Accession numbers
The accession numbers of the targeted genes by the SaCas9 toolset in this study are: PDS: Os03g0184000; DL: Os03g0215200; NAL1: Os04g0615000; SLR1: Os03g0707600; IPA1: Os08g0509600; TAC1: Os09g0529300; Ehd1: Os10g0463400; Pi‐d2: Os06g0494100; OsMKK6: Os01g051010 0; OsMPK3: Os03g0285800; Pi‐ta: Os12g0281300; OsSPL17: Os09g0491532; GL2: Os02g0701300; OsGRF3: Os04g0600900; Wx: Os06g0133000.
The next‐generation sequencing data of mutation genotyping could be achieved at NCBI BioProject ID PRJNA496604.
Conflict of interest
The authors declare no conflict of interest.
Dear Editor,
The CRISPR‐Streptococcus pyogenes Cas9 (SpCas9) system offers a rapid, simple and flexible genome editing approach. However, the targeting scope of the SpCas9 system is limited by the canonical NGG PAM. To broaden the editing range of the CRISPR system, several Cas orthologs recognized different PAM were isolated from diverse microbes and were engineered as powerful genome editing tools in eukaryotic cells (Murovec et al., 2017). One Cas9 ortholog, Staphylococcus aureus Cas9 (SaCas9), has a smaller size and comparable activity compared to SpCas9 (Kleinstiver et al., 2015a,b). Previous studies reported that the SaCas9 could induce highly efficient targeted mutagenesis in Arabidopsis, citrus, tobacco and rice (Murovec et al., 2017). SaCas9 recognizes a longer PAM motif (NNGRRT) than SpCas9 (Kleinstiver et al., 2015b). In mammalian cells, the PAM specificity of SaCas9 could be relaxed to NNNRRT by a KKH variant (SaCas9‐KKH, SaKKH; Kleinstiver et al., 2015a). In this study, we investigated the targeting capability of SaKKH in rice. Furthermore, we developed cytosine base editors (CBEs) and adenine base editors (ABEs) based on SaCas9 and SaKKH.
To achieve a high efficiency in monocots, we re‐optimized the codons of SaCas9 for rice. To broaden the targeting range of SaCas9, three key mutations (E782K/N968K/R105H) were simultaneously introduced to generate SaKKH (Figure 1a). The activity of SaCas9 was identified in PDS and DL genes via stably Agrobacterium‐mediated transformation of Japonica rice. The targeted mutations were determined in regenerated plants by high‐throughput tracking of mutation (Hi‐TOM) detection (Liu et al., 2018). After generating transgenic plants, 34 out of 53 lines and 28 out of 36 lines carried targeted mutations in the PDS‐T1 and DL‐T1 regions, achieving 64.2% and 77.8% mutagenesis efficiency respectively. This result confirms that SaCas9 efficiently edits crop genome. To examine the genome editing activity of SaKKH in plants, three protospacers with a NNNRRT PAM were further designed in rice PDS, DL and NAL1 genes. At the PDS‐T2 target with a TGCAGT PAM, mutations were detected in 90.6% of SaKKH lines (Figure 1b). In addition, we found that most of the mutated lines were homozygous or biallelic mutants (79.3%), further indicating the high‐mutagenesis frequency induced by SaKKH. Moreover, targeted mutations were obtained at the DL‐T2 and NAL1 sites at rates of 41.7% and 66.7% respectively.
Figure 1.
