Abstract
Like all outer membrane (OM) constituents, integral OM β-barrel proteins in Gram-negative bacteria are synthesized in the cytoplasm and trafficked to the OM, where they are locally assembled into the growing OM by the ubiquitous β-barrel assembly machine (Bam). While the identities and structures of all essential and accessory Bam components have been determined, the basic mechanism of Bam-assisted OM protein integration remains elusive. Here we review mechanistic analyses of OM β-barrel protein folding and Bam dynamics and summarize recent insights that inform a general model for OM protein recognition and assembly by the Bam complex.
INTRODUCTION
The presence in Gram-negative bacteria of an extracytoplasmic outer membrane (OM), which is distinct from the inner membrane (IM) both in constitution and in function, presents a complex topological problem, as all proteinaceous and lipidic OM components are synthesized cytoplasmically (1). In order to reach their destination in the growing OM, these components must translocate across the IM and traverse the aqueous, crowded periplasmic space. This problem is solved through a series of semi-independent and highly conserved transport pathways that coordinate the efficient delivery and integration of all OM constituents.
OM-specific lipopolysaccharide (LPS) is trafficked via a multicomponent trans-envelope protein bridge (2, 3), the LPS transport (Lpt) pathway, which terminates at an OM-integral translocase (LptDE) that incorporates free LPS into the outer leaflet of the OM (4). A transport system that enables retrograde (OM to IM) phospholipid transport has been described (5–10), but the mechanism of anterograde (IM to OM) transport is mysterious and represents an area of active investigation. Periplasmic lipoproteins, which can be anchored to either membrane via N-terminal lipid moieties, are sorted in a sequence-dependent manner to the OM via the Lol system, which extracts IM-associated lipoproteins and shuttles them to the OM via a soluble periplasmic carrier (11). Finally, integral OM β-barrel proteins (OMPs) are translocated in an unfolded form across the IM, ferried to the OM in a nonnative but folding-competent state via a diverse network of periplasmic chaperones, and integrated into the OM in a manner dependent on a ubiquitous, essential multiprotein complex known as the β-barrel assembly machine (Bam).
Despite the functional and structural heterogeneity observed across OMP families, the in vivo folding and membrane integration of all OM β-barrel proteins require Bam, an OM-associated heteromeric complex composed of BamA (itself a β-barrel protein) and a variable number of OM lipoproteins (BamB to -F) that bind to and act in concert with BamA to drive the OMP assembly process (1, 4, 12, 13). At present, definitive roles cannot yet be unambiguously assigned to the individual components, and the general mechanism of BamA-dependent OMP assembly remains elusive and controversial. Here we offer a compendious review of recent inquiry into the mechanism of OM β-barrel folding and its catalysis by the Bam complex.
Bam COMPLEX CONSTITUENTS
BamA, the central component of the Bam complex, is composed of a C-terminal β-barrel domain and an N-terminal periplasmic domain that serves as the physical hub of a functional network that includes substrates (14–18), accessory lipoproteins (19–26), periplasmic chaperones (27–29), and proteases (30). This fishhook-shaped domain is typically subdivided into five structurally homologous POTRA domains (31, 32) that may nucleate the early formation of OMP secondary structure through β-strand augmentation (26, 32, 33) and which, together with the Bam lipoproteins, form a cavernous periplasmic ring that circumscribes the vestibule of the BamA β-barrel lumen (24, 34). The OM-integral BamA β-barrel domain is atypical and can be distinguished from canonical membrane β-barrels in three fundamental ways: (i) the interstrand hydrogen bond network that seals β1 and β16 (to complete the barrel) is metastable, allowing transient destabilization of this seam and reversible lateral opening of the barrel (35–38); (ii) the β-barrel forms an unusually narrow protein-lipid interface adjacent to the seam that is thought to physically alter the local properties of the OM (37, 39–41); and (iii) a conserved, essential latching loop (L6) internally braces the BamA barrel interior and globally stabilizes the otherwise thermolabile β-barrel domain (42–46), likely compensating for the metastability at the β1-β16 seam (Fig. 1). The mechanistic implications of these features are discussed in an ensuing section.
Figure 1.

Unique features of the BamA β-barrel domain. (a) The BamA β-barrel (pink) is asymmetric, with one face forming a narrow protein-lipid interface (approximated by the dashed line) that is thought to physically alter the local properties of the bilayer (green). (b) The activated BamA β-barrel undergoes a dramatic conformational rearrangement that disrupts the continuous β-barrel structure, separates the β-strands comprising the lateral gate (β1 and β16), and exposes an aqueous channel that spans the membrane. Additionally, a highly conserved extracellular loop (L6, blue) internally braces and globally stabilizes the β-barrel domain and compensates for the instability introduced by the conformational dynamics. This image was generated using PDB structures 4K3B (left) and 5EKQ (right).
