Abstract
Background
A predominant difference between endometrial and normal cells is higher proliferation rate in the former cells which is benign. The genes of inhibitor of differentiation (ID) family play a major role in cell proliferation regulation which might be targeted by the nuclear transcription factor Y (NF-Y) for subsequent epigenetic modifications through the CCAAT box regulatory region. The present study was designed to investigate the epigenetic role of NF-Y on ID gene family in endometrial tissue of patients with endometriosis.
Materials & methods
In this case-control study, 20 patients with endometriosis and 20 normal women were examined for the relative expression of the NF-YA, NF-YB, NF-YC and ID genes by real-time PCR during the proliferative phase. The occupancy of NF-Y on CCAAT box region of ID genes was investigated using chromatin immunoprecipitation (ChIP) followed by real-time PCR.
Results
The NF-YA was over-expressed in eutopic endometrium during the proliferative phase. Although the expression level of NF-YB and NF-YC were unchanged in eutopic samples, they were remarkably higher in ectopic group (P<0.05). The ID2 and ID3 genes were up-regulated in ectopic and eutopic tissues, however ID1 and ID4 genes were down-regulated in these samples (P<0.05). The ChIP analysis revealed significant enrichment of NF-Y on regulatory regions of ID2,3 genes in eutopic group, but reduced binding level of NF-Y to the ID1,3 promoters in ectopic specimens (P<0.05).
Conclusion
The ability of NF-Y to regulate ID genes via CCAAT box region suggests the possible role of NF-Y transcription factor in epigenetic changes in endometrial tissues which may open novel avenues in finding new therapeutic strategies.
Keywords: Nuclear transcription factor Y, ID gene family, Endometriosis, Epigenetic
Background
Endometriosis is an estrogen-dependent inflammatory disease that is strongly associated with infertility and affects over 70 million women worldwide [1, 2]. Although there is no single theory of pathogenesis to entirely explain all the manifestations of endometriosis [3], Sampson’s theory of retrograde dissemination of menstrual debris has gained widespread acceptance as an explanation for the initiating steps in the pathogenesis of endometriosis [4]. There are several leading theories, including altered immunity, coelomic metaplasia, and metastatic spread attempting to explain the origin of endometriosis. Recent studies have also found genetic basis for endometriosis [5]. None of these theories fully explain the whole mechanisms associated with the development of disease and the actual cause remains unknown [6, 7]. Multiple factors including genetics, epigenetics, environmental modifications, aging, and diverse anatomical or biochemical aberrations of uterine function are also involved in the development of endometriosis [8, 9].Accumulating evidence suggest that various epigenetic aberrations may play a significant role in the initiation and progression of endometriosis [10]. Epigenetic modifications such as DNA methylation, chromatin modifications, and RNA interference refer to heritable changes in gene expression with no underlying alteration in the genetic sequence [11].
Nuclear transcription factor Y (NF-Y) is one of the transcriptional regulation factors which mediates the above-mentioned epigenetic modifications [12]. NF-Y complexes are trimeric proteins composed of NF-YA, NF-YB, and NF-YC subunits that play a crucial role in regulating eukaryotic gene expression. A heterodimer of NF-YB and NF-YC which are homologous in sequence to histonesH2B and H2A, respectively, interacts with NF-YA to form the heterotrimeric NF-Y complex [13]. All of these subunits are necessary for binding to the core CCAAT box, a cis element present in 30% of eukaryotic promoters [14, 15]. Among the various DNA binding proteins that interact with this sequence, NF-Y is the only protein which needs all the CCAAT box nucleotides to be able to bind the DNA and has an extremely specific interaction with this region [16]. NF-Y has been shown to (i) mediate the recruitment of polymerase II onto various CCAAT box-containing promoters to permit the transcriptional activation (a pioneering role in activation of transcription) [17], (ii) induce DNA compaction that facilitates promoter-enhancer interactions, and (iii) regulate several cell cycle regulatory genes which are known to be critical for expression control [18, 19]. Although, it has been proved that the NF-Y complex is involved in critical biological processes including cell growth, proliferation/apoptosis balance, tumorigenesis (the elevated levels of NF-Y is associated with breast [20], ovarian [21], prostate [22], and thyroid [23] cancers), and cell reaction to the stressors, the exact roles of NF-Y on regulatory regions of many developmental genes remain poorly understood [21, 24].
The inhibitor of differentiation (ID) gene family consisting of ID-1, ID-2, ID-3, and ID-4 isoforms are known to be dominant negative regulators of differentiation, but the positive regulators of cellular proliferation [25]. The ID helix-loop-helix (HLH) proteins lack the basic DNA binding region and are functional inhibitors of the basic helix-loop-helix (bHLH) transcription factors [26]. Recent data reported non-canonical functions for ID proteins, such as binding to Rb family and biochemical attributes including regulating cell fate, proliferation, differentiation, and migration [27, 28].
Since the exact roles of NF-Y on regulatory regions of ID genes are not still clarified, the current investigation was designed to evaluate the possible epigenetic role of NF-Y on ID gene family through CCAAT box region in endometrial tissue of endometriosis and non-endometriosis women.
Material & methods
Patients and tissue collection
Twenty patients with endometriosis (in stages III and IV), who underwent laparoscopic excision of ectopic endometrium lesions (ovarian endometrioma) were recruited for this study. The patients ranged in age from 20 to 45 years and had regular menstrual cycles. The exclusion criteria for participation were: endometrial hyperplasia, benign masses like fibroids and polyps, inflammatory diseases, autoimmune diseases, endocrine diseases, cancers, sepsis, asthma, glomerulonephritis, osteoporosis, psoriasis, myocardial infarction, and leukemia. It should be mentioned that based on the entity of the study, all analyzed samples were representing whole tissues, which are composed of a mixed population of cells (stromal, epithelial, and inflammatory).
