Abstract
Summary
The JCVI pan-genome pipeline is a collection of programs to run PanOCT and tools that support and extend the capabilities of PanOCT. PanOCT (pan-genome ortholog clustering tool) is a tool for pan-genome analysis of closely related prokaryotic species or strains. The JCVI Pan-Genome Pipeline wrapper invokes command-line utilities that prepare input genomes, invoke third-party tools such as NCBI Blast+, run PanOCT, generate a consensus pan-genome, annotate features of the pan-genome, detect sets of genes of interest such as antimicrobial resistance (AMR) genes and generate figures, tables and html pages to visualize the results. The pipeline can run in a hierarchical mode, lowering the RAM and compute resources used.
Availability and implementation
Source code, demo data, and detailed documentation are freely available at https://github.com/JCVenterInstitute/PanGenomePipeline.
1 Introduction
Since the publication of our pan-genome ortholog clustering tool (PanOCT) (Fouts et al., 2012) in 2012 there has been a tremendous increase in the number of publicly available bacterial genomes in RefSeq (e.g. from 6530 as of August, 2012 to 1,29,085 as of March, 2018), with Salmonella, Escherichia and Acinetobacter genomes increasing by factors of 43, 24 and 43, respectively. We present a pipeline with PanOCT as the centerpiece, providing additional analysis including: upstream data preparation, generation of a consensus pan-genome visualization, saturation curves, tree generation, annotation of clusters via HMM models and specified sequence databases and meta-grouping analysis. Additionally, the pipeline can be invoked in hierarchical mode, reducing the RAM and compute resources required to analyze large sets.
2 Design and implementation
The required input is a directory containing genome annotations in GenBank Flat File (.gb) format. Annotations can be on either peptide or nucleotide features. FASTA and gene-attribute files (which contain feature annotations and coordinates, defining gene neighborhood) are created from the feature tables in the .gb files. The pipeline distribution includes a script to download .gb files from NCBI. If .gb files are not available, the pipeline can run on user-supplied FASTA files and gene-attribute files.
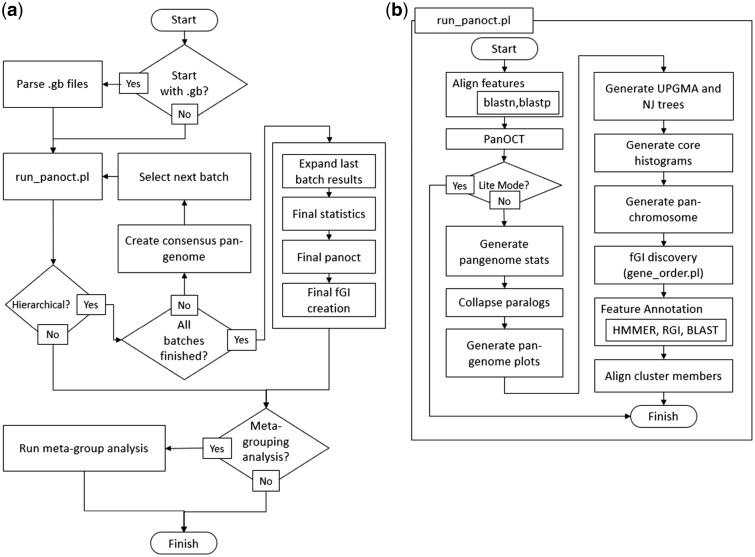
The pipeline is controlled by a wrapper script written in Perl, run_pangenome.pl [Fig. 1a]. This invokes another wrapper, run_panoct.pl [Fig. 1b], along with pre- and post-processing scripts. Run_panoct.pl controls processes used in a typical pan-genome run. Run_panoct.pl runs all-vs-all NCBI BLAST+ (Camacho et al., 2009) on the features, the results of which are used by PanOCT, along with gene-attribute files, to produce ortholog clusters of genomic features.
Fig. 1.
(a) Flowchart diagram of the entire pipeline run by run_pangenome.pl; (b) flowchart diagram of the single-batch processes by run_panoct.pl
Following PanOCT, run_panoct.pl collapses paralogs for saturation curve graphs, and generates several forms of visualizations, including UPGMA and Neighbor-Joining trees in Newick format, core gene histograms, pan-genome plots, and circular consensus pan-genome images. Clusters are annotated using HMMER3 (hmmer.org) searches comparing cluster representatives (medoids) to TIGRFAM (Haft et al., 2003) and PFAM (Sonnhammer et al., 1997) HMM libraries, RGI searches against the CARD database (McArthur et al., 2013) and BLAST+ searches against a curated set of sequences representing phage sequences and other groups of interest. These annotations can be viewed alongside flexible genomic region/island (fGR/fGI) information using PanACEA (Clarke et al., 2018).
For hierarchical PanOCT runs, a hierarchy file is supplied, describing which genomes to group together for each batch within the hierarchy. Later batches can refer to prior batches by name, incorporating their consensus pan-genomes as input. run_panoct.pl is called for each batch in the hierarchy, generating consensus pan-genome versions of FASTA and gene-attribute files for later batches to incorporate as input. Certain files from the final batch are expanded, in which the batch-based identifiers of the consensus pan-genome clusters are replaced with the members of the clusters each identifier represents. Lastly, clusters shared among predefined groups (e.g. pathogenic versus non-pathogenic, sequence type, resistant versus non-resistant) can be identified using meta-grouping analysis.
All scripts invoked from run_panoct.pl and run_pangenome.pl are available to run standalone for finer control of non-standard parameters. Many of the scripts can use an available UGE grid, but this is optional for the pipeline.
3 Summary
While the pipeline can be run on hundreds, or even thousands, of genomes, the memory used can grow to prohibitive requirements for most users. A server with 1 TB of RAM can adequately handle most pan-genome runs up to about 250–300 genomes. Using the hierarchical mode allows far higher numbers of genomes to be analyzed, decreasing the time and RAM required. The hierarchical run uses a basic divide and conquer approach to divide the n genomes into small (20–40) batches and then combine the resulting batches hierarchically in levels continuing to use the same small batches. The memory used is no more than that needed for a single small batch which scales quadratically (O(n2)) with the number of genomes in the batch. For a single batch, the compute time also scales quadratically to the number of genomes, but the divide and conquer approach scales as O(n log n).
Funding
This project has been funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Award Number U19AI110819.
Conflict of Interest: none declared.
References
- Camacho C. et al. (2009) BLAST+: architecture and applications. BMC Bioinform., 10, 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T.H. et al. (2018) PanACEA: a bioinformatics tool for the exploration and visualization of bacterial pan-chromosomes. BMC Bioinform., 19, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D.E. et al. (2012) PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res., 40, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D.H. et al. (2003) The TIGRFAMs database of protein families. Nucleic Acids Res., 31, 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A.G. et al. (2013) The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother., 57, 3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L. et al. (1997) PFAM: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]