Abstract
As microbes face changing environments, they dynamically allocate macromolecular resources to produce a particular phenotypic state. Broad “omics” data sets have revealed several interesting phenomena regarding how the proteome is allocated under differing conditions, but the functional consequences of these states and how they are achieved remain open questions. Various types of multi-scale mathematical models have been used to elucidate the genetic basis for systems-level adaptations. In this review, we outline several different strategies by which microbes accomplish resource allocation and detail how mathematical models have aided in our understanding of these processes. Ultimately, such modeling efforts have helped elucidate the principles of proteome allocation and hold promise for further discovery.
Introduction
Microbes face transiently changing environments that require the expression of new proteins and the dilution or degradation of others. To adapt to these environmental changes, cells preferentially allocate these macromolecular resources to achieve certain objectives, a process typically referred to as “resource allocation.” The total amount and allocation of these proteins is fundamentally limited by constraints such as enzyme kinetics, cell size, and nutrient availability [1–4]. Therefore, microbes are regularly under selection pressure to optimize their resource allocation.
The macromolecular state of a cell can be measured using “omics” technologies, allowing insights into how resource allocation changes in a given condition. Omics data have revealed a highly skewed distribution of macromolecular resource allocation. For example, the most abundant 190 proteins in E. coli are estimated to account for about 60% of the total protein mass [5]. The functional consequences of such skewed macromolecular compositions—and how microbes regulate their state—are open questions. Over the past few years, studies that integrate omics and mathematical modeling have increased our knowledge of how microbes allocate macromolecular resources and of the genetic basis of these allocation strategies.
In this review, we summarize the current understanding of microbial resource allocation based on recent omics measurements from the perspective of biochemical networks. We discuss how computational models have been used to elucidate the functional significance of a cellular state and how these functions are linked to a genetic basis. We close with perspectives on promising directions for future modeling studies and the potential for examining resource allocation in the context of human health.
Proteome pre-allocation provides fitness benefits at a cost
The most direct way for microbes to alter the proteome is to synthesize proteins as needed. The maximum translation rate in E. coli is 16–20 amino acids per second per ribosome [6–8], implying synthesis in ~15 seconds for a copy of protein. However, protein abundances range from ~1 to >100,000 copies per cell [9], and ribosome abundances from ~7,000 to >70,000 per cell [7]. Therefore, during a nutrient shift where hundreds of thousands of additional protein copies can be needed [9], cells must utilize efficient strategies to dynamically allocate expression machinery resources. One strategy to minimize the delay of protein synthesis is to constitutively express proteins even when they are not immediately beneficial. This pre-allocation strategy incurs the cost of using up expression machinery that could be used to express immediately useful proteins, and a metabolic (energetic) cost of expression. Combined omics and modeling analyses have been used to test the hypothesis of pre-allocation.
In E. coli, up to half of expressed protein mass potentially provides no immediate benefit for a given growth condition [10]. Even when grown on glucose minimal medium, at least 13% of the proteins expressed confer no immediate fitness benefit based on ribosomal profiling and transposon mutagenesis [11]. Genome-scale model computations suggested that pre-allocating the E. coli proteome toward alternative carbon sources may provide a fitness benefit when alternative carbon sources are encountered [10].
Pre-allocation also applies to expressing more expression machinery than immediately needed to ensure fast expression rates when needed in a new environment. For example, when growing E. coli under feast-famine cycles, growth recovery during the feast phase was maximized by strategically allocating a ribosomal protein reserve [12].
These results suggest that omics data contain information both on the immediate response to the current environment and the regulatory program shaped by the organism’s evolutionary history. Computational models help to distinguish environment-specific response from pre-programmed responses shaped by evolutionary history.
Hierarchical regulation of resource allocation robustly improves fitness
Cellular metabolism has long been recognized to be regulated through a hierarchical network of regulatory processes [13]. The slow processes of transcriptional regulation and post-translational modifications act together with fast metabolite-level allosteric regulation to control metabolic fluxes [14,15]. Metabolites also modify transcription factors (TFs), leading to a coupling between metabolism and transcriptional regulation.
Recently, there has been increasing recognition of the importance of growth rate-associated global transcriptional regulation for resource allocation. For example, a recent study [16] showed that over 90% of transcriptional changes in 100 E. coli genes across 26 environments could be explained by a surprisingly small number of metabolite-TF interactions along with global regulation. In response to nutrient shifts, E. coli was found to use a global proteome reallocation strategy [17] instead of a theoretically optimal strategy of sequentially de-bottlenecking the rate limiting enzymes [18]. This apparently conservative strategy was hypothesized to be robust by confining metabolic bottlenecks to central precursors that drive global regulatory control [17].