BamA is the central catalyst of OMP insertion, but the partner lipoprotein BamD also plays an apparently essential role in the process (13, 14, 16, 17, 20, 47–51). BamD is a solenoid protein thought to serve as a generic receptor for substrate OMPs through recognition of the β-signal, a semidegenerate C-terminal peptide motif common to all prokaryotic and eukaryotic OMPs (14, 52–60). BamD plays a critical role in the binding and OM localization of OMPs (including BamA), although BamB, a β-propeller protein, has been proposed to perform an overlapping function for some Bam substrates (17, 26, 47). Additionally, commensurate with its role as an OMP receptor, BamD has been implicated in regulation of the conformational dynamics and activity of BamA during the OMP assembly cycle, tentatively linking the recognition and binding of nascent OMP C-termini to conformational changes in BamA that enable OM insertion (14, 48, 61, 62).
The remaining Bam lipoproteins (BamB, BamC, BamE, and the BamC-like α-proteobacterial protein BamF) are variably conserved, are not central to the mechanism of Bam-catalyzed OMP assembly, and instead play accessory roles that enhance the efficiency of the process for at least a subset of OMPs (13, 63). BamCE associate indirectly with BamA via BamD and play an adjunctive role in the regulation of BamA dynamics and function, potentially by stabilizing the interaction between BamA and BamD following each round of OMP assembly (20, 21, 47, 49, 61, 64–67). BamB interacts with BamA in a BamD-independent manner through direct contacts with multiple POTRA domains (20–22, 25, 26) and has been shown to contribute significantly to the efficiency of substrate assembly for certain OMPs (17, 47, 68–75). We have proposed that BamB (together with the OMP chaperone SurA) influences substrate flux to ensure streamlined assembly of both high-abundance targets (e.g., OmpA and porins) and low-abundance, high-priority targets (e.g., LptDE) (28). Intriguing recent work has also uncovered a role for BamB in the sequestration of Bam complexes into tightly clustered “assembly precincts” that are proposed to accelerate the assembly and multimerization of abundant OMP species (75).
Bam AS A FOLDING CHAPERONE AND CATALYST OF OM PROTEIN INSERTION
A wide variety of OM β-barrel proteins can autonomously fold in hydrophobic environments in accordance with Anfinsen’s dogma. Decades of in vitro studies using model OMPs have revealed the following key observations (Fig. 2):
Figure 2.
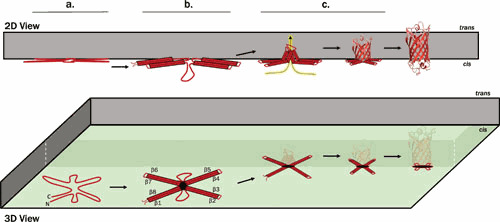
Proposed unassisted folding model for a membrane β-barrel protein. (a) A nascent eight-stranded OMP rapidly adsorbs to the cis surface (green) of the lipid bilayer, where hydrophobic lipid-facing side chains begin to penetrate into the membrane and β-strands assume a cloverleaf-like circular arrangement according to their relative position in the folded protein. (b) β-hairpins begin to form as the trans ends of the TM β-strands, oriented toward the center of the cloverleaf, plunge into the lipid phase. (c) Hydrogen bonds form between neighboring β-hairpins as they enter the membrane, stabilizing the native fold in concert with membrane insertion. The highlighted dashed line indicates the proposed path of the leading (trans) ends of the β-strands.
In the appropriate hydrophobic context, OMPs spontaneously and rapidly fold into extremely stable membrane integral species (76–89).
Native OM phospholipids impose a kinetic barrier to OMP assembly (a critical phenomenon that prevents the lethal assembly of OM β-barrels into the IM) (40, 41, 90–92).
The kinetics of OMP assembly can be dramatically accelerated by altering physical properties of the membrane so as to induce local defects (e.g., bilayer thinning or increased curvature) (39, 54, 93, 94).
OMP folding is a concerted process in which barrel formation and insertion happen concurrently and in which all β-strands integrate simultaneously rather than sequentially (79, 95–99).
