Abstract
Nuclear factor-Ys (NF-Ys) were previously shown to have important regulatory impacts in different developmental and physiological process. However, in barley the function of the NF-Y genes at system levels is not well known. To identify barley NF-Ys, Arabidopsis and wheat NF-Y protein sequences were retrieved and the BLAST program along with the hidden Markov model were used. Multiple sequence alignments of identified NF-Ys were constructed using ClustalW. Expression patterns of the NF-Ys at different physiological and developmental conditions were also surveyed based on microarray datasets in public databases and subsequently co-expression network were constructed. Validation of in silico expression analysis was performed by real-time qPCR under salt stress condition. In total, 23 barley NF-Ys (8 NF-YA, 11 NF-YB and 4 NF-YC) were identified. Based on the sequence homology, the subunits of the NF-Y complex were divided into three to five groups. Structural analysis highlighted the conserved domains of HvNF-YA, HvNF-YB and HvNF-YC. Co-expression network analysis indicated the potential functions of HvNF-Ys in photosynthesis, starch biosynthesis and osmotic stress tolerance. The results of qRT-PCR also confirmed the HvNF-Ys roles in adaptation responses of barley to salt stress. We identified some potential candidate genes which could be used for improvements of cereals tolerance to salinity stress.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-00637-1) contains supplementary material, which is available to authorized users.
Keywords: Nuclear factor Y, Phylogeny, Co-expression network, qRT-PCR, Salt stress
Introduction
Nuclear factor Y (NF-Y) is a heterotrimeric transcription factor family that binds the CCAAT element (CBF) to regulate gene expression (Quach et al. 2015). It includes three subfamilies; NF-YA, NF-YB, and NF-YC (Mantovani 1999). Each of them shows high level of sequence conservation in their central regions (Mantovani 1999). This conserved central region of the NF-YA subunit contains interaction and DNA binding domains. Interaction domain is required for contact with the NF-YB, NF-YC heterodimer, whereas the DNA-binding domain is involved in the DNA-binding site recognition (Xing et al. 1994).
In plants, each of the NF-YA, NF-YB and NF-YC proteins are encoded by a larger gene family (Liang et al. 2012; Panahi et al. 2015; Malviya et al. 2016; Ren et al. 2016). Arabidopsis and wheat encodes 36 and 37 NF-Y subunits, respectively (Siefers et al. 2009). In rice genome at least 10, 12 and 8 genes encode NF-YA, NF-YB and NF-YC proteins, respectively (Thirumurugan et al. 2008) and rapeseed (Brassica napus L.) harbors 14, 14 and 5 NF-YA, NF-YB, and NF-YC genes, respectively (Xu et al. 2014).
Plant growth and development is influenced by various abiotic stresses such as salinity (Panahi et al. 2012). Salinity is one of the major factors limiting production of crop plants worldwide, therefore enhancement of plants tolerance to salt stress is one of the main goals of plant breeders and biotechnologist in recent decades. Underlying physiological and molecular mechanisms of salt tolerance remain to be elucidated and this is the major drawback for the enhancement of plants tolerance to salt stress condition (Panahi and Mohammadi 2018). Under salt stress, some physiological events including increased respiration rate, membrane instability, and failure in the maintenance of turgor pressure are observed (Babu et al. 2012). To ameliorate these effects, some processes are coordinated and triggered in plants to maintain cellular osmolality (Panahi et al. 2012). Transcription factors are mediated genetic networks rewiring in stress condition (Panahi et al. 2015). Therefore, identification of these factors not only will be provide insight to understanding the molecular mechanism of stress responses, but also it will be suitable for identification of candidate genes to improve the stress tolerance of crops by gene manipulation approaches.
The functions of NF-Ys in diverse developmental process such as embryogenesis, nitrogen fixing nodule development, flowering and photosynthesis have been suggested (Mu et al. 2013; Panahi et al. 2015). Moreover, increasing number of evidences advocate the role of transcription factor NF-Ys in osmotic stress responses. High throughput transcriptome profiling in leaves of wheat revealed that drought stress down regulates TaNF-YA1 transcription (Stephenson et al. 2007). Additionally, it has been reported that over expression of NFY-A3 and NF-Y5 enhanced drought tolerance in soybean and Arabidopsis, respectively (Ni et al. 2013). Increased tolerance to drought stress was observed under overexpression of AtNF-YB1 and ZmNF-YB2 in Arabidopsis and Zea mays, respectively (Nelson et al. 2007).
Barley (Hordeum vulgare), member of the grass family is regarded as a salt-tolerant crop. Moreover as a diploid and inbreeding, it has been considered as a model for understanding the function and regulation of complex traits such as salt stress responses in Triticeae (I.B.G.S. 2012).
Although previous studies highlighted the importance of plant NF-Ys in wide range process, yet the molecular pathways of these transcription factors remains unclear. In this study, we characterized the function of three NF-Y families, their phylogenetic relationships, co-expression networks and expression under salt stress.