However, growth rate-dependent regulation is not always dominant and appears to be context specific. A recent study subjected E. coli to a transient nutrient stress by starvation or by switching to a lower quality carbon source [19]. The study revealed a central role for proteome allocation in triggering the “persister” phenotype, a metabolically active but non-growing state with increased antibiotic tolerance [20]. Analysis of time-course proteomics from the nutrient-stressed cells (including persisters) and proteomics from other stress conditions (pH, temperature, and osmotic) revealed that proteome allocation was mainly driven by ppGpp-mediated regulation rather than a global growth rate effect [19]. Interestingly, a recent modeling study [21] showed that the optimal control strategy for E. coli to dynamically allocate resources during environmental changes involves an iterative on-off control strategy that resembles the structure of ppGpp-mediated regulation of ribosomal RNA transcription [22]. It thus appears that resource allocation under a variety of stresses may be mediated through overlapping mechanisms that are distinct from those of unstressed conditions.
Laboratory evolution aids in understanding the genetic basis of cellular resource allocation
Adaptive laboratory evolution (ALE) is an experimental method of serially passaging cells under a selection pressure. The outcome of ALE is a set of strains possessing adaptive mutations. ALE has now been automated [23], enabling large-scale production of evolved strains, followed by phenotyping and system-level characterization by DNA re-sequencing, RNA-Seq, 13C-metabolic flux analysis, etc. ALE has been used to reveal the genetic basis of growth rate-selection under various conditions: different carbon sources [24], thermal stress [25], osmotic and chemical stress [26,27], oxidative stress [27], and gene knockouts [28].
Multiple studies have connected the systems-level adaptations in an evolved strain to a genetic basis. For example, strains of E. coli were evolved for fast aerobic growth on glucose minimal media, yielding frequent key mutations in genes including rpoB and hns. Despite the potentially broad effects of mutations in these global regulators, the strains showed little change in intracellular metabolic pathway usage. Rather, the mutations enabled higher fluxes for glucose uptake, oxygen uptake, and central carbon metabolism [29]. Interestingly, TCA cycle enzymes have been reported to be transcriptionally repressed under similar selection pressure, yet TCA cycle metabolic flux did not decrease [29]. This result suggests potential nonlinearities between transcriptome abundance, proteome allocation, and flux capacity. A genome-scale model-based analysis further suggests that this nonlinearity arises in part from the flexibility of metabolic states that support optimal growth under these conditions [29]. Specifically, simulations showed that growth at 99% of the computed maximum rate could be supported by TCA cycle fluxes at 19% of the glucose uptake rate (GUR) but also as low as 8% of the GUR.
A recent study performed ALE under dynamic switching carbon sources, which led to either generalists or co-existing specialist subcommunities [30]. Transcriptomics and genome-scale modeling showed that the evolution of generalists versus specialists could be explained by the distance between computed metabolic states for two substrates [30]. Indeed, generalists evolved when the alternating substrates were metabolically “closer” and specialists evolved when metabolic states were more “distant.”
Network models capture the molecular basis of cellular resource allocation strategies
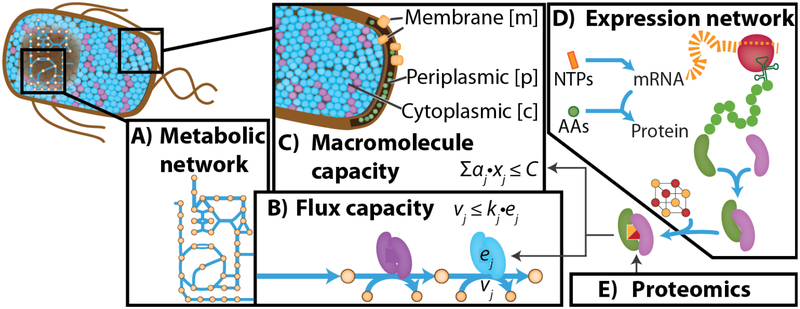
Biochemical network models, including genome-scale models of metabolism (M-models) [31,32] have traditionally been used to compute resource allocation at the level of small molecules and currency metabolites, such as NAD(P)(H) and ATP. GEMs were then extended to account for macromolecular resource allocation along two paths of development: (i) phenomenological (coarse-grained), and (ii) multi-scale (fine-grained) extensions to GEMs (Fig. 1). Here, we briefly review each category of models, followed by the new knowledge they have contributed.
Fig 1 |.