The fact that β-barrel folding and insertion occur spontaneously implies a role for Bam in accelerating the intrinsic folding kinetics of OMPs, akin to classical folding chaperones. However, alternative pathways for in vivo folding have been surmised that involve the formation of transient chimeric BamA:OMP barrels (37, 100) or elongated substrate β-sheets nucleated at BamA β1 (101) as necessary intermediates. These models have explanatory power but are problematic from a thermodynamic perspective and require unnecessary invention of a distinct and strictly BamA-dependent folding pathway for β-barrel proteins (102). A recent biochemical analysis of mitochondrial β-barrel insertion by the BamA homolog Sam50 yielded observations consistent with a β-strand exchange model (100); however, the use of nonnative, truncated substrates complicates interpretation, and the available evidence does not rule out more parsimonious alternatives. We argue that existing evidence better comports with a view of Bam as an OM-adjacent Anfinsen cage that positions client proteins for OM insertion, prevents aggregation, degradation, and off-pathway misfolding, and accelerates the native OMP folding reaction. Numerous studies have established the role of lipid bilayer defects in the acceleration of β-barrel folding kinetics, and BamA has been shown to give rise to such defects (36, 37, 39–41, 103). These observations inform a simple model in which Bam complexes effectively localize client proteins to OM “entry points” generated by the local defects imposed by BamA itself, removing the primary barrier to rapid OMP folding and enabling OM biogenesis on physiologically relevant timescales (15, 36, 90, 104).
The importance of the BamA lateral gate is a matter of ongoing debate. Artificial locking of this seam is lethal in vivo (34, 105) and moderately impairs OmpT folding kinetics into proteoliposomes (38), but it has no effect on the in vitro assembly of OmpA or OmpX, both small OMPs with minimal loop structure (36, 41). This incongruity might be reconciled if certain Bam substrates do not require opening of the gate whereas others do, such as those with large barrels or extensive hydrophilic extracellular loops (45, 106), which could avoid the entropic penalty associated with membrane translocation by traversing the OM through the hydrophilic lumen of the open BamA barrel, with the gate serving as a transient “slit” allowing passage of loops attached to transmembrane β-strands that are integrating into the lipid phase. This bears some resemblance to the mechanism of polytopic IM protein assembly by the SecYEG insertase/translocase, in which transmembrane segments diffuse into the membrane adjacent to a lateral gate and periplasmic loops are translocated through the activated, ungated SecYEG pore (107–109). However, it is also clear that specific loops from certain substrates are buried within the lumen of the barrel during folding, potentially scaffolding barrel formation and driving the maturation process, likely rendering an assisted loop translocation process dispensable.
The requirement for both rapid OM growth and a reliably impermeable OM represents an intriguing paradox and raises the possibility that local defects induced by BamA form only when OMP insertion is imminent, as a constitutively “open” complex would likely generate membrane instability and allow indiscriminate diffusion across the OM. This paradox implies tight regulation of Bam activity and a concerted mechanism for Bam activation that is linked to substrate recognition (14, 16, 48, 62). Consistent with this notion, there is mounting evidence that the BamA barrel exists in equilibrium between two conformations: a “closed” state in which extracellular loops form a dome that stabilizes a thermostable and complete β-barrel, and an “open” state in which extracellular loops reorient and the N-terminal β-strands of the barrel are dramatically wrenched outward, begetting a thermolabile, incomplete β-barrel with an exposed aqueous pore (21, 34, 37). This equilibrium can apparently be altered through mutations in L6 that prevent loop latching and destabilize the barrel (42, 43, 110), or through mutations in the BamCDE subcomplex that are presumed to slow the restoration of the closed conformation following a round of OMP assembly (61). Together with observations linking barrel-proximal POTRA 5, lipoproteins, and L6 to conformational dynamics within the BamA barrel (14, 38, 44, 61, 62, 64, 65, 111, 112), these findings extend the mechanistic model to include a role for nascent OMPs as homotropic allosteric activators of Bam that initiate an activation cascade resulting in transient opening of the BamA barrel and concomitant OM integration of folding OMPs.
A MODEL FOR BamA-ASSISTED β-BARREL FOLDING
In light of the emerging mechanistic details reviewed above, the in vivo β-barrel assembly process can be summarized as follows (Fig. 3). Unfolded OMPs are transferred from periplasmic chaperones to Bam, where they are scaffolded through direct interaction with the POTRAs and one or more Bam lipoproteins (18, 23, 24, 26). β-strands of substrate OMPs may be organized circumferentially according to their relative positions in the final folded β-barrel structure (Fig. 2, item b) (18, 113–115). Recognition of the β-signal by BamD triggers a conformational rearrangement in the complex that includes rotation of the POTRAs away from the BamA barrel lumen and/or adoption of an open barrel conformation (14, 21, 34). Local perturbation of the OM, induced by BamA and exacerbated by conformational dynamics within the activated β-barrel domain, allows rapid, spontaneous folding and simultaneous OM insertion of OMP substrates along the native folding pathway, with long, hydrophilic loops potentially threaded through the BamA barrel lumen during OMP folding via the lateral slit. Although the precise translocation path of BamA substrates remains undefined, the orientation of the periplasmic ring-like Bam apparatus relative to the OM raises the possibility that substrates are funneled through a proteinaceous aperture adjacent to the BamA barrel that could serve as both a substrate exit channel and a site of OM insertion (24). Following substrate release, the closed conformation of the BamA barrel is restored through conformational dynamics among the Bam lipoproteins and BamA, priming the complex for an ensuing round of assembly (61, 116). We anticipate that the availability of in vitro reconstitution systems, comprehensive panels of informative mutants, high-resolution imaging techniques, sensitive biophysical assays and models, and an abundance of structural data will enable a detailed analysis of the Bam catalytic mechanism and the potential variations on the generic theme presented here for diverse OMP substrates.
