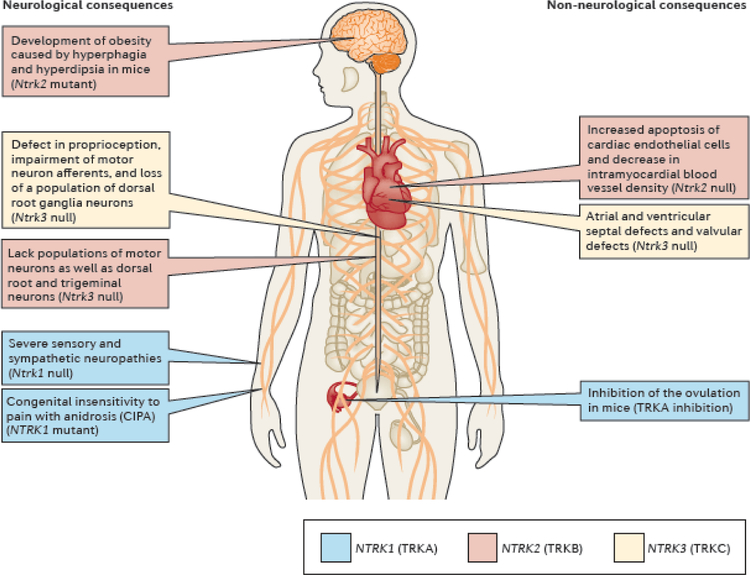
Consideration of the consequences of decreases or loss of TRK function in preclinical studies or in human disease is important to understanding the potential drug-related adverse effects of TRK inhibition in children and adults. Moreover, one must be cognizant of the fact that, while NTRK1 expression seems to be limited to visceral sensory ganglia of neural crest origin, NTRK2 and NTRK3 are widely expressed in both the central and peripheral nervous systems26,45. Accordingly, Ntrk1-null mice lack most sympathetic neurons, do not display nociceptive and temperature sensations, and die within a month after birth owing to severe sensory and sympathetic neuropathies131. By contrast, Ntrk2-knockout mice lack particular populations of motor neurons as well as dorsal root and trigeminal ganglia neurons; these mice die perinatally because they fail to eat132. Ntrk3-null mice also have deficits in the number and function of motor neurons and lack a population of dorsal root ganglia neurons, resulting in abnormal movements and posture22,133 (FIG. 6). Interestingly, defects in tissues other than the nervous system are also observed in Ntrk-null animals. For example, homozygous disruption of the Ntrk2 gene results in increased apoptosis of endothelial cells and decreased number of intramyocardial blood vessels, whereas targeted deletion of all TrkC isoforms in mice leads to severe cardiac deficiencies, including atrial, ventricular, and valvular defects that lead to animal death in the early postnatal period134,135 (FIG. 6). Consistent with this observation, TRK-mediated signalling is not exclusively confined to the nervous system as it has also been described to have roles in non-neuronal tissues, such as the vasculature, ovaries, and immune system136–139 (FIG. 6). Notably, the observation of transient but extensive upregulation of Ntrk1 expression in periovulatory follicules during the first preovulatory surge of gonadotropins in rats, with TRK inhibition at this time resulting in prevention of ovulation137, warrants caution regarding the use of TRK inhibitors in girls during puberty. Moreover, the finding that BDNF–TRKB axis has a role in mouse oocyte development into pre-implantation embryos, with TRK inhibition suppressing progression of zygotes into blastocysts139, hints at potential implications of TRK inhibition on pregnancy.
Fig. 6 |. Consequences of loss, decreased activity, or inhibition of TRK.
Genetic or pharmacological disruption of TRK signalling can cause a variety of neurological and non-neurological changes. These phenotypic effects deriving from impairments in TRK protein function were described in preclinical animal models or in patients with genetic conditions resulting in a decrease or loss of TRK activity. Consequences of loss of Ntrk1/NTRK1 (TRKA) include sensory and sympathetic neuropathies, insensitivity to pain, anhidrosis, and impairments in ovulation (blue boxes). Ntrk2/NTRK2 (TRKB) deficiency is associated with hyperphagia, the loss of specific populations of neurons, cardiac defects, and memory loss (red boxes). Similarly, loss of Ntrk3/NTRK3 (TRKC) results in defects in proprioception, a lack of certain populations of neurons, and cardiac dysfunction (yellow boxes). Consistent with these findings, paresthesias, weight gain, cognitive disturbance, and dizziness have been reported in patients treated with TRK inhibitors10,11.