Materials and methods
Identification of barley NF-Y family members
To identify barley NF-Ys, Arabidopsis and wheat NF-Y protein sequences were retrieved and BLAST program with an E-value cut-off of E−10 were used to identify HvNF-Y using the H. vulgare genomes in ensemble Plants database (http://plants.ensembl.org/Hordeum_vulgare/Tools/Blast?db=core). In addition, hidden Markov model (HMM) for the NF-Y genes was constructed using HMMER package version 3.0 (Eddy 1998). MUSCLE algorithm was used for multiple alignments based on default parameters in Ugene software (Okonechnikov et al. 2012). Subsequently, the created HMM profiles were used to perform HMM search (http://hmmer.janelia.org/search/hmmsearch) against the Barley genome (I.B.G.S. 2012). By combination the results of two strategies, the final list of HvNF-Y genes were retrieved.
Multiple alignments and phylogenetic analysis
Multiple sequence alignments of identified NF-Ys in barley were constructed using ClustalW (Thompson et al. 2002). The Neighbor-Joining trees were constructed using evolutionary distance coefficient implemented in MEGA5.0 program (Tamura et al. 2011) with the 1000 bootstrap replications.
Expression pattern under different development stages and abiotic stresses condition
The expression data of the HvNF-Y gene family under different development stages (Array accession number BB3 in PLEXdb database) and abiotic stress including cold (short term cold stress and Long term stress, accession number GSE27822 and GSE27821 respectively), drought (GSE6990) and ABA (early and late responses, GSE10328) for Morex cultivar of barley was retrieved from the Gene Expression Omnibus (GEO). The expression pattern of all NF-Y genes was presented as heatmap with a color scale indicating log2 expression values. The heatmap was constructed using Euclidean distance and complete linkage algorithm implemented in heat mapper (http://www.heatmapper.ca/) server (Babicki et al. 2016).
Identification of correlated genes and network construction
High-throughput transcriptome data sets were retrieved from the PLEXdb and GEO databases. 1200 samples were used for the expression analysis and construction of the co expression-network. Raw data were normalized using the MAS 5.0 algorithm in R (“affy” package). Co-expression network were constructed based on weighted correlations algorithm in R (“WGCNA” package).The optimal threshold of the PCC was set at 0.75. Cytoscape software (version 2.8.3) was used to construct the co-expression networks. To predict the biological function of the HvNF-Y proteins, co-expressed genes were mapped into biological pathways by the BLAST to the KEGG database (Kanehisa et al. 2014).
Plant material and stress treatment
Barley seeds (Morex cultivar) were obtained from Center of Excellence in Cereal Molecular Breeding, University of Tabriz, Tabriz, Iran. The seeds were sterilized with the 1.5% sodium hypochlorite solution for 3 min and transferred into pots containing sand. The seedlings were feed with 1/2 Hoagland’s solution and 30-days-old seedlings (planted as about 8 individuals per pot) were exposed to salt stress by adding 300 mM NaCl to the Hoagland’s solution. The treated and control plants (both shoots and roots) were sampled 6 h after salt stress treatment.
Total RNA isolation and cDNA synthesis
Total RNA was extracted using RNX™ plus kit (Cinagen, Iran). The quality and quantity of extracted RNA were examined by agarose gel electrophoresis and NanoDrop spectrophotometer, respectively. For elimination of residual genomic DNA, RNA samples were treated with an RNase-Free DNaseI (Fermentas, Lituania). cDNA was synthesized using the PrimeScript RT-PCR Kit (TaKaRa, Japan), according to the manufacturer’s instructions.
Gene expression analysis by quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was done on the Applied Biosystems 7500 device using the SYBR ExTaqTM Kit (TaKaRa, Japan). Each of 20 μl qRT-PCR reaction included 10 μl SYBR R PremixEx TaqTM (2 ×), 0.5 μl of each forward and reverse primers (10 μM), and 0.2 μl cDNA. Primer sequences of HvNF-Ys and housekeeping genes is presnetd in Supplementary Table 1. Gene expression data were normalized to 18S rRNA as housekeeping gene. Expression data were analyzed using the 2−∆∆CT method (Livack and Schmittgen 2001). To compare the gene expression between treated and normal conditions, Student’s t test was performed.
