Abstract
The nervous system represents the most complex tissue in animals. How this complexity evolved has been a challenging question to address. The explosion in single cell sequencing techniques, the development of new algorithms to cluster single cells into cell types, along with powerful tools for drawing developmental trajectories offer a unique opportunity to compare homologous cell types between species. They further permit the identification of key developmental points and transcription factors that can lead to the evolution of new cell types. At the same time, the ease of use and efficiency of CRISPR genome editing technology allow validation of predicted regulators. This promises exciting developments in the next few years in the field of neuronal evolution and development.
Introduction
Single cell sequencing gives access to transcriptomic information for every cell in any animal at nearly every developmental stage. Over the last five years, a number of computational algorithms have been generated to cluster single cell transcriptomes into different cell types in an unbiased manner [1–4], to match cell types between different species [5], and to generate developmental trajectories for these cell types [6–13]. The development of such tools offers us a unique opportunity to address the molecular developmental mechanisms that lead to the evolution of new cell types. In this review, we suggest how newly developed computational tools applied to single cell transcriptomic data can be used to study cell type evolution in neuronal systems.
How do cell types evolve?
Cell types were historically defined as a population of cells sharing the same morphology, and, when known, biochemical properties and function. In the transcriptomic era, similar transcriptomes are another feature that cells should satisfy in order to be classified as the same cell type. The inherent noise of transcription, the effect of cell cycle, as well as the response to potential stimuli can cause the transcriptome to be significantly more diverse than the morphology; two cells identical in terms of morphology and function will never have identical transcriptomes, which makes their classification into cell types more challenging.
Today, large consortia have been initiated to identify all the main cell types in human, mouse, and fly tissues thus defining ‘cell atlases’ [14]. These efforts will generate the knowledge needed to understand the extent of variability within cell types and determine whether some cell types that are thought to be distinct may actually represent elements of a larger continuum [15]. Thus, cell type classification may be seen as a taxonomy of the cells in an organism.
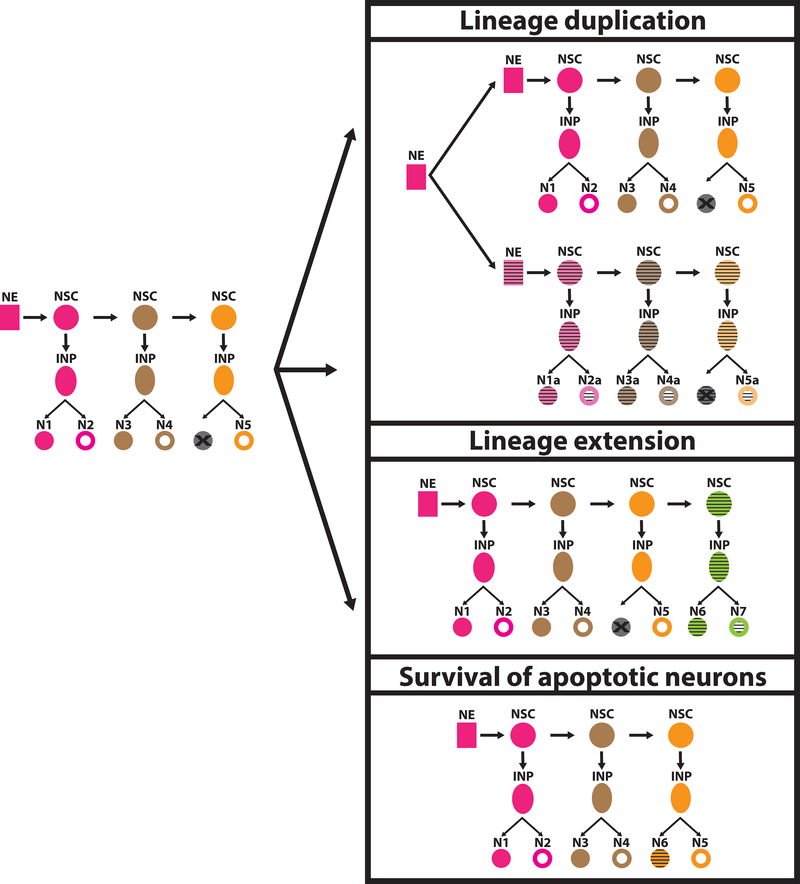
Whether cell types are distinct or form a continuum, it is important to understand how they evolved to give rise to the differences that we observe today (Figure 1). Lineage duplications or extensions can generate supernumerary cells of the same cell type that can then diverge from one another and sub-functionalize [16]. Moreover, apoptosis is a normal component of developmental processes in which cells that are not needed are purged during development [17]; their survival could underpin the evolution of new cell types [18] (Figure 1). Other ways could be envisaged for the evolution of new cell types.
