Abstract
Cell division and death can be regulated by the mechanical forces within a tissue. We study the consequences for the stability and roughness of a propagating interface, by analysing a model of mechanically-regulated tissue growth in the regime of small driving forces. For an interface driven by homeostatic pressure imbalance or leader-cell motility, long and intermediate-wavelength instabilities arise, depending respectively on an effective viscosity of cell number change, and on substrate friction. A further mechanism depends on the strength of directed motility forces acting in the bulk. We analyse the fluctuations of a stable interface subjected to cell-level stochasticity, and find that mechanical feedback can help preserve reproducibility at the tissue scale. Our results elucidate mechanisms that could be important for orderly interface motion in developing tissues.
Interfaces are ubiquitous in tissue biology, between a tissue and its environment [1–3] or between cell populations [4–8]. There is great interest in how interfaces propagate smoothly or maintain their shape in the face of cell proliferation and renewal [1, 5, 9–12], for example by line tension acting at tissue boundaries [10, 13–15].
Theoretical efforts have focused on contour instabilities in cancer [16–20], branching [21, 22] or folding [23], and wound healing [3, 24, 25]. In models that include nutrient diffusion, protruding regions access more nutrient, triggering further growth [17, 18, 26], reminiscent of the Mullins-Sekerka instability in non-living systems [27]. An epithelium-stroma interface could form undulations due to mechanical stresses from cell turnover [28, 29], while a Saffman-Taylor-like instability based on viscosity contrast has been proposed to underlie branching in the developing lung [21]. A recent cell-based simulation of imbalanced mechanically-regulated growth between two epithelia observed a stable interface, and quantified its roughness [8]. A related simulation of cells in an inert medium found finger-like protrusions, arising for higher friction in the medium relative to the cells [30, 31]. Ref. [32] calculated the steady-state surface fluctuations of a non-growing tissue maintained in its homeostatic state.
In tissue replacement, such as in the developing Drosophila abdominal epidermis [33, 34], interface propagation occurs. This may be driven by imbalances in pressure associated with cell division, and/or directed cell motility, which cause the expansion of one tissue at the other’s expense.
In this Letter, we ask whether factors that drive an interface’s propagation can also affect its stability and roughness. We are particularly interested in the consequences of mechanically-regulated cell division and death for the behaviour of interfaces. If cell number change is sensitive to mechanical forces [11, 35–44] it leads to a “homeostatic pressure” [8, 45–47] which can drive interface propagation without coherently-directed cell motility forces. Alternatively, active, directed migration is proposed to drive interface motion in wound healing [2, 48, 49] or tissue replacement [34]. We study a model of competing epithelial tissues, with an interface driven by homeostatic pressure or by directed motility, acting either at the interface (a “leader-cell” limit) or in bulk [50] [51–53]. Our results encompass also the cases of stationary interfaces maintained under constant cell renewal [9], and of a single growing tissue [1].
We find a Saffman-Taylor-like instability involving substrate friction, and a long-wavelength instability dependent on an effective viscosity of cell number change. Bulk motile forces induce an instability depending on their strength and direction in each tissue. The free boundary of a growing single tissue is generally stabilised by the mechanisms studied here. Adding a driving noise to represent, e.g., stochastic cell division, we calculate the roughness and centre-of-mass diffusion of a stably-propagating interface, and find that mechanical feedback can help preserve reproducibility at the tissue scale [1].
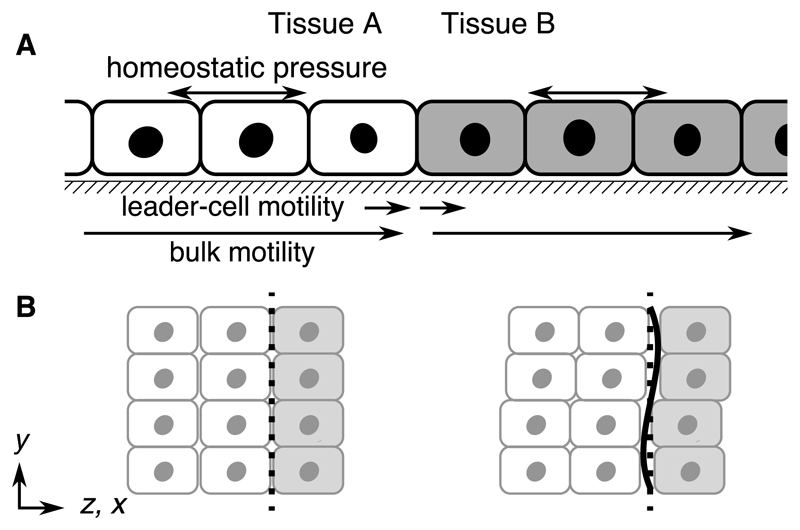
We use a 2D hydrodynamic description in terms of cell density and velocity fields. Tissues A and B cover an infinite domain, meeting at a flat interface (Fig. 1A). Since we assume a sharp interface, we state equations for a general tissue, unless decorated with A or B.
