Abstract
Urban living in modern large cities has significant adverse effects on health, increasing the risk of several chronic diseases. We focus on the two leading clusters of chronic disease, heart disease and diabetes, and develop data-driven methods to predict hospitalizations due to these conditions. We base these predictions on the patients’ medical history, recent and more distant, as described in their Electronic Health Records (EHR). We formulate the prediction problem as a binary classification problem and consider a variety of machine learning methods, including kernelized and sparse Support Vector Machines (SVM), sparse logistic regression, and random forests. To strike a balance between accuracy and interpretability of the prediction, which is important in a medical setting, we propose two novel methods: K-LRT, a likelihood ratio test-based method, and a Joint Clustering and Classification (JCC) method which identifies hidden patient clusters and adapts classifiers to each cluster. We develop theoretical out-of-sample guarantees for the latter method. We validate our algorithms on large datasets from the Boston Medical Center, the largest safety-net hospital system in New England.
Index Terms: Machine learning, Heart disease, Diabetes, Predictive analytics, Electronic Health Records, Smart health, Smart city
I. Introduction
Living in modern large cities is impacting our health in many different ways [1]. Primarily due to: (i) stress associated with fast-paced urban life, (ii) a sedentary lifestyle due to work conditions and lack of time, (iii) air pollution, and (iv) a disproportionate number of people living in poverty, urban populations face an increased risk for the development of chronic health conditions [2]. For example, according to the World Health Organization [3], ambient (outdoor air) pollution was estimated in 2012 to cause 3 million premature deaths worldwide per year; this mortality is due to exposure to small particulate matter of 10 microns or less in diameter (PM10), which cause cardiovascular, respiratory disease, and cancers. In fact, the vast majority (about 72%) of these air pollution-related premature deaths were due to ischemic heart disease and strokes.
There is an increasing percentage of the world population facing the adverse health effects of urban living. Specifically, according to the United Nations [4], 54% of the earth’s population resides in urban areas, a percentage which is expected to reach 66% by 2050. It becomes evident that the health of citizens should become an important priority in the emerging smart city agenda [5]. To that end, smart health care –“smart health” as it has been called– involves the use of ehealth and mhealth systems, intelligent and connected medical devices, and the implementation of policies that encourage health, wellness, and well-being [6]. It is estimated that by 2020 the smart city market will be worth about $1.5 trillion, with smart health corresponding to 15% of that amount [6]. Additional potential actions smart cities can adopt include ways to improve city life, reduce congestion and air pollution levels, discourage the use of tobacco products and foods high in fat and sugar which increase the risk of chronic diseases, and improve access to health care. Without overlooking the importance of all these population-level measures, our work aims at enabling personalized interventions using an algorithmic data-driven approach.
Through smart health, smart cities and governments aim at improving the quality of life of their citizens. In the state of Massachusetts, the MassHealth program –a combination of Medicaid and the Children’s Health Insurance Program–provides health insurance for 1.9 million Massachusetts residents, children in low-income households, low-wage workers, elders in nursing homes, people with disabilities, and others with very low incomes who cannot afford insurance [7], [8]. The state’s fiscal year 2018 budget includes approximately $16.6 billion for MassHealth, which is around 37% of the total state budget [8]. Clearly, this is a substantial share of the budget. Consequently, if health care costs can be lowered through smart health, more resources will become available for many other services smart cities can offer. Conversely, if other aspects of smart cities can be improved, the adverse health effects of urban living can be reduced, thus lowering health care costs. This suggests a beneficial feedback loop involving smart health and non-health-related smart city research.
Health care is also, unquestionably, an important national and global economic issue. In 2013, the United States (U.S.) spent about $3 trillion on health care, which exceeded 17% of its GDP [9]. The World Health Organization estimates that healthcare costs will grow to 20% of the U.S. GDP (nearly $5 trillion) by 2021 [10], especially with civilization diseases (or else called lifestyle diseases), like diabetes, coronary heart disease and obesity, growing.
Our goal in this paper is to explore and develop predictive analytics aiming at predicting hospitalizations due to the two leading chronic diseases: heart disease and diabetes. Prediction, naturally, is an important first step towards prevention. It allows health systems to target individuals most in need and to use (limited) health resources more effectively. We refer to [11] for a general discussion of the benefits, and some risks, associated with the use of health analytics. We seek to predict hospitalizations based on the patients’ Electronic Health Records (EHR) within a year from the time we examine the EHR, so as to allow enough lead time for prevention. What is also critical is that our methods provide an interpretation (or explanation) of the predictions. Interpretability will boost the confidence of patients and physicians in the results, hence, the chance they will act based on the predictions, and provide insight into potential preventive measures. It is interesting that interpretability is being increasingly recognized as important; for instance, recent European Union legislation [12] will enforce a citizen’s right to receive an explanation for algorithmic decisions.
Our focus on heart disease and diabetes is deliberate. Diseases of the heart have been consistently among the top causes of death. In the U.S., heart disease is yearly the cause of one in every four deaths, which translates to 610,000 people [13]. At the same time, diabetes is recognized as the world’s fastest growing chronic condition [14]. One in eleven adults has diabetes worldwide (415 million) and 12% of global health expenditures is spent on diabetes ($673 billion) [15]. In the U.S. alone, 29.1 million people or 9.3% of the population had diabetes in 2012 [16].
Our interest in hospitalizations is motivated by [17], which found that nearly $30.8 billion in hospital care cost during 2006 was preventable. Heart diseases and diabetes were the leading contributors accounting, correspondingly, for more than $9 billion, or about 31%, and for almost $6 billion, or about 20%. Clearly, even modest percentage reductions in these amounts matter.
An important enabler of our work is the increasing availability of patients’ EHRs. The digitization of patients’ records started more than two decades ago. Widespread adoption of EHRs has generated massive datasets. 87% of U.S. office-based physicians were using EHRs by the end of 2015, up from 42% in 2008 [18]. EHRs have found diverse uses [19], e.g., in assisting hospital quality management [20], in detecting adverse drug reactions [21], and in general primary care [22].
A. Contributions and Organization
Our algorithmic approach towards predicting chronic disease hospitalizations employs a variety of methods, both already well-established, as well as novel methods we introduce, tailored to solve the specific medical problem. We formulate the problem as a binary classification problem and seek to differentiate between patients that will be hospitalized in a target year and those who will not. We review related work in Section II. Section III explores baseline methods that separate the two classes of samples (patients) using a single classifier. We evaluate their performance in terms of prediction accuracy and interpretability of the model and the results. Baseline methods include linear and kernelized Support Vector Machines (SVM), random forests, and logistic regression. We also develop a novel likelihood ratio-based method, K-LRT, that identifies the K most significant features for each patient that lead to hospitalization. Surprisingly, this method, under a small value of K, performs not substantially worse than more sophisticated classifiers using all available features. This suggests that in our setting, a sparse classifier employing a handful of features can be very effective. What is more challenging is that the “discriminative” features are not necessarily the same for each patient.
Motivated by the success of sparse classifiers, in Section IV we seek to jointly identify clusters of patients who share the same set of discriminative features and, at the same time, develop per-cluster sparse classifiers using these features. Training such classifiers amounts to solving a non-convex optimization problem. We formulate it as an integer programming problem; which limits its use to rather smaller instances (training sets). To handle much larger instances we develop a local optimization approach based on alternating optimization. We establish the convergence of this local method and bound its Vapnik-Chervonenkis (VC) dimension; the latter bound leads to out-of-sample generalization guarantees.
In Section V, we provide a detailed description of the two datasets we use to evaluate the performance of the various algorithms. One dataset concerns patients with heart-related diseases and the other, patients with diabetes. The data have been made available to us from the Boston Medical Center (BMC) – the largest safety-net hospital in New England. We define the performance metrics we use in Section VI. We report and discuss our experimental settings and results in Section VII and we present our conclusions in Section VIII.
Notation
All vectors are column vectors. For economy of space, we write x = (x1, …, xdim(x)) to denote the column vector x, where dim(x) is the dimension of x. We use 0 and 1 for the vectors with all entries equal to zero and one, respectively. We denote by ℝ+ the set of all nonnegative real numbers. M ≥ 0 (resp., x ≥ 0) indicates that all entries of a matrix M (resp., vector x) are nonnegative. We use “prime” to denote the transpose of a matrix or vector and |𝒟| the cardinality of a set 𝒟. Unless otherwise specified, ‖·‖ denotes the ℓ2 norm and ‖·‖1 the ℓ1 norm.
II. Related Work
To the best of our knowledge, the problem of chronic disease hospitalization prediction using machine learning methods is novel. A closely related problem, which has received a lot of attention in the literature, is the re-hospitalization prediction, since around 20% of all hospital admissions occur within 30 days of a previous discharge. Medicare penalizes hospitals that have high rates of readmissions for some specific conditions that now include patients with heart failure, heart attack, and pneumonia. Examples of work on this problem include [23], [24], [25] and [26].
Other related problems considered in the literature are: predicting the onset of diabetes using artificial neural networks [27]; developing an intelligent system that predicts, using data-mining techniques, which patients are likely to be diagnosed with heart disease [28]; and using data-mining techniques to predict length of stay for cardiac patients (employing decision trees, SVM, and artificial neural networks) [29], or for acute pancreatitis (using artificial neural networks) [30].
We should also mention the Heritage Health Prize, a competition by Kaggle, whose goal was to predict the length of stay for patients who will be admitted to a hospital within the next year, using insurance claims data and data-mining techniques [31].
III. Baseline Methods and K-LRT
In this section we outline several baseline classification methods we use to predict whether patients will be hospitalized in a target year, given their medical history.
In medical applications, accuracy is important, but also interpretability of the predictions is indispensable [32], strengthening the confidence of medical professionals in the results. Sparse classifiers are interpretable, since they provide succinct information on few dominant features leading to the prediction [33]. Moreover, medical datasets are often imbalanced since there are much fewer patients with a condition (e.g., hospitalized) vs. “healthy” individuals (non-hospitalized). This makes it harder for supervised learning methods to learn since a training set may be dominated by negative (non-hospitalized) class samples. Sparsity, therefore, is useful in this context because there are fewer parameters in the classifier one needs to learn. In this light, we experiment with sparse versions of various classification methods and show their advantages. While harder to interpret than linear and sparse algorithms, ensemble methods that build collections of classifiers, such as random forests, can model nonlinear relationships and have been proven to provide very accurate models for common health care problems [34], including the one we study in this paper.