The eutopic endometrium samples from these patients were obtained by pipelle. All tissue samples, i.e. both eutopic (20 samples) and ectopic (20 samples) endometrium, were collected, immediately divided into two separate cryovials (one contained RNAlater (QIAGEN Ltd) for RNA extraction and the other was used in ChIP assays), and stored at − 80 °C until performing the analysis. All biopsies were performed by an experienced gynecologist at the Royan Institute for Reproductive Biomedicine.
The control group consisted of 20 women with no evidence of endometriosis and having at least one child by natural pregnancy. Endometrium samples were taken from normal women during diagnostic laparoscopy. Freshly recovered tissues also divided into two separate cryovials (one contained RNAlater (QIAGEN Ltd) for RNA extraction and the other was used in ChIP assays), and stored at − 80 °C until performing the analysis. The exclusion criteria for the women from the control group were the same as for the patients. Before sampling, the nature of the study was explained and written informed consent was obtained from all participants.
RNA isolation
Each tissue sample (50 mg approximately) preserved in RNALater was homogenized using scalpel blade and glass homogenizer. Total RNA was extracted with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Diethylpyrocarbonate (DEPC)-treated water was used for the dilution of the RNA pellet. The approximate concentration and purity of RNA was assessed by optical density [29] 260/280 ratios. To eliminate genomic DNA contamination from RNA samples, DNase I digestion was performed.
Reverse transcription (RT) and real-time polymerase chain reaction analysis
Individual RNA samples from ectopic, eutopic, and control tissue samples (n = 60) were reverse transcribed into complementary DNA (cDNA). Standard RT was carried out in reaction mixture containing 4 μLof 5X RT Buffer, 2 μL of mixed dNTPs (2 .5μm each), 1 μL of RT-MULV enzyme, 1 μL Random hexamer, 1 μL RNase Inhibitor and 1 μg RNA template. The mixture was incubated in 25 °C for 5 min, 42 °C for 60 min and 70 °C for 5 min. To study the relative mRNA expression of the NF-YA, NF-YB and NF-YC genes, real-time PCR was carried out using Step One Plus™ Real-time PCR System (Applied Biosystems International, Inc., Switzerland). Every target cDNA was co-amplified withglyceraldehyde-3-phosphate dehydrogenase (GAPDH) as endogenous control using specific primers. Real-time PCR was performed in triplicate for each sample according to the following conditions: 5 μL of SYBR Premix Ex TaqII (Takara, Japan), 1 μL of each primer (5 pmol/μL), 11 μL H2O and 2 μL cDNA (12.5 ng/μL) in a final volume of 20 μL for each reaction. The reaction conditions were 95 °C for 4 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 s and 72 °C for 30s and final extension of 72 °C for 10 min. The relative mRNA expression was calculated for each sample according to the 2−ΔΔCTandGAPDHgene was used as normalizer for the 2−ΔΔCTcalculus.All primer sets used in qRT-PCR are shown in Table 1.
Table 1.
Primer pairs used in this study
| Genes | Primer Sequences (5′-3′) | Product length (bp) |
|---|---|---|
| NF-YA | F: TTCTCCAGCAAGTTACAGTC | 183 |
| R: ACCATCATGACCATCCCT | ||
| NF-YB | F: TGCCATCAAGAGAAACGG | 151 |
| R: ACTGCTCCACCAATTCC | ||
| NF-YC | F: AGTATATCCGCTTAGCCCA | 96 |
| R: TCTGTCTGTGTAATCTGTTGAG | ||
| GAPDH | F: CTCATTTCCTGGTATGACAACGA | 122 |
| R: CTTCCTCTTGTGCTCTTGCT | ||
| ID1 | F: CCTTGCTGTTCTGAAACCC | 193 |
| R: GTGGAATGAGAGTGCGGA | ||
| ID2 | F:TGATAGACGTGCCACCTTCC | 103 |
| R: TCAGAATGAAGCCCGAGCC | ||
| ID3 | F:CACAAGATAATTCCTGACGCCA | 204 |
| R: AGTCCGCCTTTAGCCCAA | ||
| ID4 | F:CGCACGGCTCTATAAATACA | 160 |
| R:GTGTCCTAGTCACTCCCTT |
Chromatin immunoprecipitation-real-time PCR analysis
For ChIP assays, homogenized endometrial tissues were cross-linked with 1% formaldehyde and subjected to immunoprecipitation after sonication. The ChIP experiments were performed using the Orange ChIP kit (Diagenode, Belgium), according to the manufacturer’s instructions on three biological replicates. Real time PCR was used to amplify specific promoter DNA (ID1–4) bound to the immunoprecipitated histones (NF-Y) after reversing the histone-DNA cross-links (primers listed in Table 1). All the samples were amplified in triplicate. Data were represented as the percentage of input DNA associated with immunoprecipitated NF-Y relative to input chromatin. A Rabbit polyclonal antibody against NF-Y (Abcam, ab6558) was used.
Statistical analysis of real-time PCR
All the statistical analyses conducted using SPSS 16 software and values were expressed as Mean ± SEM of three separate biological experiments. The ANOVA test was performed to compare the differences between eutopic, ectopic, and control samples. Statistical significance was defined as a P value of < 0.05.