Network models of metabolism and macromolecule expression. (A) a metabolic network is reconstructed from the microbe’s annotated genome. (B) Metabolic flux (vj) is constrained by catalytic efficiency (kj) and enzyme abundance (ej). This general form can be reformulated [34]. (C) Macromolecule (xi) capacity is constrained based on its physical properties (aj) such as molecular weight and a total cell capacity (C). Macromolecule abundance can be computed using a macromolecule expression network (D), which can vary in level of detail, or from proteomics data (E). To constrain total mRNA, RNA-Seq is used instead.
Beginning with FBAwMC (flux balance analysis with molecular crowding) [33], a series of methods have been developed to extend M-models with macromolecular resource allocation constraints [34–38]. While these models do not explicitly compute the cost of expressing proteins based on the expression machinery network, they have been effective for integrating omics data to address many specific questions in different microbes: e.g., the basis of hierarchical substrate uptake [33], overflow metabolism [36,38,39], and metabolic states at sub-optimal growth rates [34]. One method extended an M-model of E. coli by integrated growth laws and proteome sector constraints [39]. The proteome sectors were derived from previous studies showing that the E. coli proteome can be partitioned into five coarse sectors whose total mass abundance correlates linearly with growth rate [5]. This coarse-grained model had been used previously to show that overflow metabolism occurs because the proteome cost of energy biogenesis by respiration exceeds that by fermentation [1].
In S. cerevisiae, an M-model extension named GECKO was developed [38]. This method constrains metabolic fluxes based on enzyme abundances that are bounded by proteomics measurements, and turnover rates obtained from BRENDA [38]. GECKO enabled prediction of new phenotypes and reduced flux variability in over 60% of metabolic reactions.
Multi-scale models of metabolism and macromolecular Expression
In a parallel effort, multi-scale models of Metabolism and macromolecular Expression (ME models) have been developed. A multi-scale biological model has been defined as one that includes components from two or more levels of biological organization (length scales) or processes occurring much faster than others (time scales) while conserving information across scales [40–42]. ME models integrate reconstructions of the macromolecule expression machinery with metabolism, enabling simultaneous computation of these two temporally and organizationally distinct but coupled processes [43,44]. These models compute steady states of the integrated system using linear or nonlinear optimization; therefore, they face technical challenges (i.e., ill-conditioning [45–47]) that are distinct from dynamic multi-scale models (e.g., stiff systems of differential equations [41]) or agent-based models [48]. ME models account for up to 80% of protein mass under fast growth conditions [44]. Protein complex stoichiometry is available for 95% of complexes in the E. coli ME model, iOL1650 [49]. ME models have been expanded to account for protein translocation [4], the proteostasis network, and temperature dependence of protein activity and stability [50]. ME models have been used to improve in silico strain performance predictions [51] and for a variety of fundamental studies. We highlight some of these studies below.
ME models have been used to identify a core set of proteins that must be expressed (resources must be allocated) to sustain microbial growth by simulating growth across 333 conditions [52] Methods have been developed to calibrate ME models to improve prediction accuracy. One method defined constraints on the mass fraction of proteome sectors, and calibrated these sectors using proteomics from 15 carbon sources [9] to improve growth rate and flux predictions [53]. Further refinement of ME model parameters was achieved using an optimization pipeline that integrated proteomics, RNA-Seq, and fluxomics [54]. This pipeline enabled a genome-wide estimation of in vivo enzyme turnover rates, based on simulated reaction fluxes and measured protein abundances across four growth conditions. Intriguingly, 284 estimated turnover rates corresponding to high-flux metabolic reactions were relatively invariant across growth conditions [54]. This result suggests that once accurately measured, the turnover rates for these reactions may be applicable across many conditions. Prioritizing the measurement of these relatively invariant kinetic parameters may be one way to approach the larger challenge of measuring the entire “kinetome” (i.e., kinetics of all enzymes) [55].
ME models have enabled deeper investigation of limitation in micronutrients or stress responses. A ME model of E. coli correctly predicted shifts in fermentation versus oxidative phosphorylation pathways in response to iron availability [56]. This simulation provided a possible explanation for why Fur regulates acnA (a TCA cycle enzyme) in a dual-mode manner of direct activation under iron-replete conditions and indirect repression under iron starvation. In a study of acid stress response in E. coli, a ME model was used to compute the fitness benefit of three alternative stress relief systems [57]. The model predicted that the active proton transporter, ndh-cbo system was more efficient than gadABC or adiC, which was consistent with up-regulation of ndh-cbo under acid stress.