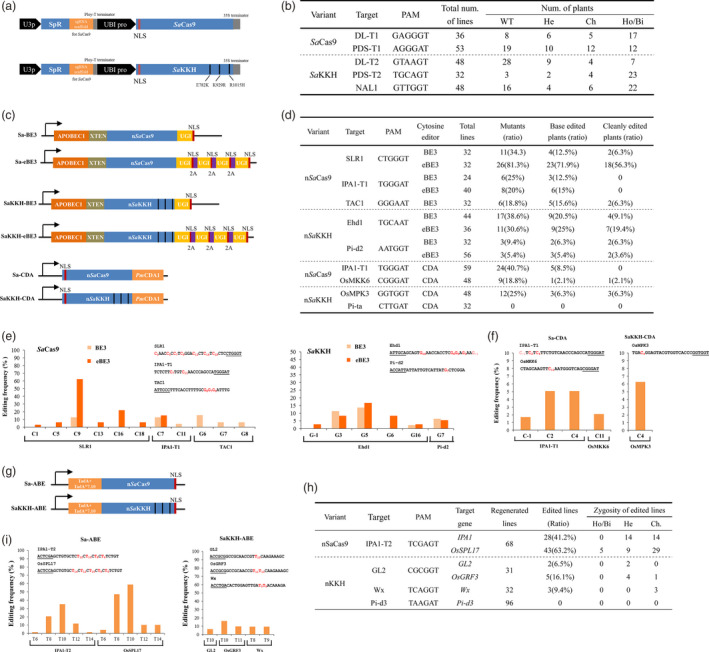
Rice genome editing generated by SaCas9 toolset. (a) Schematic illustration of the sgRNA and Cas9 expression cassettes of plant SaCas9 systems. To express sgRNA, a 21 bp protospacer sequence was inserted downstream of the rice U3 promoter (U3p) to replace the spectinomycin resistance gene (SpR). A maize ubiquitin promoter (UBI pro) was used to express SaCas9 or Sa KKH. (b) Mutations induced by SaCas9 and Sa KKH in regenerated rice plants analysed by Hi‐TOM assay with a 5% threshold (http://www.hi-tom.net/hi-tom). WT, wild‐type sequence in the target region; He, heterozygous mutation; Ch, chimeric mutation; Ho/Bi, homozygous or biallelic mutation. (c) Schematic illustration of SaCas9‐CBEs. (d) Cytosine editing induced by SaCas9 base editors in regenerated rice plants. The regenerated plants with exclusive C‐to‐T base conversions were considered as cleanly edited plants. (e) Frequencies of the base editing induced by SaCas9‐BE3 base editors at different C(G)s in the target sequence. The PAM sequence is underlined, and the targeted bases and positions in the protospacer are labeled in red. Sa‐eBE3 was not tested at the TAC1 target. (f) Frequencies of the base conversions induced by SaCas9‐CDA base editors at different C(G)s in the target sequence in regenerated plant populations. (g) Schematic illustration of SaCas9‐ABE base editors. (h) Adenine editing induced by SaCas9 base editors in regenerated rice plants. (i) Frequencies of the A‐to‐G conversion induced by SaCas9 ABEs at different A(T)s in the target sequence.
A series of CBE tools was recently developed by fusing cytosine deaminase to SpCas9 nickase (nSpCas9; Komor et al., 2016; Nishida et al., 2016). At first, we constructed SaCas9 CBEs by attaching the optimized sequence of rat APOBEC1 to the 5′ terminus and the optimized sequence of uracil glycosylase inhibitor (UGI) to the 3′ terminus of nSaCas9 (D10A) and nSaKKH(D10A) nickase, leading to Sa‐BE3 and SaKKH‐BE3 respectively. To increase the editing product purity, we separately fused three copies of UGI to the C terminus of Sa‐BE3 and SaKKH‐BE3, generating Sa‐eBE3 and SaKKH‐eBE3 (Figure 1c). A protospacer was designed to edit the C290 in SLR1 gene, using Sa‐BE3 or Sa‐eBE3. We found 11 out 40 Sa‐BE3 lines had mutations in the target region. Among them, 4 lines (10% frequency) carried base conversions, which were located at position 9 of the protospacer (the nucleotide at the 5′ end is position 1; Figure 1d,e). The ratio of base editing in SLR1 induced by Sa‐eBE3 was much higher (Fisher's exact test, P < 0.05). We found 81.3% of the regenerated lines had targeted mutations, and 71.9% of the lines were edited. More than half of the lines had only clean C‐to‐T conversion(s) in the target region (designated as clean editing). In addition, we found homozygous or biallelic clean base editing in 10 of 32 lines, further indicating the extraordinary activity of Sa‐eBE3 in this target. The base conversion at position 9 has highest