The last method we present in this section is an adaptation of a likelihood ratio test, designed to induce sparsity of the features used to make a prediction. All but the last method fall into the category of discriminative learning algorithms, while the last one is a generative algorithm. Discriminative algorithms directly partition the input space into label regions without modeling how the data are generated, while generative algorithms assume a model that generates the data, estimate the model’s parameters and use it to make classification decisions. Our experiment results show that discriminative methods are likely to give higher accuracy, but generative methods provide more interpretable models and results [35], [36]. This is the reason we experiment with methods from both families and the trade-off between accuracy and interpretability is observed in our results.
A. Radial Basis Function (RBF), Linear & Sparse Linear SVM
An SVM is an efficient binary classifier [37]. The SVM training algorithm seeks a separating hyperplane in the feature space, so that data points from the two different classes reside on different sides of that hyperplane. We can calculate the distance of each input data point from the hyperplane. The minimum over all these distances is called margin. The goal of SVM is to find the hyperplane that has the maximum margin. In many cases, however, data points are neither linearly nor perfectly separable. So called soft-margin SVM, tolerates misclassification errors and can leverage kernel functions to “elevate” the features into a higher dimensional space where linear separability is possible (kernelized SVMs) [37].
Given our interest in interpretable, hence sparse, classifiers we formulate a Sparse version of Linear SVM (SLSVM) as follows. We are given training data xi ∈ ℝD and labels yi ∈ {−1, 1}, i = 1, …, n, where xi is the vector of features for the ith patient and yi = 1 (resp., yi = −1) indicates that the patient will (resp., not) be hospitalized. We seek to find the classifier (β, β0), β ∈ ℝD, β0 ∈ ℝ, by solving:
| (1) |
where ξi is a misclassification penalty. The first term in the objective has the effect of maximizing the margin. The second objective term minimizes the total misclassification penalty. The last term, ‖β‖1, in the objective, imposes sparsity in the feature vector β, thus allowing only a sparse subset of features to contribute to the classification decision. The parameters C and ρ are tunable parameters that control the relative importance of the misclassification and the sparsity terms, respectively, compared to each other and, also, the margin term. When ρ = 0, the above formulation yields a standard linear SVM classifier.
A linear SVM finds a linear hyperplane in the feature space and can not handle well cases where a nonlinear separating surface between classes is more appropriate. To that end, kernel functions are being used that map the features to a higher dimensional space where a linear hyperplane would be applicable. In the absence of the sparse-inducing ℓ1-norm term, kernelized SVMs use as a kernel for some feature mapping function ϕ and solve an optimization problem that is based on the dual problem to (1) to find an optimal (β, β0). In our application, we will employ the widely used Radial Basis Function (RBF) K(xi, xj) = exp(−‖xi − xj‖2/2σ2) [38] as the kernel function in our experiments.
B. Random Forests
Bagging (or bootstrap aggregating) is a technique for reducing the variance of an estimated predictor by averaging many noisy but approximately unbiased models. A random forest is an ensemble of de-correlated trees [39]. Each decision tree is formed using a training set obtained by sampling (with replacement) a random subset of the original data. While growing each decision tree, random forests use a random subset of the set of features (variables) at each node split. Essentially, the algorithm uses bagging for both trees and features. Each tree is fully grown until a minimum size is reached, i.e., there is no pruning. While the predictions of a single tree are highly sensitive to noise in its training set, the average of many trees is not, as long as the trees are not correlated. Bagging achieves de-correlating the trees by constructing them using different training sets. To make a prediction at a new sample, random forests take the majority vote among the outputs of the grown trees in the ensemble. Random forests run very efficiently for large datasets, do not have the risk of overfitting (as, e.g., AdaBoost [40], a boosting method) and can handle datasets with imbalanced classes. The number of trees in the ensemble is selected through cross-validation.
C. Sparse Logistic Regression
Logistic Regression (LR) [41] is a linear classifier widely used in many classification problems. It models the posterior probability that a patient will be hospitalized as a logistic function of a linear combination of the input features, with parameters θ that weigh the input features and an offset θ0. The parameters of the model are selected by maximizing the log-likelihood using a gradient method. For the test samples, decisions are made by thresholding the log-likelihood ratio of the positive (hospitalized) class over the negative class. Logistic regression is popular in the medical literature because it predicts a probability of a sample belonging to the positive class. Here, we use an ℓ1-regularized (sparse) logistic regression [33], [42], [43], which adds an extra penalty term proportional to ‖θ‖1 in the log-likelihood. The motivation is to induce sparsity, effectively “selecting” a sparse subset of features. More specifically, we solve the following convex problem using a gradient-type method:
| (2) |
where the likelihood function is given by
and λ is a tunable parameter controlling the sparsity term. Setting λ = 0, we obtain a standard logistic regression model.
D. K-Likelihood Ratio Test
The Likelihood Ratio Test (LRT) is a naive Bayes classifier and assumes that individual features (elements) of the feature vector x = (x1, …, xD) are independent random variables [44]. The LRT algorithm empirically estimates the distribution p(xj|y) of each feature j for the hospitalized and the non-hospitalized class. Given a new test sample z = (z1, z2, ⋯, zD), LRT calculates the two likelihoods p(z|y = 1) and p(z|y = −1) and then classifies the sample by comparing the ratio
to a threshold. In our variation of the method, which we will call K-LRT,1 instead of taking into account the ratios of the likelihoods of all features, we consider only the K features with the largest ratios. We consider only the largest ratios because they correspond to features with a strong hospitalization “signal.” On the other hand, we do not consider features with the smallest ratios because they could be due to the imbalance of the dataset which has much more non-hospitalized than hospitalized patients.
The optimal K can be selected using cross-validation from a set of pre-defined values, that is, as the value with the best classification performance in a validation set. The purpose of this “feature selection” is again sparsity, that is, to identify the K most significant features for each individual patient. Thus, each patient is actually treated differently and this algorithm provides interpretability as to why a specific classification decision has been made for each individual patient.
IV. Joint Clustering and Classification (JCC)
In this section, we introduce a novel Joint Clustering and Classification method. The motivation comes from the success of K-LRT, which we will see in Section VII. Since K-LRT selects a sparse set of features for each patient, it stands to reason that there would be clusters of patients who share the same features. Moreover, since K-LRT uses the K largest likelihood ratios, feature selection is more informative for patients that are hospitalized (positive class). This is intuitive: patients are hospitalized for few underlying reasons while non-hospitalized patients appear “normal” in all features associated with a potential future hospitalization.
To reflect this reasoning, we consider a classification problem in which the positive class consists of multiple clusters, whereas negative class samples form a single cluster. It is possible to extend our framework and consider a setting where clustering is applied to both the positive and the negative class. However, because our results are satisfactory and to avoid further increasing complexity, we do not pursue this direction in this work. We assume that for each (positive class) cluster there is a sparse set of discriminative dimensions, based on which the cluster samples are separated from the negative class. Fig. 1 provides an illustration of this structure. The different clusters of patients are naturally created based on age, sex, race or different diseases. From a learning perspective, if the hidden positive groups are not predefined and we would like to learn an optimal group partition in the process of training classifiers, the problem could be viewed as a combination of clustering and classification. Furthermore, with the identified hidden clusters, the classification model becomes more interpretable in addition to generating accurate classification labels. A preliminary theoretical framework for JCC appeared in our conference paper [45], but without containing all detailed proofs of the key technical results and with very limited numerical evaluation.
Fig. 1.
The positive class contains two clusters and each cluster is linearly separable from the negative class.
A. An integer programming formulation
We next consider a joint cluster detection and classification problem under a Sparse Linear SVM (SLSVM) framework. Let and be the D-dimensional positive and negative class data points (each representing a patient), and , the corresponding labels, where i ∈ {1, 2, …, N+} and j ∈ {1, 2, …, N−}. Assuming L hidden clusters in the positive class, we seek to discover: (a) the L hidden clusters (denoted by a mapping function l(i) = l, l ∈ {1, 2, …, L}), and (b) L classifiers, one for each cluster. Let Tl be a parameter controlling the sparsity of the classifier for each cluster l. We formulate the Joint Clustering and Classification (JCC) problem as follows:
| (3) |
In the above formulation, the margin between the two classes in cluster l is equal to 2/‖βl‖, hence the first term in the objective seeks to maximize the margin. The variables represent misclassification penalties for the positive and negative data points, respectively. The first constraint limits the ℓ1 norm of βl to induce a sparse SVM for each cluster. The second (resp., third constraint) ensures that the positive (resp., negative) data points end up on the positive (resp. negative) side of the hyperplane; otherwise a penalty of (resp., ) is imposed; these misclassification penalties are minimized at optimality. We use different misclassification penalties for the positive and negative data points to accommodate a potential imbalance in the training set between available samples; typically, we have more negative (i.e., not hospitalized) samples. Notice that the misclassification costs of the negative samples are counted L times because they are drawn from a single distribution and, as a result, they are not clustered but simply copied into each cluster. The parameters λ− and λ+ control the weights of costs from the negative and the positive samples.
As stated, problem (3) is not easily solvable as it combines the cluster allocation decisions (i.e., deciding the cluster assignment l(i) for each sample i) with the determination of the SVM hyperplanes. One approach to solve JCC is shown below, where we transform the problem into a mixed integer programming problem (MIP) by introducing binary indicator variables to represent the cluster assignment in JCC (each positive sample can only be assigned to one cluster):
| (4) |
where zil = 1 when l(i) = l and 0 otherwise (binary variables describing the cluster assignments) and M is a large positive real number. The following proposition establishes the equivalence between formulations (4) and (3). The proof can be found in Appendix A.
Proposition IV.1
The MIP formulation (4) is equivalent to the original JCC formulation (3).
In order to obtain better clustering performance, we introduce a penalty term in the objective function seeking to minimize the intra-cluster distances between samples, that is, making samples in the same cluster more similar to each other. This term takes the form: , where
For σi1i2 to comply with this definition, we need to impose the constraint
The MIP approach presented above comes in a compact form, solves jointly the clustering and the classification problem, and exhibits good performance on small-scale problems. However, there are no general polynomial-time algorithms for solving MIPs, thus, making it problematic for large datasets that are most common in practice. This motivates us to develop the following Alternating Clustering and Classification (ACC) approach, which does not suffer from these limitations.
B. An alternating optimization approach
The idea behind ACC is to alternately train a classification model and then re-cluster the positive samples, yielding an algorithm which scales well and also, as we will see, comes with theoretical performance guarantees.
