Abstract
Background
The delta-like non-canonical Notch ligand 1 (DLK1)-maternally expressed 3(MEG3) locus (DLK1-MEG3 locus) plays a critical role in the maintenance and differentiation of hematopoietic stem cells. Accumulating evidence implicates the imprinted genes from this locus, DLK1 and MEG3, in the development and progression of acute myeloid leukemia (AML). However, the contribution of this locus to the treatment response of patients and their survival is unknown.
Methods
DNA methylation of select CG dinucleotide-containing amplicons (CpG sites) within the DLK1-MEG3 locus and within differentially methylated regions of other imprinted loci was assessed in the mononuclear cells of 45 AML patients by combined bisulfite restriction analysis. Methylation results were compared with patient response to first-round induction therapy and overall survival. Multivariable analysis was employed to identify independent prognostic factors for patient overall survival in AML.
Results
Increased methylation at CpG sites within the MEG3 promotor region was observed in AML patients having longer overall survival. In addition, patients with shorter overall survival had increased expression of DLK1 and MEG3, and methylation at the MEG3-DMR CpG site inversely correlated with MEG3 expression. Multivariable analysis revealed that methylation at CG9, a non-imprinted CpG site within the MEG3 promotor region which contains a CCCTC-binding factor (CTCF)-binding DNA sequence, is an independent prognostic factor for the overall survival of AML patients.
Conclusions
The results of our pilot study underscore the importance of the DLK1-MEG3 locus in AML development and progression. We identify CG9 methylation as an independent prognostic factor for AML patient survival, which suggests that distinct miRNA signatures from the DLK1-MEG3 locus could reflect varying degrees of cell stemness and present novel opportunities for personalized therapies in the future. These data provide a foundation for future studies into the role of higher-order chromatin structure at DLK1-MEG3 in AML.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0643-z) contains supplementary material, which is available to authorized users.
Keywords: DLK1-MEG3, Imprinting, Leukemia, Cancer stem cells, miRNAs
Introduction
Genomic imprinting is an epigenetic process governed by complementary chromatin structures inherited from the mother and father. With the help of long non-coding RNAs (lncRNAs), differential covalent modifications of maternally and paternally inherited DNA and histone proteins control certain gene dosages in a parent-of-origin-specific manner [1]. The human genome contains more than 100 such imprinted genes which cluster around CG-rich regions of DNA. These regions, known as differentially methylated regions (DMRs), exhibit unique patterns of methylation at CG cytosine residues based on the parental origin of the chromosome. The temporo-spatial dosage of imprinted genes governed by genomic imprinting is integral to proper growth and development, and its dysregulation is found in several developmental abnormalities [2] and malignancies such as leukemia [3].
Embryonic and postnatal growth is under the control of a select group of coregulated imprinted genes which comprise an imprinted gene network (IGN) [4, 5]. Genes that belong to the IGN are highly expressed during embryonic and early postnatal development, but they are downregulated during maturation as the somatic growth rate decelerates [4]. Interestingly, targeted deletion of the lncRNA H19, a member of the IGN, results in the overexpression of other IGN genes [6–8] and postnatal overgrowth [8], both of which normalize upon transgenic H19 re-expression [6, 7]. Similarly, a pivotal work in oocyte fusion revealed that genomic imprinting at the paternally imprinted Igf2-H19, which results in H19 overexpression in bimaternal embryos, prevents the growth and development of bimaternal mice [9], highlighting Igf2-H19 manipulation as a master switch which allows for parthenogenesis. However, it was soon realized that a second paternally imprinted locus housing IGN genes, Dlk1-Meg3, was also responsible for the restricted growth and development of bimaternal embryos [10], and a high efficiency of bimaternal mouse generation was later achieved via genetic manipulation mimicking somatic imprinting of both loci [11].
The IGF2-H19 and DLK1-MEG3 loci each contain oppositely imprinted tandem sets of IGN genes. The IGF2-H19 locus encodes the growth-promoting protein-coding insulin-like growth factor 2 (IGF2) and lncRNA H19, and similarly, the DLK1-MEG3 locus encodes the growth-related protein-coding delta-like non-canonical Notch ligand 1 (DLK1) and tumor suppressor lncRNA MEG3. Both loci also house growth-related microRNAs (miRNAs), and genomic imprinting may be similarly orchestrated for these loci by a single intergenic DMR found within each locus [12]. Interestingly, high expression of several IGN genes, including those located within Igf2-H19 and Dlk1-Meg3, is restricted to hematopoietic stem cells (HSCs) in the adult [4, 13]. Moreover, the Igf2-H19 and Dlk1-Meg3 loci are uniquely important to HSC fate, as they harbor IGN genes which are required for fetal hematopoiesis [14] and miRNAs which govern HSC quiescence [15] and stemness [16].
Accumulating evidence suggests that a direct lineage can be drawn between primordial germ cells (PGCs) and adult HSCs [17], suggesting that HSCs retain PGC-like epigenetic programming for quiescence and stemness maintenance. In this regard, recent studies found that the IGF2-H19 and DLK1-MEG3 loci are involved in the stemness maintenance, proliferation, and tumorigenic potential of embryonal carcinoma (EC) cells [18–20], the cancer stem cells responsible for teratoma formation [21], implicating these loci in the malignant transformation of PGCs to EC cells during PGC maturation. Similarly, aberrant imprinting and expression of IGN genes including those from the IGF2-H19 and DLK1-MEG3 loci occurs in leukemias, leading to the hypothesis that very small embryonic-like stem cells (VSELs), as the pluripotent stem cells atop the adult stem cell hierarchy, may adopt a malignant fate along their differentiation route to HSCs via improper epigenetic transitions [3]. Thus, VSELs potentially represent both the adult stem cell linking PGCs to HSCs [17] as well as a source of preleukemic HSCs, where epigenetic control over IGF2-H19 and DLK1-MEG3 is critical to the maintenance of a healthy HSC compartment.
Hypomethylation of the IGF2-H19 DMR with concomitant-elevated H19 expression is characteristic to VSELs and hypothesized to control their quiescence [22]. In support of this, VSEL expansion strategies are accompanied by de novo methylation of the IGF2-H19 DMR and the emergence of differentiated cells [22, 23]. Interestingly, H19 overexpression is important for leukemic cell proliferation, and DMR methylation-independent H19 overexpression predicts poor survival in AML [24]. Similarly, DLK1 overexpression prevents leukemic cell differentiation [25], and DLK1 is overexpressed in AML via aberrant methylation of an upstream insulator region [26]. On the other hand, MEG3 inhibits leukemogenesis [27] and leukemic cell proliferation [28], but its expression is decreased in AML samples [27, 29] and inversely correlates with promotor methylation [29]. Furthermore, the DLK1-MEG3 miRNA mega-cluster is known to regulate cell stemness [16, 30, 31] and self-renewal [16, 32], and the expression of these miRNAs correlates with methylation at CpG sites throughout this locus in acute promyelocytic leukemia (APL) [33]. Taken together, in contrast to the methylation-independent expression of H19 [24], it appears likely that aberrant methylation within DLK1-MEG3 underlies gene dysregulation from this locus and represents a source of potential prognostic factors for AML survival.
To this end, we assessed methylation at select CpG sites within DLK1-MEG3 (Fig. 1a) and imprinted gene expression from this locus in the peripheral blood mononuclear cells of AML patients. We then compared our results with patient outcomes to probe for independent markers of prognostic value. Based on correlations between CpG site methylation and miRNA expression from this locus in APL, in addition to the roles for this locus in HSC stemness maintenance and leukemic cell proliferation, we hypothesized that patients’ response to chemotherapy and overall survival would depend on CpG site methylation and imprinted gene expression from the DLK1-MEG3 locus.