Figure 1: Evolution of development of neuronal cell types.
Different events may lead to the evolution of new neuronal cell types. Duplication of a lineage doubles the amount of neurons that are generated from it; the duplicated neurons can then diversify from the ancestral ones and acquire new functions. The same is true in the case of lineage extensions, where the production of more neurons generates potential for cell speciation. Another alternative is the survival of apoptotic neurons that can then acquire distinct identities. New cell types are indicated by the horizontal stripes.
Neuro: Single cell mRNA sequencing of neuronal cell types
The nervous system consists of a large number of different cell types that are tightly interconnected to form functional circuits. Understanding how this amazing diversity has evolved under the constraints imposed by connectivity is still defying our technical abilities. The newly developed massively parallel single cell mRNA sequencing (scRNAseq) techniques have allowed researchers to read the transcriptomes of all cell types in neuronal tissues. Three main approaches are used to sequence single neurons: a) droplet-based techniques (Drop-seq [1] and 10xGenomics [19]) allow for the parallel sequencing of a large number of single cells at a relatively low depth, b) plate-based methods and Fluidigm [20] are used to sequence many fewer cells at increased sequencing depth per cell, and c) split-pool combinatorial barcoding [21] can be used to increase the number of single cells that can be sequenced in a single experiment. Moreover, single-nucleus techniques have also been beneficial for sequencing neuronal types in vertebrates, avoiding lengthy digestions with proteases for tissues that are complex and hard to dissociate [22]. After acquiring the transcriptome of each cell, clustering algorithms, such as density clustering, k-nearest neighbor algorithms, etc, allow the grouping of single cells into clusters of transcriptionally similar units. Finally, cluster annotation (correspondence of each cluster with a cell type) has to be performed a posteriori and relies on identifying markers that separate the clusters from each other and annotating the cell types that express these markers. A good previous knowledge of the tissue and its morphologically and functionally-defined cell types is thus very beneficial. Using these transcriptomic techniques, many different neuronal tissues have been sequenced at the single cell level over the last two years, greatly increasing our understanding of cell type diversity and development [2,3,22–33].
Evo: How to assign homologous cell types using single cell sequencing
One of the major difficulties in studying cell type evolution is to identify homologous cell types between different animals. A major issue in comparing transcriptomes between animals is assigning orthologous genes [34,35] and normalizing their expression across species [36]. A number of different approaches have been proposed to homologize cell types from one animal to another (Figure 2):
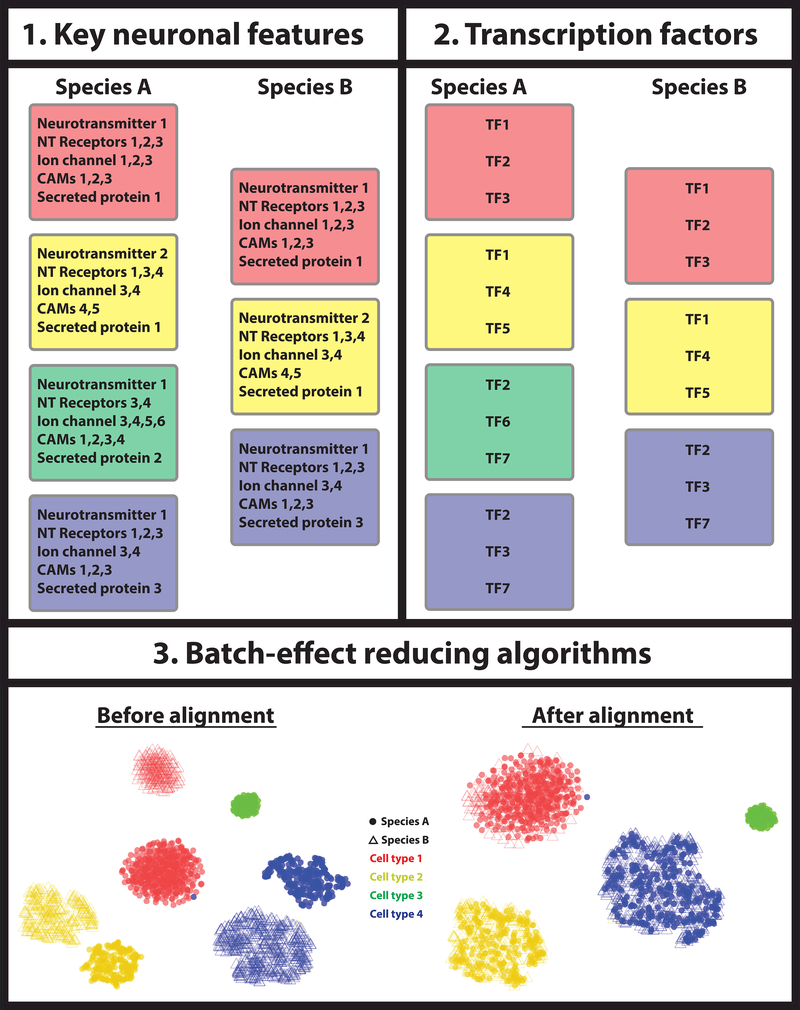
Figure 2: Identification of homologous cell types.