Figure 3.

Proposed Bam-assisted folding model. (a) Nascent OMPs (red), maintained in a folding-competent state by periplasmic chaperones (green), are transferred to the Bam complex (blue). (b) Client proteins associate with multiple epitopes on Bam, potentially stimulating formation of early β-structure and orienting circularly-arranged β-strands/hairpins toward the presumptive substrate exit pore. Recognition of conserved OMP motifs triggers a conformational change in BamA that exposes the barrel lumen and destabilizes the lateral gate, further perturbing the local membrane environment and generating an OM integration path for OMP substrates. (c) The Bam complex prevents aggregation, protects substrates from proteolysis, and lowers the kinetic barrier to OM integration to enable rapid OMP folding along the native pathway. (d) OMPs spontaneously fold into the locally destabilized membrane, with the exposed BamA lumen potentially accommodating the folding barrel and/or secreted extracellular domains of client proteins. (e) Release of substrate from the complex prompts restoration of the closed, inert state of the complex to enable an ensuing round of assembly.
ACKNOWLEDGMENTS
The authors thank fellow lab members for suggestions and critical reading of the manuscript. T.J.S. was supported by a grant from NIGMS (R35GM118024).
Contributor Information
Dante P. Ricci, Department of Early Research, Achaogen, Inc., South San Francisco, CA 94080
Thomas J. Silhavy, Department of Molecular Biology, Princeton University, Princeton, NJ 08544
Maria Sandkvist, Department of Microbiology and Immunology, University of Michigan, Ann Arbor, Michigan.
Eric Cascales, CNRS Aix-Marseille Université, Mediterranean Institute of Microbiology, Marseille, France.
Peter J. Christie, Department of Microbiology and Molecular Genetics, McGovern Medical School, Houston, Texas
REFERENCES
- 1.Konovalova A, Kahne DE, Silhavy TJ. 2017. Outer membrane biogenesis. Annu Rev Microbiol 71:539–556. 10.1146/annurev-micro-090816-093754. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okuda S, Freinkman E, Kahne D. 2012. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338:1214–1217. 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D. 2018. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359:798–801. 10.1126/science.aar1886. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botos I, Noinaj N, Buchanan SK. 2017. Insertion of proteins and lipopolysaccharide into the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 372:20160224. 10.1098/rstb.2016.0224. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC,Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. 10.1073/pnas.1601375113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. 10.1073/pnas.0903229106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. 10.1038/s41564-017-0046-x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Chong Z-S, Woo W-F, Chng S-S. 2015. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol 98:1133–1146. 10.1111/mmi.13202. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Powers MJ, Trent MS. 2018. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci U S A 115:E8518–E8527. 10.1073/pnas.1806714115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285.e17. 10.1016/j.cell.2017.03.019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. 10.1146/annurev-micro-090110-102859. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Ranava D, Caumont-Sarcos A, Albenne C, Ieva R. 2018. Bacterial machineries for the assembly of membrane-embedded β-barrel proteins. FEMS Microbiol Lett 365:fny087. 10.1093/femsle/fny087. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Anwari K, Webb CT, Poggio S, Perry AJ, Belousoff M, Celik N, Ramm G, Lovering A, Sockett RE, Smit J, Jacobs-Wagner C, Lithgow T. 2012. The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol Microbiol 84:832–844. 10.1111/j.1365-2958.2012.08059.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, Kahne D. 2018. Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc Natl Acad Sci U S A 115:2359–2364. 10.1073/pnas.1711727115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. 2016. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci U S A 113:8717–8722. 10.1073/pnas.1604100113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagan CL, Wzorek JS, Kahne D. 2015. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A 112:2011–2016. 10.1073/pnas.1415955112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagan CL, Westwood DB, Kahne D. 2013. Bam lipoproteins assemble BamA in vitro. Biochemistry 52:6108–6113. 10.1021/bi400865z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ieva R, Tian P, Peterson JH, Bernstein HD. 2011. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci U S A 108:E383–E391. 10.1073/pnas.1103827108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vuong P, Bennion D, Mantei J, Frost D, Misra R. 2008. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol 190:1507–1517. 10.1128/JB.01477-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. 10.1111/j.1365-2958.2006.05211.