Results
Identification, multiple alignments and phylogenetic analyses of HvNF-Y genes
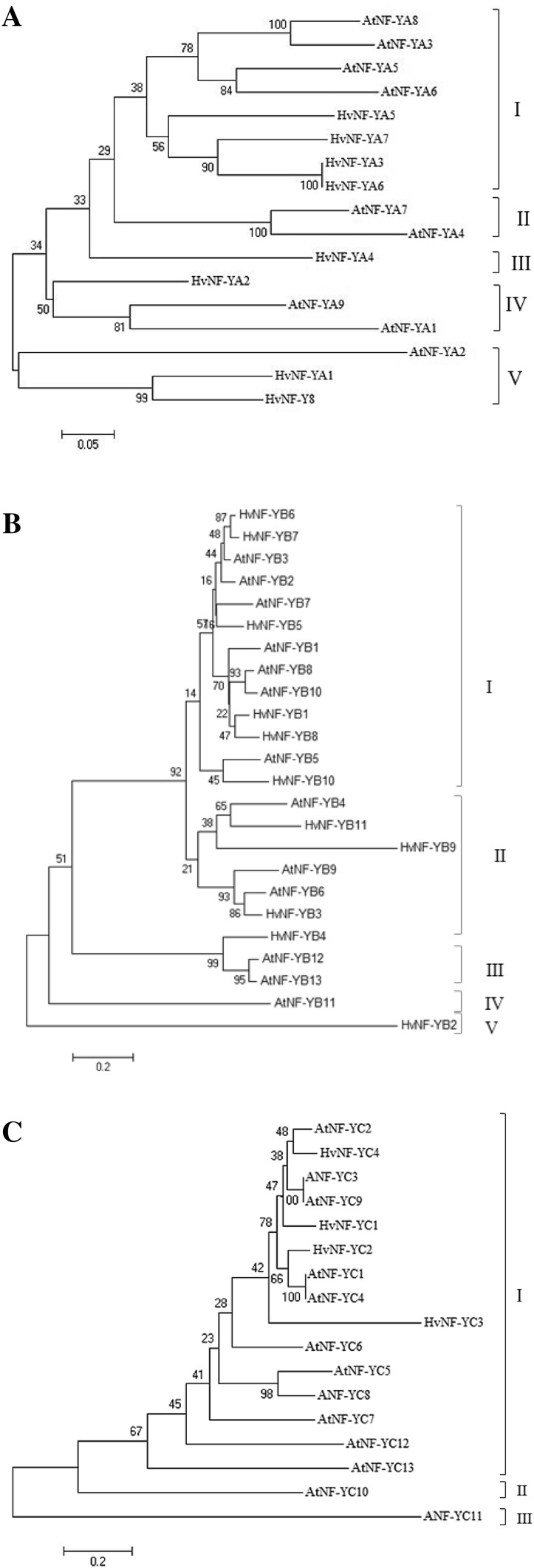
We used wheat and Arabidopsis NF-Y protein sequences as queries to search barley NF-Y genes. In total, 23 unique NF-Y sequences belonging to the three subgroups including 8 NF-YA, 11 NF-YB and 4 NF-YC genes were identified (Table 1). To understand the evolutionary relationship among individual HvNF-Ys, phylogenetic analysis was carried out (Fig. 1a–c). Based on the obtained tress, HvNF-YAs were clustered into five discrete clades (I–V). Clade I consisted of HvNF-YA 3, 5, 6 and 7 as well as AtNF-YA 3, 5, 6, 8 (Fig. 1a). The HvNF-YB subunit was clustered into five distinct groups (I–V) (Fig. 1b). In this grouping, HvNF-YB 1, 5, 7, 8 and 10 and correspondence Arabidopsis NF-YBs (AtNF-YBs 1, 5, 7, 8 and 10) were grouped in same clade. As shown in the Fig. 1c, all-YCs were grouped in the clade I.
Table 1.
List of identified NF-Ys in barley
| NF-YC | NF-YB | NF-YA | |||
|---|---|---|---|---|---|
| Gene ID | NF-Y name | Gene ID | NF-Y name | Gene ID | NF-Y name |
| MLOC_5869 | NF-YA1 | MLOC_5897 | NF-YB1 | BAJ98529 | NF-YC1 |
| MLOC_24655 | NF-YA2 | MLOC_70684 | NF-YB2 | BAK03493 | NF-YC2 |
| MLOC_36554 | NF-YA3 | MLOC_72428 | NF-YB3 | MLOC_11547 | NF-YC3 |
| MLOC_52315 | NF-YA4 | MLOC_36682 | NF-YB4 | MLOC_53118 | NF-YC4 |
| AK368372 | NF-YA5 | MLOC_36944 | NF-YB5 | ||
| MLOC_62730 | NF-YA6 | MLOC_57782 | NF-YB6 | ||
| MLOC_67781 | NF-YA7 | MLOC_75867 | NF-YB7 | ||
| MLOC_76757 | NF-YA8 | MLOC_7755 | NF-YB8 | ||
| MLOC_79487 | NF-YB9 | ||||
| AK357982 | NF-YB10 | ||||
| MLOC_36879 | NF-YB11 | ||||
Fig. 1.