Given cluster assignments l(i) for all positive training samples i, the JCC problem (3) can be decoupled into L separate quadratic optimization problems, essentially solving an SVM training problem per cluster. Our alternating optimization approach, summarized in Algorithms 1–2, consists of two major modules: (i) training a classifier for each cluster and (ii) re-clustering positive samples given all the estimated classifiers.
The process starts with an initial (e.g., random or using some clustering algorithm) cluster assignment of the positive samples and then alternates between the two modules. Algorithm 1 orchestrates the alternating optimization process; given samples’ assignment to clusters, it obtains the optimal per cluster SLSVM classifiers and calls the re-clustering procedure described in Algorithm 2.
Algorithm 2 uses the computed L classifiers and assigns a positive sample i to the cluster l whose classification hyperplane is the furthest away from the sample i, that is, whose classifier better separates sample i from the negative class. Notice that the re-clustering of the positive samples is based on 𝒞, a subset of {1, …, D}, which is a set of selected features that allows us to select which features are important in cluster discrimination so that the identified clusters are more interpretable. In a notational remark, we denote (resp., x𝒞) as the projection of the D-dimensional feature vector (resp., x) on the subset 𝒞. We also impose the constraint (5) in Algorithm 2, which is necessary for proving the convergence of ACC.
Algorithm 1.
ACC Training
| Initialization: |
| Randomly assign positive class sample i to cluster l(i), for i ∈ {1, …, N+} and l(i) ∈ {1, …, L}. |
| repeat |
| Classification Step: |
| Train an SLSVM classifier for each cluster of positive samples combined with all negative samples. Each classifier is the outcome of a quadratic optimization problem (cf. (11)) and provides a hyperplane perpendicular to βl and a corresponding optimal objective value Ol. |
| Re-clustering Step: |
| Re-cluster the positive samples based on the classifiers βl and update the l(i)’s. |
| until no l(i) is changed or ΣlOl is not decreasing. |
Finally, Algorithm 3 describes how ACC classifies new samples not used in training. Specifically, it assigns a new sample to the cluster whose classifier is furthest away from that sample and uses the classifier of that cluster to make the classification decision.
Algorithm 2.
Re-clustering procedure given classifiers
| Input: positive samples , classifiers βl, current cluster assignment which assigns sample i to cluster l(i). | ||
| for all i ∈ {1, …, N+} do | ||
| for all l ∈ {1, …, L} do | ||
| calculate the projection of positive sample i onto the classifier for cluster l using only elements in a feature set 𝒞: ; | ||
| end for | ||
update cluster assignment of sample i from l(i) to
, subject to
| ||
| end for |
Algorithm 3.
ACC Testing
| for each test sample x do |
| Assign it to cluster . |
| Classify x with βl*. |
| end for |
C. ACC performance and convergence guarantees
In this subsection, we rigorously prove ACC convergence and establish out-of-sample (in a “test” set not seen during training) performance guarantees. While theoretical, such results are important because (i) they establish that ACC will converge to a set of clusters and a classifier per cluster and (ii) characterize the number of samples needed for training, as well as (iii) bound out-of-sample classification performance in terms of the in-sample performance.
We first present a result that suggests a favorable sample complexity for SLSVM compared to the standard linear SVM. Suppose that SLSVM for the l-th cluster yields Ql < D nonzero elements of βl, thus, selecting a Ql-dimensional subspace of features used for classification. The value of Ql is controlled by the parameter Tl (cf. (4)).
As is common in the learning literature [46], we draw independent and identically distributed (i.i.d.) training samples from some underlying probability distribution. Specifically, we draw N− negative samples from some distribution 𝒫0 and positive samples for cluster l from some distribution , where the total number of positive and negative samples used to derive the classifier of cluster l is . Let denote the expected training error rate and Rl the expected test error (out-of-sample) for the classifier of cluster l under these distributions. The proof of the following result is in Appendix B. We note that e in (6) is the base of the natural logarithm.
Theorem IV.2
For a specific cluster l, suppose that the corresponding sparse linear SVM classifier lies in a Ql-dimensional subspace of the original D-dimensional space. Then, for any ε > 0 and δ ∈ (0, 1), if the sample size Nl satisfies
| (6) |
it follows that with probability no smaller than 1 − δ, .
Theorem IV.2 suggests that if the training set contains a number of samples roughly proportional to (Ql+log(1/δ))/ε2, then we can guarantee with probability at least 1 − δ an out-of-sample error rate ε-close to the training error rate. In other words, sparse SVM classification requires samples proportional to the effective dimension of the sparse classifier and not the (potentially much larger) dimension D of the feature space.
Next we establish that the ACC training algorithm converges. The proof is given in Appendix C. As a remark on convergence, it is worth mentioning that the values λ+ and λ− should be fixed across all clusters to guarantee convergence.
Theorem IV.3
The ACC training algorithm (Alg. 1) converges for any set 𝒞.
The following theorem establishes a bound on the VC-dimension of the class of decision functions produced by ACC training. As we will see, this bound will then lead to out-of-sample performance guarantees. To state the result, let us denote by ℋ the family of clustering/classification functions produced by ACC training. The proof of the following theorem is in Appendix D.
Theorem IV.4
The VC-dimension of ℋ is bounded by
Theorem IV.4 implies that the VC-dimension of ACC-based classification grows linearly with the dimension of data samples and polynomially (between quadratic and cubic) with the number of clusters. Since the local (per cluster) classifiers are trained under an ℓ1 constraint, they are typically defined in a lower dimensional subspace. At the same time, the clustering function also lies in a lower dimensional space 𝒞. Thus, the “effective” VC-dimension could be smaller than the bound in Theorem IV.4.
An immediate consequence of Thm. IV.4 is the following corollary which establishes out-of-sample generalization guarantees for ACC-based classification and is based on a result in [47] (see also Appendix B). To state the result, let N = N+ + N− the size of the training set. Let RN denote the expected training error rate and R the expected test error (out-of-sample) of the ACC-based classifier.
Corollary IV.5
For any ρ ∈ (0, 1), with probability at least 1 − ρ it holds:
V. The Data
The data we use to evaluate the various methods we presented come from the Boston Medical Center (BMC). BMC is the largest safety-net hospital in New England and with 13 affiliated Community Health Centers (CHCs) provides care for about 30% of Boston residents. The data integrate information from hospital records, information from the community health centers, and some billing records, thus forming a fairly rich and diverse dataset.
The study is focused on patients with at least one heart-related diagnosis or procedure record in the period 01/01/2005–12/31/2010 or a diagnosis record of diabetes mellitus between 01/01/2007–12/31/2012. For each patient in the above set, we extract the medical history (demographics, hospital/physician visits, problems, medications, labs, procedures and limited clinical observations) for the period 01/01/2001–12/31/2010 and 01/01/2001–12/31/2012, correspondingly, which includes relevant medical factors from which we will construct a set of patient features. Data were available both from the hospital EHR and billing systems. Table I shows the ontologies, along with the number of factors and some examples corresponding to each of the heart patients. Similarly, Table II shows the ontologies with some examples for the diabetic patients. In these tables, ICD9 (International Classification of Diseases, 9th revision) [48], CPT (Current Procedural Terminology) [49], LOINC (Logical Observation Identifiers Names and Codes) [50], and MSDRG (Medicare Severity-Diagnosis Related Group) [51] are commonly used medical coding systems for diseases, procedures, laboratory observations, and diagnoses, respectively.
TABLE I.
Medical Factors in the Heart Diseases Dataset.
| Ontology | Number of Factors |
Examples |
|---|---|---|
| Demographics | 4 | Sex, Age, Race, Zip Code |
| Diagnoses | 22 | e.g., Acute Myocardial Infarction (ICD9: 410), Cardiac Dysrhythmias (ICD9: 427), Heart Failure (ICD9: 428), Acute Pulmonary Heart Disease (ICD9: 415), Diabetes Mellitus with Complications (ICD9: 250.1–250.4, 250.6–250.9), Obesity (ICD9: 278.0) |
| Procedures CPT | 3 | Cardiovascular Procedures (including CPT 93501, 93503, 93505, etc.), Surgical Procedures on the Arteries and Vein (including CPT 35686, 35501, 35509, etc.), Surgical Procedures on the Heart and Pericardium (including CPT 33533, 33534, 33535) |
| Procedures ICD9 | 4 | Operations on the Cardiovascular System (ICD9: 35–39.99), Cardiac Stress Test and pacemaker checks (ICD9: 89.4), Angiocardiography and Aortography (ICD9: 88.5), Diagnostic Ultrasound of Heart (ICD9: 88.72) |
| Vitals | 2 | Diastolic Blood Pressure, Systolic Blood Pressure |
| Lab Tests | 4 | CPK (Creatine phosphokinase) (LOINC:2157-6), CRP Cardio (C-reactive protein) (LOINC:30522-7), Direct LDL (Low-density lipoprotein) (LOINC:2574-2), HDL (High-Density Lipoprotein) (LOINC:9830-1) |
| Tobacco | 2 | Current Cigarette Use, Ever Cigarette Use |
| Visits to the ER | 1 | Visits to the Emergency Room |
| Admissions | 17 | e.g., Heart Transplant or Implant of Heart Assist System (MSDRG: 001, 002), Cardiac Valve and Other Major Cardiothoracic procedures (MSDRG: 216–221), Coronary Bypass (MSDRG: 231–234), Acute Myocardial Infarction (MSDRG: 280–285), Heart Failure and Shock (MSDRG: 291–293), Cardiac Arrest (MSDRG: 296–298), Chest Pain (MSDRG: 313), Respiratory System related admissions (MSDRG: 175–176, 190–192) |
TABLE II.