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Bakelar J, Buchanan SK, Noinaj N. 2016. The structure of the β-barrel assembly machinery complex. Science 351:180–186. 10.1126/science.aad3460. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Zhan LH, Hou HF, Gao ZQ, Xu JH, Dong C, Dong YH. 2016. Structural basis for the interaction of BamB with the POTRA3-4 domains of BamA. Acta Crystallogr D Struct Biol 72:236–244. 10.1107/S2059798315024729. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Bergal HT, Hopkins AH, Metzner SI, Sousa MC. 2016. The structure of a BamA-BamD fusion illuminates the architecture of the β-barrel assembly machine core. Structure 24:243–251. 10.1016/j.str.2015.10.030. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. 2016. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196. 10.1038/nsmb.3181. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Jansen KB, Baker SL, Sousa MC. 2015. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine. J Biol Chem 290:2126–2136. 10.1074/jbc.M114.584524. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuck A, Schleiffer A, Clausen T. 2011. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol 406:659–666. 10.1016/j.jmb.2011.01.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol 77:1153–1171. 10.1111/j.1365-2958.2010.07280.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci DP, Schwalm J, Gonzales-Cope M, Silhavy TJ. 2013. The activity and specificity of the outer membrane protein chaperone SurA are modulated by a proline isomerase domain. mBio 4:e00540-13. 10.1128/mBio.00540-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel GJ, Kleinschmidt JH. 2013. The lipid bilayer-inserted membrane protein BamA of Escherichia coli facilitates insertion and folding of outer membrane protein A from its complex with Skp. Biochemistry 52:3974–3986. 10.1021/bi400103t. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Narita S, Masui C, Suzuki T, Dohmae N, Akiyama Y. 2013. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 110:E3612–E3621. 10.1073/pnas.1312012110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. 2010. Structure and flexibility of the complete periplasmic domain of BamA:the protein insertion machine of the outer membrane. Structure 18:1492–1501. 10.1016/j.str.2010.08.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964. 10.1126/science.1143993. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Remaut H, Waksman G. 2006. Protein-protein interaction through beta-strand addition. Trends Biochem Sci 31:436–444. 10.1016/j.tibs.2006.06.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. 2016. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69. 10.1038/nature17199. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Lundquist K, Bakelar J, Noinaj N, Gumbart JC. 2018. C-terminal kink formation is required for lateral gating in BamA. Proc Natl Acad Sci U S A 115:E7942–E7949. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doerner PA, Sousa MC. 2017. Extreme dynamics in the BamA β-barrel seam. Biochemistry 56:3142–3149. 10.1021/acs.biochem.7b00281. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501:385–390. 10.1038/nature12521. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. 2016. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865. 10.1038/ncomms12865. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danoff EJ, Fleming KG. 2015. Membrane defects accelerate outer membrane β-barrel protein folding. Biochemistry 54:97–99. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. 2014. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci U S A 111:5878–5883. 10.1073/pnas.1322473111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiffrin B, Calabrese AN, Higgins AJ, Humes JR, Ashcroft AE, Kalli AC, Brockwell DJ, Radford SE. 2017. Effects of periplasmic chaperones and membrane thickness on bama-catalyzed outer-membrane protein folding. J Mol Biol 429:3776–3792. 10.1016/j.jmb.2017.09.008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonard-Rivera M, Misra R. 2012. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol 194:4662–4668. 10.1128/JB.00825-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwyer RS, Ricci DP, Colwell LJ, Silhavy TJ, Wingreen NS. 2013. Predicting functionally informative mutations in Escherichia coli BamA using evolutionary covariance analysis. Genetics 195:443–455. 10.1534/genetics.113.155861. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoma J, Sun Y, Ritzmann N, Müller DJ. 2018. POTRA domains, extracellular lid, and membrane composition modulate the conformational stability of the β barrel assembly factor BamA. Structure 26:987–996.e3. 10.1016/j.str.2018.04.017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Wzorek JS, Lee J, Tomasek D, Hagan CL, Kahne DE. 2017. Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. Proc Natl Acad Sci U S A 114:2598–2603. 10.1073/pnas.1616576114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartmann J-B, Zahn M, Burmann IM, Bibow S, Hiller S. 2018.Sequence-specific solution NMR assignments of the β-barrel insertase BamA to monitor its conformational ensemble at the atomic level. J Am Chem Soc 140:11252–11260. 