Various diseases can be caused either by loss-of-function mutations in NTRK genes or impairments in the synthesis of neurotrophins. For example, loss-of-function NTRK1 mutations have been identified in patients with congenital insensitivity to pain with anhidrosis (CIPA)140,141 (FIG. 6), a hereditary sensory and autonomic neuropathy. Patients with this rare inherited disorder have an impaired ability to sense differences in temperature or feel pain. Impairments in the BDNF–TRKB axis have been shown to cause hyperphagia and consequent obesity, in both mice and humans, in addition to defects in memory, learning, and nociception in humans142,143 (FIG. 6). Moreover, mice with impairments in the synthesis of BDNF develop severe ataxia owing to decreased levels of BDNF in the cerebellum, which is associated with reduced levels of γ-aminobutyric acid (GABA) and fewer, smaller inhibitory synapses in cerebellar neurons144.
When considering the adverse events of larotrectinib and entrectinib, it should be noted that these drugs have a favourable overall safety profile compared with other TKIs. Specifically, off-target adverse effects typically observed at high frequency, and often at high severity (grade ≥3) with the use of other TKIs (such as fatigue, nausea, diarrhoea, constipation, vomiting, and increased serum transaminase levels)102,104 have been seen at relatively lower frequencies and were mostly grade 1 or 2 in severity in the trials of entrectinib and larotrectinib10,11 (TABLE 1). Accordingly, dose reductions were infrequent and occurred in only 15% of patients treated with either TRK inhibitor10,11.
Occasional on-target adverse effects have been observed with the use of TRK inhibitors in the clinic, consistent with the consequences of gene knockout and loss-of-function mutations detailed above. These included paresthesias, dizziness (potentially associated with decreased proprioception), and weight gain10,11 (TABLE 1). Moreover, a dose-limiting toxicity of grade 3 cognitive disturbance was noted with entrectinib during the dose-escalation phase of the STARTRK-1 phase I trial11. These adverse events were reversible in patients for whom dose modification was required11. Finally, in select patients, pain flares have been noted with treatment discontinuation, consistent with the fact that TRK pathway inhibitors, such as monoclonal antibodies against NGF or TRKA, are hypothesized to decrease pain and were initially developed for this purpose145,146.
Fortunately, on the basis of published reports, the frequency of moderate-to-severe on-target adverse events mediated by TRK inhibition is low (TABLE 1) but will best be estimated after longer follow-up durations, especially given the durability of response achieved in patients with NTRK fusion-positive cancers. Treatment guidelines will need to be developed to manage these adverse effects. Beyond dose modification, potential supportive medications include gabapentin and pregabalin for paraesthesias and peripheral neuropathy, weight-modifying agents (for example, liraglutide) for substantial weight gain not managed by diet and exercise, and mecilizine or midodrine for positional dizziness. Notably, however, the true utility of these interventions in the management of the toxicities of TRK inhibition has not yet been established and requires further study. Non-narcotic and/or narcotic pain medication could be used in patients with pain flare associated with drug discontinuation.
Conclusions
NTRK fusions are drivers of a wide variety of adult and paediatric cancers. These oncogenes are either highly enriched in select tumour types or infrequently found in subsets of other cancers, including common tumours. Comprehensive nucleic acid-based profiling and complementary IHC assays can be used to detect these fusions in the clinic. The first-generation TRK inhibitors larotrectinib and entrectinib have demonstrated histology-agnostic and age-independent activity in adult and paediatric patients with diverse cancers harbouring NTRK fusions. Acquired resistance to TRK inhibitor therapy remains an ongoing challenge, but on-target resistance mediated by the acquisition of NTRK kinase domain mutations can be abrogated by the use of second-generation TRK inhibitors. The toxicological consequences of TRK inhibition include on-target neurological adverse events, although larotrectinib and entrectinib have a favourable overall safety profile compared with many other TKIs and are, therefore, amenable to chronic dosing.
Key points.
NTRK fusions, encoding TRK fusion proteins, are oncogenic drivers of a wide variety of adult and paediatric tumours, supporting a basket trial approach to drug development.
These alterations are found at high frequencies (up to or greater than 90%) in rare cancer types (secretory breast carcinoma, mammary analogue secretory carcinoma, cellular or mixed congenital mesoblastic nephroma, and infantile fibrosarcoma) and at lower frequencies (commonly <1%) in a range of other tumour types.
NTRK fusions are clinically actionable: first-generation TRK tyrosine kinase inhibitors (larotrectinib or entrectinib) result in histology-agnostic responses in both adult and paediatric patients with NTRK fusion-positive cancers.
Resistance to TRK inhibition can be mediated by the acquisition of NTRK kinase domain mutations, including solvent-front and gatekeeper mutations; second-generation TRK inhibitors have been developed to overcome these mechanisms of resistance.
First-generation TRK inhibitors are generally well-tolerated and, with consideration of the biological roles of TRK receptors in normal development and adulthood, the occasional on-target adverse effects are predictable.
Acknowledgements
We would like to thank Andrew Drilon, Sandra Misale, and Chandra Verma for assisting with the creation and design of the figures in this article. The work of the authors is supported by Cycle for Survival and the National Institutes of Health awards P30 CA008748 and R01CA226864. E.C. is a recipient of a MSK Society Scholar Prize. Given the space limitations of this review, the authors apologize for their inability to cite everyone who has contributed to this field of inquiry.