Three different approaches can be used to match homologous cell types between different species: 1) one can use key features that define each neuronal type, e.g. the expression of specific neurotransmitter, neurotransmitter receptors, ion channels, cell adhesion molecules (CAMs), secreted proteins etc. If the different species’ cell types express the same functional molecules, they are most likely homologous. 2) Alternatively, one can select a core regulatory complex of transcription factors and compare their expression between cell types of different species. Homologous cell types should express the same transcription factor fingerprint to regulate their phenotypic characteristics. 3) Finally, one could apply batch effect correction algorithms before comparing the different cell types between the two species, with the assumption that the effect of their independent evolution resembles a technical batch effect.
1). Identification of key features that endow each neuronal type with its functional characters.
One approach to compare neuronal cell types between different species is to uncover a collective set of effector genes that are directly responsible for the specific neuronal features of each cell type and to compare their expression across species. Such features could be neurotransmitter identity, neurotransmitter receptors, ion channels, cell adhesion molecules, etc (Figure 2). This necessarily relies on identifying orthologous genes, which can be a difficult task in the case of large gene families, such as neurotransmitter receptors that often have many different subunits. To overcome this problem, scmap [37] uses an unsupervised approach to select features that are representative of the underlying biological differences [38], which are then used to project data sets from one experiment onto a reference data set of another. Such approach was shown to reliably match pancreatic cell clusters coming from different single cell sequencing studies [37]. It remains to be shown whether scmap can match cell clusters from distinct species.
2). Identification of a core regulatory complex of transcription factors that defines a cell type.
Transcription factors control the morphology, physiology, molecular characteristics, and hence the identity of a neuron. Therefore, the unique combination of transcription factors can be used to define cell types more precisely than the combination of effector genes that are often shared by multiple cell types. One way to compare cell types between different species would be to define a cell type-specific transcription factor identity (core regulatory complex or CoRC [16]) and use this for assigning homologous cell types (Figure 2).
This could be done either in a supervised or unsupervised manner. If the tissue of interest has been studied extensively, one could select transcription factors that are known to specify different neuronal features, such as the case of terminal selectors in C. elegans [39]. Alternatively, one could use unsupervised models to detect combinations of transcription factor present in single cell clusters and then select those that appear to define a cluster in different species.
3). Use of batch-effect reducing algorithms.