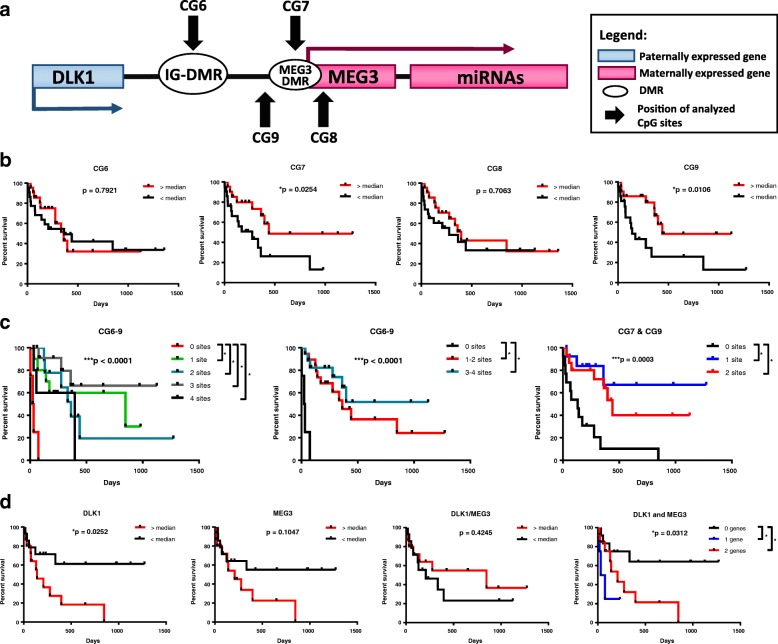
Fig. 1.
Survival of AML patients based on DLK1-MEG3 CpG site methylation and imprinted gene expression in AML MNCs. a Schematic representation of the human imprinted DLK1-MEG3 locus. b Analysis of AML patients’ survival depending on median methylation values for particular CpG site amplicons using the Mantel-Cox test. *p < 0.05. Sample counts for “< median”: CG6 n = 21, CG7 n = 20, CG8 n = 22, and CG9 n = 20; and for “> median”: CG6 n = 22, CG7 n = 21, CG8 n = 23, and CG9 n = 21. c Analysis of AML patients’ survival depending on pooled CG6–CG9 or CG7 and CG9 amplicon median methylation values using the Mantel-Cox test. Lines represent the number of CpG site amplicons out of 4 (CG6–9) or 2 (CG7 and CG9) analyzed which exhibited increased methylation relative to their respective median values. ***p < 0.001 and Bonferroni post hoc analysis *p < 0.05. Sample counts for CG6–9 (0–4 sites): 0 site n = 3, 1 site n = 10, 2 sites n = 9, 3 sites n = 13, and 4 sites n = 5; for CG6–9 (0, 1–2, 3–4 sites): 0 site n = 3, 1–2 sites n = 19, 3–4 sites n = 18; and for CG7 and 9: 0 site n = 12, 1 site n = 14, and 2 sites n = 15. d Mantel-Cox analysis of AML patients’ survival depending on median DLK1, MEG3, and DLK1/MEG3 and pooled DLK1 and MEG3 expression ratio values obtained by RT-qPCR. Red lines represent patients with increased expression of DLK1 and MEG3 and an increased DLK1/MEG3 ratio. For pooled expression results, the Bonferroni post hoc analysis was used and lines represent the number of genes which exhibited increased expression relative to their respective median values. *p < 0.05. Sample counts for “> median” expression: DLK1, MEG3, and DLK1/MEG3 n = 14; and for “<median” expression: DLK1, MEG3, and DLK1/MEG3 n = 15
Methods
Study design and description of patient samples
This investigational pilot study was designed to evaluate the impact of imprinting within DLK1-MEG3 locus on survival and response to treatment of patients with acute non-promyelocytic leukemia (AML). Forty-five patients with newly diagnosed AML were included in the study. Both patients with acute promyelocytic leukemia, due to the specific biology and a different outcome, as well as AML patients who received corticosteroids at the beginning of the treatment course, were not included in the study. Diagnoses were established following the WHO classification system [34]. Blood counts and flow cytometry were performed to confirm the presence of blastic cells, whereas cytogenetic and molecular studies, including the FISH study (AML1/ETO, CBFß/MYH11, and MLLT3-MLL and frequently mutated genes FLT3-ITD, NPM1, and CEBPA), were performed to determine the risk group, according to the WHO recommendation. On the basis of the above, patients were classified as follows (Table 1): 7 (15.6%) patients had good risk (1 patient with t(8;21), 4 with inv. [16]/t(16;16), and 1 with mutated CCAAT/enhancer-binding protein alpha (CEBPAmut)), 10 (22.2%) subjects had 1st intermediate risk (diploid karyotype features with 2 having both mutated nucleophosmin (NPM1mut) and internal tandem duplication of FMS-like tyrosine kinase 3 (FLT3-ITD) and 7 with FLT3-ITD without NMP1mut), 18 (17.8%) patients had 2nd intermediate risk (3 patients with t(9;11) and 7 with different abnormalities not assigned to either good or bad risk group), and 20 subjects (44.4%) were classified as unfavorable risk group with del(5q), del(7q), or complex (≥ 3) abnormalities. Patient information is listed in Table 1.
Table 1.
AML patient information
| Parameter | Characteristic | Value | |
|---|---|---|---|
| General Information | Number of patients | 45 | |
| Mean (range) age in years | 48.7 (19–65) | ||
| Mean (± SD) white blood cell count (G/l) | 45.01 ± 65.95 | ||
| Mean (range) of blastic cells in the peripheral blood (%) | 56 (0–97) | ||
| Mean (range) of blastic cells in the bone marrow (%) | 65 (20–97) | ||
| Mean (± SD) lactate dehydrogenase (U/L) | 504.4 ± 344.4 | ||
| AML subtypes based on WHO classification (n,(%)) | AML with recurrent genetic abnormalities | t(8;21)(q22;q22);(AML1/ETO) | 1 (2.2%) |
| inv [16](p13;q22) or t(16;16)(p13;q22);(CBFβ/MYH11) | 4 (8.9%) | ||
| t(9;11); MLLT3-MLL | 2 (4.4%) | ||
| AML with multilineage dysplasia without antecedent MDS | 3 (6.7%) | ||
| AML (therapy-related) | 0 (0%) | ||
| AML (not otherwise categorized; n = 35) | AML (minimally differentiated) | 4 (8.9%) | |
| AML (without maturation) | 8 (17.8%) | ||
| AML (with maturation) | 13 (28.9%) | ||
| Acute myelomonocytic leukemia (AMMoL) | 8 (17.8%) | ||
| AMMoL with eosinophilia | 0 (0.0%) | ||
| Acute monocytic leukemia | 2 (4.4%) | ||
| Acute erythroid leukemia | 0 (0.0%) | ||
| Acute megakaryoblastic leukemia | 0 (0.0%) | ||
| Mutations | FLT3-ITD/NPM1mut/CEBPAmut | 9/3/1 | |
| Induction therapy outcome | Complete remission after 1st induction | 24 | |
| Complete remission after 2nd induction | 6 | ||
| Complete remission after 3rd induction | 3 | ||
| Mortality (1st/2nd/3rd induction/consolidation) | 5/4/3/0 | ||
| Post-consolidation treatment | Allogenic hematopoietic stem cell transplant | 30 | |
| Maintenance | 3 | ||
| Risk | Favorable risk | 7 (15.6%) | |
| Intermediate risk I | 10 (22.2%) | ||
| Intermediate risk II | 8 (17.8%) | ||
| Unfavorable risk | 20 (44.4%) | ||
NPM1mut mutated nucleophosmin, CEBPAmut CCAAT/enhancer-binding protein alpha, FLT3-ITD internal tandem duplication of FMS-like tyrosine kinase 3
All patient samples were collected with the approval of the Ethics Committee at the Medical University of Bialystok and with a written informed consent form in accordance with the Declaration of Helsinki. AML patients were treated in the Hematology Department of the Medical University of Bialystok from 2008 to 2016 with 7-day induction chemotherapy regimens corresponding to the standard therapy based on the Polish Adult Leukemia Group: cytarabine was delivered as a continuous IV infusion for seven consecutive days at a dose of 200 mg/m2, while anthracycline for three consecutive days as an IV push at a dose of 50 mg/m2, and cladribine was administered for 5 days as an IV push at a dose of 5 mg/m2 (DAC schedule) [35]. Following evaluation of the induction response, non-responding patients were given re-induction protocol therapy of cytarabine, cladribine, filgrastim, and mitoxantrone (CLAG-M) and/or idarubicin, cytarabine, and etoposide (ICE) [36–38]. Consolidation therapies for patients who achieved complete remission consisted of cytarabine and mitoxantrone first, then a high dose of cytarabine, and finally either allogenic hematopoietic stem cell transplantation or maintenance therapy.