Footnotes
Competing interests
A.D. has received honoraria (as an advisory board member) from Bayer, Ignyta, Loxo Oncology, Pfizer, Roche/Genentech, and TP Therapeutics, and research funding from Loxo Oncology. M.S. has received research funding from Daiichi Sankyo and Puma Biotechnology.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Clinical Oncology thanks T. Laetsch, and two other anonymous reviewers, for their contribution to the peer review of this work.
References
- 1.Robert C et al. Improved Overall Survival in Melanoma with Combined Dabrafenib and Trametinib. The New England journal of medicine 372, 30–39, doi: 10.1056/NEJMoa1412690 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Geyer CE et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. The New England journal of medicine 355, 2733–2743, doi: 10.1056/NEJMoa064320 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Planchard D et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. The Lancet. Oncology 17, 984–993, doi: 10.1016/S1470-2045(16)30146-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. The New England journal of medicine 371, 1963–1971, doi: 10.1056/NEJMoa1406766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine 344, 783–792, doi: 10.1056/NEJM200103153441101 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Solomon BJ et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. The New England journal of medicine 371, 2167–2177, doi: 10.1056/NEJMoa1408440 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Hyman DM et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. The New England journal of medicine 373, 726–736, doi: 10.1056/NEJMoa1502309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyman DM et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 35, 2251–2259, doi: 10.1200/JCO.2017.73.0143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty KT et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine 363, 809–819, doi: 10.1056/NEJMoa1002011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drilon A et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. The New England journal of medicine 378, 731–739, doi: 10.1056/NEJMoa1714448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drilon A et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372–001 and STARTRK-1). Cancer discovery 7, 400–409, doi: 10.1158/2159-8290.CD-16-1237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulciani S et al. Oncogenes in solid human tumours. Nature 300, 539–542 (1982). [DOI] [PubMed] [Google Scholar]
- 13.Martin-Zanca D, Hughes SH & Barbacid M A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature 319, 743–748, doi: 10.1038/319743a0 (1986). [DOI] [PubMed] [Google Scholar]
- 14.Martin-Zanca D, Oskam R, Mitra G, Copeland T & Barbacid M Molecular and biochemical characterization of the human trk proto-oncogene. Molecular and cellular biology 9, 24–33 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein R, Jing SQ, Nanduri V, O’Rourke E & Barbacid M The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 65, 189–197 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Kaplan DR, Hempstead BL, Martin-Zanca D, Chao MV & Parada LF The trk proto-oncogene product: a signal transducing receptor for nerve growth factor. Science 252, 554–558 (1991). [DOI] [PubMed] [Google Scholar]
- 17.Klein R, Parada LF, Coulier F & Barbacid M trkB, a novel tyrosine protein kinase receptor expressed during mouse neural development. The EMBO journal 8, 3701–3709 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamballe F, Klein R & Barbacid M trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell 66, 967–979 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Davies AM et al. Neurotrophin-4/5 is a mammalian-specific survival factor for distinct populations of sensory neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 13, 4961–4967 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soppet D et al. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell 65, 895–903 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Deinhardt K & Chao MV Trk receptors. Handbook of experimental pharmacology 220, 103–119, doi: 10.1007/978-3-642-45106-5_5 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Snider WD Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell 77, 627–638 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Lemmon MA & Schlessinger J Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134, doi: 10.1016/j.cell.2010.06.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arevalo JC et al. A novel mutation within the extracellular domain of TrkA causes constitutive receptor activation. Oncogene 20, 1229–1234, doi: 10.1038/sj.onc.1204215 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Zaccaro MC, Ivanisevic L, Perez P, Meakin SO & Saragovi HU p75 Co-receptors regulate ligand-dependent and ligand-independent Trk receptor activation, in part by altering Trk docking subdomains. The Journal of biological chemistry 276, 31023–31029, doi: 10.1074/jbc.M104630200 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Barbacid M The Trk family of neurotrophin receptors. Journal of neurobiology 25, 1386–1403, doi: 10.1002/neu.480251107 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Huang EJ & Reichardt LF Trk receptors: roles in neuronal signal transduction. Annual review of biochemistry 72, 609–642, doi: 10.1146/annurev.biochem.72.121801.161629 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Clary DO & Reichardt LF An alternatively spliced form of the nerve growth factor receptor TrkA confers an enhanced response to neurotrophin 3. Proceedings of the National Academy of Sciences of the United States of America 91, 11133–11137 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodeur GM et al. Trk receptor expression and inhibition in neuroblastomas. Clinical cancer research : an official journal of the American Association for Cancer Research 15, 3244–3250, doi: 10.1158/1078-0432.CCR-08-1815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacconelli A et al. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer cell 6, 347–360, doi: 10.1016/j.ccr.2004.09.011 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Tacconelli A, Farina AR, Cappabianca L, Gulino A & Mackay AR Alternative TrkAIII splicing: a potential regulated tumor-promoting switch and therapeutic target in neuroblastoma. Future oncology 1, 689–698, doi: 10.2217/14796694.1.5.689 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Strohmaier C, Carter BD, Urfer R, Barde YA & Dechant G A splice variant of the neurotrophin receptor trkB with increased specificity for brain-derived neurotrophic factor. The EMBO journal 15, 3332–3337 (1996). [PMC free article] [PubMed] [Google Scholar]
- 33.Boeshore KL, Luckey CN, Zigmond RE & Large TH TrkB isoforms with distinct neurotrophin specificities are expressed in predominantly nonoverlapping populations of avian dorsal root ganglion neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 4739–4747 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luberg K, Wong J, Weickert CS & Timmusk T Human TrkB gene: novel alternative transcripts, protein isoforms and expression pattern in the prefrontal cerebral cortex during postnatal development. Journal of neurochemistry 113, 952–964, doi: 10.1111/j.1471-4159.2010.06662.x (2010). [DOI] [PubMed] [Google Scholar]
- 35.Valenzuela DM et al. Alternative forms of rat TrkC with different functional capabilities. Neuron 10, 963–974 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Stoilov P, Castren E & Stamm S Analysis of the human TrkB gene genomic organization reveals novel TrkB isoforms, unusual gene length, and splicing mechanism. Biochemical and biophysical research communications 290, 1054–1065, doi: 10.1006/bbrc.2001.6301 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Tsoulfas P, Stephens RM, Kaplan DR & Parada LF TrkC isoforms with inserts in the kinase domain show impaired signaling responses. The Journal of biological chemistry 271, 5691–5697 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Esteban PF et al. A kinase-deficient TrkC receptor isoform activates Arf6-Rac1 signaling through the scaffold protein tamalin. The Journal of cell biology 173, 291–299, doi: 10.1083/jcb.200512013 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohira K et al. A truncated tropomyosin-related kinase B receptor, T1, regulates glial cell morphology via Rho GDP dissociation inhibitor 1. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 1343–1353, doi: 10.1523/JNEUROSCI.4436-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham ME & Greene LA A function-structure model for NGF-activated TRK. The EMBO journal 17, 7282–7293, doi: 10.1093/emboj/17.24.7282 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichardt LF Neurotrophin-regulated signalling pathways. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 361, 1545–1564, doi: 10.1098/rstb.2006.1894 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minichiello L et al. Point mutation in trkB causes loss of NT4-dependent neurons without major effects on diverse BDNF responses. Neuron 21, 335–345 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Postigo A et al. Distinct requirements for TrkB and TrkC signaling in target innervation by sensory neurons. Genes & development 16, 633–645, doi: 10.1101/gad.217902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wai DH et al. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene 19, 906–915, doi: 10.1038/sj.onc.1203396 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Barbacid M, Lamballe F, Pulido D & Klein R The trk family of tyrosine protein kinase receptors. Biochimica et biophysica acta 1072, 115–127 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Levi-Montalcini R The nerve growth factor: thirty-five years later. Bioscience reports 7, 681–699 (1987). [DOI] [PubMed] [Google Scholar]
- 47.Cohen S, Levi-Montalcini R & Hamburger V A Nerve Growth-Stimulating Factor Isolated from Sarcom as 37 and 180. Proceedings of the National Academy of Sciences of the United States of America 40, 1014–1018 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frade JM & Barde YA Nerve growth factor: two receptors, multiple functions. BioEssays : news and reviews in molecular, cellular and developmental biology 20, 137–145, doi: (1998). [DOI] [PubMed] [Google Scholar]
- 49.Patel TD, Jackman A, Rice FL, Kucera J & Snider WD Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron 25, 345–357 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Teng HK et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 5455–5463, doi: 10.1523/JNEUROSCI.5123-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahadeo D, Kaplan L, Chao MV & Hempstead BL High affinity nerve growth factor binding displays a faster rate of association than p140trk binding. Implications for multi-subunit polypeptide receptors. The Journal of biological chemistry 269, 6884–6891 (1994). [PubMed] [Google Scholar]