Experimental results are affected by technical sources of variation, known as “batch-effect”, which may compromise data analysis. One could consider the differences in expression between the same cell type in different species as the batch effect of an experiment performed by evolution. In this case, an approach to compare cell types from different species would be to apply batch effect correction algorithms before aligning the two datasets (Figure 2). A number of batch-effect correction methods have recently been developed [5,40,41]; however, only one such algorithm has been used to integrate data from different species [5]. It relies on identifying a shared gene correlation structure that is conserved between the data sets. It can then spot cells that cannot be well described by this shared structure, which can be used to identify cell types that are nonoverlapping between the data sets. It then aligns the data sets into a conserved lowdimensional space, using nonlinear ‘warping’ algorithms to normalize for differences in feature scale, in which one can perform comparative analysis to identify changes in population density or gene expression. This is an active area of research and, as new data from different species are being generated, new algorithms will emerge.
The techniques described above could allow for the identification of homologous cell types, as well as taxon-specific cell types. Such an example is the homology between neo-endometrial stromal fibroblasts (neo-ESFs), which are found in eutherian mammals, and paleo-ESFs found in marsupial mammals. Neo-ESFs are evolutionarily related to decidual stromal cells, which are unique to eutherian mammals and are important for the immune tolerance following embryo implantation [42]. How do taxon-specific cell types, such as decidual stromal cells, arise during development? Were they gained in one lineage or were they lost in the other? What are the molecular mechanisms that led to their evolution?
Devo: How to define developmental trajectories
Neuronal cell type diversity is established during development. To recognize the potential for the generation of a new cell type during evolution (“cell speciation”), it is important to know the cell lineage of the tissue of interest in different species to then be able to discover differences in these lineages. Extensive research in model organisms such as worms, flies and mice, has provided a significant breadth of information regarding the development of neuronal tissues. In the absence of such information in an animal of interest, one can use a number of different recently developed techniques that rely on sequential CRISPR-generated mutations and single cell sequencing and allow the reconstruction of cell lineages during development.
scGESTALT [44], ScarTrace [46], and LINNAEUS [45] rely on the use of CRISPR for generating different marks in multiple target loci that have been integrated in the genome. The inherent variability of the targeting and repair process introduces different mutations at these loci in different branches of the cell lineage. The mutated targets can then be read in adult cells alongside the cell-specific transcriptome using single cell sequencing; the transcriptome allows for the identification of the cell type, while the specific CRISPR-induced mutations are used to reconstruct the lineage tree, in ways similar to the reconstruction of phylogenetic trees based on specific DNA sequences. This has enabled the reconstruction of neuronal lineages in zebrafish, both in larva and adult brains. Moreover, the ease-of-use of CRISPR and efficiency in diverse species [47] provides the opportunity to apply such lineage tracing methods beyond the established model organisms.
Developmental cell lineages are extremely useful to understand the development of cell types, but coupling them with the knowledge of the molecules that drive the differentiation of sister cells after each division is necessary to understand how each cell type is generated. A large number of trajectory inference (TI) methods have been developed over the last 5 years to probe developmental trajectories, and predict the critical genes at each bifurcation along the trajectory [6–13].
TI methods typically consist of a dimensionality reduction step and a trajectory-building step. The first step performs dimensionality reduction (PCA, ICA, t-SNE, and diffusion maps), clustering, and/or graph building to represent the cells in a reduced feature space, while maintaining the main characteristics of the data. In the second step, the cells are ordered along the simplified space in order to find a path through a series of nodes. Depending on the algorithm, the user may input different priors such as the initial cell or node or the topology of the trajectory [7].
Although most of the developed algorithms are able to handle multiple bifurcations, it is clear that the more complex the tissue of interest, the harder the reconstruction of the developmental trajectories of its constituent cells. Neuronal tissues are among the most complex ones, consisting of a great variety of neuronal cell types. While its complexity is what makes the neuronal tissue attractive for studying how diversity develops and evolves, it is this same complexity that makes it extremely difficult to study. Improving the depth of the sequencing in single cells and the number of available single cell transcriptomes are two factors that can facilitate the reconstruction of reliable developmental trajectories. Moreover, new developmental trajectory algorithms are being developed to accommodate leaps in the development of single cell sequencing technology and the constantly increasing amount of available data.