Fig. 1.
A) Side-on schematic of competing epithelial tissues. Each tissue is described with coarse-grained fields of cell density ρ, stress σij and velocity vi. Each has an elastic modulus χ (Eq. 3), substrate friction ξ (Eq. 4), and division/death responsive to strain on a timescale τ (Eq. 2). B) Top-down view of the tissues illustrating an interface contour fluctuation. We use a 2D description, with z a coordinate parallel to x in the comoving frame of the interface.
We begin with continuity of the areal cell density, ρ,
| (1) |
where vi is the velocity field and
| (2) |
is an expansion of the net division/death rate about the homeostatic density ρd, with τ a characteristic timescale [45]. The ρ units of each tissue are independent, so we set ρd ≡ 1. We consider a linearised, isotropic elastic stress,
| (3) |
where σh is a tissue’s homeostatic stress, χ its elastic modulus and Δρ ≡ ρ − 1. The homeostatic pressure imbalance is −Δσh ≡ −(σhA − σhB). The quantity χτ is an effective bulk viscosity for cell number change [21]; on a timescale τ, a tissue loses its elastic character as cells are lost or created in response to elastic stress [32, 54]. Based on parameter estimates (see supplement [50]), we neglect viscous stresses anticipating [47, 50]. Force balance expresses the tissue velocity as
| (4) |
for substrate friction ξ and a density of active motility forces fi = δixf directed normal to the interface. “Leader-cell” motility at the interface gives an effective contribution to Δσh [50]. We thus take f in Eq. 4 as uniform in a given tissue, to account for “bulk” directed motility forces, as may arise from cryptic lamellipodia away from tissue edges [51–53].
Moving steady state
We first solve for the steady propagation of a flat interface (cf. Refs. [8, 47]). The comoving coordinate is z ≡ x − V0t, with V0 the velocity of the interface at z0 = 0, propagating in z. Assuming driving forces small enough that nonlinear terms in Δρ, v can be neglected in Eq. 1, we write
| (5) |
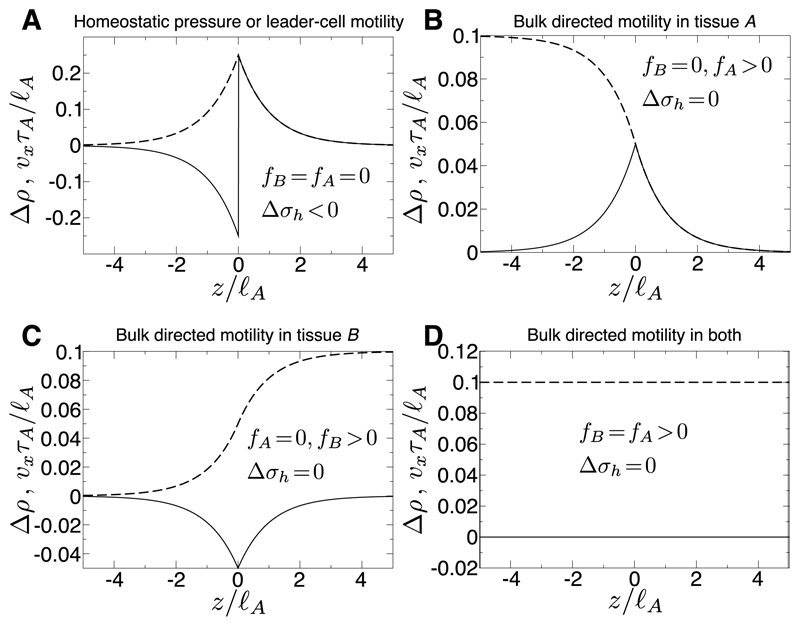