Medical Factors in the Diabetes Dataset.
| Ontology | Examples |
|---|---|
| Demographics | Sex, Age, Race, Zip Code |
| Diagnoses | e.g., Diabetes mellitus with complications, Thyroid disorders, Hypertensive disease, Pulmonary heart disease, Heart failure, Aneurysm, Skin infections, Abnormal glucose tolerance test, Family history of diabetes mellitus |
| Procedures (CPT or ICD9) | e.g., Procedure on single vessel, Insertion of intraocular lens prosthesis at time of cataract extraction, Venous catheterization, Hemodialysis, Transfusion of packed cells |
| Admissions | e.g., Diabetes (with and without) complications, Heart failure and shock, Deep Vein Thrombophlebitis, Renal failure, Chest pain, Chronic obstructive pulmonary disease, Nutritional. & misc metabolic disorders, Bone Diseases & Arthropathies, Kidney & urinary tract infections, Acute myocardial infarction, O.R. procedures for obesity, Hypertension |
| Service by Department | Inpatient (admit), Inpatient (observe), Outpatient, Emergency Room |
We note that some of the diagnoses and admissions in Table I are not directly heart-related, but may be good indicators of a heart problem. Also, as expected, many of the diagnoses and procedures in Table II are direct complications due to diabetes. Diabetes-related admissions are not trivially identifiable, and are revealed through the procedure described in Subsection V-B. Overall, our heart dataset contains 45,579 patients and our diabetes dataset consists of 33,122 patients after preprocessing, respectively. Among these patients, 3,033 patients in the heart dataset and 5,622 patients in the diabetes dataset are labeled as hospitalized in a target year. For each dataset we randomly select 60% of the patients for training and keep the remaining 40% of the patients for testing.
Our objective is to leverage past medical factors for each patient to predict whether she/he will be hospitalized or not during a target year which, as we explain below, could be different for each patient.
In order to organize all the available information in a uniform way for all patients, some preprocessing of the data is needed to summarize the information over a time interval. Details will be discussed in Subsections V-A and V-B. We will refer to the summarized information of the medical factors over a specific time interval as features.
Each feature related to diagnoses, procedures (CPT), procedures (ICD9) and visits to the Emergency Room (ER) is an integer count of such records for a specific patient during the specific time interval. Zero indicates the absence of any record. Blood pressure and lab tests features are continuous valued. Missing values are replaced by the average of values of patients with a record at the same time interval. Features related to tobacco use are indicators of current- or past-smoker in the specific time interval. Admission features contain the total number of days of hospitalization over the specific time interval the feature corresponds to. Admission records are used both to form the admission features (past admission records) and in order to calculate the prediction variable (existence of admission records in the target year). We treat our problem as a classification problem and each patient is assigned a label: 1 if there is a heart-related (or diabetes-related) hospitalization in the target year and −1 otherwise.
A. Heart Data Preprocessing
In this section we discuss several data organization and preprocessing choices we make for the heart dataset. For each patient, a target year is fixed (the year in which a hospitalization prediction is sought) and all past patient records are organized as follows.
Summarization of the medical factors in the history of a patient: After exploring multiple alternatives, an effective way to summarize each patient’s medical history is to form four time blocks for each medical factor. Time blocks 1, 2, and 3 summarize the medical factors over one, two, and three years before the target year, whereas the 4th block summarizes all earlier patient records. For tobacco use, there are only two features, indicating whether the patient is currently smoking and whether he/she has ever smoked. After removing features with zero standard deviation, this process results in a vector of 212 features for each patient.
Selection of the target year: As a result of the nature of the data, the two classes are highly imbalanced. When we fix the target year for all patients to be 2010, the number of hospitalized patients is about 2% of the total number of patients, which does not yield enough positive samples for effective training. Thus, and to increase the number of hospitalized patient examples, if a patient had only one hospitalization throughout 2007–2010, the year of hospitalization is set as the target year for that patient. If a patient had multiple hospitalizations, a target year between the first and the last hospitalization is randomly selected.
Setting the target time interval to be a year: After testing several options, a year appears to be an appropriate time interval for prediction. Shorter prediction windows increase variability and do not allow sufficient time for prevention. Moreover, given that hospitalization occurs roughly uniformly within a year, we take the prediction time interval to be a calendar year.
Removing noisy samples: Patients who have no records before the target year are impossible to predict and are thus removed.
B. Identifying Diabetes-Related Hospitalizations
Identifying the hospitalizations that occur mainly due to diabetes is not a trivial task, because for financial reasons (i.e., higher reimbursement) many diabetes-related hospitalizations are recorded in the system as other types of admissions, e.g., heart-related. Therefore, as a first step, we seek to separate diabetes-related admissions from all the rest. To that end, we consider all patients with at least one admission record between 1/1/2007 and 12/31/2012. From this set, patients with at least one diabetes mellitus record during the same period are assigned to the diabetic population, while the rest are assigned to the non-diabetic population.
We list the union of all unique admission types for both populations (732 unique types). The total number of admission samples for the diabetic and non-diabetic populations are N1 = 47, 352 and N2 = 116, 934, respectively. For each type of admission d, each admission sample can be viewed as the outcome of a binary random variable that takes the value 1, if the hospitalization occurs because of this type of admission, and 0, otherwise. Thus, we can transform the two sets of admission records for the two populations into binary (0/1) sequences. By (statistically) comparing the proportions of d in the two populations, we can infer whether admission d was caused mainly by diabetes or not.
To that end, we will utilize a statistical hypothesis test comparing sample differences of proportions. Suppose we generate two sets of admissions 𝒮1 and 𝒮2 of size N1 and N2 drawn from the diabetic and the non-diabetic patient populations, respectively. Consider a specific admission type d and suppose that it appears with probability p1, out of all possible admission types in 𝒮1. Similarly, a type d admission appears with probability p2 in 𝒮2. Given now the two sets of admissions from diabetics (𝒮1) and non-diabetics (𝒮2), let P1 and P2 be the corresponding sample proportions of type d admissions. We want to statistically compare P1 and P2 and assess whether a type d admission is more prevalent in 𝒮1 vs. 𝒮2. Consider as the null hypothesis the case where p1 = p2, i.e., a type d admission is equally likely in the two populations. Under the null hypothesis, the sampling distribution of differences in proportions is approximately normally distributed, with its mean and standard deviation given by
where p = (N1P1 + N2P2)/(N1 + N2) is used as an estimate of the probability of a type d admission in both populations and q = 1 − p. By using the standardized variable z = (P1 − P2)/(σP1 − P2) we can assess if the results observed in the samples differ markedly from the results expected under the null hypothesis. We do that using the single sided p-value of the statistic z. The smaller the p-value is, the higher the confidence we have in the alternative hypothesis or equivalently in the fact that the diabetic patients have higher chance of getting admission records of type d than the non-diabetic ones (since we consider the difference P1 − P2). We list admission types in increasing order of p-value and we set a threshold of p-value ≤ α = 0.0001; admission types with p-value less than α are considered to be attributed to diabetes.2 Examples of diabetes-related admissions are shown in Table II.
C. Diabetes Data Preprocessing
The features are formed as combinations of different medical factors (instead of considering the factors as separate features) that better describe what happened to the patients during their visits to the hospital. Specifically, we form triplets that consist of a diagnosis, a procedure (or the information that no procedure was done), and the service department. An example of a complex feature (a triplet) is the diagnosis of ischemic heart disease that led to an adjunct vascular system procedure (procedure on single vessel) while the patient was admitted to the inpatient care. Clearly, since each category can take one of several discrete values, a huge number of combinations should be considered. Naturally, not all possible combinations occur, which reduces significantly the total number of potential features that describe each patient. Also for each patient, we extract information about the diabetes type over their history and demographics including age, gender and race. Next, we present several data organization and preprocessing steps we take. For each patient, a target year is fixed and all past patient records are organized as follows.
Forming the complex features: We create a diagnoses-procedures indicator matrix to keep track of which diagnosis occurs with which procedure. The procedures that are not associated with any diabetes-related diagnosis are removed. Procedures in the dataset are listed in the most detailed level of the ICD9 coding system [48] or the CPT coding system [49]. We group together procedures that belong to the same ICD9/CPT family, resulting in 31 categories (out of 2004 in total).
Summarization of the complex features in the history of a patient: We use the same approach as with heart diseases: we form four time blocks for each medical factor with all corresponding records summarized over one, two, three years before the target year, and a fourth time block containing averages of all the earlier records. This produces a 9, 402-dimensional vector of features characterizing each patient.
Reducing the number of features: We remove all the features that do not contain enough information for a significant amount of the population (less than 1% of the patients), as they could not help us generalize. This leaves 320 medical and 3 demographical features.
Identifying the diabetes type: The ICD9 code for diabetes is assigned to category 250 (diabetes mellitus). The fifth digit of the diagnosis code determines the type of diabetes and whether it is uncontrolled or not stated as uncontrolled. Thus, we have four types of diabetes diagnoses: type II, not stated as uncontrolled (fifth digit 0); type I, not stated as uncontrolled (fifth digit 1), type II or unspecified type, uncontrolled (fifth digit 2) and type I, uncontrolled (fifth digit 3). Based on these four types, we count how many records of each type each patient had in the four time blocks before the target year, thus adding 16 new features for each patient.
Setting the target time interval to a calendar year: Again, as with heart diseases, we seek to predict hospitalizations in the target time interval of a year starting on the 1st of January and ending on the 31st of December.
Selection of the target year: As a result of the nature of the data, the two classes are highly imbalanced. To increase the number of hospitalized patient examples, if a patient had only one hospitalization throughout 2007–2012, the year of hospitalization will be set as the target year. If a patient had multiple hospitalizations, a target year between the first and the last hospitalizations will be randomly selected. 2012 is set as the target year for patients with no hospitalization, so that there is as much available history for them as possible. By this policy, the ratio of hospitalized patients in the dataset is 16.97%.
Removing patients with no record: Patients who have no records before the target year are removed, since there is nothing on which a prediction can be based. The total number of patients left is 33,122.
Splitting the data into a training set and a test set randomly: As is common in supervised machine learning, the population is randomly split into a training and a test set. Since from a statistical point of view, all the data points (patients’ features) are drawn from the same distribution, we do not differentiate between patients whose records appear earlier in time than others with later time stamps. A retrospective/prospective approach appears more often in the medical literature and is more relevant in a clinical trial setting, rather than in our algorithmic approach. What is critical in our setting is that for each patient prediction we make (hospitalization/non-hospitalization in a target year), we only use that patients’ information before the target year.
VI. Performance Evaluation
Typically, the primary goal of learning algorithms is to maximize the prediction accuracy or equivalently minimize the error rate. However, in the specific medical application problem we study, the ultimate goal is to alert and assist patients and doctors in taking further actions to prevent hospitalizations before they occur, whenever possible. Thus, our models and results should be accessible and easily explainable to doctors and not only machine learning experts. With that in mind, we examine our models from two aspects: prediction accuracy and interpretability.