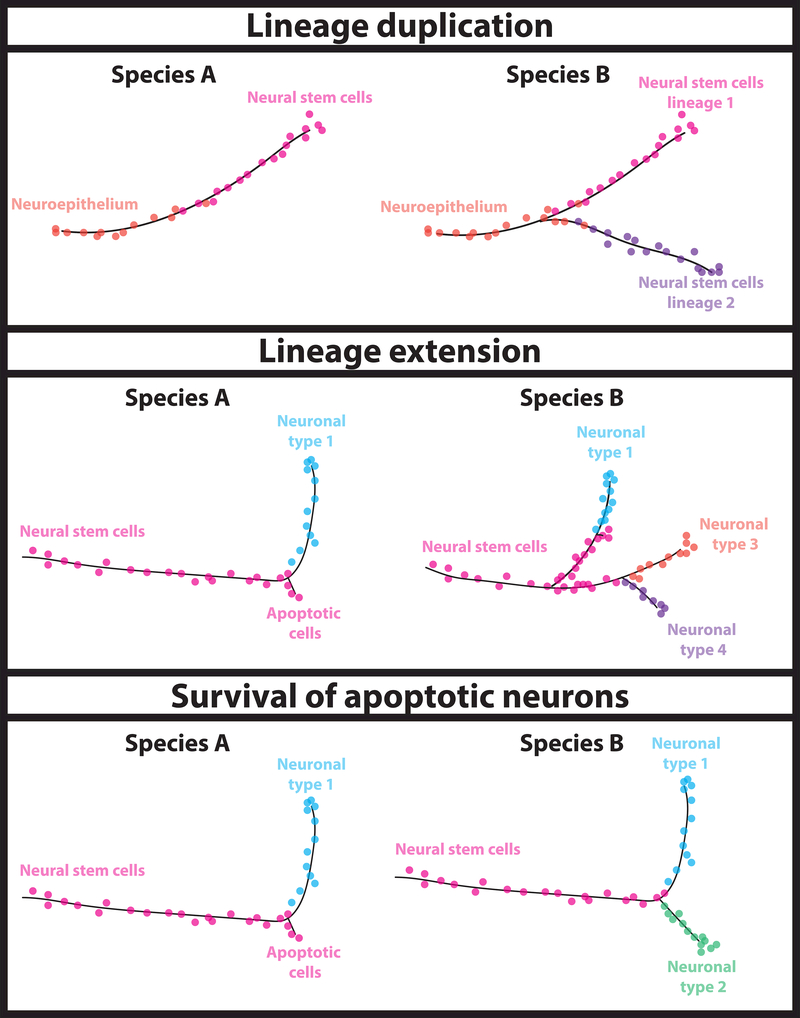
Once the developmental trajectories of a tissue in different species are defined, it becomes possible to figure out how cell types that are unique in one animal evolved. Differences in the developmental trajectories can pinpoint key splits in the trajectory of evolutionary related cells, indicate the mechanisms that led to the evolution of new cell types, and suggest transcription factors that differ between species and may have been involved in this split (Figure 3). But what is the significance of these regulators? How do they contribute to the generation of a new cell type? What are the genetic changes that led to the evolution of a new cell type?
Figure 3: Comparison of developmental trajectories between species.
After inferring developmental trajectories for the cell types of interest in two different species, one can compare these trajectories to identify the developmental mechanisms that led to the evolution of the new cell type. Based on the developmental trajectory, one can distinguish between cases of lineage duplication and extension, as well as instances where cells that undergo apoptosis in one species survive in the other, and define key splits in the trajectory of evolutionary related cells. Studying the differential expression of genes along the trajectory, one can identify putative transcription factors that differ between species and may have been involved in the key splits.
CRISPR-induced mutations present the opportunity to test such regulators. Once the regulator that allows for the differentiation of a newly evolved cell type from its evolutionary related cell type is determined, one can look for genomic changes in its cis elements that may have changed its expression. One can then use CRISPR to exchange enhancers and try to force the generation of this cell type in a species where it doesn’t exist. This opens up other questions (e.g. the identification of cis and trans elements that influence the expression of genes in specific cell types), which can be addressed using available single cell chromatin accessibility techniques [48,49], and can give us a holistic understanding of how a new neuronal cell type evolved.