The evolutional cladograms of the NF-Y proteins HvNF-YA (a), HvNF-YB (b), and HvNF-YC (c) genes generated by ClustalW using neighbor joining algorithm implemented in the MEGA software
Expression patterns of HvNF-Ys under different developmental stages and abiotic stresses condition
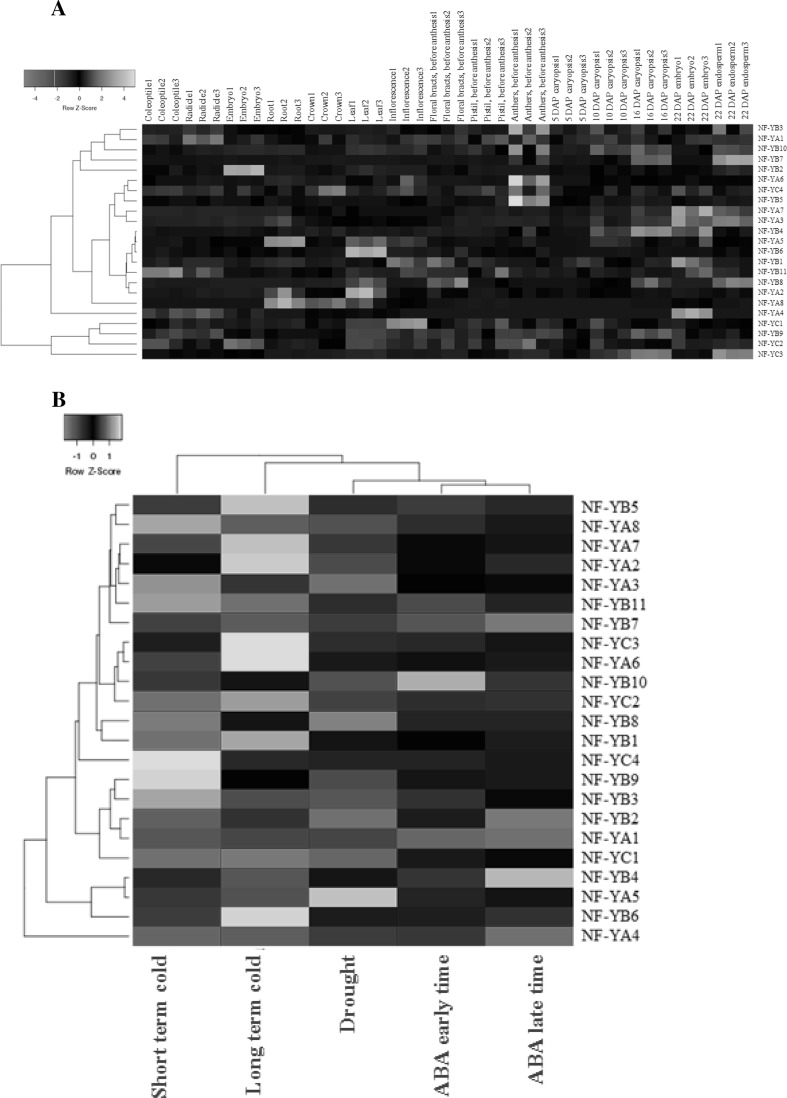
To examine the transcript accumulation of HvNF-Y genes in the entire barley life cycle, their expression profiles was analyzed at the 15 barley developmental stages. To infer the functional relationship of 23 HvNF-Y genes, hierarchical cluster was performed based on their expression levels (Fig. 2a). Based on expression patterns, the HvNF-Y genes were classified into three major groups.\ Groups A containing 5 genes including HvNF-YB3, HvNF-YA1, HvNF-YB10, HvNF-YB7and HvNF-YB2. Group B consisted of 13 genes which were further divided into three subgroups. In subgroup B1, HvNF-YA6, HvNF-YC4 and HvNF-YB5 showed high expression in the anthers before full anthesis. HvNF-YA7 and HvNF-YA3 showed high expression in embryos (22 day after pollination; Fig. 2a). HvNF-YA8 showed high expression in the roots, crown and shoots, whereas HvNF-YA2 showed high expression in the root and leafs. High expressions of HvNF-YAs were also detected in the coleoptile, radicals and embryos. Group C contained HvNF-YC1, HvNF-YB9, HvNF-YC2 and HvNF-YC3.
Fig. 2.
Expression patterns of HvNF-Y genes in various barley tissues (a) and abiotic stresses (b). Hierarchical clustering displays the expression profiles of the 23 HvNF-Y genes. Cluster analyses were carried out with Euclidean distances and the hierarchical cluster method of the “complete linkage”. The color bar at the base represents log2 expression values: green, low expression; black, medium expression; red, high expression
Transcriptome analysis of HvNF-Y genes under different abiotic stresses were shown in the Fig. 2b. The results revealed that HvNF-YA1 gene expressed in similar pattern under ABA and drought treatments condition and high expression of HvNF-YA4 gene was observed under ABA, drought and cold stress conditions. As shown in the Fig. 2b, HvNF-YA3, HvNF-YA2, HvNF-YA7 and HvNF-YA8 expressed at similar pattern under drought stress condition and expression of HvNF-YC3 gene was higher under ABA and drought stress conditions (Fig. 2b).