Combined bisulfite restriction analysis
Genomic DNA (gDNA) from whole peripheral blood mononuclear cells (MNCs) of AML patients and from human cord blood MNCs (CBMNCs; Cleveland Cord Blood Center, Cleveland, OH) was purified using the DNA Mini Kit (Qiagen, Germantown, MD) and subjected to bisulfite treatment using the EpiTect Bisulfite Kit (Qiagen). Sequences of bisulfite-treated genomic DNA (BSgDNA) were amplified using specific oligo primers and the following thermocycler conditions: for AmpliTaq DNA Polymerase (Applied Biosystems, Waltham, MA)—(95 °C for 2 min, annealing temperature for 1 min, 72 °C for 1 min) × 1 cycle, (95 °C for 30 s, annealing temperature for 1 min, 72 °C for 1 min) × cycle number, and (72 °C for 10 min) × 1 cycle; for AmpliTaq Gold DNA Polymerase (Applied Biosystems)—(95 °C for 8 min) × 1 cycle, (95 °C for 2 min, annealing temperature for 1 min, 72 °C for 1 min) × 2 cycles, (95 °C for 30 s, annealing temperature for 1 min, 72 °C for 1 min) × cycle number, and (72 °C for 10 min) × 1 cycle. Primer sequences for each locus are listed in Table 2. The annealing temperatures and cycle numbers for each amplicon are listed in Table 3. Primer binding locations are listed in Table 4.
Table 2.
Primer list for PCR of BSgDNA
| Locus | 1st PCR primer pair (5′-3′) | 2nd PCR primer air (5′-3′) |
|---|---|---|
| IGF2-H19 ICR | F–AGGTGTTTTAGTTTTATGGATGATGG [47] R–TCCCATAAATATCCTATTCCCAAATAACC [47] |
F–TGTATAGTATATGGGTATTTTTGGAGGTTT [47] R–TCCCATAAATATCCTATTCCCAAATAACC [47] |
| DLK1-MEG3 IG-DMR (CG6) | F–TGGGAATTGGGGTATTGTTTATAT R–AAACAATTTAACAACAACTTTCCTC |
F–GTTAAGAGTTTGTGGATTTGTGAGAAATG [39] R–CTAAAAATCACCAAAACCCATAAAATCAC [39] |
| MEG3 DMR (CG7) | F–TTATTTTTTTGAATAATAAGAGAAAGTATG R–CTCATTTCTCTAAAAATAATTAACC |
F–TTATTTTTTTGAATAATAAGAGAAAGTATG R–CCCCAAATTCTATAACAAATTACT [39] |
| MEG3 promotor (CG9) | F–TGAGGAAGTAGGGGTTTATAGAGAG R–AACCCTACAACCCCACAAAA [39] |
F–GGAGAGTGGGGTTTATTGTGAA [39] R–AACCCTACAACCCCACAAAA [39] |
| MEG3 intragenic (CG8) | F–GTTTGAGATTTGTTGGGTATTT [39] R–AATTTAACTAACAAATCACAAATATTAACT |
F–GTTTGAGATTTGTTGGGTATTT [39] R–AATTTAACTAACAAATCACAAATATTAACT |
| ZAC DMR | F–GGGGTAGTYGTGTTTATAGTTTAGTA [48] R–CRAACACCCAAACACCTACCCTA [48] |
F–GGGGTAGTYGTGTTTATAGTTTAGTA [48] R–CRAACACCCAAACACCTACCCTA [48] |
| PEG1 DMR | F–TTGTTGGTTAGTTTTGTAYGGTT [47] R–AAAAATAACACCCCCTCCTCAAAT [47] |
F–TTGTTGGTTAGTTTTGTAYGGTT [47] R–CCCAAAAACAACCCCAACTC [47] |
| PEG3 DMR | F–AAAAGGTATTAATTATTTATAGTTTGGT [49] R–AAAAATATCCACCCTAAACTAATAA [49] |
F–AAAAGGTATTAATTATTTATAGTTTGGT [49] R–AAAAATATCCACCCTAAACTAATAA [49] |
Table 3.
PCR conditions for amplifying BSgDNA
| Locus | 1st PCR condition | 2nd PCR condition | COBRA | ||||
|---|---|---|---|---|---|---|---|
| Polymerase | Anealing temperature (°C) | Cycles | Polymerase | Anealing temperature (°C) | Cycles | Restriction enzyme | |
| IGF2-H19 DMR | AmpliTaq | 55 | 35 | AmpliTaq | 55 | 35 | BstUI |
| DLK1-MEG3 IG-DMR (CG6) | AmpliTaq | 55 | 35 | AmpliTaq | 55 | 35 | BstUI |
| MEG3 DMR (CG7) | GoldTaq | 57 | 40 | GoldTaq | 57 | 40 | TaqI |
| MEG3 promotor (CG9) | AmpliTaq | 57 | 35 | AmpliTaq | 57 | 35 | TaqI |
| MEG3 intragenic (CG8) | AmpliTaq | 55 | 35 | AmpliTaq | 55 | 35 | BstUI |
| ZAC DMR | GoldTaq | 55 | 35 | GoldTaq | 55 | 38 | BstUI |
| PEG1 DMR | AmpliTaq | 55 | 35 | AmpliTaq | 55 | 38 | TaqI |
| PEG3 DMR | AmpliTaq | 55 | 35 | AmpliTaq | 55 | 35 | BstUI |
Table 4.
Nucleotide coordinates for primers and DLK1-MEG3 CTCF-binding sites
| Locus | Chromosome | Nucleotide coordinates |
|---|---|---|
| IGF2-H19 ICR | 11 | 1999842–2000072 |
| DLK1-MEG3 IG-DMR (CG6) | 14 | 100810848–100811276 |
| MEG3 DMR (CG7) | 14 | 100825668–100825999 |
| MEG3 promotor (CG9) | 14 | 100823942–100824184 |
| MEG3 intragenic (CG8) | 14 | 100828129–100828322 |
| ZAC DMR | 6 | 144132370–144132521 |
| PEG1 DMR | 7 | 130492229–130492444 |
| PEG3 DMR | 19 | 56840361–56840682 |
| DLK1-MEG3 CTCF site B | 14 | 100824015–100824074 |
| DLK1-MEG3 CTCF site D | 14 | 100825725–100825784 |
| DLK1-MEG3 CTCF site F | 14 | 100828145–100828204 |
Combined bisulfite restriction analysis (COBRA) of each amplicon was carried out on each amplicon via restriction enzyme digestion using either TaqI or BstUI (New England Biolabs, Ipswich, MA) using the following thermocycler conditions: 60 °C for 2 h for BstUI and 65 °C for 2 h for TaqI. The digested amplicons were separated by electrophoresis in agarose gels, and densitometric analysis of ethidium bromide-labeled bands was performed on photographed agarose gels using ImageJ software (National Institutes of Health). The specific COBRA conditions for each amplicon are listed in Table 3. Additional file 1: Figure S1 illustrates the quantitation method used for COBRA and includes all gels used for COBRA analyses. The median (range) values of methylation of studied CpG sites are presented in Table 5.