Very recently, the ME model of E. coli was extended to include part of the cytoplasmic proteostasis network, and to account for temperature-dependent protein stability and activity [50]. The model, called FoldME, recapitulated changes in growth rate as a function of temperature in three different media conditions. Additionally, the model computed the change in abundances for the approximately 450 proteins that were predicted to be expressed at a given temperature (ranging between 28°C to 45°C). Despite the lack of high-quality protein thermostability data for the majority of the 1,554 modeled proteins, temperature-dependent growth rate predictions were robust against parameter uncertainty [50].
Macromolecular expression networks have also been reconstructed for other organisms besides E. coli and T. maritima. A genome-scale model of the plant pathogen Ralstonia solanacearum was recently reconstructed, and it included macromolecule biosynthesis and secretion, host interaction, and DNA modification [58]. The macromolecule network included 135 reactions for macromolecule biosynthesis, and 165 reactions for secretion processes. The model could compute the cost of synthesizing virulence factors, including exopolysaccharides.
A genome-scale model of Bacillus subtilis integrated 72 cellular processes including metabolism, protein translation and folding, ribosome maturation, and flagella synthesis [37]. The model included 614 reactions, 467 metabolites and ions, and 672 protein-coding genes, and was solved using a new method called Resource Balance Analysis (RBA) [59]. While the metabolic and expression machinery networks are less detailed than a typical ME model, the RBA model parameters could be calibrated using proteomics data, enabling quantitative prediction of protein abundance in new environments. RBA showed that protein allocation for most cellular processes in B. subtilis agrees well with the growth rate maximization objective but that a few processes integrate more complex objectives such as stress or survival [37].
Models can identify simple biological principles and connect them back to biomolecular mechanisms
Mathematical models aid knowledge expansion by distilling complex data into simple principles that may be transferable to other biological systems or contexts. Examples include phenomenological models that describe bacterial growth laws that dictate the allocation of proteome sectors [5,60]https://paperpile.com/c/ZDFrmo/j1wl+3H60. Computational models of microbes have been increasing in detail, with detailed models such as the whole-cell model of Mycoplasma genitalium [61] and ME models for T. maritima [62] and E. coli [43,44] being examples of detailed biomolecular and biochemical network models. We posit that such models help to reveal principles from complex interactions and data with the added benefit of connecting these principles back to biomolecular mechanisms and specific genes that can be validated using omics.
For example, Chen et al. [50] simulated growth, metabolism and response of the chaperone network under thermal stress and recovered a simple, linear “growth law.” This linear relationship described the mass fraction of ribosomes, molecular chaperones, and metabolic enzymes as a function of temperature and growth rate. The study additionally showed that thermosensitivity of growth was attributable not to a few rate-limiting enzymes but to many enzymes. Thus, in addition to computing the global consequences of thermal stress, the model also could enumerate plausible biomolecular responses.
Conclusions and Outlook
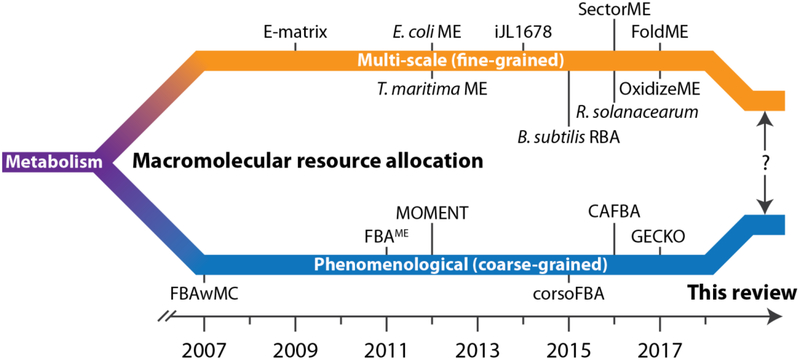
Our current understanding of cellular resource allocation has expanded toward a deeper appreciation of the coupling between regulatory processes across multiple biological scales. Fast metabolite-level regulation feeds back into slow transcriptional regulation, which in turn is hierarchically organized into general growth-associated regulation, global transcriptional programs, and local regulons. Advances in omics technologies and accumulation of multi-omics data have provided readouts of cellular state at these multiple biological scales. More than ever, there is a need for multi-scale network models that integrate these scales. Genome-scale models have risen to this challenge from two directions (Fig. 2). One is developing multi-scale reconstructions of macromolecular expression networks that are integrated with metabolism. This approach, while powerful, is time-consuming and has led to reconstructions for two different organisms so far. Research to accelerate reconstruction of these networks is ongoing [63]. The second approach is extending existing metabolic reconstructions with coarse-grained (phenomenological) constraints and variables that incorporate macromolecular abundance and catalytic efficiency. Based on these advances, we suggest two parallel, near-term efforts.