Results
The expression analysis of NF-YA, NF-YB and NF-YC genes, and ID gene family
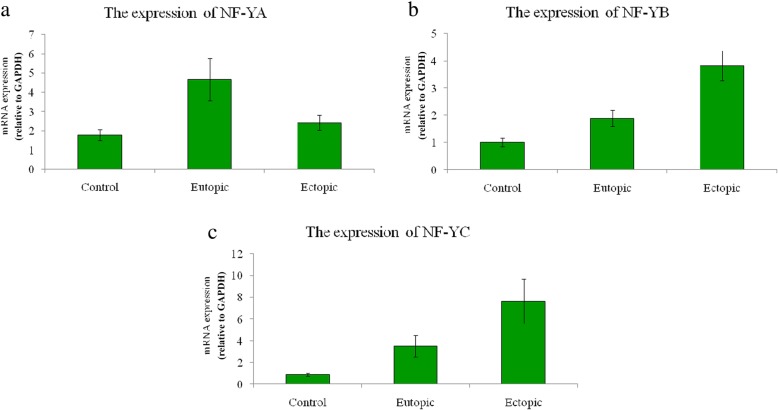
To quantify the relative expression levels of NF-Y (NF-YA, NF-YB, and NF-YC) and ID (ID1–ID4) genes in ectopic, eutopic, and control groups, quantitative real-time PCR was performed (Figs. 1 and 2) (Table 2) during the proliferative phase. As illustrated in Fig. 1a, the expression level of NF-YA is increased in eutopic endometrium in comparison to control (3.39 ± 1.09 versus 1.77 ± 0.29) (P = 0.006) (Table 2). In addition, the expression of NF-YA in eutopic tissues was greater than the ectopic samples (2.41 ± 0.38) (P = 0.039). The expression level of NF-YB and NF-YC had no remarkable changes in eutopic tissue (1.88 ± 0.3 and 3.51 ± 0.97, respectively) in relation to control (1.01 ± 0.15 and 0.88 ± 0.13, respectively) (P = 0.277 and P = 0.397, respectively) (Table 2). On the other hand, their expression significantly increased in ectopic group (NF-YB: 3.81 ± 0.55, and NF-YC: 7.63 ± 2.05) during the proliferative phase compared to that of control (P = 0.000 and P = 0.002, respectively) (Table 2). NF-YB and NF-YC gene expression changes in the ectopic tissues were significant compared to eutopic tissues (Fig. 1b and c).
Fig. 1.
The expression profile of NF-Y subunits in endometriosis patients vs. control group in proliferative phase. Although the NF-YA overexpression in eutopic tissue samples (n = 20) is evident (p = 0.006), there is no remarkable change in its expression level in ectopic specimens (n = 20) compared to normal group (n = 20) (p = 0.730). The NF-YB and NF-YC were up-regulated in ectopic tissues (p = 0.000 and 0.002, respectively), while remained unchanged in eutopic samples (p = 0.277 and 0.397, respectively). (The measures are provided as mean ± SEM)
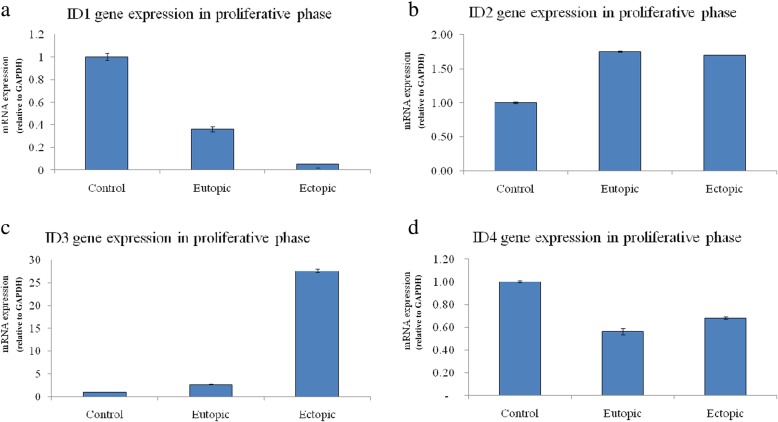
Fig. 2.
The expression profile of ID genes in endometriosis patients vs. control group in proliferative phase. The ID1 and ID4 expression levels were reduced in both ectopic (n = 20) (p = 0.001 and 0.015, respectively) and eutopic (n = 20) (p = 0.025 and 0.019, respectively) samples in comparison with control specimens (n = 20). On the other hand, ID2 and ID3 were overexpressed in ectopic (p = 0.0006 and 0.005, respectively) and eutopic tissues (p = 0.012 and 0.004, respectively). (The measures are presented as mean ± SEM)
Table 2.