Conclusions
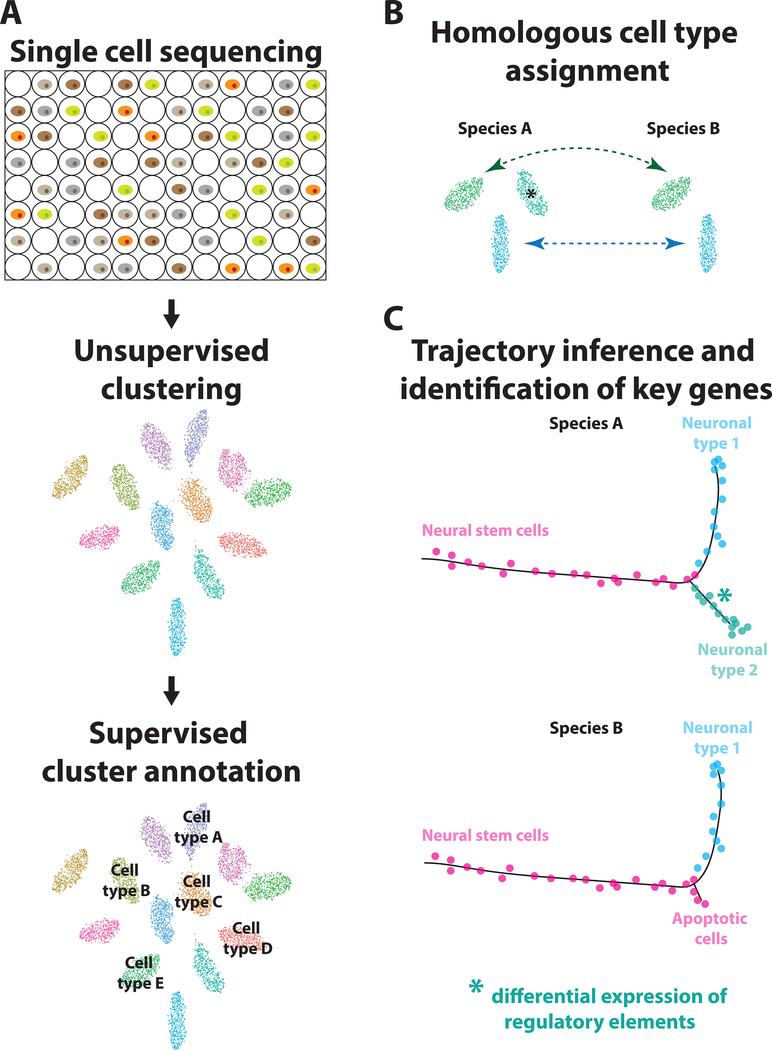
We present here how new single cell sequencing technologies and available algorithms can be used to probe neuronal cell type evolution in three steps (Figure 4): a) single cell sequencing of the same tissue in two different species, clustering, and cluster annotation, b) identification of homologous cell types, as well as cell types that are unique in one species, c) trajectory inference for the development of these cell types, comparison of the trajectories in the two species, and identification and validation of candidate transcription factors that may be causing these different trajectories. This will give us the opportunity to address evo-devo questions in a thorough manner, switching from candidate genes to unbiased large-scale sequencing and from model to non-model organisms. As discussed here, important challenges remain, which will soon be possible to overcome with the fast improvements of single-cell sequencing technologies and computational algorithms. Cells are the product of their evolutionary and developmental history, which is reflected in their transcriptome. By reading their history, we will be able to reverse-engineer it to guide successful in vitro differentiation of pluripotent stem cells towards specific neuronal types.
Figure 4: Overview of technologies and algorithms to probe neuronal cell type evolution.