Table 5.
The median (range) values of methylation of studied CpG sites and chosen gene expression
| Analysis | AML patient MNCs Median (range) [number of patient] |
Control MNCs Median (range) [number of sample] |
|---|---|---|
| Methylation (%) | ||
| CG6 | 88.28 (5.00–99.53) [44] | 60.89 (48.75–70.89) [9] |
| CG7 | 74.45 (32.65–97.69) [42] | 60.21 (24.98–72.70) [9] |
| CG8 | 91.91 (30.43–99.65) [44] | 65.65 (60.11–82.36) [9] |
| CG9 | 55.34 (7.86–91.73) [42] | 46.53 (39.23–60.13) [8] |
| ZAC | 49.24 (18.96–71.97) [42] | 62.91 (46.50–78.97) [8] |
| IGF2-H19 | 70.20 (40.97–99.56) [45] | 78.12 (68.64–86.20) [9] |
| PEG3 | 67.18 (15.95–98.10) [42] | 67.41 (41.82–79.71) [9] |
| PEG1 | 56.29 (23.08–95.13) [45] | 66.74 (57.85–68.91) [9] |
| Expression (∆Ct) | ||
| DLK1 (× 10−7) | 50.98 (0.88–161,542.28) [29] | |
| MEG3 (× 10− 6) | 34.189 (1.36–19,339.85) [29] | |
| DLK1/MEG3 (× 10− 2) | 10.31 (0.20–1121.10) [29] | |
Real-time quantitative PCR
Total RNA was purified from whole peripheral blood MNCs of AML patients using TRIZOL (Life Technologies, Waltham, MA), and 2500 ng of RNA was transcribed to cDNA using Superscript VILO (Life Technologies). Gene expression analysis was carried out at least twice in duplicate using 2 ng of cDNA, 12.5 μL SYBR Select Master Mix (Applied Biosystems), and 150 nM forward and reverse primer sequences in 25 μL reaction mixtures. Real-time quantitative PCR (RT-qPCR) was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems), and results were quantified using the ΔΔCt method. The following amplification conditions were carried out for real-time quantitative PCR (RT-qPCR) of each reaction mixture: 95 °C (15 s), 45 cycles at 95 °C (15 s), and 60 °C (1 min). B2M was used as a control gene. Primer sequences for each gene are listed in Table 6. The median (range) values of the chosen gene expression are presented in Table 5.
Table 6.
Primer list for RT-qPCR
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| DLK1 | GCGAGGATGACAATGTTTGCA | GGTTCTCCACAGAGTCCGTGAA |
| MEG3 | ATCCCGGACCCAAGTCTTCT | CCACATTCGAGGTCCCTTCC |
| B2M | TGACTTTGTCACAGCCCAAGATA | AATGCGGCATCTTCAAACCT |
Statistical analysis
GraphPad Prism 7 (GraphPad, La Jolla, CA), GraphPad InStat 3 (GraphPad), and SPSS software (IBM Corporation, Armonk, NY) were used for the statistical analysis. The Mantel-Cox test was used to analyze patient survival data with use of the Bonferroni post hoc analysis noted where appropriate (GraphPad Prism). Normal distribution of samples was tested using D’Agostino-Pearson normality test (GraphPad Prism). Individual gene expression, methylation, risk group, FMS-like tyrosine kinase 3 (FLT3) mutation, and patient first-round induction therapy response data were analyzed using the Mann-Whitney test or Kruskal-Wallis test and presented as mean ± SEM (GraphPad Prism). Analyses based on nucleophosmin 1 (NPM1) and CCAAT/enhancer-binding protein alpha (CEBPA) mutation status were omitted due to the limited number of patients. Pooled methylation data was analyzed with gene expression data using the Kruskal-Wallis test and presented as mean ± SEM (GraphPad Prism). Correlation data were obtained using Spearman’s rank-order correlation (GraphPad Prism) and the point-biserial correlation (SPSS Software). SPSS software was used for the Cox regression analysis (both univariable and multivariable analyses) and for the testing of a Cox proportional hazard assumption. Multicollinearity of samples was analyzed using GraphPad InStat software. All tests were performed as two-sided tests, and differences between sample sets were considered significant for p < 0.05.
Results
Increased methylation at the MEG3 promotor region is found in AML patients with better overall survival
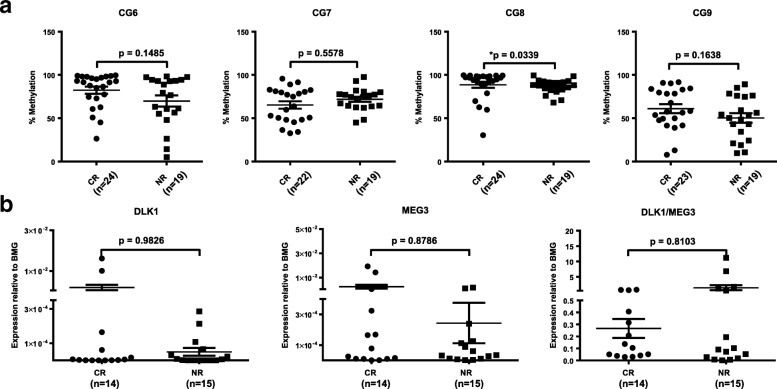
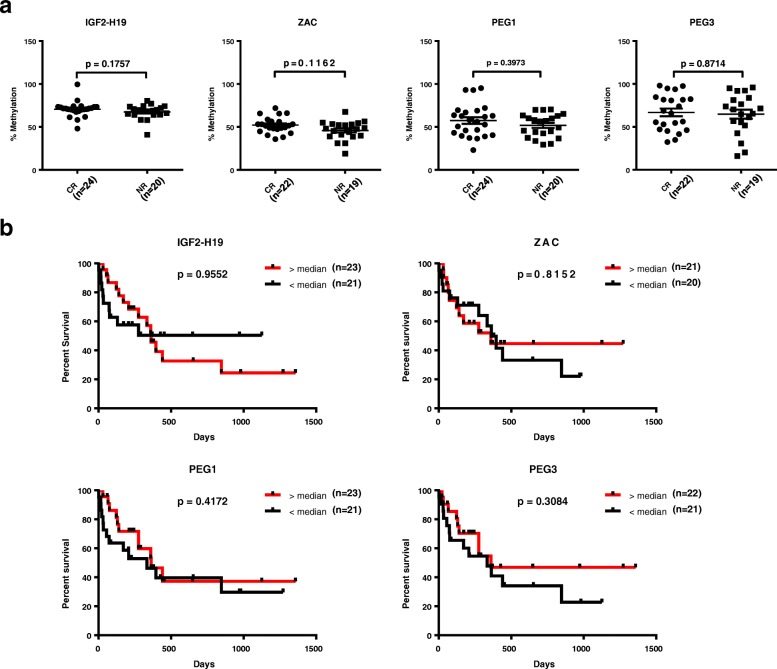
To investigate the relationship between methylation at the DLK1-MEG3 locus and patient outcomes in AML, we assessed the methylation of four CpG sites within this locus (Fig. 1a) in AML patient peripheral blood mononuclear cells (MNCs) and compared our results with patient overall survival (OS) and first-round induction therapy response (IR). Previous methylation analyses of 9 CpG sites within DLK1-MEG3 (CG1–9) in uniparental disomy patients revealed that CG6 and CG7, but not CG8 or CG9, are paternally imprinted [39]. In addition, at least seven CTCF-binding sites exist within DLK1-MEG3 (sites A–G) [40], and three of these CTCF-binding sites are located within CG7 (site D), CG8 (site F), and CG9 (site B) (Table 4). Together, these studies allowed us to analyze one confirmed non-imprinted CpG site which contains no known CTCF-binding site (CG6), one imprinted CpG site which contains a CTCF-binding site (CG7), and two non-imprinted CpG sites which each contain one CTCF-binding site (CG8, CG9). Our COBRA analyses of CG6–9 revealed that patients with increased methylation at CpG sites within the MEG3 promotor region (CG7 and CG9) had significantly longer OS (Fig. 1a, b). We also utilized Spearman’s rank-order correlation to determine the correlation between methylation at CG6–9. The correlation coefficient (rho) indicates the strength and direction of the relationship between two variables, where − 1 and 1 indicate strong correlations (negative and positive, respectively) and 0 indicates a lack of correlation. Our analysis indicated that methylation at CG6 and CG8 was correlated (rho = 0.4121, p = 0.0060) but did not impact patient OS (Fig. 1b). In addition, slightly increased methylation at CG8 was observed in patients who achieved complete remission following first-round induction therapy (Fig. 2a). Interestingly, by pooling the methylation results at each CpG site within the DLK1-MEG3 locus for each patient, we determined that patients with increased methylation at this locus had significantly longer OS than patients with lower methylation at this locus (Fig. 1c). Moreover, we found no significant contributions from the IGF2-H19, ZAC, PEG1, or PEG3 locus on the IR (Fig. 3a) or OS (Fig. 3b) of AML patients. Taken together, these data highlight CpG sites within the MEG3 promotor region as potential prognostic factors for AML patient OS and suggest that DLK1-MEG3 imprinted gene dysregulation may explain this phenomenon.