The prediction accuracy is captured in two metrics: the false alarm rate (how many patients were predicted to be in the positive class, i.e., hospitalized, while they truly were not) and the detection rate (how many patients were predicted to be hospitalized while they truly were). In the medical literature, the detection rate is often referred to as sensitivity and the term specificity is used for one minus the false alarm rate. Two other terms that are commonly used are the recall rate, which is the same as the detection rate, and the precision rate, which is defined as the ratio of true positives (hospitalizations) over all the predicted positives (true and false). For a binary classification system, the evaluation of the performance is typically illustrated with the Receiver Operating Characteristic (ROC) curve, which plots the detection rate versus the false alarm rate at various threshold settings. To summarize the ROC curve and be able to compare different methods using only one metric, we will use the Area Under the ROC Curve (AUC). An ideal classifier achieves an AUC equal to 1 (or 100%), while a classifier that makes random classification decisions achieves an AUC equal to 0.5 (or 50%). Thus, the “best” (most accurate) classification method will be the one that achieves the highest AUC.
For the heart study we conduct, we will also generate the ROC curve based on patients’ 10-year risk of general cardiovascular disease derived by the Framingham Heart Study (FHS) [52]. FHS is a seminal study on heart diseases that has developed a set of risk factors for various heart problems. The 10-year risk we are using is the closest to our purpose and has been widely used. It uses the following features (predictors): age, diabetes, smoking, treated and untreated systolic blood pressure, total cholesterol, High-Density Lipoprotein (HDL), and BMI (Body Mass Index) which can be used to replace lipids in a simpler model. We calculate this risk value (which we call the Framingham Risk Factor-FRF) for every patient and make the classification based on this risk factor only. We also generate an ROC curve by applying random forests just to the features involved in FRF. The generated ROC curve serves as a baseline for comparing our methods to classifiers that are based on features suggested only by medical intuition.
For the diabetes study, we also consider baseline classifiers that are based only on features commonly considered by physicians. More specifically, the features we select are: age, race, gender, average over the entire patient history of the hemoglobin A1c, or HbA1c for short (which measures average blood sugar concentrations for the preceding two to three months), and the number of emergency room visits over the entire patient history. All these features are part of a 3-year risk of diabetes metric in [53]. We apply random forests to just these features to obtain a baseline to compare our methods against.
Let us also note that we will compare our new algorithm ACC to SVMs (linear and RBF), and two other hierarchical approaches that combine clustering with classification, to which we refer as Cluster-Then-Linear-SVM (CT-LSVM) and Cluster-Then-Sparse-Linear-SVM (CT-SLSVM). Specifically, CT-LSVM first clusters the positive samples (still based on the feature set 𝒞) with the widely used k-means method [39], then copies negative samples into each cluster, and finally trains classifiers with linear SVM for each cluster. The only difference between algorithm CT-SLSVM and CT-LSVM is that CT-SLSVM adopts sparse linear SVM in the last step.
Notice that ACC implements an alternating procedure while CT-LSVM and CT-SLSVM do not. With only one-time clustering, CT-LSVM and CT-SLSVM create unsupervised clusters without making use of the negative samples, whereas ACC is taking class information and classifiers under consideration so that the clusters also help the classification.
VII. Experimental Results
In this section, we will present experimental results on the two datasets for all methods we have presented so far, in terms of both accuracy and interpretability.
For SVM, tuning parameters are the misclassification penalty coefficient C (cf. (1)) and the kernel parameter σ; we used the values {0.3, 1, 3} and {0.5, 1, 2, 7, 15, 25, 35, 50, 70, 100}, respectively. Optimal values of 1 and 7, respectively, were selected by cross-validation.
For K-LRT, we quantize the data as shown in Table III. After experimentation, the best performance of K-LRT is achieved by setting k = 4.
TABLE III.
Quantization of Features.
| Features | Levels of quantization |
Comments |
|---|---|---|
| Sex | 3 | 0 represents missing information |
| Age | 6 | Thresholds at 40, 55, 65, 75 and 85 years old |
| Race | 10 | |
| Zip Code | 0 | Removed due to its vast variation |
| Tobacco (Current and Ever Cigarette Use) | 2 | Indicators of tobacco use |
| Diastolic Blood Pres-sure (DBP) | 3 | Level 1 if DBP < 60mmHg, Level 2 if 60mmHg ≤ DBP ≤ 90mmHg and Level 3 if DBP > 90mmHg |
| Systolic Blood Pressure (SBP) | 3 | Level 1 if SBP < 90mmHg, Level 2 if 90mmHg ≤ SBP ≤ 140mmHg and Level 3 if SBP > 140mmHg |
| Lab Tests | 2 | Existing lab record or Non-Existing lab ecord in the specific time period |
| All other dimensions | 7 | Thresholds are set to 0.01%, 5%, 10%, 20%, 40% and 70% of the maximum value of each dimension |
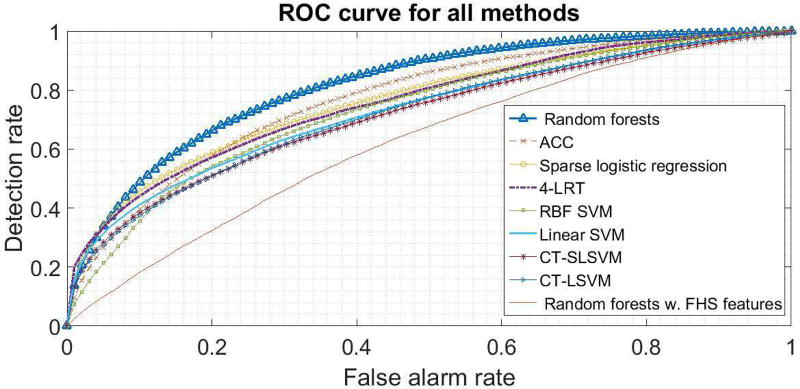
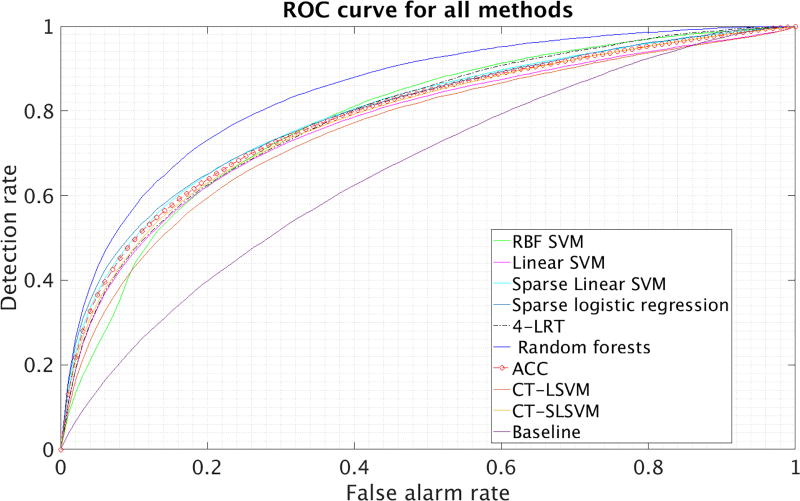
In Figures 2 and 3, we present the ROC curves of all methods, for a particular random split of the data into a training and test set. In Tables IV and V, we present the average (avg.) and the standard deviation (std) of the AUC over 10 different splits of the data into a training and a test set. In these tables, Lin. and RBF SVM correspond to SVM with a linear and an RBF kernel, respectively. Sparse LR corresponds to sparse logistic regression (cf. Sec. III-C). FRF 10-yr risk corresponds to thresholding the Framingham 10-year risk and random forests on FRF features simply trains a random forest on the features used in the Framingham 10-year risk. We also report the baseline diabetes method we presented in subsection VI in the last row of Table V.
Fig. 2.
ROC curves for the heart data.
Fig. 3.
ROC curves for diabetes data.
TABLE IV.
Prediction accuracy (AUC) on heart data.
| Settings | avg. AUC | std AUC |
|---|---|---|
| ACC, L = 1 (SLSVM) | 76.54% | 0.59% |
| ACC, L = 2 | 76.83% | 0.87% |
| ACC, L = 3 | 77.06% | 1.04% |
| ACC, L = 4 | 75.14% | 0.92% |
| ACC, L = 5 | 75.14% | 1.00% |
| ACC, L = 6 | 74.32% | 0.87% |
| 4-LRT | 75.78% | 0.53% |
| Lin. SVM | 72.83% | 0.51% |
| RBF SVM | 73.35% | 1.07% |
| sparse LR | 75.87% | 0.67% |
| CT-LSVM (L = 2) | 71.31% | 0.76% |
| CT-SLSVM (L = 2) | 71.97% | 0.73% |
| random forests | 81.62% | 0.37% |
| FRF 10-yr risk | 56.48% | 1.09% |
| random forests on FRF features | 62.20% | 1.13% |
TABLE V.
Prediction Accuracy (AUC) on diabetes data.
| Settings | avg. AUC | std AUC |
|---|---|---|
| ACC, L = 1 (SLSVM) | 79.24% | 0.52% |
| ACC, L = 2 | 78.55% | 0.41% |
| ACC, L = 3 | 78.53% | 0.41% |
| ACC, L = 4 | 78.46% | 0.35% |
| ACC, L = 5 | 78.36% | 0.36% |
| ACC, L = 6 | 78.18% | 0.50% |
| 4-LRT | 78.74% | 0.28% |
| Lin. SVM | 76.87% | 0.48% |
| RBF SVM | 77.96% | 0.27% |
| sparse LR | 78.91% | 0.38% |
| CT-LSVM (L = 2) | 75.63% | 0.50% |
| CT-SLSVM (L = 2) | 77.99% | 0.49% |
| random forests | 84.53% | 0.26% |
| random forests on selected features (baseline) | 65.77% | 0.47% |
Based on the results, random forests perform the best followed by our ACC. It is interesting that using features selected by physicians (as in FRF or the diabetes baseline method) leads to significantly inferior performance even if a very sophisticated classifier (like random forests) is being used. This suggests that the most intuitive medical features do not contain all the information that could be used in making an accurate prediction.
In terms of interpretability, with RBF SVM, the features are mapped through a kernel function from the original space into a higher-dimensional space. This, however, makes the features in the new space not interpretable. Random forests are also not easy to interpret. While a single tree classifier which is used as the base learner is explainable, the weighted sum of a large number of trees makes it relatively complicated to find the direct attribution of each feature to the final decision. LRT itself lacks interpretability, because we have more than 200 features for each sample and there is no direct relationship between prediction of hospitalization and the reasons that led to it. On the other hand, sparse linear SVM (SLSVM which coincides with ACC using L = 1 cluster), ACC, K-LRT, and sparse LR are easily interpretable because they are based on sparse classifiers involving relatively few features. ACC, in addition, clusters patients and cluster membership provides extra interpretation.