- 52.Hempstead BL, Martin-Zanca D, Kaplan DR, Parada LF & Chao MV High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature 350, 678–683, doi: 10.1038/350678a0 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Hempstead BL et al. Overexpression of the trk tyrosine kinase rapidly accelerates nerve growth factor-induced differentiation. Neuron 9, 883–896 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Bibel M, Hoppe E & Barde YA Biochemical and functional interactions between the neurotrophin receptors trk and p75NTR. The EMBO journal 18, 616–622, doi: 10.1093/emboj/18.3.616 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makkerh JP et al. p75 neurotrophin receptor reduces ligand-induced Trk receptor ubiquitination and delays Trk receptor internalization and degradation. EMBO reports 6, 936–941, doi: 10.1038/sj.embor.7400503 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eggert A, Sieverts H, Ikegaki N & Brodeur GM p75 mediated apoptosis in neuroblastoma cells is inhibited by expression of TrkA. Medical and pediatric oncology 35, 573–576 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Rabizadeh S et al. Induction of apoptosis by the low-affinity NGF receptor. Science 261, 345–348 (1993). [DOI] [PubMed] [Google Scholar]
- 58.Geiger TR, Song JY, Rosado A & Peeper DS Functional characterization of human cancer-derived TRKB mutations. PloS one 6, e16871, doi: 10.1371/journal.pone.0016871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harada T et al. Role and relevance of TrkB mutations and expression in non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 17, 2638–2645, doi: 10.1158/1078-0432.CCR-10-3034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchetti A et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Human mutation 29, 609–616, doi: 10.1002/humu.20707 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Miranda C, Mazzoni M, Sensi M, Pierotti MA & Greco A Functional characterization of NTRK1 mutations identified in melanoma. Genes, chromosomes & cancer 53, 875–880, doi: 10.1002/gcc.22200 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Tomasson MH et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 111, 4797–4808, doi: 10.1182/blood-2007-09-113027 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reuther GW, Lambert QT, Caligiuri MA & Der CJ Identification and characterization of an activating TrkA deletion mutation in acute myeloid leukemia. Molecular and cellular biology 20, 8655–8666 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eggert A et al. Expression of the neurotrophin receptor TrkA down-regulates expression and function of angiogenic stimulators in SH-SY5Y neuroblastoma cells. Cancer research 62, 1802–1808 (2002). [PubMed] [Google Scholar]
- 65.Lagadec C et al. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene 28, 1960–1970, doi: 10.1038/onc.2009.61 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Lange AM & Lo HW Inhibiting TRK Proteins in Clinical Cancer Therapy. Cancers 10, doi: 10.3390/cancers10040105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajan N et al. Dysregulated TRK signalling is a therapeutic target in CYLD defective tumours. Oncogene 30, 4243–4260, doi: 10.1038/onc.2011.133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakagawara A, Azar CG, Scavarda NJ & Brodeur GM Expression and function of TRK-B and BDNF in human neuroblastomas. Molecular and cellular biology 14, 759–767 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans AE et al. Antitumor activity of CEP-751 (KT-6587) on human neuroblastoma and medulloblastoma xenografts. Clinical cancer research : an official journal of the American Association for Cancer Research 5, 3594–3602 (1999). [PubMed] [Google Scholar]
- 70.Evans AE et al. Effect of CEP-751 (KT-6587) on neuroblastoma xenografts expressing TrkB. Medical and pediatric oncology 36, 181–184, doi: (2001). [DOI] [PubMed] [Google Scholar]
- 71.Minturn JE et al. Phase I trial of lestaurtinib for children with refractory neuroblastoma: a new approaches to neuroblastoma therapy consortium study. Cancer chemotherapy and pharmacology 68, 1057–1065, doi: 10.1007/s00280-011-1581-4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaishnavi A, Le AT & Doebele RC TRKing down an old oncogene in a new era of targeted therapy. Cancer discovery 5, 25–34, doi: 10.1158/2159-8290.CD-14-0765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schram AM, Chang MT, Jonsson P & Drilon A Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nature reviews. Clinical oncology 14, 735–748, doi: 10.1038/nrclinonc.2017.127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coulier F, Martin-Zanca D, Ernst M & Barbacid M Mechanism of activation of the human trk oncogene. Molecular and cellular biology 9, 15–23 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knezevich SR, McFadden DE, Tao W, Lim JF & Sorensen PH A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nature genetics 18, 184–187, doi: 10.1038/ng0298-184 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Hechtman JF et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. The American journal of surgical pathology 41, 1547–1551, doi: 10.1097/PAS.0000000000000911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh D et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235, doi: 10.1126/science.1220834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borrello MG et al. The oncogenic versions of the Ret and Trk tyrosine kinases bind Shc and Grb2 adaptor proteins. Oncogene 9, 1661–1668 (1994). [PubMed] [Google Scholar]