Fig. 2.
Differences in DLK1-MEG3 CpG site methylation and imprinted gene expression in AML MNCs based on patient response to first-round induction therapy. a Mann-Whitney analysis of AML patients’ response to treatment depending on the methylation values for particular CpG site amplicons. CR, complete remission; NR, non-responders. *p < 0.05. b Mann-Whitney analysis of AML patients’ response to treatment depending on DLK1, MEG3, and DLK1/MEG3 expression ratio values obtained by RT-qPCR. CR, complete remission; NR, non-responders
Fig. 3.
Patient response to first-round induction therapy and overall survival based on DMR methylation at imprinted gene network loci in AML MNCs. a Mann-Whitney analysis of AML patients’ response to treatment depending on the methylation values for particular imprinted gene loci DMR amplicons. CR, complete remission; NR, non-responders. b Analysis of AML patients’ survival depending on the median methylation values for particular imprinted gene loci DMR amplicons using the Mantel-Cox test
Increased expression of imprinted genes at the DLK1-MEG3 locus is found in AML patients with worse overall survival
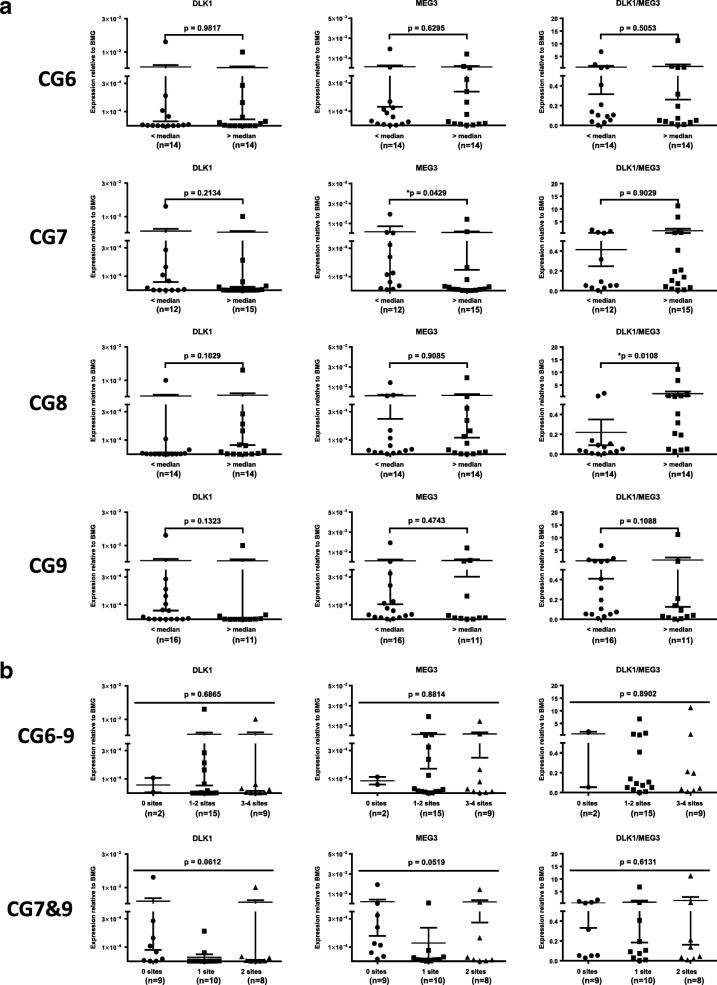
To gain insight into the relationship between DLK1-MEG3 imprinted gene expression and AML patient outcomes, we probed DLK1 and MEG3 expression in AML MNCs and found no significant differences in the expression of these genes based on IR (Fig. 2b). As mentioned before, DLK1 overexpression enhances the proliferation of leukemic cells [41], and we found increased DLK1 expression in patients with significantly shorter OS (Fig. 1d). Interestingly, no significant differences in DLK1 expression were found based on individual (Fig. 4a) or pooled (Fig. 4b) CpG site methylation, consistent with the known mechanism of DLK1 regulation in AML [26]. On the other hand, we observed a trend for shorter OS in patients with increased expression of the tumor suppressor [27] MEG3 (Fig. 1d). Our pooled results for DLK1 and MEG3 expression also revealed that patients with increased expression of DLK1, MEG3, or both genes had significantly shorter OS than patients with lower expression of these genes (Fig. 1d). We found significant differences in MEG3 expression based on CG7 methylation (Fig. 4a) and a significant correlation between them (rho = − 0.3846, p = 0.0476), despite our survival results for CG7 methylation (Fig. 1b). DLK1 expression also correlated with MEG3 expression (rho = 0.7163, p < 0.0001) and the DLK1/MEG3 expression ratio (rho = 0.6227, p = 0.0003). In addition, significant differences in the DLK1/MEG3 expression ratio were found based on CG8 methylation (Fig. 4a) along with a significant correlation between them (rho = 0.4532, p = 0.0154). No significant differences in the MEG3 expression or DLK1/MEG3 expression ratio were found based on pooled methylation results (Fig. 4b). Collectively, these data indicate that DLK1 is a potential prognostic factor for AML patient OS and the tumor-suppressing abilities of MEG3 may be overwhelmed by DLK1 expression and/or CpG site-specific signatures of its downstream miRNAs [33].
Fig. 4.