where vz ≡ vx − V0. The resulting propagating steady state is derived in the supplement [50] by matching the tissues’ stress and velocity at the interface. Density perturbations ∝ e±z/ℓ decay from the interface (Fig. 2) governed by each tissue’s hydrodynamic length Their sign (see Eq. S2 [50]) depends on −Δσh, and on Δvf ≡ fA/ξA − fB/ξB, a difference in “bare” velocities f/ξ associated to the bulk directed motilities. In Fig. 2A, the growing tissue has decreased density so, by Eq. 2, is proliferative near the interface, while the shrinking tissue has increased density so undergoes net apoptosis near the interface. In Fig. 2B, both tissues are apoptotic near the interface. The steady interface velocity,
| (6) |
is, for fA = fB = 0 and ξB = ξA, that found in Ref. [8]. To justify ignoring nonlinear terms, we require that stresses from homeostatic pressure, motility and interface line tension γ are small relative to the tissues’ elastic moduli: |Δσh| ≪ χ, |f/ξ| ≪ ℓ/τ, γ/ℓ ≪ χ. Much stronger stresses would lead to nonlinear responses and, eventually, tissue rupture [47, 50, 55].
Fig. 2.
A) Steady state density perturbation profile Δρ (solid line) and velocity vx (dashed) for tissues A (z < 0) and B (z > 0). Parameters: homeostatic pressure difference −Δσh = 0.5χA, frictions ξB = ξA, elastic moduli χB = χA, division/death timescales τB = τA, bulk motility forces fA = fB = 0. B) As A, but with Δσh = 0, fA = 0.1χA/ℓA. C) As A, but with Δσh = 0, fB = 0.1χA/ℓA. D) As A, but with Δσh = 0, fA = fB = 0.1χA/ℓA. In this particular case the tissue moves uniformly and the density perturbation cancels to zero.
Interface stability
In the supplement [50], starting from Fourier and Laplace transforms y → q and t → s of Eq. 5, we perturb the propagating steady state calculated above, to find the fate of an interface fluctuation (Fig. 1B) δz0 = ϵ(t) cos(qy), where ϵ(0) = ϵ0. For line tension γ ≥ 0 (e.g., increased myosin at heterotypic junctions [10] or a supracellular actin cable [56]),
| (7) |
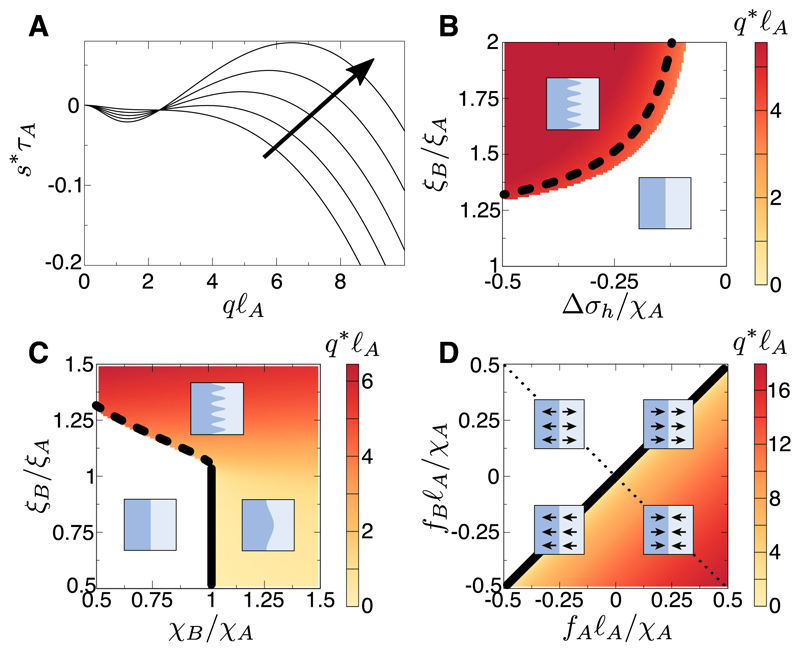
where δσ is the deviation from stress σ of the propagating steady state. The dominant growth rate is denoted s*(q), with s* < 0, > 0 indicating stability or instability (in the applicable parameter regime we do not find complex poles, so treat s* as real [50]). Dispersion relations (e.g., Fig. 3A) maximised over q yield phase diagrams (Fig. 3B,C,D) of the most-unstable wavenumber q*. We approximate the dispersion relation in limits of q [50] to find the analytic criteria discussed below.