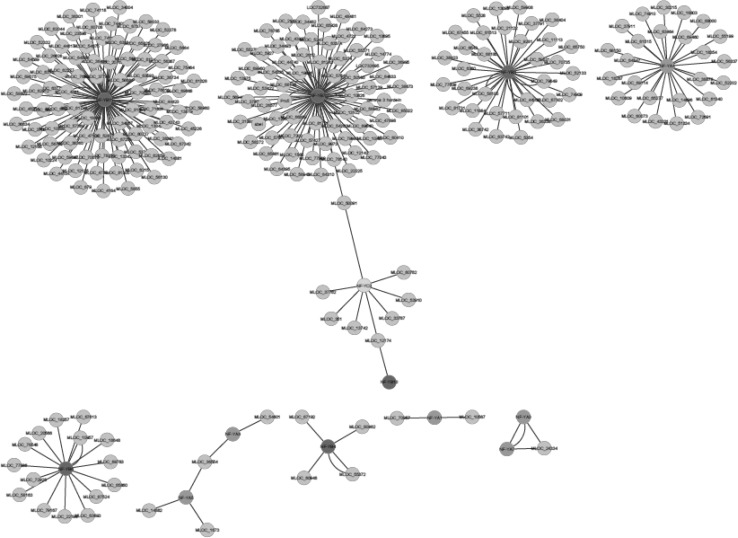
Co-expression networks and functional analysis
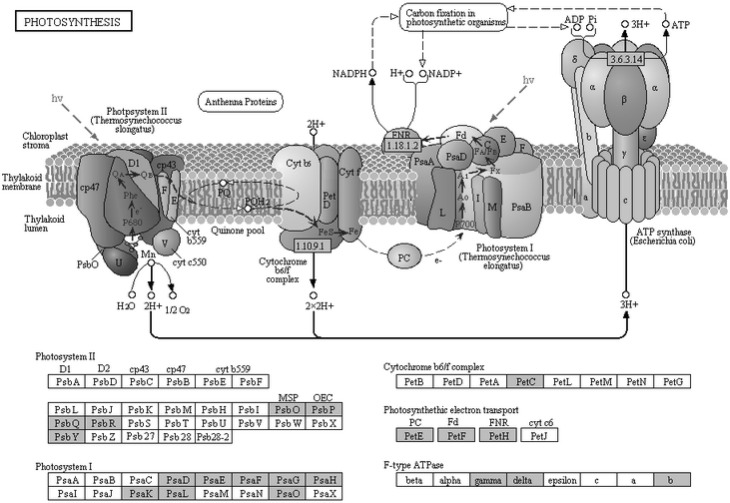
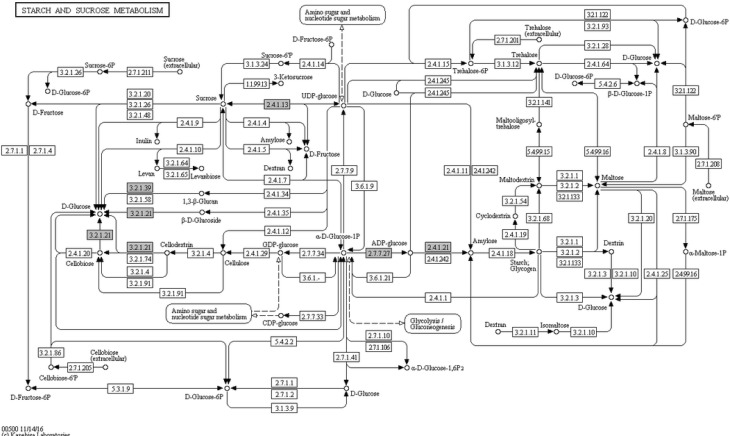
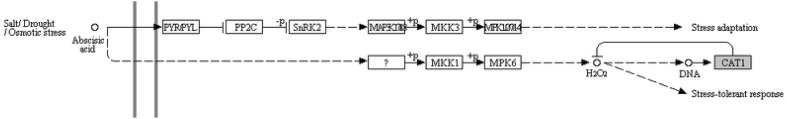
Further functional analysis of HvNF-Ys was done using construction of co-expression network (Fig. 3). Subsequently for functional annotation, co-expressed genes were mapped to KEGG database. Based on cut off of P ≤ 0.05, photosynthesis, starch, sucrose metabolism and salt stress responding pathways were enriched (Figs. 4, 5, 6). In photosynthesis pathway, five components of photosystem II (PSII) were co-expressed with the HvNF-Ys. Other pathways correlated with the HvNF-Y expression were sucrose and starch pathways. The correspondence genes for sucrose and starch metabolism were sucrose synthase (EC 2.4.1.13), 4-alpha-d-glucan glucohydrolase (EC 3.2.1.3), beta-glucosidase (EC 3.2.1.21), starch synthase (EC 2.4.1.21), glucose-1-phosphate adenylyl transferase (EC 2.7.7.27) (Fig. 5).
Fig. 3.
Co-expression networks of HvNF-Y genes. Green, pink and yellow color nodes represent the NF-YB, NF-YC and NF-YA, respectively
Fig. 4.
Photosynthesis pathway, as adapted from the Kyoto Encyclopedia of Genes and Genomes (KEGG). Co-expressed genes identified are highlighted by green boxes
Fig. 5.
Starch and Sucrose Metabolism pathway, adapted from the Kyoto Encyclopedia of Genes and Genomes (KEGG). Co-expressed genes identified in the present study highlighted by green boxes
Fig. 6.
Salt/Drought/Osmotic signaling pathway, adapted from the Kyoto Encyclopedia of Genes and Genomes (KEGG). Co-expressed genes identified in the present study highlighted by the green box
Expression analysis of NF-Ys under salt stress
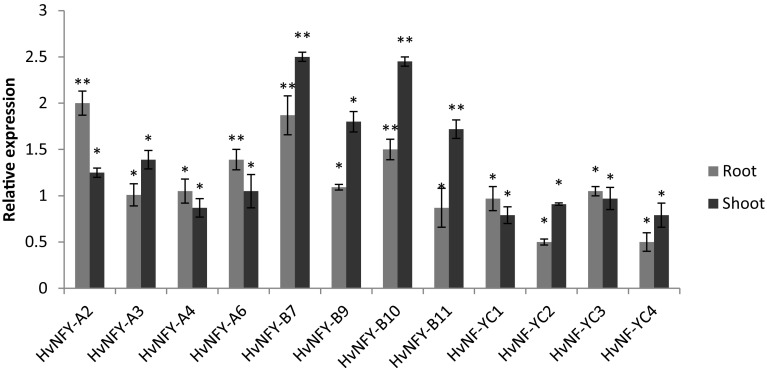
The expression response of 12 selected HvNF-Ys (8 NF-YA and 4 NF-YC) under salt stress condition (in both root and shoot parts) was validated using qRT-PCR. As shown in a Fig. 7, expression of the selected HvNF-Ys was changed in response to salt stress. HvNF-YA1 and HvNF-YA6 showed the highest differences in salt treated roots (Fig. 7).