Expression analysis of genes encoding NFY complex subunits and ID gene family members and the evaluation of NFY complex incorporation on ID genes upstream regions in normal and endometrial ectopic and eutopic tissue specimens
| Tissue | Gene | qPCR | P-value | ChIP-seq | P-value |
|---|---|---|---|---|---|
| Eutopic | NF-YA | 3.39 ± 1.09 | 0.006 | ||
| NF-YB | 1.88 ± 0.3 | 0.277 | |||
| NF-YC | 3.51 ± 0.97 | 0.397 | |||
| ID1 | 0.36 ± 0.02 | 0.025 | 0.34 ± 0.045 | 0.195 | |
| ID2 | 1.75 ± 0.01 | 0.012 | 0.45 ± 0.088 | 0.014 | |
| ID3 | 2.67 ± 0.09 | 0.004 | 0.30 ± 0.037 | 0.025 | |
| ID4 | 0.56 ± 0.03 | 0.019 | |||
| Ectopic | NF-YA | 2.41 ± 0.38 | 0.730 | ||
| NF-YB | 3.81 ± 0.55 | 0.000 | |||
| NF-YC | 7.63 ± 2.05 | 0.002 | |||
| ID1 | 0.05 ± 0.03 | 0.001 | 0.05 ± 0.016 | 0.005 | |
| ID2 | 1.7 ± 0.00 | 0.0006 | 0.20 ± 0.076 | 0.799 | |
| ID3 | 27.54 ± 0.4 | 0.005 | 0.58 ± 0.02 | 0.025 | |
| ID4 | 0.68 ± 0.01 | 0.015 | |||
| Normal | NF-YA | 1.77 ± 0.29 | |||
| NF-YB | 1.01 ± 0.15 | ||||
| NF-YC | 0.88 ± 0.13 | ||||
| ID1 | 1 ± 0.03 | 0.25 ± 0.032 | |||
| ID2 | 1 ± 0.01 | 0.14 ± 0.008 | |||
| ID3 | 1 ± 0.03 | 0.19 ± 0.034 | |||
| ID4 | 1 ± 0.01 |
In proliferative phase, the expression level of ID1, ID2, ID3, and ID4 genes in control, ectopic, and eutopic groups were assessed (Fig. 2a-c) (Table 2). The results indicated the down-regulation of ID1 and ID4 genes in both ectopic (ID1: 0.05 ± 0.03, and ID4: 0.68 ± 0.01) (P = 0.001, and P = 0.015) and eutopic (ID1: 0.36 ± 0.02, ID4: 0.56 ± 0.03) (P = 0.025, and P = 0.019) tissues in proliferative phase (Fig. 2a, d) (Table 2). However, the expression level of ID2 and ID3 was observed to be increased in ectopic (ID2: 1.7 ± 0.00, ID3:27.54 ± 0.4) (P = 0.0006, and P = 0.005) and eutopic (ID2: 1.75 ± 0.01, ID3: 2.67 ± 0.09) (P = 0.012, and P = 0.004) tissues (Fig. 2b, c) (Table 2).
NF-Y incorporation on regulatory regions of ID genes in ectopic and eutopic tissues of women with endometriosis
To gain insight into NF-Y-mediated transcriptional regulation of ID genes in endometriosis, we investigated the occupancy of NF-Y complex on CCAAT regulatory region and/ or its complementary sequence in eutopic, ectopic, and normal tissues during the proliferative phase using ChIP analysis followed by real time PCR.
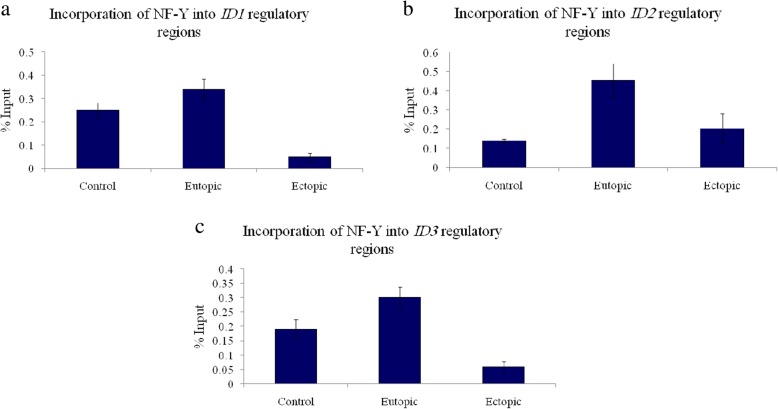
The ChIP analysis of the eutopic samples revealed significant enrichment of NF-Y on the CCAAT-containing regions of ID2, and ID3 promoters in the proliferative phase (ID2: 0.45 ± 0.088, and ID3: 0.30 ± 0.037) in comparison with the normal samples (ID2: 0.14 ± 0.008, and ID3: 0.19 ± 0.034) (P = 0.014 and P = 0.025, respectively) (Fig. 3b, c) (Table 2). However, there was no remarkable difference in NF-Y binding level to the regulatory regions of ID1gene between eutopic (ID1: 0.34 ± 0.045) and control (ID1: 0.25 ± 0.032) tissues (P = 0.195) (Fig. 3a) (Table 2).
Fig. 3.
Incorporation of NF-Y complex on the regulatory regions of a: ID1, b: ID2, and c: ID3 genes in endometriosis patients vs. control group in proliferative phase. The data shows statistically significant NFY complex enrichment on ID2 (p = 0.014) and ID3 (p = 0.025) promoters in eutopic group (n = 20), but notable reduction in NFY binding to ID1 (p = 0.005) and ID3 (p = 0.025) upstream regions in ectopic group (n = 20) in comparison with normal group (n = 20). (The measures represent mean ± SEM)
Comparing the level of NF-Y binding to the upstream regions of ID genes in ectopic and control specimens in proliferative phase, it was found that the binding level to the ID1 and ID3 CCAAT boxes was decreased in ectopic tissue samples (ID1: 0.05 ± 0.016, and ID3: 0.58 ± 0.02) (P = 0.005 and P = 0.025, respectively) (Fig. 3a, c) (Table 2). In ectopic group, a slight increment of binding to the ID2 upstream regions (ID2: 0.20 ± 0.076) was observed which was not statistically significant (P = 0.799) (Fig. 3b) (Table 2).