Fig. 3.
A) Example numerically-determined dispersion relations. Parameters: frictions ξB = 1.5ξA, moduli χB = 0.5χA, division/death timescales τB = τA, bulk motility forces fA = fB = 0, line tension γ = 0.001ℓAχA. The homeostatic pressure imbalance/leader-cell motility parameter varies, Δσh = −0.1χA, −0.2χA, …, −0.5χA, in the direction of the arrow, crossing a “type Is” instability transition [57]. B) Phase diagram in (Δσh, ξB) of the most unstable wavenumber q* (white if no instability), using χB = 0.5χA, τB = τA, fA = fB = 0, γ = 0.001ℓAχA. The dashed line indicates the approximation of the type Is transition line by Eq. 8. C) Phase diagram in (χB, ξB) using Δσh = −0.5χA, τB = τA, fA = fB = 0, γ = 0.001ℓAχA. The meaning of the dashed line is as in C, whereas the solid line indicates the type IIs transition (Eq. 9). D) Phase diagram in bulk directed motilities (fA, fB), with other parameters ξB = ξA, χB = χA, τB = τA, γ = 0.001ℓAχA. The black line is the transition approximated by Eq. 10. The dotted line is V0 = 0, with the upper half-space V0 > 0 (tissue A growing) and the lower V0 < 0. In each quadrant, cartoons illustrate the direction of the bulk motilities.
Interface driven by homeostatic pressure or leader-cell motility
We first discuss growth of tissue A driven by Δσh < 0 (Fig. 3A,B,C), without bulk directed motility (fA = fB = 0). Analytic dispersion relations [50] show that, for strong enough Δσh, the interface is unstable if tissue B has greater friction ξB > ξA or effective viscosity χBτB > χAτA. Instability criteria (given Δσh < 0) are
| (8) |
and
| (9) |
where Eq. 8 is approximate [50]. Two types of transition arise. Fig. 3A and Eq. 8 show a “type Is” transition in the Cross-Hohenberg classification [57], where an intermediate band qmin < q < qmax, qmin ≠ 0 becomes unstable (“s” indicates that the instabilities found are stationary, not oscillatory). Fig. S2A [50] and Eq. 9 show a “type IIs” transition, with unstable band 0 < q < qmax and onset at q → 0. Then, one expects near threshold that the characteristic wavelength scales with system size. Eqs. 8, 9 are combined with phase diagrams of q* in Fig. 3B,C.
Interface driven by bulk directed motility
We now consider the case Δσh = 0, with bulk directed motility forces fA ≠ 0, fB ≠ 0. Instability occurs when Δvf ≡ fA/ξA − fB/ξB [50] satisfies
| (10) |
Fig. S2B [50] shows dispersion relations crossing the type IIs transition of Eq. 10. The phase diagram in Fig. 3D shows that a static interface (V0 = 0), marginally stable for f = 0 [50], can be stable or unstable depending on the direction of fA, fB.
Single tissue
The free boundary of a growing single tissue (e.g., epithelium invading empty substrate [2]), is stabilised by the mechanisms studied here (Eqs. S20–S22 [50]). Protrusion formation is often observed in wound-healing, via a number of proposed mechanisms we have not included [3, 24, 25, 58].
A stable interface subject to noise
Interface propagation and maintenance takes place in the presence of stochasticity in cell divisions, motilities, material parameters, etc. In the supplement [50] we model this with i) a driving noise in Eq. 5, ∂tΔρ+∂zvz +∂yvy = −(1/τ)Δρ+k where 〈k(z, y, t)k(z′, y′, t′)〉 = DA(B)δ(z−z′)δ(y−y′)δ(t−t′), corresponding to a contribution of random cell division; or (ii) a noisy motile force contribution to Eq. 4, 〈fi(z, y, t)fj(z, y, t)〉 = Dfδ(y − y′)δ(z − z′)δ(t − t′)δij. Noisy motility could arise, e.g., from ‘swirling’ patterns [3], provided that the correlation length of these patterns is small compared to other length scales discussed. We focus here on cell division noise, but find qualitatively similar results for noise on the motile force [50].