Fig 2 |.
Developmental paths of genome-scale resource allocation models. Models or algorithms listed are: FBAwMC [33], FBAME (Membrane Economics) [36], MOMENT [35], corsoFBA [34], CAFBA [39], GECKO [38], E-matrix [64], E. coli ME [43,44], T. maritima ME [62], iJL1678 (E. coli ME with the protein translocation network) [4], B. subtilis RBA [37], R. solanacearum [58], SectorME [53], FoldME [50], and OxidizeME [65].
First, there is a need to continue detailed reconstruction of non-metabolic processes. For the E. coli ME model, ~20% of protein mass is not modeled for rapidly growing cells [44]. Approximately half of this mass is allocated toward proteostasis and stress response [10]. The recent study of E. coli thermosensitivity has taken first steps toward a detailed reconstruction of the proteostasis network and thermal stress response [50]. Reconstructing periplasmic protein homeostasis and response to additional key stresses (oxidative, acid, osmotic, etc.) are likely to be achievable in the near future.
Second, we suggest a systematic interplay between detailed macromolecule expression reconstructions and coarse-grained resource allocation models as an attractive direction for future modeling studies. In principle, a model’s scope and scale should match the requirements (biological question and available data) of a specific study but this selection process is not trivial and often iterative [48]. ME models currently provide highly detailed reconstructions of expression networks integrated with metabolism. While reliable methods are now available to solve these multi-scale and nonlinear models [46,47], they are still computationally expensive. Methods for metabolic network reduction [66–68] may be adopted to macromolecular networks to reduce complexity. Additionally, methods to systematically coarse-grain selected parts of complex models into phenomenological constraints having few parameters may benefit future studies [69–71].
Cellular resource allocation has applications for microbial cell factory design [51] and also in the context of infectious disease. For example, recent evidence suggests that the persister cellular state, which contribute to chronic infection, may be regulated by a limitation in certain metabolic fluxes that triggers resource reallocation from growth to stress protection [19,72]. Another study used M-models to show that the environment alters the cost-benefit tradeoff of antibiotic resistance mutations through a change in metabolic state [73]. Additionally, antibiotic lethality by certain bactericidal antibiotics directly or indirectly affect macromolecular processes but the mechanisms of lethality are not fully known [74]. Future modeling studies of cellular resource allocation may thus provide new directions for therapeutic interventions against infectious disease.
Highlights.
Microbes allocate macromolecular machinery to perform different tasks
Proteome pre-allocation and hierarchical regulation give fitness benefits at a cost
Examining evolution helps elucidate the genetic basis of cellular resource allocation
Macromolecular resource allocation can be modeled by coarse-grained constraints
Detailed reconstructions of macromolecular expression networks are growing in number
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health Grants U01 GM102098 and R01 GM057089, and Novo Nordisk Foundation Grant NNF10CC1016517.
Source of support
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health Grants U01 GM102098 and R01 GM057089, and Novo Nordisk Foundation Grant NNF10CC1016517.
Footnotes
Conflict of interest
The authors declare no competing financial interests.
References
The full reference list is below in the next section. The following references are annotated as being of special (*) or outstanding (**) interest:
- 1.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T: Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 2015, 528:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T: Need-based activation of ammonium uptake in Escherichia coli. Mol Syst Biol 2012, 8:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vazquez A, Beg QK, Demenezes M a, Ernst J, Bar-Joseph Z, Barabási A-L, Boros LG, Oltvai ZN: Impact of the solvent capacity constraint on E. coli metabolism. BMC Syst Biol 2008, 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu JK, O’Brien EJ, Lerman J a, Zengler K, Palsson BO, Feist AM: Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale. BMC Syst Biol 2014, 8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui S, Silverman JM, Chen SS, Erickson DW, Basan M, Wang J, Hwa T, Williamson JR: Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol Syst Biol 2015, 11:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, Williamson JR, Fredrick K, Wang Y-P, Hwa T: Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol 2016, 2:16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milo R, Jorgensen P, Moran U, Weber G, Springer M: BioNumbers--the database of key numbers in molecular and cell biology. Nucleic Acids Res 2010, 38:D750–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bremer H, Dennis PP: Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus 2008, 3. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M: The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 2015, in press.** - A quantitative E. coli proteomics dataset measuring >2300 proteins (55% of E. coli genes) measured under 22 experimental conditions. Provides a resource for validating and calibrating large-scale resource allocation models.