Eventually, NF-Y incorporation on the ID1, ID2 and ID3 genes in eutopic group was remarkably greater than the ectopic group (P = 0.000, P = 0.045, and P = 0.000, respectively) (Fig. 3). However, the incorporation of NF-Y was not identified on ID4 gene.
Discussion
Endometriosis is a benign progressive disease with endometrial lesions leading to infertility in women of reproductive age [30]. According to the recent investigations, endometriosis is caused by several factors and the epigenetic plays a major role in development of the disease [10, 31, 32].
The NF-Y is one of the factors responsible for epigenetic changes. This factor facilitates the interaction between promoter and the enhancers by binding to the CCAAT BOX regulatory region through affecting the DNA condensation. This is a crucial process in the expression regulation of genes with this motif [13, 33]. Any changes in NF-Y (whether its expression or DNA binding properties) is regarded as a disease causing factor [13].
The current study revealed the up-regulation of NF-YB and NF-YC genes in ectopic tissue of endometriosis patients in the proliferative phase of the menstrual cycle which was significant in comparison to the control group. The NF-YA gene expression was remarkably increased in the endometrium of endometriosis patients (eutopic tissue) than that of the control group and the augmentation in this group was higher than the ectopic group.
Previous studies demonstrated that the cell growth and proliferation in endometriosis patients is increased in the proliferative phase [34]. It was also declared that some signaling pathways, related to the increased cell division and survival, are more activated in endometriotic cells of these patients [35]. Furthermore, the eutopic and ectopic endometriums have less differentiated cells which results in decreased levels of apoptosis in such tissues [35]. NF-Y regulates the balance between proliferation and differentiation states through the recruitment of RNA polymerase II and communication with transcription factors and the enzymes binding to the promoter regions of cell cycle-regulated genes [13, 36, 37].
In present study, the binding level of NF-Y factor to the regulatory regions of ID1, ID2, and ID3 genes in endometrium of the control and patient groups was compared in the proliferative phase in order to epigenetic assessments.
The ID family genes (ID1, ID2, ID3, and ID4) are major regulators of two important cell processes: the proliferation and differentiation. These genes play important roles in signaling pathways, cell fate determination, cell death, tumorigenesis, cell cycle and others as a result of their ability to restrain the differentiation and stimulate the proliferation [38–40]. An interesting feature of ID family genes (which led us to select them for the current research) is the existence of CCAAT regulatory region or its complementary sequence (ATTGG) in their promoter which affects the regulated binding of NF-Y to this region. It is worth to mention that among all the members of ID gene family, the ID4 is deficient in the presence of CCAAT sequence in its upstream region to be occupied by the NFY complex. The binding of NF-Y protein to the ID gene promoter implies that such connection is necessary for transcription initiation in the cell division process [33].
Regarding the results from quantitative comparison of ID1, ID2, ID3, and ID4 gene expression between the control and endometriosis groups in the proliferative phase, the ID2 and ID3 gene expression in both ectopic and eutopic tissues were significantly higher than the control group and the expression level of ID1 and ID4 genes was decreased in the ectopic and also eutopic tissue.
Among the factors affecting the expression of ID gene family, c-Myc is a reasonable candidate for being responsible for ID gene family expression changes in endometriosis patients. C-Myc, a multifunctional nuclear phosphoprotein, participates in the cell cycle and apoptosis processes as a transcription factor regulating the expression of certain genes [41]. C-Myc contains a binding site for ID genes’ promoter which has more affinity for ID2 promoter than others. However, it was clarified that ID3 promoter is also a target for this protein in recent years [42, 43]. The previous investigations indicated that c-Myc factor increases the expression of these genes. The studies revealed that the expression of c-Myc as a regulating factor for cell cycle is increased in the eutopic and ectopic tissues of endometriosis patients in the proliferative phase in comparison to normal people. The elevated level of c-Myc expression in endometriosis patients seems to be related to the estrogen level increment and also TGF-β expression reduction [44]. Furthermore, it has formerly been disclosed that in endometrial tissues, there is an elevated level of ID2 gene expression negatively affected by increased levels of TGF-β [45]. This expression pattern exhibits conformity to the expression of ID2 and ID3 in current study. Regarding the similar expression patterns, it can be interpreted that c-Myc family is responsible for ID2 and ID3 up-regulation in eutopic and ectopic tissues in the proliferative phase.
Moreover, the helix-loop-helix (HLH) domain is conserved in all the members of ID family but the C- and N-terminal regions are different which provides the possibility of interaction between ID genes and different factors resulting in various consequences [46]. The hematopoietic stem cell studies indicated that each member of ID family participates in a certain phase of hematopoietic cell evolution. For example, once the cell is stimulated by inducing cytokines like Interleukine-3, Interleukine-6, and Erythropoietin, the ID1 gene is over-expressed suggesting the role ID1 plays in proliferation and pluripotency maintenance of hematopoietic stem cells. ID1 gene also participates in cell fate determination in T cell or NK cell differentiation pathway but the expression of ID2 is not notably increased in this phase and is significantly elevated in final stages (granulocyte differentiation) instead [47].
The above mentioned differences also lead to various ID family activities in different cells and tissues. For example, the over-expression of ID1 gene in fetus cerebral cortex results in neural cell proliferation and ID4 gene is necessary for mammary gland development [48–50].