Excluding the q = 0 mode (discussed below), we calculate the correlation function 〈δz0(y + y′, t + t′)δz0(y, t)〉 of an interface in the stable parameter regime. The saturation (late-time) roughness as the system size in y, L⊥, becomes large, is
| (11) |
The dependence on L⊥ is as in 1-dimensional Edwards-Wilkinson deposition [59]. The positive denominator is expanded in the applicable regime of small Δvf, γ, Δσh. Roughness can be reduced by three now-familiar mechanisms: line tension γ > 0; stabilising effective viscosity contrast χAτA > χBτB, Δσh < 0; stabilising bulk motilities Δvf < 0. For identical tissues without line tension, the “interface” is an arbitrary line in the tissue: Eq. 11 then diverges, i.e., the roughness grows indefinitely. Identical tissues with line tension yield so that mechanical regulation (i.e., smaller τ) reduces boundary roughness. This is true also for a single tissue,
| (12) |
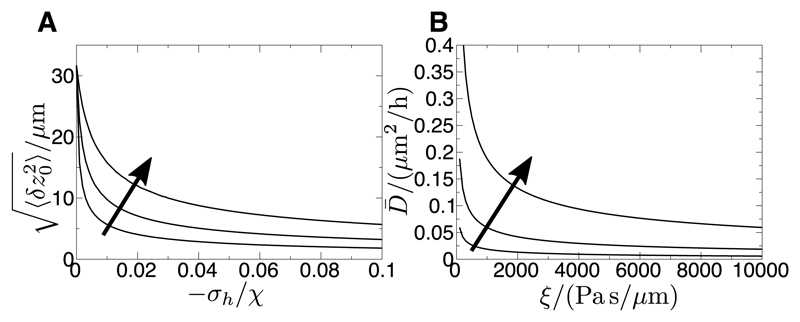
where if the tissue is growing (σh < 0) the roughness is decreased. Fig. 4A shows this behaviour quantitatively for estimated physical values of the parameters [50].
Fig. 4.
A) Root-mean-squared interface deviation for a single tissue using example physical parameters (Eq. 12). The homeostatic pressure/leader-cell motility parameter σh is varied, and the arrow shows increasing friction ξ = 103, 104, 105 Pa s/µm. Other parameters: χ = 104 Pa µm, τ = 3.6 × 104 s, γ = 103 pN, L⊥ = 1000 µm, D = 0.4 µm2/h. B) Interface centre-of-mass diffusion coefficient for a single tissue (Eq. 14) using example physical parameters. The friction ξ is varied, and the arrow shows increasing elastic modulus χ = 103, 104, 105 Pa µm. Other parameters: σh = 0, τ = 3.6 × 104 s, γ = 103 pN, L⊥ = 1000 µm, D = 0.4 µm2/h.
The q = 0 mode leads to an effective diffusion coefficient for the interface centre-of-mass,
| (13) |
and for a single tissue,
| (14) |
The behaviour of Eq. 14 for varying friction ξ and elastic modulus χ is shown in Fig. 4B. These equations control the accumulating uncertainty in tissue size as the interface centre-of-mass progressively diffuses away from its noise-free trajectory. A larger friction coefficient leads to more precise growth (Fig. 4B) but decreases the velocity (Eq. 6), which suggests trade-offs might be necessary to optimise the speed and precision of growth.
Discussion
Given experimental evidence of mechanically-regulated cell number change [11, 35–43], models of the type used here are widely studied [8, 31, 45–47]. There is much interest in mechanisms of boundary maintenance between cell populations [10, 56, 60]. Recent simulations showed how the topology of cellular interactions can stabilise anomalously smooth interfaces [13], while experiments suggest that interface maintenance is not only local but is connected to mechanical waves and jamming processes deep within neighbouring tissues [61]. Our results add to this picture, showing that mechanically-regulated cell number change within in the tissue bulk can exert an important influence on the properties of interfaces. We have shown how the forces driving overall interface propagation can also generate instabilities, and affect the response of interfaces to cell-level stochasticity.