Our modified LRT, K-LRT, is particularly interpretable and it is surprising that such a simple classifier has strong performance. It highlights the top K features that lead to the classification decision. These features could be of help in assisting physicians reviewing the patient’s EHR profile and formulating hospitalization-prevention strategies. To provide an example of intuition that can be gleaned from this information, we consider the heart disease dataset and in Table VI we present the features highlighted by 1-LRT. We remind the reader that in 1-LRT, each test patient is essentially associated with a single feature. For each feature j, we (i) count how many times it was selected as the primary feature in the test set, and (ii) calculate the average likelihood ratio p(zj|y = 1)/p(zj|y = −1) over all test patients. We normalize both quantities (i) and (ii) to have zero mean and variance equal to 1. The average of these two normalized quantities is treated as the importance score of the feature j. We rank the importance scores and report the top 10 features in Table VI. In the table, CPK stands for creatine phosphokinase, an enzyme which, when elevated, it indicates injury or stress to the heart muscle tissue, e.g., as a result of a myocardial infarction (heart attack). It is interesting that in addition to heart-related medical factors, utilization features such as lab tests and emergency room visits, contribute to the classification decision. This is likely the reason why our methods, which use the entirety of the EHR, perform much better than the Framingham-based methods.
TABLE VI.
Top 10 significant features for 1-LRT.
| 1-LRT Importance Score |
1-LRT Feature Name |
|---|---|
| 10.50 | Admission of heart failure, 1 year before the target year |
| 9.71 | Age |
| 6.23 | Diagnosis of heart failure, 1 year before the target year |
| 5.43 | Admission with other circulatory system diagnoses, 1 year before the target year |
| 4.38 | Diagnosis of heart failure, 2 years before the target year |
| 4.16 | Diagnosis of hematologic disease, 1 year before the target year |
| 3.45 | Diagnosis of diabetes mellitus w/o complications, 1 year before the target year |
| 3.40 | Symptoms involving respiratory system and other chest symptoms, 1 year before the target year |
| 3.24 | visit to the Emergency Room, 1 year before the target year |
| 3.13 | Lab test CPK, 1 year before the target year |
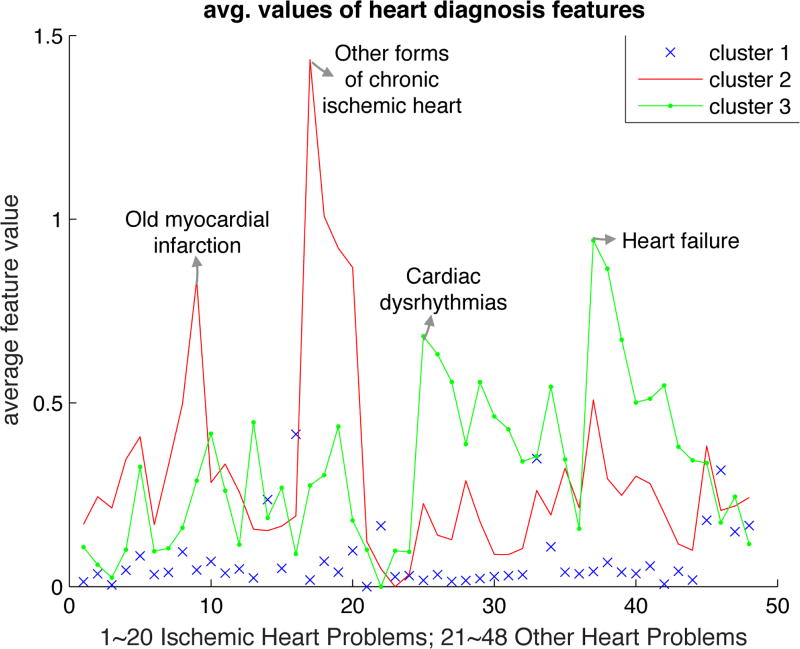
To interpret the clusters generated by ACC for the heart study (for the case L = 3 which yields the best performance), we plot in Figure 4 the mean value over each cluster of each element in the feature vector x𝒞. The 3 clusters are well-separated. Cluster 2 contains patients with other forms of chronic ischemic disease (mainly coronary atherosclerosis) and myocardial infarction that had occurred sometime in the past. Cluster 3 contains patients with dysrhythmias and heart failure. Cardiologists would agree that these clusters contain patients with very different types of heart disease. Finally, Cluster 1 contains all other cases with some peaks corresponding to endocardium/pericardium disease. It is interesting, and a bit surprising, that ACC identifies meaningful clusters of heart-disease even though it is completely agnostic of medical knowledge.
Fig. 4.
Average feature values in each cluster (L = 3) for the heart diseases dataset.
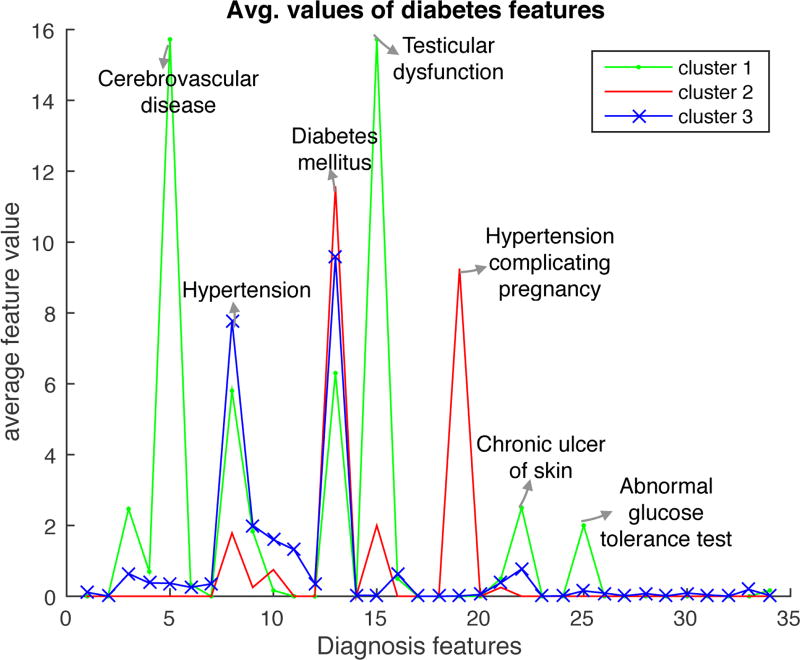
In the diabetes dataset, best ACC performance is obtained for L = 1 (a single cluster). Still, it is of interest to examine whether meaningful clusters emerge for L > 1. We plot again in Figure 5 the mean value over each cluster of each element in the feature vector, using as “diagnostic” features the subset of features which have a correlation larger than 0.01 with the labels in the training set. This is done for a single repetition of the experiment and L = 3, yielding interesting clusters and highlighting the interpretative power of ACC. We observe that Cluster 1 contains diabetes patients with chronic cerebrovascular disease, skin ulcers, hypertension, an abnormal glucose tolerance test, and other complications as a result of diabetes. Cluster 2 contains patients with diabetes complicating pregnancy. Cluster 3 contains patients with less acute disease, combining diabetes with hypertension. The feature values of these three clusters clearly separate from the feature values in the negative class.
Fig. 5.
Average feature values in each cluster (L = 3) for the diabetes dataset.
VIII. Conclusions
In this paper, we focused on the challenge of predicting future hospitalizations for patients with heart problems or diabetes, based on their Electronic Health Records (EHRs). We explored a diverse set of methods, namely kernelized, linear and ℓ1-regularized linear Support Vector Machines, ℓ1-regularized logistic regression and random forests. We proposed a likelihood ratio test-based method, K-LRT, that is able to identify the K most significant features for each patient that lead to hospitalization.
Our main contribution is the introduction of a novel joint clustering and classification method that discovers hidden clusters in the positive samples (hospitalized) and identifies sparse classifiers for each cluster separating the positive samples from the negative ones (non-hospitalized). The joint problem is non-convex (formulated as an integer optimization problem); still we developed an alternating optimization approach (termed ACC) that can solve very large instances. We established the convergence of ACC, characterized its sample complexity, and derived a bound on VC dimension that leads to out-of-sample performance guarantees.
For all the methods we proposed, we evaluated their performance in terms of classification accuracy and interpretability, an equally crucial criterion in the medical domain. Our ACC approach yielded the best performance among methods that are amenable to an interpretation (or explanation) of the prediction.
Our findings highlight a number of important insights and opportunities by offering a more targeted strategy for “at-risk” individuals. Our algorithms could easily be applied to care management reports or EHR-based prompts and alerts with the goal of identifying individuals who might benefit from additional care management and outreach. Depending on available resources and economic considerations, a medical facility can select a specific point on the ROC curve to operate at. This is equivalent to selecting a tolerable maximum false positive (alarm) rate, or, equivalently, a minimum specificity. Because costs associated with preventive actions (such as tests, medications, office visits) are orders of magnitude lower than hospitalization costs, one can tolerate significant false alarm rates and still save a large amount of money in preventable hospitalization costs. To get a sense of this difference, the average cost per hospital stay in the U.S. was $9,700 in 2010 [54], with some heart related hospitalizations costing much more on average (e.g., $18,200 for Acute Myocardial Infarction). In contrast, an office visit costs on the order of $200, tests like an ECG or an echo on the order of $100–$230, and a 90-day supply of common medication (hypertension or cholesterol) no more than $50. Clearly, even a small fraction of prevented hospitalizations can lead to large savings. Our methods can be seen as enabling such prevention efforts.
Acknowledgments
This work was supported in part by the National Science Foundation (NSF) under Grants CNS-1645681, CCF-1527292, IIS-1237022, and IIS-172499; by the Army Research Office (ARO) under Grant W911NF-12-1-0390; by the National Institutes of Health (NIH) under Grant 1UL1TR001430 to the Clinical & Translational Science Institute at Boston University; and by the Boston University Digital Health Initiative.