frequency, while the editing also occurred at positions 1, 5, 9, 13, 16 and 18 (Figure 1e), suggesting the Sa‐eBE3 have a broad editing window. The miR156 binding region in the IPA1 gene was also used to design a sgRNA (IPA‐T1). Screening of 24 Sa‐BE3 lines and 40 Sa‐eBE3 lines detected 2 and 6 edited lines respectively. Moreover, the 3′ splicing point of the 4th intron of rice TAC1 was targeted by Sa‐BE3. The base editing of G (s) was obtained in 15.6% regenerated plants. To test the editing activity of SaKKH BE3s, the protospacer with TGCAAT and AATGGT was designed to target the G655 and G1383 in Ehd1 and Pi‐d2 genes respectively. We found that 20.5% and 6.3% of SaKKH‐BE3 plants have targeted base conversions in the Ehd1 and Pi‐d2 target region respectively (Figure 1D). Furthermore, we noticed that the editing efficiency at the Ehd1 target was increased to 25% by the SaKKH‐eBE3 vector. In addition, the clean editing frequency induced by SaKKH‐eBE3 is increased to 19.4% from the 9.1% generated by SaKKH‐BE3. The SaKKH‐eBE3‐induced base conversion ratio remained as low as 5.4% at the Pi‐d2 target, which may be caused by the inconvenient GC context for the rat APOBEC1 activity. The base conversions in the Ehd1 target were occurred in Gs at position 3, 5, 6, 16 in the 21 bp target sequence and the G immediately 5′ upstream of the protospacer (position ‐1), while the editing on the Pi‐d2 target were only detected in the G at position 7 (Figure 1e). Very recently, a similar SaKKH‐BE3 was constructed and assembled into the pRCBEsakkh‐OsU6sa vector (CBE‐P5; Hua et al., 2018b). The editing activity of CBE‐P5 was not observed at the target in the rice PMS1 gene (Hua et al., 2018b), suggesting insufficient activity of SaKKH‐BE3. However, the mutagenesis efficiency introduced by the SaKKH‐BE3 in this study reached as high as 38.6%. Because the activity of CBE‐P5 was examined at only one site (Hua et al., 2018b), the editing efficiency of the two systems cannot be directly compared based on current results.
To expand the editing scope of the Petromyzon marinus cytidine deaminase 1 (PmCDA1) ‐based CBE in plant (Shimatani et al., 2017), the nSaCas9 or nSaKKH nickase was separately assembled with a PmCDA1 domain, which was designated Sa‐CDA or SaKKH‐CDA respectively (Figure 1c). Two targets were selected and examined for each CDA base editor. For Sa‐CDA, the IPA‐T1 protospacer of Sa‐BE3 was used. In 59 regenerated plants, base editing was observed in 5 lines (Figure 1d). The substitutions occurred at the position 2 and 4 of the protospacer and the position ‐1 out of the protospacer (Figure 1f). At another target site in OsMKK6, one cleanly edited line out of 48 screened regenerated plants was obtained. For SaKKH‐CDA, protospacers were designed to edit the TEY domain of OsMPK3 and the C2753 of Pi‐ta. At the OsMPK3 target, 3 out 48 lines carried a clean C‐to‐T conversion, all of which were located at position 4; while no mutation was detected in the Pi‐ta target with a CTTGAT PAM.