10.1021/jacs.8b03220. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Misra R, Stikeleather R, Gabriele R. 2015. In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the β-barrel assembly machine of Escherichia coli. J Mol Biol 427:1061–1074. 10.1016/j.jmb.2014.04.021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. 2012. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109:3487–3491. 10.1073/pnas.1201362109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikora AE, Wierzbicki IH, Zielke RA, Ryner RF, Korotkov KV, Buchanan SK, Noinaj N. 2018. Structural and functional insights into the role of BamD and BamE within the β-barrel assembly machinery in Neisseria gonorrhoeae. J Biol Chem 293:1106–1119. 10.1074/jbc.RA117.000437. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahoney TF, Ricci DP, Silhavy TJ. 2016. Classifying β-barrel assembly substrates by manipulating essential Bam complex members. J Bacteriol 198:1984–1992. 10.1128/JB.00263-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossiter AE, Leyton DL, Tveen-Jensen K, Browning DF, Sevastsyanovich Y, Knowles TJ, Nichols KB, Cunningham AF, Overduin M, Schembri MA, Henderson IR. 2011. The essential β-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J Bacteriol 193:4250–4253. 10.1128/JB.00192-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N. 2008. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 132:1011–1024. 10.1016/j.cell.2008.01.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D. 2009. Signals in bacterial beta-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci U S A 106:2531–2536. 10.1073/pnas.0807830106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iyer BR, Zadafiya P, Vetal PV, Mahalakshmi R. 2017. Energetics of side-chain partitioning of β-signal residues in unassisted folding of a transmembrane β-barrel protein. J Biol Chem 292:12351–12365. 10.1074/jbc.M117.789446. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paramasivam N, Habeck M, Linke D. 2012. Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not? BMC Genomics 13:510. 10.1186/1471-2164-13-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozjak-Pavlovic V, Ott C, Götz M, Rudel T. 2011. Neisserial Omp85 protein is selectively recognized and assembled into functional complexes in the outer membrane of human mitochondria. J Biol Chem 286:27019–27026. 10.1074/jbc.M111.232249. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller JEN, Papic D, Ulrich T, Grin I, Schütz M, Oberhettinger P, Tommassen J, Linke D, Dimmer KS, Autenrieth IB, Rapaport D. 2011. Mitochondria can recognize and assemble fragments of a beta-barrel structure. Mol Biol Cell 22:1638–1647. 10.1091/mbc.e10-12-0943. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4:e377. 10.1371/journal.pbio.0040377. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albrecht R, Zeth K. 2011. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem 286:27792–27803. 10.1074/jbc.M111.238931. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. 2011. Crystal structure of BamD: an essential component of the β-barrel assembly machinery of gram-negative bacteria. J Mol Biol 409:348–357. 10.1016/j.jmb.2011.03.035. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rigel NW, Ricci DP, Silhavy TJ. 2013. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc Natl Acad Sci U S A 110:5151–5156. 10.1073/pnas.1302662110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCabe AL, Ricci D, Adetunji M, Silhavy TJ. 2017. Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J Bacteriol 199:e00373-17. 10.1128/JB.00373-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb CT, Heinz E, Lithgow T. 2012. Evolution of the β-barrel assembly machinery. Trends Microbiol 20:612–620. 10.1016/j.tim.2012.08.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Tellez R, Jr, Misra R. 2012. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J Bacteriol 194:317–324. 10.1128/JB.06192-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. 2012. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol 194:1002–1008. 10.1128/JB.06426-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 104:6400–6405. 10.1073/pnas.0701579104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim KH, Kang H-S, Okon M, Escobar-Cabrera E, McIntosh LP, Paetzel M. 2011. Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry 50:1081–1090. 10.1021/bi101659u. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. 2007. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol 189:446–454. 10.1128/JB.01103-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charlson ES, Werner JN, Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol 188:7186–7194. 10.1128/JB.00571-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317. 10.1016/j.cell.2005.02.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Hsieh P-F, Hsu C-R, Chen C-T, Lin T-L, Wang J-T. 2016. The Klebsiella pneumoniae YfgL (BamB) lipoprotein contributes to outer membrane protein biogenesis, type-1 fimbriae expression, anti-phagocytosis, and in vivo virulence. Virulence 7:587–601. 