Biographies

Theodora Brisimi received her Diploma (five-year degree) in 2011 from the National Technical University of Athens, Athens, Greece. She is currently a Ph.D. student in the Department of Electrical and Computer Engineering, Boston University, Boston, MA, USA, working on developing and applying new techniques in machine learning with applications to healthcare and smart cities. Her current research focus is on developing classification methods to predict future hospitalizations of patients based on their Electronic Health Records (EHR) history. She has also been involved in Boston’s Street Bump Project, work in collaboration with the City of Boston, to detect fixable bumps on city streets. Ms. Brisimi has industrial experience, working during the summer of 2014 as an intern at Mitsubishi Electric Research Labs and during the summer of 2015 as an intern at Palo Alto Research Center, a Xerox company.

Tingting Xu received her B.S. degree from China University of Mining and Technology, Beijing, China, in 2011, and the M.S. degree from the Key Laboratory of Systems and Control, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, China, in 2014. She is currently a Ph.D. candidate at the Division of Systems Engineering, Boston University, Boston, MA, USA. Her research interests lie in the fields of systems and control, game theory, machine learning and medical informatics.

Taiyao Wang obtained the M.S. degree in complex systems and control from the Institute of Systems Science, Chinese Academy of Sciences, Beijing, China, in June 2014. Since September 2014 he has been with the Division of Systems Engineering, Boston University, Boston, MA, USA, where he is pursuing the Ph.D. degree in systems engineering. His research interests include optimization, applied probability and statistics, machine learning, and systems and control with applications in healthcare and biology.

Wuyang Dai received the Ph.D. in electrical and computer engineering from the Boston University, Boston, MA, USA, in 2015. During his Ph.D. studies, his research focus was on designing machine learning algorithms with applications in medical informatics. He is now a data scientist with Adobe Systems, Inc. His current work is mainly on customer predictive analytics based on meta data, including static profiles and dynamic usage data.

William G. Adams received the M.D. degree from the Columbia University College of Physicians and Surgeons, New York, NY, in 1987. He was a resident at Childrens Hospital, Boston, MA, in 1990, and a fellow in Epidemiology and Public Health, Centers for Disease Control, Atlanta, GA, in 1993. He is currently an epidemiologist, medical informatician, and practicing pediatrician at Boston Medical Center (BMC), Boston, MA. He is also Director of BUCTSI Clinical Research Informatics for Boston University and Director of Community Health Informatics for the Boston HealthNet – an urban integrated health delivery system. His primary research has been, and continues to be, focused on developing and evaluating information technology (IT)-based solutions for improving the quality of health and healthcare for urban populations, particularly children. His focuses include the child health EHR, state-wide health registries and decision support, patient-centered health IT, and clinical data warehousing for research and practice.

Ioannis Ch. Paschalidis (M’96–SM’06–F’14) received the M.S. and Ph.D. degrees both in electrical engineering and computer science from the Massachusetts Institute of Technology (MIT), Cambridge, MA, USA, in 1993 and 1996, respectively. In September 1996 he joined Boston University where he has been ever since. He is a Professor at Boston University with appointments in the Department of Electrical and Computer Engineering, the Division of Systems Engineering, and the Department of Biomedical Engineering. He is the Director of the Center for Information and Systems Engineering (CISE). He has held visiting appointments with MIT and Columbia University, New York, NY, USA. His current research interests lie in the fields of systems and control, networking, applied probability, optimization, operations research, computational biology, and medical informatics.
Dr. Paschalidis is a recipient of the NSF CAREER award (2000), several best paper and best algorithmic performance awards, and a 2014 IBM/IEEE Smarter Planet Challenge Award. He was an invited participant at the 2002 Frontiers of Engineering Symposium, organized by the U.S. National Academy of Engineering and the 2014 U.S. National Academies Keck Futures Initiative (NAFKI) Conference. He is the inaugural Editor-in-Chief of the IEEE Transactions on Control of Network Systems.
Appendix A
Proof of Proposition IV.1
Proof
Let and be the optimal objective values of problems (3) and (4).
Given any feasible solution to the JCC problem (3): l(i), βl, , ∀l, i, and , a feasible solution to the MIP problem is:
and βl, remain the same as in the JCC solution.
The feasibility of the constructed MIP solution is verified as follows. Notice that except for the 2nd constraint in the MIP formulation (4) (the big-M constraint), all other constraints can be easily verified to be satisfied by the constructed MIP solution. For the big-M constraint, if zil = 1, then M Σk≠l zik = 0, and the big-M constraint holds since . If, however, zil = 0, then M Σk≠l zik = M, and the big-M constraint also holds (trivially).
The above two feasible solutions have the same objective values, and this equality holds for any feasible solution to the JCC problem, hence we can conclude that .
Next, we prove that each optimal solution to the MIP problem satisfies when zil = 0. Note that when zil = 0, M Σk≠l zik = M, and the big-M constraint becomes , which will always hold since M is a large enough number. Therefore, to minimize the objective, the optimal solution should select the smallest feasible , i.e., .
Given an optimal solution to the MIP problem, a corresponding feasible solution to JCC problem is: if zil = 1, then , and l(i) = l; and all other variables retain their values in the MIP solution. Since the two solutions have the same objective cost, it follows .
Appendix B
Proof of Theorem IV.2
Proof
To simplify notation we drop the cluster index l. We will use a result from [47]. We note that the family of linear classifiers in a D-dimensional space has VC-dimension D + 1 ([35]). Let 𝒢 be a function family with VC-dimension D + 1. Let RN (g) denote the training error rate of classifier g on N training samples randomly drawn from an underlying distribution 𝒫. Let R(g) denote the expected test error of g with respect to 𝒫. The following theorem from [47] is useful in establishing our result.
Theorem B.1 ([47])
If the function family 𝒢 has VC-dimension D + 1, then the probability
| (7) |
for any function g ∈ 𝒢 and ρ ∈ (0, 1).
For the given ε in the statement of Theorem IV.2, select large enough N such that
or
| (8) |
It follows from Thm. B.1,
| (9) |
In our setting, the classifier g is restricted to a Q-dimensional subspace of the D-dimensional feature space. Thus, the bound in (8) holds by replacing D with Q in the right hand side and the bound in (9) holds for any such Q-dimensional subspace selected by the ℓ1-penalized optimization. Since there are possible choices for the subspace, using the union bound we obtain:
Using the bound , it follows:
| (10) |
For the given δ ∈ (0, 1) in the statement of Theorem IV.2, select small enough ρ such that
or equivalently
Using (8) (with Q replacing D), we obtain
which implies that N must be large enough to satisfy
This establishes P (R(g) − RN (g) ≥ ε) ≤ δ, which is equivalent to Theorem IV.2.
Appendix C
Proof of Theorem IV.3
Proof
At each alternating cycle, and for each cluster l, we train a SLSVM using as training samples the positive samples of that cluster combined with all negative samples. This produces an optimal value Ol for the corresponding SLSVM training optimization problem (cf. (3)) and the corresponding classifier (βl, ). Specifically, the SLSVM training problem for cluster l is:
| (11) |
Set
where l(i) maps sample i to cluster l(i), , and βl, , and are optimal solutions of (11) for each l. Let us now consider the change of Z at each iteration of the ACC training procedure.
First, we consider the re-clustering step (Alg. 2) given computed SLSVMs for each cluster. During the re-clustering step, the classifier and slack variables for negative samples are not modified. Only the get modified since the assignment functions l(i) change. When we switch positive sample i from cluster l(i) to l*(i), we can simply assign value to . Therefore, the value of Z does not change during the re-clustering phase and takes the form
Next, given new cluster assignments, we re-train the local classifiers by resolving problem (11) for each cluster l. Notice that re-clustering was done subject to the constraint in Eq. (5). Since for all positive samples, we have
The first inequality is due to being feasible for (11). The second inequality is due to and Eq. (5). Thus, by assigning to it follows that the remain feasible for problem (11). Given that the remaining decision variables do not change, forms a feasible solution of problem (11). This solution has a cost equal to Ol. Re-optimizing can produce an optimal value that is no worse. It follows that in every iteration of ACC, Z is monotonically non-increasing. Monotonicity and the fact that Z is bounded below by zero, suffices to establish convergence.
Appendix D
Proof of Theorem IV.4
Proof
The proof is based on Lemma 2 of [55]. Given an assignment of each positive sample i to cluster l(i), define L clustering functions
Hence, positive sample i is assigned to cluster arg maxl gl(i). This can be viewed as the output of (L − 1)L/2 comparisons between pairs of gl1 and gl2, where 1 ≤ l1 < l2 ≤ L. This pairwise comparison could be further transformed into a boolean function (i.e., sgn(gl1 − gl2)). Together with the L classifiers (one for each cluster), we have a total of (L + 1)L/2 boolean functions. Among all these boolean functions, the maximum VC-dimension is D + 1, because at most D features are being used as input. Therefore, by Lemma 2 of [55], the VC-dimension of the function family ℋ is bounded by , or equivalently .
Footnotes
Research partially supported by the NSF under grants CNS-1645681, CCF-1527292, IIS-1237022, and IIS-1724990, by the ARO under grant W911NF-12-1-0390, by the NIH under grant 1UL1TR001430 to the Clinical & Translational Science Institute at Boston University, and by the Boston University Digital Health Initiative.
K-LRT was first proposed in [44] and was applied only to a heart-disease dataset.
Apart from selecting a small-value α, we also ensure that the cumulative fraction of patients that are potentially labeled as belonging to the hospitalized class is not too small, so that the dataset is not highly imbalanced.
References
- 1.Stahl G. Health impacts of living in a city. Global Citizen. 2015 Oct 31; [Online]. Available: https://www.globalcitizen.org/en/content/health-impacts-of-living-in-a-city/
- 2.Yach D, Hawkes C, Gould CL, Hofman KJ. The global burden of chronic diseases: overcoming impediments to prevention and control. Jama. 2004;291(21):2616–2622. doi: 10.1001/jama.291.21.2616. [DOI] [PubMed] [Google Scholar]
- 3.Ambient (outdoor) air quality and health. Fact sheet, World Health Organization; Sep, 2016. [Online]. Available: http://www.who.int/mediacentre/factsheets/fs313/en/ [Google Scholar]
- 4.World’s population increasingly urban with more than half living in urban areas. Report on World Urbanization Prospects, United Nations Department of Economic and Social Affairs. 2014 Jul; http://www.un.org/en/development/desa/news/population/world-urbanization-prospects-2014.html.
- 5.Steep M. Smart cities improve the health of their citizens. Forbes. 2016 Jun 27; [Online]. Available: https://www.forbes.com/sites/mikesteep/2016/06/27/can-smart-cities-improve-the-health-of-its-citizens/#45b7bd9e3957.