- 10.O’Brien EJ, Utrilla J, Palsson BO: Quantification and Classification of E. coli Proteome Utilization and Unused Protein Costs across Environments. PLoS Comput Biol 2016, 12:e1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price MN, Wetmore KM, Deutschbauer AM, Arkin AP: A Comparison of the Costs and Benefits of Bacterial Gene Expression. PLoS One 2016, 11:e0164314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori M, Schink S, Erickson DW, Gerland U, Hwa T: Quantifying the benefit of a proteome reserve in fluctuating environments. Nat Commun 2017, 8:1225.* - Showed using phenotype, total RNA, total proteome measurements and an empirical model that growth recovery during the feast phase of feast-famine cycles is maximized by strategically allocating a ribosomal protein reserve.
- 13.ter Kuile BH, Westerhoff HV: Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett 2001, 500:169–171. [DOI] [PubMed] [Google Scholar]
- 14.Chubukov V, Gerosa L, Kochanowski K, Sauer U: Coordination of microbial metabolism. Nat Rev Microbiol 2014, 12:327–340. [DOI] [PubMed] [Google Scholar]
- 15.Gerosa L, Sauer U: Regulation and control of metabolic fluxes in microbes. Curr Opin Biotechnol 2011, 22:566–575. [DOI] [PubMed] [Google Scholar]
- 16.Kochanowski K, Gerosa L, Brunner SF, Christodoulou D, Nikolaev YV, Sauer U: Few regulatory metabolites coordinate expression of central metabolic genes in Escherichia coli. Mol Syst Biol 2017, 13:903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson DW, Schink SJ, Patsalo V, Williamson JR, Gerland U, Hwa T: A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 2017, 551:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlov MY, Ehrenberg M: Optimal control of gene expression for fast proteome adaptation to environmental change. Proc Natl Acad Sci U S A 2013, 110:20527–20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radzikowski JL, Vedelaar S, Siegel D, Ortega ÁD, Schmidt A, Heinemann M: Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol Syst Biol 2016, 12:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litsios A, Ortega ÁD, Wit EC, Heinemann M: Metabolic-flux dependent regulation of microbial physiology. Curr Opin Microbiol 2017, 42:71–78. [DOI] [PubMed] [Google Scholar]
- 21.Giordano N, Mairet F, Gouzé J-L, Geiselmann J, de Jong H: Dynamical Allocation of Cellular Resources as an Optimal Control Problem: Novel Insights into Microbial Growth Strategies. PLoS Comput Biol 2016, 12:e1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosdriesz E, Molenaar D, Teusink B, Bruggeman FJ: How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J 2015, 282:2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feist AM, Palsson BO: What do cells actually want? Genome Biol 2016, 17:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM: Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 2015, 81:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandberg TE, Pedersen M, LaCroix RA, others: Evolution of Escherichia coli to 42 C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Biol Evol 2014, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT: Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution. Metab Eng 2017, 39:141–150. [DOI] [PubMed] [Google Scholar]
- 27.Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I: Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 2013, 9:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzmán GI, Utrilla J, Nurk S, Brunk E, Monk JM, Ebrahim A, Palsson BO, Feist AM: Model-driven discovery of underground metabolic functions in Escherichia coli. Proc Natl Acad Sci U S A 2015, 112:929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR: Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab Eng 2017, 44:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM: Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies. Appl Environ Microbiol 2017, 83.* - Adaptive laboratory evolution under dynamic switching carbon sources led to either generalists or co-existing specialist subcommunities. Transcriptomics and genome-scale modeling showed that the distance between two substrates’ metabolic states was consistent with the evolution of generalists versus specialists.
- 31.O’Brien EJ, Monk JM, Palsson BO: Using Genome-scale Models to Predict Biological Capabilities. Cell 2015, 161:971–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monk JM, Lloyd CJ, Brunk E, Mih N, Sastry A, King Z, Takeuchi R, Nomura W, Zhang Z, Mori H, et al. : iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol 2017, 35:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beg QK, Vazquez A, Ernst J, de Menezes MA, Bar-Joseph Z, Barabási A-L, Oltvai ZN: Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci U S A 2007, 104:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz A, Qutub AA: Predicting internal cell fluxes at sub-optimal growth. BMC Syst Biol 2015, 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adadi R, Volkmer B, Milo R, Heinemann M, Shlomi T: Prediction of microbial growth rate versus biomass yield by a metabolic network with kinetic parameters. PLoS Comput Biol 2012, 8:e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang K, Vemuri GN, Mahadevan R: Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol 2011, 7:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goelzer A, Muntel J, Chubukov V, Jules M, Prestel E, Nölker R, Mariadassou M, Aymerich S, Hecker M, Noirot P, et al. : Quantitative prediction of genome-wide resource allocation in bacteria. Metab Eng 2015, 32:232–243.** - Developed the constraint-based modeling method Resource Balance Analysis (RBA). RBA calibrated with proteomics data showed that resource allocation of most cellular processes in B. subtilis agrees well with the growth rate maximization objective but a few processes integrate more complex objectives such as stress or survival.