10.1080/21505594.2016.1171435. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892. 10.1126/science.1188919. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palomino C, Marín E, Fernández LÁ. 2011. The fimbrial usher FimD follows the SurA-BamB pathway for its assembly in the outer membrane of Escherichia coli. J Bacteriol 193:5222–5230. 10.1128/JB.05585-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwalm J, Mahoney TF, Soltes GR, Silhavy TJ. 2013. Role for Skp in LptD assembly in Escherichia coli. J Bacteriol 195:3734–3742. 10.1128/JB.00431-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunasinghe SD, Shiota T, Stubenrauch CJ, Schulze KE, Webb CT, Fulcher AJ, Dunstan RA, Hay ID, Naderer T, Whelan DR, Bell TDM, Elgass KD, Strugnell RA, Lithgow T. 2018. The WD40 protein BamB mediates coupling of BAM complexes into assembly precincts in the bacterial outer membrane. Cell Rep 23:2782–2794. 10.1016/j.celrep.2018.04.093. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Huysmans GHM, Radford SE, Brockwell DJ, Baldwin SA. 2007. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP. J Mol Biol 373:529–540. 10.1016/j.jmb.2007.07.072. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hou VC, Moe GR, Raad Z, Wuorimaa T, Granoff DM. 2003. Conformational epitopes recognized by protective anti-neisserial surface protein A antibodies. Infect Immun 71:6844–6849. 10.1128/IAI.71.12.6844-6849.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaturvedi D, Mahalakshmi R. 2013. Methionine mutations of outer membrane protein X influence structural stability and beta-barrel unfolding. PLoS One 8:e79351. 10.1371/journal.pone.0079351. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. 1999. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry 38:5006–5016. 10.1021/bi982465w. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Johansson MU, Alioth S, Hu K, Walser R, Koebnik R, Pervushin K. 2007. A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance. Biochemistry 46:1128–1140. 10.1021/bi061265e. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Surrey T, Schmid A, Jähnig F. 1996. Folding and membrane insertion of the trimeric β-barrel protein OmpF. Biochemistry 35:2283–2288. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Jansen C, Wiese A, Reubsaet L, Dekker N, de Cock H, Seydel U, Tommassen J. 2000. Biochemical and biophysical characterization of in vitro folded outer membrane porin PorA of Neisseria meningitidis. Biochim Biophys Acta 1464:284–298. 10.1016/S0005-2736(00)00155-3. [DOI] [PubMed] [Google Scholar]
- 83.de Cock H, Hendriks R, de Vrije T, Tommassen J. 1990. Assembly of an in vitro synthesized Escherichia coli outer membrane porin into its stable trimeric configuration. J Biol Chem 265:4646–4651. [PubMed] [PubMed] [Google Scholar]
- 84.Otzen DE, Andersen KK. 2013. Folding of outer membrane proteins. Arch Biochem Biophys 531:34–43. 10.1016/j.abb.2012.10.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Conlan S, Bayley H. 2003. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry 42:9453–9465. 10.1021/bi0344228. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Mogensen JE, Kleinschmidt JH, Schmidt MA, Otzen DE. 2005. Misfolding of a bacterial autotransporter. Protein Sci 14:2814–2827. 10.1110/ps.051628705. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kramer RA, Zandwijken D, Egmond MR, Dekker N. 2000. In vitro folding, purification and characterization of Escherichia coli outer membrane protease ompT. Eur J Biochem 267:885–893. 10.1046/j.1432-1327.2000.01073.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Pocanschi CL, Apell H-J, Puntervoll P, Høgh B, Jensen HB, Welte W, Kleinschmidt JH. 2006. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J Mol Biol 355:548–561. 10.1016/j.jmb.2005.10.060. [PubMed] [DOI] [PubMed] [Google Scholar]
- 89.Dekker N, Merck K, Tommassen J, Verheij HM. 1995. In vitro folding of Escherichia coli outer-membrane phospholipase A. Eur J Biochem 232:214–219. 10.1111/j.1432-1033.1995.tb20801.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Fleming KG. 2015. A combined kinetic push and thermodynamic pull as driving forces for outer membrane protein sorting and folding in bacteria. Philos Trans R Soc Lond B Biol Sci 370:20150026. 10.1098/rstb.2015.0026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peterson JH, Plummer AM, Fleming KG, Bernstein HD. 2017. Selective pressure for rapid membrane integration constrains the sequence of bacterial outer membrane proteins. Mol Microbiol 106:777–792. 10.1111/mmi.13845. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grabowicz M, Koren D, Silhavy TJ. 2016. The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. mBio 7:e00312-16. 10.1128/mBio.00312-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maurya SR, Chaturvedi D, Mahalakshmi R. 2013. Modulating lipid dynamics and membrane fluidity to drive rapid folding of a transmembrane barrel. Sci Rep 3:1989. 