Relationship between DLK1-MEG3 CpG site methylation and imprinted gene expression in AML MNCs. a Mann-Whitney analysis of DLK1 and MEG3 expression in AML MNCs based on the methylation values for particular CpG site amplicons. *p < 0.05. b Kruskal-Wallis test of DLK1 and MEG3 expression in AML MNCs based on the pooled CG6–CG9 or CG7 and CG9 amplicon median methylation results. Groups represent the number of CpG site amplicons out of 4 (CG6–9) or 2 (CG7 and CG9) analyzed which exhibited increased methylation relative to their respective median values
Multivariable analysis confirms CG9 methylation as an independent prognostic factor for survival
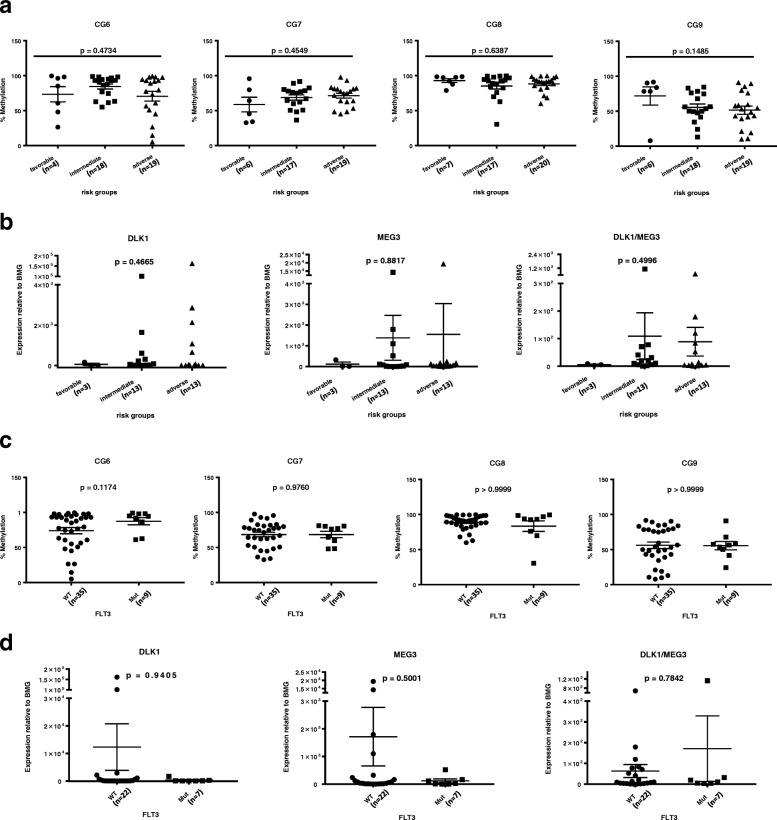
To examine the prognostic values of our methylation and expression results in AML survival, we compared them with already known survival predictors including cytogenetics (risk groups and FLT3 mutation status), white blood count (WBC), lactate dehydrogenase (LDH) level, and age. Analyses based on nucleophosmin (NPM1) and CCAAT/enhancer-binding protein alpha (CEBPA) mutation status were omitted due to the limited number of patients. Significant correlations were observed between CG7 methylation and age (rho = − 0.3088, p = 0.0466) and between patient WBC and LDH level (rho = 0.4497; p = 0.0022). No significant differences in CpG site methylation or imprinted gene expression were found based on the patient risk group (Fig. 5a, b) or FLT3 mutation status (Fig. 5c, d). Univariable analysis performed for all patients confirmed that risk group, IR, and LDH level are strong predictors of OS and identified CG7 methylation, CG9 methylation, and DLK1 expression as new possible predictors of OS. Multivariable Cox proportional hazard models carried out independently for significant methylation (CG7 and CG9) and expression (DLK1) factors indicated that only CG9 methylation can be considered a prognostic factor for survival independent of risk group, IR, and LDH level (Table 7). In total, these data identify CG9 methylation specifically as an independent prognostic factor for AML patient OS and suggest that miRNAs mediate this phenomenon.
Fig. 5.
Differences in DLK1-MEG3 CpG site methylation and imprinted gene expression based in AML MNCs based on patient risk group and FLT3 mutation status. Kruskal-Wallis (a) or Mann-Whitney (c) analysis of methylation values for particular CpG site amplicons depending on AML patients’ risk groups (a) or presence of FLT3 mutation (c). WT, wild-type; Mut, mutated. Kruskal-Wallis (b) or Mann-Whitney (d) analysis of DLK1 and MEG3 and the DLK1/MEG3 expression ratio in MNCs from AML patients based on their risk group (b) or presence of FLT3 mutation (d). WT, wild-type; Mut, mutated
Table 7.
Statistical analyses of overall survival
| Overall survival | |||
|---|---|---|---|
| HR | 95% confidence interval | p value | |
| Univariate analysis | |||
| Demographic | |||
| Sex (male vs female) | 0.887 | 0.396–1.987 | 0.770 |
| Age (< 60 vs 60+) | 1.461 | 0.625–3.414 | 0.381 |
| Cytogenetic/clinical factors | |||
| Risk groups (favorable vs intermediate vs adverse) | 2.325 | 1.183–4.569 | 0.014 |
| FLT3 (WT vs mutated) | 1.282 | 0.474–3.466 | 0.624 |
| Response (NR vs CR) | 0.213 | 0.089–0.509 | 0.001 |
| WBC count (low vs high) | 1.168 | 0.514–2.650 | 0.711 |
| LDH level (low vs high) | 0.242 | 0.101–0.584 | 0.002 |
| Methylation | |||
| CG6 (low vs high) | 0.894 | 0.386–2.068 | 0.793 |
| CG7 (low vs high) | 0.380 | 0.157–0.917 | 0.031 |
| CG8 (low vs high) | 0.762 | 0.335–1.731 | 0.516 |
| CG9 (low vs high) | 0.331 | 0.136–0.805 | 0.015 |
| Expression | |||
| DLK1 (low vs high) | 3.212 | 1.096–9.413 | 0.033 |
| MEG3 (low vs high) | 2.294 | 0.819–6.421 | 0.114 |
| DLK1/MEG3 (low vs high) | 0.668 | 0.247–1.808 | 0.428 |
| Multivariate analysis | |||
| Methylation | |||
| CG7 (low vs high) | 0.533 | 0.185–1.538 | 0.245 |
| CG9 (low vs high) | 0.294 | 0.092–0.935 | 0.038 |
| Expression | |||
| DLK1 (low vs high) | 1.758 | 0.467–6.615 | 0.404 |
Low or high refers to values either lower or higher than median calculated for all AML patients included in this study. HR hazard rate, FLT3 FMS-like tyrosine kinase 3, NR non-responders, CR complete remission
Discussion
Multiple CpG sites within DLK1-MEG3 house CCCTC-binding factor (CTCF)-binding DNA sequences [26, 40] which govern the expression of coding [26] and non-coding RNAs (ncRNAs) [33] from this locus. Our investigation into the relationship between select CpG site methylation and AML patient outcomes identified increased methylation at CG7 and CG9 in patients with longer OS. Interestingly, CG7 and CG9 both contain CTCF-binding DNA sequences [40] and are located within the MEG3 promotor region, where increased methylation was previously found to predict poor OS for AML patients [42]. This disparity is likely explained by differences in methodology and CpG site selection given that, similar to our results, increased methylation at a region containing CG7 was also observed in longer-living patients with MDS or AML with myelodysplasia-related changes (AML-MR) [43]. Furthermore, our data for DMRs within other IGN loci uncovered no differences in patient outcomes based on their methylation, highlighting the significance of our observation for CG7 and CG9 in survival prediction. Thus, the imprinted CG7 and non-imprinted CG9 [44] are potential prognostic factors for AML patient OS, and future investigations may reveal CpG site methylation-dependent [33] miRNA signatures from this locus which promote cell stemness [16, 30, 31] and self-renewal [30].