The expression of ID1, ID2, and at a lesser amount the ID3 genes were also investigated in several studies including prostate cancer researches and the relationship between their over-expression and cancer incidence was clearly perceived. On the other hand, some studies declared that the reduction of ID4 gene expression is responsible for prostate cancer development. Other studies clarified that the ID4 gene expression in normal breast and stomach tissues is highly elevated in comparison to cancerous tissues denoting the tumor suppressive role for this protein [51, 52].
The binding level of NF-Y protein complex to the promoter regions of ID1, ID2, and ID3 genes was displayed through ChIP-Real-Time-PCR and according to the results; it was considerably increased in eutopic group of endometriosis patients in proliferative phase.
The function of NF-Y transcription factor is regulated by post-translational molecular mechanisms. The regulation of NF-Y is accomplished at protein level and through the NF-YA subunit [16, 53, 54]. The protein level of NF-Y is increased and/ or decreased depending on the cell status. It shows that the binding level of NF-Y protein fluctuates in cells and the regulation through NF-YA subunit prevents the whole complex from binding to the DNA.
The results indicated that the NF-YA is significantly over-expressed in eutopic tissue of endometriosis patients and the binding level of NF-Y complex to the promoter regions of ID genes in eutopic tissue is elevated as well. This is significant as the DNA binding of NF-Y complex is accomplished through NF-YA subunit and it is the only subunit possessing the DNA binding domain which without this domain the complex is not able to bind the CCAAT motif [55].
Comparing the binding level of NF-Y complex to the promoter regions of ID1, ID2, and ID3 genes with quantitative data from expression of ID1, ID2, and ID3 genes in control and endometriosis groups in proliferative phase, it was revealed that the level of NF-Y complex binding to the ID2 promoter region in endometrium of endometriosis women (the eutopoic tissue) in proliferative phase of the menstrual cycle is augmented in comparison with the control group which is accompanied by over-expression of this gene in the endometrium and endometrial lesions of endometriosis patients in proliferative phase.
Moreover, a reduction in binding level of NF-Y to the ID1 gene promoter was observed in endometrial lesions (ectopic tissue) in proliferative phase compared to the control group.
In contrast, the NF-Y binding to the ID3 promoter region in the endometrial lesions (ectopic tissue) in proliferative phase is significantly decreased compared to the control group which is contradictory to ID3 gene expression data in the proliferative phase.
The studies demonstrated that CCAAT box sequence (the sequence which NF-Y binds to) is one of the most common elements in the upstream of eukaryotic promoters. In addition to the CCAAT box motif, there are other conserved regions in class II eukaryotic promoters including GC-box and TATA-box [56]. Likewise, in addition to NF-Y protein, there are other proteins comprising HSP-CBF, Y-BOX FACTORS, CTF/NF-1, and C/EBF to bind to this site of DNA [55].
Therefore, the involvement of other conserved regulatory regions and/ or other regulatory elements in the process of ID3 gene expression regulation in endometrial lesions (ectopic tissue) of endometriosis patients have a great likelihood. It was also determined that the genes without TATA box are more dependent to their CAT box region [12, 16].
The ID2 gene has two CAT box regions and no TATA box, while the ID1 and ID3 genes have only one CAT box [33]. As the TATA box lacking genes are intensively dependent to their CAT box region, the role of NF-Y protein in ID2 gene regulation and epigenetic in endometriosis patients is more significant than its binding to the ID1 and ID3 genes.
Conclusion
The altered levels of NF-YA, NF-YB, and NF-YC expression resulted in some changes in the expression of ID gene family in both ectopic and eutopic tissues of endometriosis patients in proliferative phase. Furthermore, the incorporation of NF-Y complex on CCAAT box region of ID1, ID2, and ID3 promoters was highly enhanced in endometriosis patients. Thus, it can be suggested that NF-Y transcription factor has regulatory role on ID gene family through CCAAT box region and is responsible for epigenetic changes in endometrial tissues of endometriosis patients.
However, more investigations are required to clarify the different underlying molecular regulatory mechanisms in eutopic and ectopic endometriums in endometriosis patients which may open novel avenues in understanding of endometriosis pathophysiology and give rise to novel therapeutic strategies.
Acknowledgements
We are thakful to all the participants of the study.