- 38.Sánchez BJ, Zhang C, Nilsson A, Lahtvee P-J, Kerkhoven EJ, Nielsen J: Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol Syst Biol 2017, 13:935.* - Proteomics was used with enzyme turnover values from BRENDA to constrain a genome-scale model of S. cerevisiae metabolism.
- 39.Mori M, Hwa T, Martin OC, De Martino A, Marinari E: Constrained Allocation Flux Balance Analysis. PLoS Comput Biol 2016, 12:e1004913.** - Developed a modeling method (Constrained Allocation Flux Balance Analysis) integrating growth laws and coarse-grained proteome sector constraints into a genome-scale model of metabolism.
- 40.Southern J, Pitt-Francis J, Whiteley J, Stokeley D, Kobashi H, Nobes R, Kadooka Y, Gavaghan D: Multi-scale computational modelling in biology and physiology. Prog Biophys Mol Biol 2008, 96:60–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dada JO, Mendes P: Multi-scale modelling and simulation in systems biology. Integr Biol 2011, 3:86. [DOI] [PubMed] [Google Scholar]
- 42.Qu Z, Garfinkel A, Weiss JN, Nivala M: Multi-scale modeling in biology: How to bridge the gaps between scales? Prog Biophys Mol Biol 2011, 107:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiele I, Fleming RMT, Que R, Bordbar A, Diep D, Palsson BO: Multiscale Modeling of Metabolism and Macromolecular Synthesis in E. coli and Its Application to the Evolution of Codon Usage. PLoS One 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Brien EJ, Lerman JA, Chang RL, Hyduke DR, Palsson BØ: Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol 2013, 9:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Fleming RMT, Thiele I, Saunders MA: Robust flux balance analysis of multiscale biochemical reaction networks. BMC Bioinformatics 2013, 14:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma D, Yang L, Fleming RMT, Thiele I, Palsson BO, Saunders MA: Reliable and efficient solution of genome-scale models of Metabolism and macromolecular Expression. Sci Rep 2017, 7:40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Ma D, Ebrahim A, Lloyd CJ, Saunders MA, Palsson BO: solveME: fast and reliable solution of nonlinear ME models. BMC Bioinformatics 2016, 17:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu JS, Bagheri N: Multi-class and multi-scale models of complex biological phenomena. Curr Opin Biotechnol 2016, 39:167–173. [DOI] [PubMed] [Google Scholar]
- 49.Brunk E, Mih N, Monk J, Zhang Z, O’Brien EJ, Bliven SE, Chen K, Chang RL, Bourne PE, Palsson BO: Systems biology of the structural proteome. BMC Syst Biol 2016, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen K, Gao Y, Mih N, O’Brien EJ, Yang L, Palsson BO: Thermosensitivity of growth is determined by chaperone-mediated proteome reallocation. Proc Natl Acad Sci U S A 2017, 114:11548–11553.** - A ME model of E. coli was expanded to include the cytoplasmic proteostasis network and temperature-dependent enzyme activity and stability to show that thermosensitivity of growth is determined by chaperone-mediated proteome reallocation.
- 51.King ZA, O’Brien EJ, Feist AM, Palsson BO: Literature mining supports a next-generation modeling approach to predict cellular byproduct secretion. Metab Eng 2017, 39:220–227. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Tan J, O’Brien EJ, Monk JM, Kim D, Li HJ, Charusanti P, Ebrahim A, Lloyd CJ, Yurkovich JT, et al. : Systems biology definition of the core proteome of metabolism and expression is consistent with high-throughput data. Proc Natl Acad Sci U S A 2015, 112:10810–10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Yurkovich JT, Lloyd CJ, Ebrahim A, Saunders MA, Palsson BO: Principles of proteome allocation are revealed using proteomic data and genome-scale models. Sci Rep 2016, 6:36734.* - Proteomics data was used to calibrate the allocation of proteome sectors for a ME model of E. coli to improve growth rate and metabolic flux predictions
- 54.Ebrahim A, Brunk E, Tan J, O’Brien EJ, Kim D, Szubin R, Lerman JA, Lechner A, Sastry A, Bordbar A, et al. : Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun 2016, 7:13091.* - A multi-omics pipeline to estimate kinetic parameters in ME models. Found that enzyme turnover rates were typically condition invariant.