Pioneering studies in bimaternal mouse models uncovered a requirement for the balanced expression of growth-promoting DLK1 and growth-suppressing MEG3 in embryonal growth [10] and a more specific requirement for Dlk1 in fetal hematopoiesis [14]. Accumulating evidence suggests the opposing roles of these genes on growth and development are mirrored in leukemias, where DLK1 maintains cell stemness [25] and enhances the proliferation of leukemic cells [41] and MEG3 suppresses leukemogenesis [27] and leukemic cell proliferation [28]. By analyzing the expression of DLK1 and MEG3 in AML patient MNCs, we determined that increased expression of these genes is found in patients with worse overall survival. While this is consistent with the growth-promoting effects of DLK1 on leukemic cells, our data for MEG3 alludes to the presence of underlying miRNA signatures from this locus which may overcome the tumor-suppressing abilities of MEG3. In support of this, the negative correlation we observed between CG7 methylation and MEG3 expression is consistent with the recent work of Yao et al. [29] in AML patients from a Hainan population and supported by work by Merkerova et al. which observed increased MEG3 expression in MDS/AML-MR patients with shorter OS [43]. Taken together, our data emphasizes the overlapping roles of paternally [25] and maternally expressed [16, 30] genes from the DLK1-MEG3 locus in stemness maintenance and proliferation as potential drivers of leukemia progression.
Several lines of evidence point to the dysregulation of the DLK1-MEG3 locus as a key component of leukemia development and progression. DLK1 expression is elevated in the CD34+ cells and MNCs of myelodysplastic syndrome (MDS) patients and MNCs of AML patients, and increased levels of DLK1 are found in MDS patient sera [41]. MEG3 expression is similarly decreased in AML patients [29], and methylation at CpG sites within the DLK1-MEG3 locus is likely responsible for the abnormal expression of these genes [26, 29]. We therefore sought to delineate the unique contribution(s) of the DLK1-MEG3 locus to AML patient OS, and univariable analyses validated our observations that CG7 methylation, CG9 methylation, and DLK1 expression are potential prognostic factors for survival of this disease. The multivariable analysis further clarified these results to reveal CG9 methylation as an independent prognostic factor for AML patient OS. We found this particularly interesting because, unlike CG6 or CG7, CG9 is not imprinted [44]. We also found no correlations between CG9 and patient biomarkers or gene expression. Thus, we hypothesize that the prognostic value of CG9 methylation can be explained by its regulation of the DLK1-MEG3 miRNA mega-cluster [33] via the chromatin-modifying CTCF protein [40]. In addition, emerging evidence points to small nucleolar RNAs (snoRNAs) from the DLK1-MEG3 locus as potential mediators of leukemic cell proliferation [45] and differentiation [46]. As a pilot study, we recognize the limitations of a 45-patient cohort. However, the results of our CpG site selection strategy add to the mounting evidence that methylation at CTCF-binding sites is responsible for chromatin dysregulation of the entire DLK1-MEG3 locus, and our data suggest that increased methylation at CG9 specifically exerts a protective role in AML. Future studies with large patient cohorts are necessary to delineate the CTCF-mediated chromatin organization at DLK1-MEG3 and its role in AML development and prognosis.
Conclusions
Taken together, our screenings of imprinted loci methylation and gene expression in AML patient MNCs highlight a unique role for the DLK1-MEG3 locus in AML patient prognosis. To our knowledge, we are the first to compare methylation at confirmed DLK1-MEG3 CpG sites with survival in a relatively large cohort of AML patients comprised of multiple AML subtypes. We identify methylation at the non-imprinted CpG site CG9 as a novel independent prognostic factor for survival in AML patients which implicates CTCF-mediated ncRNA regulation as a key determinant of AML patient survival. In particular, considering that the DLK1-MEG3 locus is the home to over 40 miRNAs, many of which exhibit functional redundancies [16, 30], the assessment of unique miRNA signatures from this locus for their prognostic values represents a valuable pursuit. Future studies involving large patient cohorts are required to confirm our findings for CG9 and further clarify the prognostic impact of the role of chromatin organization at DLK1-MEG3 in AML.
Additional file
Scheme of densitometry analysis. a Equal amounts of PCR product (treated and untreated with restriction enzyme, RE) were separated on a gel, and a picture of the gel was captured. Step (1) Selection of bands for analysis. Step (2) Intensity of the bands is translated to the surface of the corresponding peaks using ImageJ software. Step (3) Analysis of the surface of the peaks corresponding to particular bands and re-calculation of arbitrary units to percentages. In analysis 1, the surface of the “unmethylated” band from the re-treated sample is compared with the total amount of PCR product that was used for digestion. In analysis 2, the surface of the “unmethylated” band is compared to the total surface of all peaks in the digested sample. The mean of the methylation from both analyses is then used for further studies. Gels are shown for the COBRA analysis of CG6 (b), CG7 (c), CG8 (d), CG9 (e), ZAC (f), IGF2-H19 (g), PEG1 (h), and PEG3 (i). (PDF 1340 kb)
Acknowledgements
Not applicable
Funding
This work was supported by NIH grants 2R01 DK074720 and R01HL112788, the Stella and Henry Endowment, and the Harmonia NCN grant UMO-2014/14/M/NZ3/00475 to MZR and by a grant from the University of Louisville School of Medicine to GS.
Availability of data and materials
Not applicable.
Authors’ contributions
ZPS, MZR, and GS contributed to the study conception and design. GS contributed to the development of methodology. LB, JK, MLW, KL, and MM acquired and managed the patients, collected the samples, and prepared the cDNA and DNA for analysis. ZPS and GS contributed to the methylation and expression analysis. ZPS, GS, and LB contributed to the analysis and interpretation of data. ZPS, LB, MZR, and GS contributed to the writing, review, and/or revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patient samples were collected with the approval of the Ethics Committee at the Medical University of Bialystok and with a written informed consent form in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mariusz Z. Ratajczak, Phone: 502-852-1788, Email: mzrata01@louisville.edu
Gabriela Schneider, Phone: 502-852-1203, Email: g0schn01@louisville.edu.
References
- 1.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6(2). 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed]
- 2.Sanchez-Delgado M, Riccio A, Eggermann T, Maher ER, Lapunzina P, Mackay D, et al. Causes and consequences of multi-locus imprinting disturbances in humans. Trends Genet. 2016;32(7):444–455. doi: 10.1016/j.tig.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Benetatos L, Vartholomatos G. Imprinted genes in myeloid lineage commitment in normal and malignant hematopoiesis. Leukemia. 2015;29(6):1233–1242. doi: 10.1038/leu.2015.47. [DOI] [PubMed] [Google Scholar]
- 4.Lui JC, Finkielstain GP, Barnes KM, Baron J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol. 2008;295(1):R189–R196. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 7.Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc Natl Acad Sci U S A. 2013;110(51):20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 9.Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428(6985):860–864. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Kumagai T, Kawahara M, Ogawa H, Hiura H, Obata Y, et al. Regulated expression of two sets of paternally imprinted genes is necessary for mouse parthenogenetic development to term. Reproduction. 2006;131(3):481–488. doi: 10.1530/rep.1.00933. [DOI] [PubMed] [Google Scholar]
- 11.Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, et al. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol. 2007;25(9):1045–1050. doi: 10.1038/nbt1331. [DOI] [PubMed] [Google Scholar]