Funding
Not applicable.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Abbreviations
- bHLH
Basic helix-loop-helix
- c DNA
Complementary DNA
- ChIP
Chromatin immunoprecipitation
- DEPEC
Diethylpyrocarbonate
- HLH
Helix-loop-helix
- ID
Inhibitor of differentiation
- NF-Y
Nuclear transcription factor Y
- RT
Reverse transcription
Authors’ contributions
ShA: Data collection, Data analysis and interpretation, Drafting the manuscript; MA: Data collection; FR: Critical revision of the manuscript; RA: Final approval of the manuscript; PA: Data collection; MSh: Study design and Supervising the research. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by Ethical Committee of Royan Institute for Reproductive Biomedicine. Informed consent was obtained from all the participants for being included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bulun SE. Mechanisms of disease endometriosis. Th e new england journal o f medicine. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 2.Nothnick W, Alali Z. Recent advances in the understanding of endometriosis: the role of inflammatory mediators in disease pathogenesis and treatment. F1000Research. 2016;5. 10.12688/f1000research.7504.1. [DOI] [PMC free article] [PubMed]
- 3.Sourial S, Tempest N, Hapangama DK. Theories on the pathogenesis of endometriosis. International journal of reproductive medicine. 2014;2014. [DOI] [PMC free article] [PubMed]
- 4.Sampson JA. Endometrial carcinoma of the ovary, arising in endometrial tissue in that organ. Arch Surg. 1925;10(1):1–72. [Google Scholar]
- 5.Pavone ME, Lyttle BM. Endometriosis and ovarian cancer: links, risks, and challenges faced. Int J Women's Health. 2015;7:663. doi: 10.2147/IJWH.S66824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acién P, Velasco I. Endometriosis: a disease that remains enigmatic. ISRN obstetrics and gynecology 2013;2013. 10.1155/2013/242149. [DOI] [PMC free article] [PubMed]
- 7.Kumar R, Clerc A-C, Gori I, Russell R, Pellegrini C, Govender L, et al. Lipoxin A4 prevents the progression of de novo and established endometriosis in a mouse model by attenuating prostaglandin E2 production and estrogen signaling. PLoS One. 2014;9(2):e89742. doi: 10.1371/journal.pone.0089742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forte A, Cipollaro M, Galderisi U. Genetic, epigenetic and stem cell alterations in endometriosis: new insights and potential therapeutic perspectives. Clin Sci. 2014;126(2):123–138. doi: 10.1042/CS20130099. [DOI] [PubMed] [Google Scholar]
- 9.Shao R, Cao S, Wang X, Feng Y, Billig H. The elusive and controversial roles of estrogen and progesterone receptors in human endometriosis. Am J Transl Res. 2014;6(2):104–113. [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S-W. Epigenetics of endometriosis. Mol Hum Reprod. 2009;15(10):587–607. doi: 10.1093/molehr/gap064. [DOI] [PubMed] [Google Scholar]
- 11.Nasu K, Kawano Y, Tsukamoto Y, Takano M, Takai N, Li H, et al. Aberrant DNA methylation status of endometriosis: epigenetics as the pathogenesis, biomarker and therapeutic target. J Obstet Gynaecol Res. 2011;37(7):683–695. doi: 10.1111/j.1447-0756.2011.01663.x. [DOI] [PubMed] [Google Scholar]
- 12.Ly LL, Yoshida H, Yamaguchi M. Nuclear transcription factor Y and its roles in cellular processes related to human disease. Am J Cancer Res. 2013;3(4):339–346. [PMC free article] [PubMed] [Google Scholar]
- 13.Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47(1):29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- 14.Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8(24):4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- 15.Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152(1):132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26(5):1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shilovskiĭ I, Mazurov D, Khaitov M. The development of vector constructions for respiratory syncitial virus (RSV) P-gene silencing. Patologicheskaia fiziologiia i eksperimental’naia terapiia. 2009;(4):11–6. [PubMed]
- 18.Guerra RF, Imperadori L, Mantovani R, Dunlap DD, Finzi L. DNA compaction by the nuclear factor-Y. Biophys J. 2007;93(1):176–182. doi: 10.1529/biophysj.106.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolfini D, Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y&quest. Cell Death & Differentiation. 2013;20(5):676–685. doi: 10.1038/cdd.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niida A, Smith AD, Imoto S, Tsutsumi S, Aburatani H, Zhang MQ, et al. Integrative bioinformatics analysis of transcriptional regulatory programs in breast cancer cells. BMC bioinformatics. 2008;9(1):404. doi: 10.1186/1471-2105-9-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garipov A, Li H, Bitler BG, Thapa RJ, Balachandran S, Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol Cancer Res 2013;11(4):360-369. [DOI] [PMC free article] [PubMed]
- 22.Calvo A, Perez-Stable C, Segura V, Catena R, Guruceaga E, Nguewa P, et al. Molecular characterization of the Gγ-globin-tag transgenic mouse model of hormone refractory prostate cancer: comparison to human prostate cancer. Prostate. 2010;70(6):630–645. doi: 10.1002/pros.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvatore G, Nappi TC, Salerno P, Jiang Y, Garbi C, Ugolini C, et al. A cell proliferation and chromosomal instability signature in anaplastic thyroid carcinoma. Cancer Res. 2007;67(21):10148–10158. doi: 10.1158/0008-5472.CAN-07-1887. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Zhao H, Wang L, Wang Y, Guo X, Xu B. The animal nuclear factor Y: an enigmatic and important heterotrimeric transcription factor. Am J Cancer Res. 2018;8(7):1106–1125. [PMC free article] [PubMed] [Google Scholar]
- 25.Chaudhary J, Sadler-Riggleman I, Ague JM, Skinner MK. The helix-loop-helix inhibitor of differentiation (ID) proteins induce post-mitotic terminally differentiated Sertoli cells to re-enter the cell cycle and proliferate. Biol Reprod. 2005;72(5):1205–1217. doi: 10.1095/biolreprod.104.035717. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole PJ, Inoue T, Emerson L, Morrison IE, Mackie AR, Cherry RJ, et al. Id proteins negatively regulate basic helix-loop-helix transcription factor function by disrupting subnuclear compartmentalization. J Biol Chem. 2003;278(46):45770–45776. doi: 10.1074/jbc.M306056200. [DOI] [PubMed] [Google Scholar]
- 27.Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20(58):8317. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- 28.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14(2):77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhary J, Johnson J, Kim G, Skinner MK. Hormonal Regulation and Differential Actions of the Helix-Loop-Helix Transcriptional Inhibitors of Differentiation (Id1, Id2, Id3, and Id4) in Sertoli Cells. Endocrinology. 2011;142(5):1727–1736. doi: 10.1210/endo.142.5.8134. [DOI] [PubMed] [Google Scholar]
- 30.Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 31.Samadieh Y, Favaedi R, Ramezanali F, Afsharian P, Aflatoonian R, Shahhoseini M. Epigenetic dynamics of HOXA10 gene in infertile women with endometriosis. Reprod Sci. 2018;26(1):1933719118766255. doi: 10.1177/1933719118766255. [DOI] [PubMed] [Google Scholar]
- 32.Hosseini E, Mehraein F, Shahhoseini M, Karimian L, Nikmard F, Ashrafi M, et al. Epigenetic alterations of CYP19A1 gene in cumulus cells and its relevance to infertility in endometriosis. J Assist Reprod Genet. 2016;33(8):1105–1113. doi: 10.1007/s10815-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeinvaziri F, Shahhoseini M. Epigenetic role of CCAAT box-binding transcription factor NF-Y on ID gene family in human embryonic carcinoma cells. IUBMB Life. 2015;67(11):880–887. doi: 10.1002/iub.1443. [DOI] [PubMed] [Google Scholar]
- 34.Wingfield M, Macpherson A, Healy DL, Rogers PA. Cell proliferation is increased in the endometrium of women with endometriosis. Fertil Steril. 1995;64(2):340–346. doi: 10.1016/s0015-0282(16)57733-4. [DOI] [PubMed] [Google Scholar]
- 35.Giudice LC, Evers JL, Healy DL. Endometriosis: science and practice: John Wiley & Sons; 2012.