10.1038/srep01989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Horne JE, Radford SE. 2016. A growing toolbox of techniques for studying β-barrel outer membrane protein folding and biogenesis. Biochem Soc Trans 44:802–809. 10.1042/BST20160020. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleinschmidt JH. 2015. Folding of β-barrel membrane proteins in lipid bilayers—unassisted and assisted folding and insertion. Biochim Biophys Acta 1848:1927–1943. 10.1016/j.bbamem.2015.05.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Danoff EJ, Fleming KG. 2017. Novel kinetic intermediates populated along the folding pathway of the transmembrane β-barrel OmpA. Biochemistry 56:47–60. 10.1021/acs.biochem.6b00809. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huysmans GHM, Baldwin SA, Brockwell DJ, Radford SE. 2010. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A 107:4099–4104. 10.1073/pnas.0911904107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tamm LK, Hong H, Liang B. 2004. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta 1666:250–263. 10.1016/j.bbamem.2004.06.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Thoma J, Bosshart P, Pfreundschuh M, Müller DJ. 2012. Out but not in: the large transmembrane β-barrel protein FhuA unfolds but cannot refold via β-hairpins. Structure 20:2185–2190. 10.1016/j.str.2012.10.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Höhr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N, Wiedemann N. 2018. Membrane protein insertion through a mitochondrial β-barrel gate. Science 359:eaah6834. 10.1126/science.aah6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schiffrin B, Brockwell DJ, Radford SE. 2017. Outer membrane protein folding from an energy landscape perspective. BMC Biol 15:123. 10.1186/s12915-017-0464-5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Plummer AM, Fleming KG. 2016. From chaperones to the membrane with a BAM! Trends Biochem Sci 41:872–882. 10.1016/j.tibs.2016.06.005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fleming PJ, Patel DS, Wu EL, Qi Y, Yeom MS, Sousa MC, Fleming KG, Im W. 2016. BamA POTRA domain interacts with a native lipid membrane surface. Biophys J 110:2698–2709. 10.1016/j.bpj.2016.05.010. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Michalik M, Orwick-Rydmark M, Habeck M, Alva V, Arnold T, Linke D. 2017. An evolutionarily conserved glycine-tyrosine motif forms a folding core in outer membrane proteins. PLoS One 12:e0182016. 10.1371/journal.pone.0182016. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22:1055–1062. 10.1016/j.str.2014.05.008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hagan CL, Silhavy TJ, Kahne D. 2011. β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem 80:189–210. 10.1146/annurev-biochem-061408-144611. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Botte M, Zaccai NR, Nijeholt JLÀ, Martin R, Knoops K, Papai G, Zou J, Deniaud A, Karuppasamy M, Jiang Q, Roy AS, Schulten K, Schultz P, Rappsilber J, Zaccai G, Berger I, Collinson I, Schaffitzel C. 2016. A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci Rep 6:38399. 10.1038/srep38399. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li L, Park E, Ling J, Ingram J, Ploegh H, Rapoport TA. 2016. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531:395–399. 10.1038/nature17163. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dalbey RE, Wang P, Kuhn A. 2011. Assembly of bacterial inner membrane proteins. Annu Rev Biochem 80:161–187. 10.1146/annurev-biochem-060409-092524. [PubMed] [DOI] [PubMed] [Google Scholar]
- 110.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y. 2014. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 28:2677–2685. 10.1096/fj.13-248450. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Noinaj N, Gumbart JC, Buchanan SK. 2017. The β-barrel assembly machinery in motion. Nat Rev Microbiol 15:197–204. 10.1038/nrmicro.2016.191. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sinnige T, Weingarth M, Daniëls M, Boelens R, Bonvin AMJJ, Houben K, Baldus M. 2015. Conformational plasticity of the POTRA 5 domain in the outer membrane protein assembly factor BamA. Structure 23:1317–1324. 10.1016/j.str.2015.04.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Albenne C, Ieva R. 2017. Job contenders: roles of the β-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol Microbiol 106:505–517. 10.1111/mmi.13832. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Pavlova O, Peterson JH, Ieva R, Bernstein HD. 2013. Mechanistic link between β barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A 110:E938–E947. 10.1073/pnas.1219076110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ieva R, Skillman KM, Bernstein HD. 2008. Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol 67:188–201. 10.1111/j.1365-2958.2007.06048.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Plummer AM, Fleming KG. 2015. BamA alone accelerates outer membrane protein folding in vitro through a catalytic mechanism. Biochemistry 54:6009–6011. 10.1021/acs.biochem.5b00950. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