- 6.Frost & Sullivan. Strategic Opportunity Analysis of the Global Smart City Market. 2016 http://www.egr.msu.edu/~aesc310-web/resources/SmartCities/Smart%20City%20Market%20Report%202.pdf.
- 7.Learn about MassHealth coverage types for individuals, families, and people with disabilities. 2017 [Online]. Available: https://www.mass.gov/service-details/learn-about-masshealth-coverage-types-for-individuals-families-and-people-with.
- 8.Wagman N. What is the actual state cost of MassHealth in 2018? Massachusetts Budget And Policy Center; 2017. [Online]. Available: http://massbudget.org/report_window.php?loc=What-Is-the-Actual-State-Cost-of-MassHealth-in-2018.html. [Google Scholar]
- 9.Kayyali B, Knott D, Van Kuiken S. The big-data revolution in US health care: Accelerating value and innovation. McKinsey & Company. 2013:1–13. [Google Scholar]
- 10.What is driving US health care spending? America’s unsustainable health care cost growth. Bipartisan Policy Center; Washington, DC: 2012. [Google Scholar]
- 11.Paschalidis IC. How machine learning is helping us predict heart disease and diabetes. Harvard Business Review. 2017 May 30; [Online]. Available: https://hbr.org/2017/05/how-machine-learning-is-helping-us-predict-heart-disease-and-diabetes.
- 12.EU general data protection regulation. EU Regulation 2016/679, 2018. [Online]. Available: https://en.wikipedia.org/wiki/General_Data_Protection_Regulation.
- 13.Centers for Disease Control and Prevention. Heart Disease Facts. 2015 www.cdc.gov/heartdisease/facts.htm.
- 14.Diabetes globally. Diabetes Australia; 2017. [Online]. Available: https://www.diabetesaustralia.com.au/diabetes-globally. [Google Scholar]
- 15.International Diabetes Federation. Diabetes Atlas. 2015 www.diabetesatlas.org/component/attachments/?task=download&id=116.
- 16.Centers for Disease Control and Prevention. National diabetes statistics report: estimates of diabetes and its burden in the United States. 2014 www.cdc.gov/diabetes/pdfs/data/2014-report-acknowledgments.pdf.
- 17.Jiang HJ, Russo CA, Barrett ML. Nationwide frequency and costs of potentially preventable hospitalizations, 2006. 2009 [Online]. Available: http://www.hcup-us.ahrq.gov/reports/statbriefs/sb72.jsp. [PubMed]
- 18.Office of the National Coordinator for Health Information Technology. Office-based Physician Electronic Health Record Adoption, Health IT Quick-Stat# 50. 2016 dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php.
- 19.Ludwick DA, Doucette J. Adopting electronic medical records in primary care: lessons learned from health information systems implementation experience in seven countries. International Journal of Medical Informatics. 2009;78(1):22–31. doi: 10.1016/j.ijmedinf.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Takeda H, Matsumura Y, Nakajima K, Kuwata S, Zhenjun Y, Shanmai J, Qiyan Z, Yufen C, Kusuoka H, Inoue M. Health care quality management by means of an incident report system and an electronic patient record system. International Journal of Medical Informatics. 2003;69(2):285–293. doi: 10.1016/s1386-5056(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 21.Hannan TJ. Detecting adverse drug reactions to improve patient outcomes. International Journal of Medical Informatics. 1999;55(1):61–64. doi: 10.1016/s1386-5056(99)00020-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang SJ, Middleton B, Prosser LA, Bardon CG, Spurr CD, Carchidi PJ, Kittler AF, Goldszer RC, Fairchild DG, Sussman AJ, et al. A cost-benefit analysis of electronic medical records in primary care. The American Journal of Medicine. 2003;114(5):397–403. doi: 10.1016/s0002-9343(03)00057-3. [DOI] [PubMed] [Google Scholar]
- 23.Hosseinzadeh A, Izadi MT, Verma A, Precup D, Buckeridge DL. Assessing the predictability of hospital readmission using machine learning. IAAI. 2013 [Google Scholar]
- 24.Zolfaghar K, Meadem N, Teredesai A, Roy SB, Chin S-C, Muckian B. Big Data, 2013 IEEE International Conference on. IEEE; 2013. Big data solutions for predicting risk-of-readmission for congestive heart failure patients; pp. 64–71. [Google Scholar]
- 25.Strack B, DeShazo JP, Gennings C, Olmo JL, Ventura S, Cios KJ, Clore JN. Impact of hba1c measurement on hospital readmission rates: analysis of 70,000 clinical database patient records. BioMed research international. 2014;2014 doi: 10.1155/2014/781670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caruana R, Lou Y, Gehrke J, Koch P, Sturm M, Elhadad N. Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; 2015. Intelligible models for healthcare: Predicting pneumonia risk and hospital 30-day readmission; pp. 1721–1730. [Google Scholar]
- 27.Pradhan M, Sahu RK. Predict the onset of diabetes disease using artificial neural network (ann) International Journal of Computer Science & Emerging Technologies (E-ISSN: 2044-6004) 2011;2(2) [Google Scholar]
- 28.Palaniappan S, Awang R. Intelligent heart disease prediction system using data mining techniques. Computer Systems and Applications, 2008. AICCSA 2008. IEEE/ACS International Conference on; IEEE; 2008. pp. 108–115. [Google Scholar]
- 29.Hachesu PR, Ahmadi M, Alizadeh S, Sadoughi F. Use of data mining techniques to determine and predict length of stay of cardiac patients. Healthcare informatics research. 2013;19(2):121–129. doi: 10.4258/hir.2013.19.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pofahl WE, Walczak SM, Rhone E, Izenberg SD. Use of an artificial neural network to predict length of stay in acute pancreatitis. The American Surgeon. 1998;64(9):868. [PubMed] [Google Scholar]
- 31.Heritage health prize. Heritage Provider Network; 2011. [Google Scholar]
- 32.Vellido A, Martín-Guerrero JD, Lisboa PJ. ESANN. Vol. 12. Citeseer; 2012. Making machine learning models interpretable; pp. 163–172. [Google Scholar]
- 33.Lee S-I, Lee H, Abbeel P, Ng AY. Efficient l1 regularized logistic regression. AAAI. 2006;6:401–408. [Google Scholar]
- 34.Casanova R, Saldana S, Chew EY, Danis RP, Greven CM, Ambrosius WT. Application of random forests methods to diabetic retinopathy classification analyses. PLoS One. 2014;9(6):e98587. doi: 10.1371/journal.pone.0098587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vapnik VN. The nature of statistical learning theory. New York, NY, USA: Springer-Verlag New York, Inc; 1995. [Google Scholar]
- 36.Ng AY, Jordan MI. On discriminative vs. generative classifiers: A comparison of logistic regression and naive bayes. Advances in neural information processing systems. 2002:841–848. [Google Scholar]
- 37.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 38.Scholkopf B, Sung K-K, Burges CJ, Girosi F, Niyogi P, Poggio T, Vapnik V. Comparing support vector machines with gaussian kernels to radial basis function classifiers. Signal Processing, IEEE Transactions on. 1997;45(11):2758–2765. [Google Scholar]
- 39.Friedman J, Hastie T, Tibshirani R. The elements of statistical learning. Vol. 1 Springer series in statistics Springer; Berlin: 2001. [Google Scholar]
- 40.Freund Y, Schapire RE. European conference on computational learning theory. Springer; 1995. A desicion-theoretic generalization of on-line learning and an application to boosting; pp. 23–37. [Google Scholar]
- 41.Bishop CM. Pattern recognition and machine learning. Springer; 2006. [Google Scholar]
- 42.Pudil P, Novovičová J, Kittler J. Floating search methods in feature selection. Pattern recognition letters. 1994;15(11):1119–1125. [Google Scholar]
- 43.Brisimi TS, Cassandras CG, Osgood C, Paschalidis IC, Zhang Y. Sensing and classifying roadway obstacles in smart cities: The Street Bump system. IEEE Access. 2016;4:1301–1312. [Google Scholar]
- 44.Dai W, Brisimi TS, Adams WG, Mela T, Saligrama V, Paschalidis IC. Prediction of hospitalization due to heart diseases by supervised learning methods. International Journal of Medical Informatics. 2014 doi: 10.1016/j.ijmedinf.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu T, Brisimi TS, Wang T, Dai W, Paschalidis IC. A joint sparse clustering and classification approach with applications to hospitalization prediction; Decision and Control (CDC), 2016 IEEE 55th Conference on; 2016. pp. 4566–4571. [Google Scholar]
- 46.Cristianini N, Shawe-Taylor J. An introduction to support vector machines and other kernel-based learning methods. Cambridge university press; 2000. [Google Scholar]
- 47.Bousquet O, Boucheron S, Lugosi G. Advanced lectures on machine learning. Springer; 2004. Introduction to statistical learning theory,” in; pp. 169–207. [Google Scholar]
- 48.International classification of diseases, 9th revision (icd9) World Health Organization; 1999. [Online]. Available: www.who.int/classifications/icd/en/ [Google Scholar]
- 49.Current Procedural Terminology (CPT) American Medical Association; 2014. [Online]. Available: www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/coding-billing-insurance/cpt.page. [Google Scholar]
- 50.Logical observation identifiers names and codes (LOINC) Regenstrief Institute. 2017 [Online]. Available: https://loinc.org/
- 51.Medicare severity-diagnosis related group (MS-DRG) Centers for Medicare and Medicaid Services; 2017. [Online]. Available: https://www.cms.gov/icd10manual/version30-fullcode-cms/P0001.html. [Google Scholar]
- 52.D’ Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General cardiovascular risk profile for use in primary care the Framingham heart study. Circulation. 2008;117(6):743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 53.McAllister DA, Hughes KA, Lone N, Mills NL, Sattar N, McKnight J, Wild SH. Stress hyperglycaemia in hospitalised patients and their 3-year risk of diabetes: a scottish retrospective cohort study. PLoS medicine. 2014;11(8):e1001708. doi: 10.1371/journal.pmed.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfuntner A, Wier LM, Steiner C. Costs for hospital stays in the united states, 2010. Agency for Healthcare Research and Quality, Tech. Rep., 2013, statistical Brief. 146 [PubMed] [Google Scholar]
- 55.Sontag ED. Neural Networks and Machine Learning. Springer; 1998. VC dimension of neural networks; pp. 69–95. [Google Scholar]