- 55.Nilsson A, Nielsen J, systems BP-C, 2017 undefined: Metabolic Models of Protein Allocation Call for the Kinetome. ElsevierPaperpile [date unknown], [DOI] [PubMed] [Google Scholar]
- 56.Seo SW, Kim D, Latif H, O’Brien EJ, Szubin R, Palsson BO: Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 2014, 5:4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo SW, Kim D, O’Brien EJ, Szubin R, Palsson BO: Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun 2015, 6:7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peyraud R, Cottret L, Marmiesse L, Gouzy J, Genin S: A Resource Allocation Trade-Off between Virulence and Proliferation Drives Metabolic Versatility in the Plant Pathogen Ralstonia solanacearum. PLoS Pathog 2016, 12:e1005939.** - A genome-scale reconstruction of R. solanacearum metabolism, macromolecule biosynthesis and secretion, host interaction, and DNA modification. The model could compute the cost of synthesizing virulence factors, including exopolysaccharides.
- 59.Goelzer A, Fromion V: Resource allocation in living organisms. 2017, doi: 10.1042/BST20160436. [DOI] [PubMed] [Google Scholar]
- 60.Barenholz U, Keren L, Segal E, Milo R: A Minimalistic Resource Allocation Model to Explain Ubiquitous Increase in Protein Expression with Growth Rate. PLoS One 2016, 11:e0153344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karr JR, Sanghvi JC, MacKlin DN, Gutschow MV., Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW: A whole-cell computational model predicts phenotype from genotype. Cell 2012, 150:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerman J a., Hyduke DR, Latif H, Portnoy V a., Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K, et al. : In silico method for modelling metabolism and gene product expression at genome scale. Nat Commun 2012, 3:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lloyd CJ, Ebrahim A, Yang L, King ZA, Catoiu E, O’Brien EJ, Liu JK, Palsson BO: COBRAme: A Computational Framework for Models of Metabolism and Gene Expression. 2017, doi: 10.1101/106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiele I, Jamshidi N, Fleming RMT, Palsson BØ: Genome-Scale Reconstruction of Escherichia coli’s Transcriptional and Translational Machinery: A Knowledge Base, Its Mathematical Formulation, and Its Functional Characterization. PLoS Comput Biol 2009, 5:e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Mih N, Yurkovich JT, Park JH, Seo S, Kim D, Monk JM, Lloyd CJ, Tan J, Gao Y, et al. : Multi-scale model of the proteomic and metabolic consequences of reactive oxygen species. bioRxiv 2017, doi: 10.1101/227892. [DOI] [Google Scholar]
- 66.Ataman M, Hernandez Gardiol DF, Fengos G, Hatzimanikatis V: redGEM: Systematic reduction and analysis of genome-scale metabolic reconstructions for development of consistent core metabolic models. PLoS Comput Biol 2017, 13:e1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Röhl A, Bockmayr A: A mixed-integer linear programming approach to the reduction of genome-scale metabolic networks. BMC Bioinformatics 2017, 18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erdrich P, Steuer R, Klamt S: An algorithm for the reduction of genome-scale metabolic network models to meaningful core models. BMC Syst Biol 2015, 9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirschner D, Pienaar E, Marino S, Linderman JJ: A review of computational and mathematical modeling contributions to our understanding of Mycobacterium tuberculosis within-host infection and treatment. Curr Opin Syst Biol 2017, 3:170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cilfone NA, Kirschner DE, Linderman JJ: Strategies for efficient numerical implementation of hybrid multi-scale agent-based models to describe biological systems. Cell Mol Bioeng 2015, 8:119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirschner DE, Hunt CA, Marino S, Fallahi-Sichani M, Linderman JJ: Tuneable resolution as a systems biology approach for multi-scale, multi-compartment computational models. Wiley Interdiscip Rev Syst Biol Med 2014, 6:289–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Radzikowski JL, Schramke H, Heinemann M: Bacterial persistence from a system-level perspective. Curr Opin Biotechnol 2017, 46:98–105. [DOI] [PubMed] [Google Scholar]
- 73.Zampieri M, Enke T, Chubukov V, Ricci V, Piddock L, Sauer U: Metabolic constraints on the evolution of antibiotic resistance. Mol Syst Biol 2017, 13:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang JH, Bening SC, Collins JJ: Antibiotic efficacy-context matters. Curr Opin Microbiol 2017, 39:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]