Adenine base editors convert A:T to G:C in the target site (Gaudelli et al., 2017). To increase the range of ABEs in plants, a rice codon‐optimized TadA‐XTEN‐TadA*7.10 fragment was assembled into the nSaCas9 (D10A) or nSaKKH (D10A) nickase, resulting Sa‐ABE and SaKKH‐ABE, respectively (Figure 1g). A protospacer (IPA‐T2) that simultaneously targeted two genomic sites in IPA1 and OsSPL17 was selected for testing Sa‐ABE. The editing occurred in IPA1 and OsSPL17 with 41.2% and 63.2% efficiency respectively (Figure 1h). In both targets, the Cs at positions 8 and 10 were edited with higher frequency (Figure 1i). Other than the targeted base conversions, an undesired mutation (a 35 nt deletion) was found in only one plant at the OsSPL17 site, indicating high‐editing purity of Sa‐ABE. Moreover, 21 lines (31.9%) were edited at both sites, suggesting the capability of multiplexed editing with Sa‐ABE. This result is similar to the editing generated by the ABE‐P2 in a recent report (Hua et al., 2018a). We notice that the SaKKH‐ABE (ABE‐P5) was also recently reported with relatively lower (0%–6.5%) efficiency compared to Sa‐ABE in plant (Hua et al., 2018b). We further examined the activity of the SaKKH‐ABE with three independent protospacers with NNHRRT PAMs. For the protospacers targeting the Wx and Pi‐d3 gene, the targeting efficiency was 0% and 9.4% respectively. In addition, The GL2 protospacer simultaneously targeted GL2 and OsGRF3 genes. We found that only 6.5% of plants (2 out of 31 lines) carried A‐to‐G conversions in the GL2 gene, while the editing frequency reached 16.1% (five edited lines) in OsGRF3, suggesting the SaKKH‐ABE may have potential to achieve efficient editing in some targets.
In this study, we developed a series of mutagenesis and base editing tools using SaCas9 and its derivative for plant genome editing. These tools expand the scope of genome editing to targets with a NNNRRT PAM. It is desired to simultaneously perform different editing events, such as base editing and targeted mutation, in a single plant. Using the SaCas9 tools, this purpose would be robustly achieved by co‐transforming or stacking the previously established SpCas9 systems. Taking these results together, we established a highly efficient and wildly adaptive SaCas9 toolset that may advance plant research and accelerate crop improvement.
Acknowledgments
This work was funded by the Genetically Modified Breeding Major Projects (No. 2016ZX08010‐002‐008) and the National Natural Science Foundation of China (No. 31701405).
Contributor Information
Xiuqing Zhang, Email: xiuqingzhang@cau.edu.cn.
Pengcheng Wei, Email: Weipengcheng@gmail.com.
References
- Gaudelli, N.M. , Komor, A.C. , Rees, H.A. , Packer, M.S. , Badran, A.H. , Bryson, D.I. and Liu, D.R. (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. , Yuan, F. , Wang, D. and Zhu, J.‐K. (2018a) Precise A·T to G·C base editing in the rice genome. Molecular Plant, 11, 627–630. [DOI] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. and Zhu, J.‐K. (2018b) Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 10.1111/pbi.12993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Nguyen, N.T. , Topkar, V.V. , Zheng, Z. and Joung, J.K. (2015a) Broadening the targeting range of Staphylococcus aureus CRISPR‐Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Topkar, V.V. , Nguyen, N.T. , Zheng, Z. , Gonzales, A.P.W. et al. (2015b) Engineered CRISPR‐Cas9 nucleases with altered PAM specificities. Nature, 523, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2018) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. bioRxiv. [DOI] [PubMed]
- Murovec, J. , Pirc, Ž. and Yang, B. (2017) New variants of CRISPR RNA‐guided genome editing enzymes. Plant Biotechnol. J. 15, 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K. , Arazoe, T. , Yachie, N. , Banno, S. , Kakimoto, M. , Tabata, M. , Mochizuki, M. et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353, aaf8729. [DOI] [PubMed] [Google Scholar]
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441. [DOI] [PubMed] [Google Scholar]