- 79.Miranda C, Greco A, Miele C, Pierotti MA & Van Obberghen E IRS-1 and IRS-2 are recruited by TrkA receptor and oncogenic TRK-T1. Journal of cellular physiology 186, 35–46, doi: (2001). [DOI] [PubMed] [Google Scholar]
- 80.Ranzi V et al. The signaling adapters fibroblast growth factor receptor substrate 2 and 3 are activated by the thyroid TRK oncoproteins. Endocrinology 144, 922–928, doi: 10.1210/en.2002-221002 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Jin W et al. Cellular transformation and activation of the phosphoinositide-3-kinase-Akt cascade by the ETV6-NTRK3 chimeric tyrosine kinase requires c-Src. Cancer research 67, 3192–3200, doi: 10.1158/0008-5472.CAN-06-3526 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Tognon CE et al. A tripartite complex composed of ETV6-NTRK3, IRS1 and IGF1R is required for ETV6-NTRK3-mediated membrane localization and transformation. Oncogene 31, 1334–1340, doi: 10.1038/onc.2011.323 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Drilon A et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Annals of oncology : official journal of the European Society for Medical Oncology 27, 920–926, doi: 10.1093/annonc/mdw042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vaishnavi A et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nature medicine 19, 1469–1472, doi: 10.1038/nm.3352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vaishnavi A et al. EGFR Mediates Responses to Small-Molecule Drugs Targeting Oncogenic Fusion Kinases. Cancer research 77, 3551–3563, doi: 10.1158/0008-5472.CAN-17-0109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen S et al. A new ETV6-NTRK3 cell line model reveals MALAT1 as a novel therapeutic target - a short report. Cellular oncology 41, 93–101, doi: 10.1007/s13402-017-0356-2 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Cook PJ et al. Somatic chromosomal engineering identifies BCAN-NTRK1 as a potent glioma driver and therapeutic target. Nature communications 8, 15987, doi: 10.1038/ncomms15987 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roberts KG et al. ETV6-NTRK3 induces aggressive acute lymphoblastic leukemia highly sensitive to selective TRK inhibition. Blood, doi: 10.1182/blood-2018-05-849554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Witte K et al. TFG-1 function in protein secretion and oncogenesis. Nature cell biology 13, 550–558, doi: 10.1038/ncb2225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amatu A, Sartore-Bianchi A & Siena S NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO open 1, e000023, doi: 10.1136/esmoopen-2015-000023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halalsheh H et al. Dramatic bone remodeling following larotrectinib administration for bone metastasis in a patient with TRK fusion congenital mesoblastic nephroma. Pediatric blood & cancer, e27271, doi: 10.1002/pbc.27271 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Davis JL et al. Infantile NTRK-associated Mesenchymal Tumors. Pediatric and developmental pathology : the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society 21, 68–78, doi: 10.1177/1093526617712639 (2018). [DOI] [PubMed] [Google Scholar]
- 93.Laetsch TW et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. The Lancet. Oncology 19, 705–714, doi: 10.1016/S1470-2045(18)30119-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tognon C et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer cell 2, 367–376 (2002). [DOI] [PubMed] [Google Scholar]
- 95.Taylor J et al. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. The Journal of clinical investigation, doi: 10.1172/JCI120787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benayed R et al. Comprehensive detection of targetable fusions in lung adenocarcinomas by complementary targeted DNAseq and RNAseq assays. Journal of Clinical Oncology 36, 12076–12076, doi: 10.1200/JCO.2018.36.15_suppl.12076 (2018). [DOI] [Google Scholar]
- 97.Drilon A et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer discovery 7, 963–972, doi: 10.1158/2159-8290.CD-17-0507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Russo M et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer discovery 6, 36–44, doi: 10.1158/2159-8290.CD-15-0940 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Rudzinski ER et al. Pan-Trk Immunohistochemistry Identifies NTRK Rearrangements in Pediatric Mesenchymal Tumors. The American journal of surgical pathology 42, 927–935, doi: 10.1097/PAS.0000000000001062 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Menichincheri M et al. Discovery of Entrectinib: A New 3-Aminoindazole As a Potent Anaplastic Lymphoma Kinase (ALK), c-ros Oncogene 1 Kinase (ROS1), and Pan-Tropomyosin Receptor Kinases (Pan-TRKs) inhibitor. Journal of medicinal chemistry 59, 3392–3408, doi: 10.1021/acs.jmedchem.6b00064 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Doebele RC et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer discovery 5, 1049–1057, doi: 10.1158/2159-8290.CD-15-0443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kazandjian D et al. Benefit-Risk Summary of Crizotinib for the Treatment of Patients With ROS1 Alteration-Positive, Metastatic Non-Small Cell Lung Cancer. The oncologist 21, 974–980, doi: 10.1634/theoncologist.2016-0101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Malik SM et al. U.S. Food and Drug Administration approval: crizotinib for treatment of advanced or metastatic non-small cell lung cancer that is anaplastic lymphoma kinase positive. Clinical cancer research : an official journal of the American Association for Cancer Research 20, 2029–2034, doi: 10.1158/1078-0432.CCR-13-3077 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Singh H et al. U.S. Food and Drug Administration Approval: Cabozantinib for the Treatment of Advanced Renal Cell Carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 330–335, doi: 10.1158/1078-0432.CCR-16-1073 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Shamroe CL & Comeau JM Ponatinib: A new tyrosine kinase inhibitor for the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. The Annals of pharmacotherapy 47, 1540–1546, doi: 10.1177/1060028013501144 (2013). [DOI] [PubMed] [Google Scholar]
- 106.Karimi-Shah BA & Chowdhury BA Forced vital capacity in idiopathic pulmonary fibrosis--FDA review of pirfenidone and nintedanib. The New England journal of medicine 372, 1189–1191, doi: 10.1056/NEJMp1500526 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Zou HY et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer research 67, 4408–4417, doi: 10.1158/0008-5472.can-06-4443 (2007). [DOI] [PubMed] [Google Scholar]
- 108.Christensen JG et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 6, 3314–3322, doi: 10.1158/1535-7163.MCT-07-0365 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Cui JJ et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). Journal of medicinal chemistry 54, 6342–6363, doi: 10.1021/jm2007613 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Bergethon K et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 30, 863–870, doi: 10.1200/JCO.2011.35.6345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bowles DW, Kessler ER & Jimeno A Multi-targeted tyrosine kinase inhibitors in clinical development: focus on XL-184 (cabozantinib). Drugs Today (Barc) 47, 857–868, doi: 10.1358/dot.2011.47.11.1688487 (2011). [DOI] [PubMed] [Google Scholar]
- 112.O’Hare T et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell 16, 401–412, doi: 10.1016/j.ccr.2009.09.028 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fuse MJ et al. Mechanisms of Resistance to NTRK Inhibitors and Therapeutic Strategies in NTRK1-Rearranged Cancers. Molecular cancer therapeutics 16, 2130–2143, doi: 10.1158/1535-7163.MCT-16-0909 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Hilberg F et al. BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res 68, 4774–4782, doi: 10.1158/0008-5472.CAN-07-6307 (2008). [DOI] [PubMed] [Google Scholar]
- 115.Mok S, Duffy C, Du R & Allison JP Abstract A146: Increase antitumor activity of immunotherapy by blocking colony stimulating factor 1 receptor and tropomyosin receptor kinase. Cancer Immunology Research 4, A146–A146, doi: 10.1158/2326-6066.imm2016-a146 (2016). [DOI] [Google Scholar]
- 116.Patwardhan PP, Ivy KS, Musi E, de Stanchina E & Schwartz GK Significant blockade of multiple receptor tyrosine kinases by MGCD516 (Sitravatinib), a novel small molecule inhibitor, shows potent anti-tumor activity in preclinical models of sarcoma. Oncotarget 7, 4093–4109, doi: 10.18632/oncotarget.6547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith KM et al. Anti-Tumor Activity of Entrectinib, a Pan-TRK, ROS1 and ALK Inhibitor, in ETV6-NTRK3-Positive Acute Myeloid Leukemia. Molecular Cancer Therapeutics, doi: 10.1158/1535-7163.mct-17-0419 (2017). [DOI] [PubMed] [Google Scholar]
- 118.Kiga M et al. Preclinical characterization and antitumor efficacy of DS-6051b, a novel, orally available small molecule tyrosine kinase inhibitor of ROS1 and NTRKs. European Journal of Cancer 69, S35–S36, doi: 10.1016/S0959-8049(16)32687-9 (2016). [DOI] [Google Scholar]
- 119.Smith BD et al. Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2. Mol Cancer Ther 14, 2023–2034, doi: 10.1158/1535-7163.MCT-14-1105 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Smith BD et al. Abstract 790: Altiratinib is a potent inhibitor of TRK kinases and is efficacious in TRK-fusion driven cancer models. Cancer Research 75, 790–790, doi: 10.1158/1538-7445.am2015-790 (2015). [DOI] [Google Scholar]
- 121.Kawada I et al. Dramatic antitumor effects of the dual MET/RON small-molecule inhibitor LY2801653 in non-small cell lung cancer. Cancer Res 74, 884–895, doi: 10.1158/0008-5472.CAN-12-3583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yan SB et al. MET-targeting antibody (emibetuzumab) and kinase inhibitor (merestinib) as single agent or in combination in a cancer model bearing MET exon 14 skipping. Invest New Drugs, doi: 10.1007/s10637-017-0545-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Konicek BW et al. Merestinib (LY2801653) inhibits neurotrophic receptor kinase (NTRK) and suppresses growth of NTRK fusion bearing tumors. Oncotarget 9, 13796–13806, doi: 10.18632/oncotarget.24488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yan SB et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investigational new drugs 31, 833–844, doi: 10.1007/s10637-012-9912-9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Katayama R et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Science translational medicine 4, 120ra117, doi: 10.1126/scitranslmed.3003316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Awad MM, Engelman JA & Shaw AT Acquired resistance to crizotinib from a mutation in CD74-ROS1. The New England journal of medicine 369, 1173, doi: 10.1056/NEJMc1309091 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Kim S et al. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 8, 415–422, doi: 10.1097/JTO.0b013e318283dcc0 (2013). [DOI] [PubMed] [Google Scholar]