To understand how neuronal cell types evolve, one can combine available single-cell sequencing techniques with powerful algorithms: A) Use massive parallel sequencing technologies to sequence every single cell in the tissue of interest in different species, clustering algorithms to group single cells in transcriptionally similar clusters and then annotate the clusters based on the expression of specific markers. B) Assign homologous cell types between different species (arrows) and identify cell types that are unique in one of them (asterisk). C) Draw developmental trajectories of species-specific cell types and their evolutionary sister cells, compare the trajectories between the two species, and identify key genes that may have played a causal role in the evolution of these species-specific cell types. These genes can be then interrogated for their role using CRISPR driven knock-outs.
Highlights.
-
–
New sequencing technologies can define the transcriptome of any single cell type
-
–
Algorithms can identify homologous cell types in different species
-
–
New computational tools exist to infer developmental trajectories
-
–
It is possible to pinpoint the role of transcription factors in the evolution of neuronal cell types
-
–
It is a unique time to address developmental mechanisms that guide neural evolution
Acknowledgements
The authors would like to thank Michalis Averof and members of the Desplan lab for critical reading and comments on the manuscript. This work was supported by the National Institutes of Health [R01 EY13012]. NK was supported by an EMBO long-term fellowship (365–2014) and a postdoctoral HFSP fellowship (LT000122/2015-L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. : Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015, 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 3.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. : Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 2016, 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Z, Mich JK, Ku S, Menon V, Krostag AR, Martinez RA, Furchtgott L, Mulholland H, Bort S, Fuqua MA, et al. : A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell Stem Cell 2017, 20:120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.••.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R: Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018, 36:411–420. This paper introduces the first algorithm to try to match cell populations across different species using single cell RNA-Seq datasets. The authors were able to align single cell RNA-Seq datasets from human and mouse pancreatic islets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannoodt R, Saelens W, Saeys Y: Computational methods for trajectory inference from single-cell transcriptomics. Eur J Immunol 2016, 46:2496–2506. [DOI] [PubMed] [Google Scholar]
- 7.••.Saelens W, Cannoodt R, Todorov H, Saeys Y: A comparison of single-cell trajectory inference methods: towards more accurate and robust tools. bioRxiv 2018. This paper compares more than 50 available trajectory inference methods, highlighting their advantages and limitations and providing guidelines for researchers who want to choose the appropriate method for their system. [DOI] [PubMed] [Google Scholar]
- 8.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL: The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014, 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ: Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 2016, 13:845–848. [DOI] [PubMed] [Google Scholar]
- 10.La Manno G, Soldatov R, Hochgerner H, Zeisel A, Petukhov V, Kastriti M, Lonnerberg P, Furlan A, Fan J, Liu Z, et al. : RNA velocity in single cells. bioRxiv 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf FA, Angerer P, Theis FJ: SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018, 19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.•.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C: Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017, 14:979–982. This paper introduces Monocle 2, currently one of the most efficient algorithms in inferring multibranch trajectories from complex tissues. Monocle 2 was used to draw the trajectory of differentiating blood cells and identify key regulators that when mutated alter cellular identity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe’er D: Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 2016, 34:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. : The Human Cell Atlas. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cembrowski MS, Menon V: Continuous Variation within Cell Types of the Nervous System. Trends Neurosci 2018. [DOI] [PubMed] [Google Scholar]
- 16.Arendt D, Musser JM, Baker CV, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, et al. : The origin and evolution of cell types. Nat Rev Genet 2016, 17:744–757. [DOI] [PubMed] [Google Scholar]
- 17.Pinto-Teixeira F, Konstantinides N, Desplan C: Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett 2016, 590:2435–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry M, Konstantinides N, Pinto-Teixeira F, Desplan C: Generation and Evolution of Neural Cell Types and Circuits: Insights from the Drosophila Visual System. Annu Rev Genet 2017, 51:501–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. : Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017, 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, et al. : Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods 2014, 11:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, et al. : Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018, 360:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.