Fig. 7.
Expression changes of HvNF-Ys in response to salt stress in roots and shoots. Their transcript levels were normalized to the 18SrRNA and then compared with control
Discussion
According to Gray et al. (2009), each protein was named with a two-letter code corresponding to Hordeum vulgare (Hv), followed by the NF-Y letter code and the family designation (NF-YA, B, or C). Finally, the identified genes were numbered based on their chromosomal positions (Cao et al. 2011).
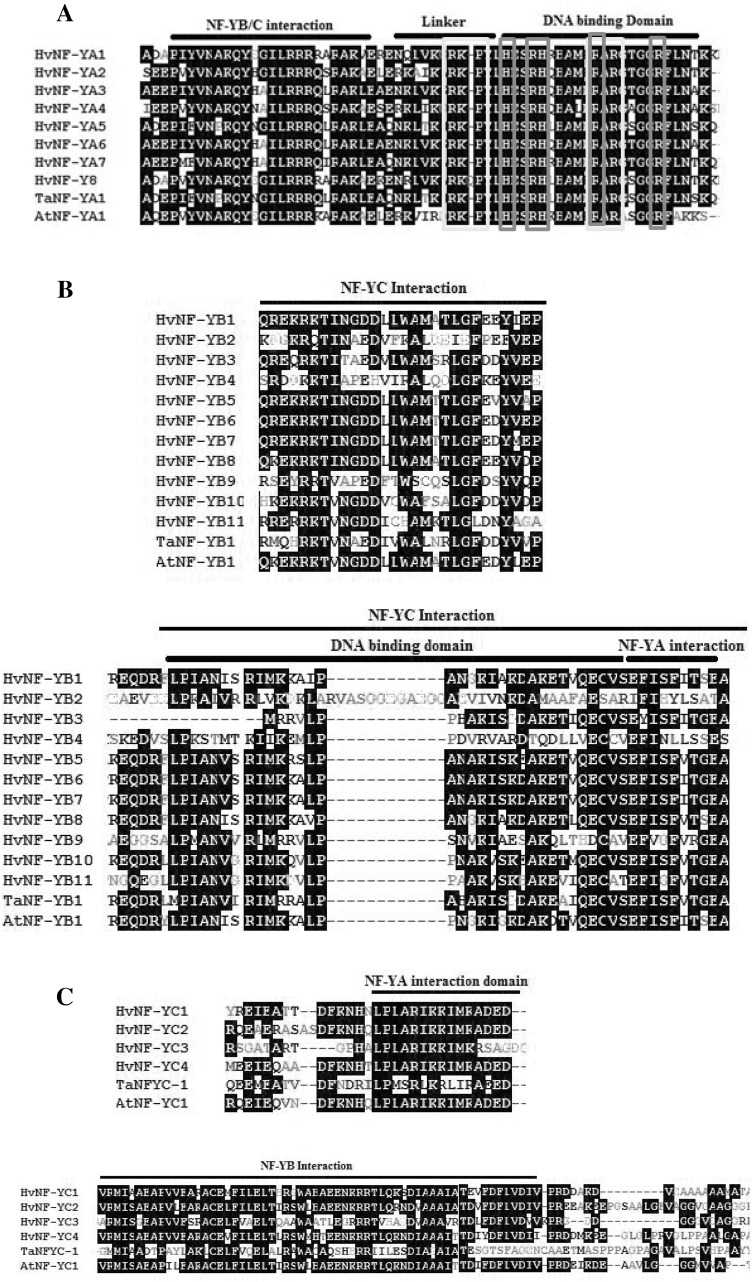
In concordance with Siefers et al. (2009) and Cao et al. (2011), in the HvNF-YA phylogeny tree, paralogus genes were equidistant from each other. This is in contrast to the HvNF-YB and HvNF-YC, where clear blocks of the conserved regions was observed (Fig. 8b, c). NF-YA proteins consisted of two conserved parts; interaction and DNA-binding domains (Fig. 8a; Hackenberg et al. 2012) which are separated with a linker. The interaction domain generally has 20 amino acids and interacts with NF-YB/NF-YC heterodimer (Mantovani 1999). Presence of three clusters of basic residues (red boxs in Fig. 8a) which serve as a nuclear localization signal is a distinct characteristic of NF-YA proteins (Thon et al. 2010).
Fig. 8.