- 128.Zhai D, Deng W, Huang J, Rogers E & Cui JJ Abstract 3161: TPX-0005, an ALK/ROS1/TRK inhibitor, overcomes multiple resistance mechanisms by targeting SRC/FAK signaling. Cancer research 77, 3161–3161, doi: 10.1158/1538-7445.am2017-3161 (2017). [DOI] [Google Scholar]
- 129.Kozaki R, Yoshizawa T, Tsukamoto K, Kato H & Kawabata K Abstract 2954A: A potent and selective TRK inhibitor ONO-5390556, shows potent antitumor activity against both TRK-rearranged cancers and the resistant mutants. Cancer research 76, 2954A–2954A, doi: 10.1158/1538-7445.am2016-2954a (2016).26980765 [DOI] [Google Scholar]
- 130.Drilon AE et al. A phase 1 study of the next-generation ALK/ROS1/TRK inhibitor ropotrectinib (TPX-0005) in patients with advanced ALK/ROS1/NTRK+ cancers (TRIDENT-1). Journal of Clinical Oncology 36, 2513–2513, doi: 10.1200/JCO.2018.36.15_suppl.2513 (2018). [DOI] [Google Scholar]
- 131.Smeyne RJ et al. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature 368, 246–249, doi: 10.1038/368246a0 (1994). [DOI] [PubMed] [Google Scholar]
- 132.Klein R et al. Targeted disruption of the trkB neurotrophin receptor gene results in nervous system lesions and neonatal death. Cell 75, 113–122 (1993). [PubMed] [Google Scholar]
- 133.Klein R et al. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. Nature 368, 249–251, doi: 10.1038/368249a0 (1994). [DOI] [PubMed] [Google Scholar]
- 134.Wagner N et al. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes & development 19, 2631–2642, doi: 10.1101/gad.346405 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tessarollo L et al. Targeted deletion of all isoforms of the trkC gene suggests the use of alternate receptors by its ligand neurotrophin-3 in neuronal development and implicates trkC in normal cardiogenesis. Proceedings of the National Academy of Sciences of the United States of America 94, 14776–14781 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kermani P & Hempstead B Brain-derived neurotrophic factor: a newly described mediator of angiogenesis. Trends in cardiovascular medicine 17, 140–143, doi: 10.1016/j.tcm.2007.03.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dissen GA et al. A role for trkA nerve growth factor receptors in mammalian ovulation. Endocrinology 137, 198–209, doi: 10.1210/endo.137.1.8536613 (1996). [DOI] [PubMed] [Google Scholar]
- 138.Kermani P et al. Neurotrophins promote revascularization by local recruitment of TrkB+ endothelial cells and systemic mobilization of hematopoietic progenitors. The Journal of clinical investigation 115, 653–663, doi: 10.1172/JCI22655 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kawamura K, Kawamura N, Mulders SM, Sollewijn Gelpke MD & Hsueh AJ Ovarian brain-derived neurotrophic factor (BDNF) promotes the development of oocytes into preimplantation embryos. Proceedings of the National Academy of Sciences of the United States of America 102, 9206–9211, doi: 10.1073/pnas.0502442102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Greco A, Villa R, Fusetti L, Orlandi R & Pierotti MA The Gly571Arg mutation, associated with the autonomic and sensory disorder congenital insensitivity to pain with anhidrosis, causes the inactivation of the NTRK1/nerve growth factor receptor. Journal of cellular physiology 182, 127–133, doi: (2000). [DOI] [PubMed] [Google Scholar]
- 141.Indo Y et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nature genetics 13, 485–488, doi: 10.1038/ng0896-485 (1996). [DOI] [PubMed] [Google Scholar]
- 142.Yeo GS et al. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nature neuroscience 7, 1187–1189, doi: 10.1038/nn1336 (2004). [DOI] [PubMed] [Google Scholar]
- 143.Xu B et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nature neuroscience 6, 736–742, doi: 10.1038/nn1073 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Richardson CA & Leitch B Phenotype of cerebellar glutamatergic neurons is altered in stargazer mutant mice lacking brain-derived neurotrophic factor mRNA expression. The Journal of comparative neurology 481, 145–159, doi: 10.1002/cne.20386 (2005). [DOI] [PubMed] [Google Scholar]
- 145.Ashraf S, Bouhana KS, Pheneger J, Andrews SW & Walsh DA Selective inhibition of tropomyosin-receptor-kinase A (TrkA) reduces pain and joint damage in two rat models of inflammatory arthritis. Arthritis research & therapy 18, 97, doi: 10.1186/s13075-016-0996-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hirose M, Kuroda Y & Murata E NGF/TrkA Signaling as a Therapeutic Target for Pain. Pain practice : the official journal of World Institute of Pain 16, 175–182, doi: 10.1111/papr.12342 (2016). [DOI] [PubMed] [Google Scholar]