- 36.Gurtner A, Fuschi P, Magi F, Colussi C, Gaetano C, Dobbelstein M, et al. NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue. PLoS One. 2008;3(4):e2047. doi: 10.1371/journal.pone.0002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceribelli M, Dolfini D, Merico D, Gatta R, Viganò AM, Pavesi G, et al. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28(6):2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 39.Yokota Y. Id and development. Oncogene. 2001;20(58):8290–8298. doi: 10.1038/sj.onc.1205090. [DOI] [PubMed] [Google Scholar]
- 40.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13(8):410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 41.Gearhart J, Pashos EE, Prasad MK. Pluripotency redux—advances in stem-cell research. N Engl J Med. 2007;357(15):1469–1472. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
- 42.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5(8):603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 43.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20(58):8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 44.Johnson MC, Torres M, Alves A, Bacallao K, Fuentes A, Vega M, et al. Augmented cell survival in eutopic endometrium from women with endometriosis: expression of c-myc, TGF-beta1 and bax genes. Reprod Biol Endocrinol. 2005;3(1):45. doi: 10.1186/1477-7827-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young VJ, Ahmad SF, Brown JK, Duncan WC, Horne AW. Id2 mediates the transforming growth factor-β1-induced Warburg-like effect seen in the peritoneum of women with endometriosis. MHR: Basic Sci Reprod Med. 2016;22(9):648–654. doi: 10.1093/molehr/gaw045. [DOI] [PubMed] [Google Scholar]
- 46.Rockman SP, Currie SA, Ciavarella M, Vincan E, Dow C, Thomas RJ, et al. Id2 is a target of the β-catenin/T cell factor pathway in colon carcinoma. J Biol Chem. 2001;276(48):45113–45119. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- 47.Kamalian L, Gosney JR, Forootan SS, Foster CS, Bao ZZ, Beesley C, et al. Increased expression of id family proteins in small cell lung cancer and its prognostic significance. Clin Cancer Res. 2008;14(8):2318–2325. doi: 10.1158/1078-0432.CCR-07-4716. [DOI] [PubMed] [Google Scholar]
- 48.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22. 3 in bladder cancer. Molecular Cancer. 2005;4(1):16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Franklyn J, Stratford A, Boelaert K, Watkinson J, Eggo M, et al. Pituitary tumor-transforming gene regulates multiple downstream angiogenic genes in thyroid cancer. J Clin Endocrinol Metabol. 2006;91(3):1119–1128. doi: 10.1210/jc.2005-1826. [DOI] [PubMed] [Google Scholar]
- 50.Mern D, Hasskarl J, Burwinkel B. Inhibition of id proteins by a peptide aptamer induces cell-cycle arrest and apoptosis in ovarian cancer cells. Br J Cancer. 2010;103(8):1237–1244. doi: 10.1038/sj.bjc.6605897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan ASW, Tsui WY, Chen X, Chu KM, Chan TL, Chan ASY, et al. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003;22(44):6946–6953. doi: 10.1038/sj.onc.1206799. [DOI] [PubMed] [Google Scholar]
- 52.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci. 2002;99(11):7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9(1):173. doi: 10.1186/1471-2407-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper CL, Newburger PE. Differential expression of id genes in multipotent myeloid progenitor cells: id-1 is induced by early-and late-acting cytokines while id-2 is selectively induced by cytokines that drive terminal granulocytic differentiation. J Cell Biochem. 1998;71(2):277–285. doi: 10.1002/(sici)1097-4644(19981101)71:2<277::aid-jcb12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 55.Matuoka K, Chen KY. Nuclear factor Y (NF-Y) and cellular senescence. Exp Cell Res. 1999;253(2):365–371. doi: 10.1006/excr.1999.4605. [DOI] [PubMed] [Google Scholar]
- 56.Stewart JJ, Fischbeck JA, Chen X, Stargell LA. Non-optimal TATA elements exhibit diverse mechanistic consequences. J Biol Chem. 2006;281(32):22665–22673. doi: 10.1074/jbc.M603237200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.