Multiple sequence alignments of HvNF-YA (a) HvNF-YB (b) and HvNF-YC (c) as generated by the ClustalW software. In NF-YB/C interaction domain NF-YB and C create ahueteroduplex on NF-YA transcription factor. The red boxes highlighted the residues that is required for nuclear targeting
It has been suggested that evolutionarily constrains in NF-YA proteins is higher than NF-YB or NF-YC proteins (Cao et al. 2011). Structural studies revealed that NF-YA subunits are specifically responsible for all physical contacts with the CCAAT sequence (Romier et al. 2003). These residues are conserved in all eight HvNF-YAs (residues in yellow box in Fig. 8a) as well as in NF-YAs in other species (Thirumurugan et al. 2008)
Our results highlighted the regulatory potential of HvNF-Ys in photosynthesis process. As presented in Fig. 4, HvNF-Ys coordinately expressed with ATP synthase, ferredoxin, subunits of PSI, PSII and cytochrome b6f. In line with our results, functional CCAAT-boxes has been previously identified in the promoters of photosynthesis-related genes, but the role of NF-Ys in photosynthesis pathway is currently not well known. It has been also shown that NF-YB and NF-YA proteins regulate some of the photosynthesis-related genes such as chlorophyll a/b-binding protein (CAB) and the small subunit of Rubisco (RBCS) in stress conditions (Miyoshi et al. 2003; Warpeha et al. 2007).
The results of co-expression network analysis were consistent with the previous studies. Systematic analysis highlighted the OsNF-YB1 roles in the grain filling (Xu et al. 2016). It has been reported that OsNF-YB1 regulates the transcription of sucrose transporters through direct binding (Xu et al. 2016) and in Vitis vinifera, NF-Ys (NF-YA1 and NF-YB7) are induced by sucrose, which hints the involvement of NF-Ys in the biosynthesis and/or transport of sucrose (Ren et al. 2016).
As shown in the Fig. 6, Cat1 (Catalase) was co-expressed with HvNF-Ys. Catalase (EC 1.11.1.6) is considered as one of the main factors in plant tolerance to environmental stress (Navabpour et al. 2013; Shahriari Ahmadi et al. 2013; Panahi et al. 2013, 2014). It is one of the most important determinants of H2O2 levels during stress conditions. It has been proposed that one molecule of catalase eliminates about six million molecules of H2O2 per minute (Gill and Tuteja 2010). Recent evidences suggest the involvement of HAP2E and NF-YC1 in the H2O2 signaling pathways (Chen et al. 2015; Thön et al. 2010). Feng et al. (2015) reported that H2O2 induces the NF-YA1 and NF-YB8 expression under stress conditions.
The previous studies demonstrated that NF-Y transcription factors play key roles in abiotic stresses (Han et al. 2013; Ni et al. 2013). Therefore, we attempted further validation of barley NF-Ys function during salt stress. Among HvNF-YA genes, HvNF-YC2 and HvNF-YC3 was strongly up-regulated under salt stress condition. Comparison of expression changes of HvNF-Ys which are defined as hub genes in the co-expression networks showed notable changes of HvNF-Y3 gene in leaves and roots under salt stress condition. The results suggested that the most of HvNF-Ys might be the putative regulators in abiotic stress responses. This is also confirmed by the expression analysis of transcriptomic data under abiotic stress condition (Fig. 2b). In line with our results, it has been hypothesized that coordination of antioxidant defense systems such the production of CAT, thioredoxin reductases (TR) (EC 1.8.1.9) are regulated by NF-Ys (Ikbal et al. 2014). However, more research is needed to confirm of this hypothesis.
Conclusion
In conclusion, barley NF-Ys comprises relatively diverse gene family involved in different process such as starch metabolism, sucrose metabolism and photosynthesis. Functional analysis of HvNF-Ys by co-expression network and real time PCR revealed involvement of HvNF-Ys in adaptation to salt stress condition that can contribute to the maintenance of plant growth and productivity under stress conditions. The results of this study offer useful foundation for functional studies of HvNF-Ys and support the potential application of HvNF-Ys in the improvement of salt tolerance in cereals via biotechnological strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thanks of Dr Jalil Fallah (Lister Institute of Microbiology) for providing the Real time PCR reagants.
Authors’ Contributions
Conceived and designed the experiments: BP, SAM. Performed the experiments: BP, HAH,SAM, Analyzed the data: BP, SAM, HAH, MZM, KR. Wrote the paper: BP, SAM, KR, HAH, MZM.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Bahman Panahi, Email: panahibahman@ymail.com, Email: b.panahi@abrii.ac.ir.
Seyyed Abolghasem Mohammadi, Email: mohammadi@tabrizu.ac.ir.