- 12.Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet. 2002;11(1):77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 13.Berg JS, Lin KK, Sonnet C, Boles NC, Weksberg DC, Nguyen H, et al. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS One. 2011;6(10):e26410. doi: 10.1371/journal.pone.0026410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Kawahara M, Kono T. Synergistic role of Igf2 and Dlk1 in fetal liver development and hematopoiesis in bi-maternal mice. J Reprod Dev. 2008;54(3):177–182. doi: 10.1262/jrd.19146. [DOI] [PubMed] [Google Scholar]
- 15.Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18(2):214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratajczak MZ. Why are hematopoietic stem cells so ‘sexy’? On a search for developmental explanation. Leukemia. 2017;31(8):1671–77. [DOI] [PMC free article] [PubMed]
- 18.Sellers ZP, Schneider G, Maj M, Ratajczak MZ. Analysis of the paternally-imprinted DLK1-MEG3 and IGF2-H19 tandem gene loci in NT2 embryonal carcinoma cells identifies DLK1 as a potential therapeutic target. Stem Cell Rev. 2018;14(6):823-36 [DOI] [PubMed]
- 19.Yang NQ, Luo XJ, Zhang J, Wang GM, Guo JM. Crosstalk between Meg3 and miR-1297 regulates growth of testicular germ cell tumor through PTEN/PI3K/AKT pathway. Am J Transl Res. 2016;8(2):1091–1099. [PMC free article] [PubMed] [Google Scholar]
- 20.Zeira E, Abramovitch R, Meir K, Even Ram S, Gil Y, Bulvik B, et al. The knockdown of H19lncRNA reveals its regulatory role in pluripotency and tumorigenesis of human embryonic carcinoma cells. Oncotarget. 2015;6(33):34691–34703. doi: 10.18632/oncotarget.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinsmith LJ, Pierce GB., Jr Multipotentiality of single embryonal carcinoma cells. Cancer Res. 1964;24(9):1544–1551. [PubMed] [Google Scholar]
- 22.Shin DM, Zuba-Surma EK, Wu W, Ratajczak J, Wysoczynski M, Ratajczak MZ, et al. Novel epigenetic mechanisms that control pluripotency and quiescence of adult bone marrow-derived Oct4(+) very small embryonic-like stem cells. Leukemia. 2009;23(11):2042–2051. doi: 10.1038/leu.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratajczak MZ, Ratajczak J, Suszynska M, Miller DM, Kucia M, Shin DM. A novel view of the adult stem cell compartment from the perspective of a quiescent population of very small embryonic-like stem cells. Circ Res. 2017;120(1):166–178. doi: 10.1161/CIRCRESAHA.116.309362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang TJ, Zhou JD, Zhang W, Lin J, Ma JC, Wen XM, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi: 10.1186/s13148-018-0486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Forman SJ, Bhatia R. Expression of DLK1 in hematopoietic cells results in inhibition of differentiation and proliferation. Oncogene. 2005;24(27):4472–4476. doi: 10.1038/sj.onc.1208637. [DOI] [PubMed] [Google Scholar]
- 26.Khoury H, Suarez-Saiz F, Wu S, Minden MD. An upstream insulator regulates DLK1 imprinting in AML. Blood. 2010;115(11):2260–2263. doi: 10.1182/blood-2009-03-212746. [DOI] [PubMed] [Google Scholar]
- 27.Lyu Y, Lou J, Yang Y, Feng J, Hao Y, Huang S, et al. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31(12):2543-51. [DOI] [PMC free article] [PubMed]
- 28.Li J, Zi Y, Wang W, Li Y. LncRNA MEG3 inhibits cell proliferation and metastasis in chronic myeloid leukemia via targeting MiR-184. Oncol Res. 2018;26(2):297-305. [DOI] [PMC free article] [PubMed]
- 29.Yao H, Duan M, Lin L, Wu C, Fu X, Wang H, et al. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget. 2017;8(11):18337–18347. doi: 10.18632/oncotarget.15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moradi S, Sharifi-Zarchi A, Ahmadi A, Mollamohammadi S, Stubenvoll A, Gunther S, et al. Small RNA sequencing reveals Dlk1-Dio3 locus-embedded microRNAs as major drivers of ground-state pluripotency. Stem Cell Reports. 2017;9(6):2081–2096. doi: 10.1016/j.stemcr.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moradi S, Braun T, Baharvand H. miR-302b-3p promotes self-renewal properties in leukemia inhibitory factor-withdrawn embryonic stem cells. Cell J. 2018;20(1):61–72. doi: 10.22074/cellj.2018.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manodoro F, Marzec J, Chaplin T, Miraki-Moud F, Moravcsik E, Jovanovic JV, et al. Loss of imprinting at the 14q32 domain is associated with microRNA overexpression in acute promyelocytic leukemia. Blood. 2014;123(13):2066–2074. doi: 10.1182/blood-2012-12-469833. [DOI] [PubMed] [Google Scholar]
- 34.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 35.Holowiecki J, Grosicki S, Giebel S, Robak T, Kyrcz-Krzemien S, Kuliczkowski K, et al. Cladribine, but not fludarabine, added to daunorubicin and cytarabine during induction prolongs survival of patients with acute myeloid leukemia: a multicenter, randomized phase III study. J Clin Oncol. 2012;30(20):2441–2448. doi: 10.1200/JCO.2011.37.1286. [DOI] [PubMed] [Google Scholar]
- 36.Bradstock KF, Matthews JP, Lowenthal RM, Baxter H, Catalano J, Brighton T, et al. A randomized trial of high-versus conventional-dose cytarabine in consolidation chemotherapy for adult de novo acute myeloid leukemia in first remission after induction therapy containing high-dose cytarabine. Blood. 2005;105(2):481–488. doi: 10.1182/blood-2004-01-0326. [DOI] [PubMed] [Google Scholar]
- 37.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 38.Jaglal MV, Duong VH, Bello CM, Al Ali NH, Padron E, Fernandez HF, et al. Cladribine, cytarabine, filgrastim, and mitoxantrone (CLAG-M) compared to standard induction in acute myeloid leukemia from myelodysplastic syndrome after azanucleoside failure. Leuk Res. 2014;38(4):443–446. doi: 10.1016/j.leukres.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40(2):237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 40.Rosa AL, Wu YQ, Kwabi-Addo B, Coveler KJ, Reid Sutton V, Shaffer LG. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosom Res. 2005;13(8):809–818. doi: 10.1007/s10577-005-1015-4. [DOI] [PubMed] [Google Scholar]
- 41.Sakajiri S, O'Kelly J, Yin D, Miller CW, Hofmann WK, Oshimi K, et al. Dlk1 in normal and abnormal hematopoiesis. Leukemia. 2005;19(8):1404–1410. doi: 10.1038/sj.leu.2403832. [DOI] [PubMed] [Google Scholar]
- 42.Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34(2):148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 43.Merkerova MD, Remesova H, Krejcik Z, Loudova N, Hrustincova A, Szikszai K, et al. Relationship between altered miRNA expression and DNA methylation of the DLK1-DIO3 region in Azacitidine-treated patients with myelodysplastic syndromes and acute myeloid leukemia with myelodysplasia-related changes. Cells. 2018;7(9). 10.3390/cells7090138. [DOI] [PMC free article] [PubMed]
- 44.Ogata T, Kagami M, Ferguson-Smith AC. Molecular mechanisms regulating phenotypic outcome in paternal and maternal uniparental disomy for chromosome 14. Epigenetics. 2008;3(4):181–187. doi: 10.4161/epi.3.4.6550. [DOI] [PubMed] [Google Scholar]
- 45.Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26(9):2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- 46.Warner WA, Spencer DH, Trissal M, White BS, Helton N, Ley TJ, et al. Expression profiling of snoRNAs in normal hematopoiesis and AML. Blood Adv. 2018;2(2):151–163. doi: 10.1182/bloodadvances.2017006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9(14):2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 48.Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, et al. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115(5):690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16(21):2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scheme of densitometry analysis. a Equal amounts of PCR product (treated and untreated with restriction enzyme, RE) were separated on a gel, and a picture of the gel was captured. Step (1) Selection of bands for analysis. Step (2) Intensity of the bands is translated to the surface of the corresponding peaks using ImageJ software. Step (3) Analysis of the surface of the peaks corresponding to particular bands and re-calculation of arbitrary units to percentages. In analysis 1, the surface of the “unmethylated” band from the re-treated sample is compared with the total amount of PCR product that was used for digestion. In analysis 2, the surface of the “unmethylated” band is compared to the total surface of all peaks in the digested sample. The mean of the methylation from both analyses is then used for further studies. Gels are shown for the COBRA analysis of CG6 (b), CG7 (c), CG8 (d), CG9 (e), ZAC (f), IGF2-H19 (g), PEG1 (h), and PEG3 (i). (PDF 1340 kb)
Data Availability Statement
Not applicable.