•.Lacar B, Linker SB, Jaeger BN, Krishnaswami S, Barron J, Kelder M, Parylak S, Paquola A, Venepally P, Novotny M, et al. : Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun 2016, 7:11022 One of the first papers where single nuclei sequencing was used to study differential neuronal responses upon neuronal activation. The authors observed a continuum of activation states and ordered them into an temporal pattern based on their transcriptomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. : Massively parallel single-nucleus RNAseq with DroNc-seq. Nat Methods 2017, 14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. : Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 2016, 166:1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croset V, Treiber CD, Waddell S: Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. : Neuronal subtypes and diversity revealed by singlenucleus RNA sequencing of the human brain. Science 2016, 352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA, et al. : Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016, 352:1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. : Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015, 18:145–153. [DOI] [PubMed] [Google Scholar]
- 29.Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Sudhof TC, Quake SR: Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep 2016, 16:1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.•.Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G: Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 2018, 360:881–888. In this paper, the authors used single cell RNA sequencing to study gene expression in reptilian brains and identify homologous interneuron classes between mammals and reptiles. [DOI] [PubMed] [Google Scholar]
- 31.Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. : A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 2018, 555:524–528. [DOI] [PubMed] [Google Scholar]
- 32.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, Aibar S, Makhzami S, Christiaens V, Bravo Gonzalez-Blas C, et al. : A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.•.Konstantinides N, Kapuralin K, Fadil C, Barboza L, Satija R, Desplan C: Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell 2018. The authors used a combiantion of single cell RNA sequencing and cell type RNA sequencing after FACS-sorting to build a cell atlas of the Drosophila optic lobes.They used these data to show that same effector genes are regulated by different transcription factors in different cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. : Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 2016, 167:566–580 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marioni JC, Arendt D: How Single-Cell Genomics Is Changing Evolutionary and Developmental Biology. Annu Rev Cell Dev Biol 2017, 33:537–553. [DOI] [PubMed] [Google Scholar]
- 36.Lun AT, Bach K, Marioni JC: Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 2016, 17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.••.Kiselev VY, Yiu A, Hemberg M: scmap: projection of single-cell RNA-seq data across data sets. Nat Methods 2018, 15:359–362. This paper describes scmap, an algorithm that can select biologically relevant features to project data sets from one experiment onto a reference data set of another. Proof of concept was performed using published data sets from human and mouse tissues. [DOI] [PubMed] [Google Scholar]
- 38.Andrews TS, Hemberg M: Modelling dropouts for feature selection in scRNASeq experiments. bioRxiv 2017. [Google Scholar]
- 39.Hobert O: A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip Rev Dev Biol 2016, 5:474–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haghverdi L, Lun ATL, Morgan MD, Marioni JC: Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018, 36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaham U, Stanton KP, Zhao J, Li H, Raddassi K, Montgomery R, Kluger Y: Removal of batch effects using distribution-matching residual networks. Bioinformatics 2017, 33:2539–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kin K, Nnamani MC, Lynch VJ, Michaelides E, Wagner GP: Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep 2015, 10:1398–1409. [DOI] [PubMed] [Google Scholar]
- 43..••. Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glazar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N: Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 2018, 360 The authors sequenced all cells of a flatworm. They developed a new algorithm to reconstruct a lineage tree, where all cell types are shown to originate from a single stem cell compartment, the neoblasts, which are known to be able to regenerate every cell type. They further identified gene expression changes that are related to tissue regeneration and cell type differentiation. [DOI] [PubMed] [Google Scholar]
- 44.•.Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF: Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 2018, 36:442–450. This is one of the three papers that reported at the same time reconstruction of developmental cell lineages using sequential CRISPR-generated mutations and single cell sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP: Simultaneous lineage tracing and cell-type identification using CRISPRCas9-induced genetic scars. Nat Biotechnol 2018, 36:469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A: Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556:108–112. [DOI] [PubMed] [Google Scholar]
- 47.Gilles AF, Averof M: Functional genetics for all: engineered nucleases, CRISPR and the gene editing revolution. Evodevo 2014, 5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J: Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.•.Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R, Pliner HA, Christiansen L, Qiu X, Steemers FJ, et al. : The cisregulatory dynamics of embryonic development at single-cell resolution. Nature 2018, 555:538–542. This paper uses single-cell combinatorial indexing assay for transposase accessible chromatin with sequencing (sci-ATAC-seq) to assess chromatin accessibility changes in the Drosophila nuclei during early embryonic development (2–12h) and identify tissue-specific regulatory elements. [DOI] [PMC free article] [PubMed] [Google Scholar]