References
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MA, Singh D, Gothandam KM. The effect of salinity on growth, hormones and mineral elements in leaf and fruit of tomato cultivar Pkm1. J Anim Plant Sci. 2012;22:159–164. [Google Scholar]
- Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt BF. Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE. 2011;6(6):e21805. doi: 10.1371/journal.pone.0021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhao Y, Zhuo C, Lu S, Guo Z. Overexpression of a NF-YC transcription factor from Bermuda grass confers tolerance to drought, salinity in transgenic rice. Plant Biotechnol J. 2015;13:482–491. doi: 10.1111/pbi.12270. [DOI] [PubMed] [Google Scholar]
- Consortium, I.B.G.S A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Feng Z-J, He G-H, Zheng W-J, et al. Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front Plant Sci. 2015;6:1142. doi: 10.3389/fpls.2015.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gray J, Bevan M, Brutnell T, Buell CR, Cone K, et al. A recommendation for naming transcription factor proteins in the grasses. Plant Physiol. 2009;149:4–6. doi: 10.1104/pp.108.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci. 2012;13:3458–3477. doi: 10.3390/ijms13033458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Tang S, An Y, Zheng DC, Xia XL, Yin WL. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance, improves water-use efficiency in Arabidopsis. J Exp Bot. 2013;64:4589–4601. doi: 10.1093/jxb/ert262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikbal FE, Hernández JA, Barba-Espín G, Koussa T, Aziz A, Faize M, et al. Enhanced salt-induced antioxidative responses involve a contribution of polyamine biosynthesis in grapevine plants. J Plant Physiol. 2014;171:779–788. doi: 10.1016/j.jplph.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Hole D, Wu J, Blake T, Wu Y. Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta. 2012;235:779–791. doi: 10.1007/s00425-011-1539-0. [DOI] [PubMed] [Google Scholar]
- Livack KJ, Schmittgen TD. Analysis of relative gene expression data using time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malviya N, Jaiswal P, Yadav D. Genome- wide characterization of Nuclear Factor Y (NF-Y) gene family of sorghum [Sorghum bicolor L. Moench]: a bioinformatics approach. Physiol Mol Biol Plants. 2016;22:33–49. doi: 10.1007/s12298-016-0349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/S0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ito Y, Serizawa A, Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003;36:532–540. doi: 10.1046/j.1365-313X.2003.01897.x. [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6:188–201. doi: 10.1093/mp/sss061. [DOI] [PubMed] [Google Scholar]
- Navabpour S, Moloudi F, Soltanloo H, et al. Catalase and Metallothionein genes expression analysis in wheat cultivars under drought stress condition. J Plant Mol Breed. 2013;1:54–68. [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82:113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- Panahi B, Mohammadi SA. Function of Alternative Splicing in plants (In Farsi) Modern Genetics J. 2018;1:1–9. [Google Scholar]
- Panahi B, Moshtaghi N, Torktaz I, Panahi A, Roy S. Homology modeling and structural analysis of NHX antiporter of Leptochloa fusca L. J Proteomics Bioinform. 2012;5:214–216. doi: 10.4172/jpb.1000238. [DOI] [Google Scholar]
- Panahi B, Shahriari Ahmadi F, Marashi H, Zare M, Moshtaghi N. Molecular cloning and expression analysis of Na+/H+ antiporter in monocot halophyte Leptochloa fusca L. NJAS-Wageningen J Life Sci. 2013;65:87–93. doi: 10.1016/j.njas.2013.05.002. [DOI] [Google Scholar]
- Panahi B, Abbaszadeh B, Taghizadeghan M, Ebrahimi E. Genome-wide survey of Alternative Splicing in Sorghum Bicolor. Physiol Mol Biol Plants. 2014;20:323–329. doi: 10.1007/s12298-014-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahi B, Mohammadi SA, Khaksefidi RE, Mehrabadi JF, Ebrahimie E. Genome-wide analysis of alternative splicing events in Hordeum vulgare: highlighting retention of intron-based splicing and its possible function through network analysis. FEBS Lett. 2015;589:3564–3575. doi: 10.1016/j.febslet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Quach TN, Nguyen HTM, Valliyodan B, JoshiT XuD, Nguyen HT. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Genet Genomics. 2015;290:1095–1115. doi: 10.1007/s00438-014-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Zhang Z, Wang Y, Li S, Liang Z. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.) BMC Genom. 2016;17:605–621. doi: 10.1186/s12864-016-2989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- Shahriari Ahmadi F, Panahi B, Marashi H, Moshtaghi N, Mirshamsi Kakhki A. Coordinate up-regulation of vacuolar Na+/H+ antiporter and V-PPase to early time salt stress in monocot halophyte Leptochloa fusca roots. J Agric Sci Technol. 2013;15:369–376. [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, IV, Tayrose G, et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol. 2007;65:77–92. doi: 10.1007/s11103-007-9200-9. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Steecher G, NeiMand Kumar S. MEGA: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genomics. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinf. 2002;1:2–3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, Brakhage AA. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010;38:1098–1113. doi: 10.1093/nar/gkp1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thön M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, et al. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010;38:1098–1113. doi: 10.1093/nar/gkp1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Zhang S, Olesen JT, Rich A, Guarente L. Subunit interaction in the CCAAT-binding heteromeric complexis mediated by a very short alpha-helix in HAP2. Proc Natl Acad Sci USA. 1994;91:3009–3013. doi: 10.1073/pnas.91.8.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lin Z, Tao Q, Liang M, Zhao G, et al. Multiple NUCLEAR FACTOR Y transcription factors respond to abiotic stress in Brassica napus L. PLoS ONE. 2014;9(10):e111354. doi: 10.1371/journal.pone.0111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JJ, Zhang XF, Xue HW. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J Exp Bot. 2016;67(22):6399–6411. doi: 10.1093/jxb/erw409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.