A Saffman-Taylor-like instability based on substrate friction [30, 31] (Eqs. 8, S17 [50]) accords with the tumour literature, where tissues with weaker cell-matrix adhesions tend to be more invasive [62]. A longer-wavelength instability (Eq. 9) occurs if the effective bulk viscosity for cell number change is smallest in the growing tissue. Cell-based simulations [8] could explore our predictions, which could in turn be extended to include, e.g., cell growth anisotropy, proposed to play a role in the stable interfaces found in Ref. [8].
The effects of bulk directed motility depend on Δvf (Fig. 3D). Repulsive migration, known to occur due to Eph and ephrin signalling [14, 60], should yield Δvf < 0, favouring stability. In Drosophila abdominal epidermis, larval epithelial cells being replaced by histoblasts are proposed to actively migrate away from the propagating interface [34]. This motility force would promote stability, presumably desirable to ensure reproducible, well-controlled tissue replacement. This Drosophila system, or model experiments [61], could be used to test our theory by perturbing, e.g., motility, substrate friction, or cell division, and observing the effect on interfaces.
We found that mechanical feedback can help to smooth a stable interface in the presence of noise, as well as determining how quickly the interface centre-of-mass diffuses away from its noise-free position. These findings are relevant to the question of how tissue-level reproducibility is achieved despite cell-level stochasticity [1].
Supplementary Material
Acknowledgments
We thank Anna Ainslie, John Robert Davis, Federica Mangione and Nic Tapon for discussions. J. J. W. acknowledges support by a Wellcome Trust Investigator award to Dr Nic Tapon (107885/Z/15/Z), and acknowledges discussions with Ruth Curtain, Claire McIlroy, Pasha Tabatabai and Ansgar Trächtler. G. S. and J. J. W. acknowledge support by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001317, FC001175), the UK Medical Research Council (FC001317, FC001175), and the Wellcome Trust (FC001317, FC001175).
References
- [1].Hong L, Dumond M, Tsugawa S, Sapala A, Routier-Kierzkowska A-L, Zhou Y, Chen C, Kiss A, Zhu M, Hamant O, Smith RS, et al. Developmental Cell. 2016;38:15. doi: 10.1016/j.devcel.2016.06.016. [DOI] [PubMed] [Google Scholar]
- [2].Ravasio A, Cheddadi I, Chen T, Pereira T, Ong HT, Bertocchi C, Brugues A, Jacinto A, Kabla AJ, Toyama Y, Trepat X, et al. Nature Communications. 2015;6:7683. doi: 10.1038/ncomms8683. EP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Basan M, Elgeti J, Hannezo E, Rappel WJ, Levine H. Proceedings of the National Academy of Sciences. 2013;110:2452. doi: 10.1073/pnas.1219937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Baker NE. Curr Biol. 2011;21:R11. doi: 10.1016/j.cub.2010.11.030. [DOI] [PubMed] [Google Scholar]
- [5].Ninov N, Martin-Blanco E. Nature Protocols. 2007;2:3074. doi: 10.1038/nprot.2007.417. [DOI] [PubMed] [Google Scholar]
- [6].Vincent J-P, Fletcher AG, Baena-Lopez LA. Nature Reviews Molecular Cell Biology. 2013;14:581. doi: 10.1038/nrm3639. [DOI] [PubMed] [Google Scholar]
- [7].Gogna R, Shee K, Moreno E. Annual review of genetics. 2015;49:697. doi: 10.1146/annurev-genet-112414-055214. [DOI] [PubMed] [Google Scholar]
- [8].Podewitz N, Jülicher F, Gompper G, Elgeti J. New Journal of Physics. 2016;18:083020. [Google Scholar]
- [9].Marianes A, Spradling AC. eLife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Landsberg KP, Farhadifar R, Ranft J, Umetsu D, Widmann TJ, Bittig T, Said A, Julicher F, Dahmann C. Current Biology. 2009;19:1950. doi: 10.1016/j.cub.2009.10.021. [DOI] [PubMed] [Google Scholar]
- [11].de la Loza MD, Thompson B. Mechanisms of Development. 2017;144(Part A):23. doi: 10.1016/j.mod.2016.10.003. [DOI] [PubMed] [Google Scholar]
- [12].Curtius K, Wright NA, Graham TA. Nature Reviews Cancer. 2017;18:19. doi: 10.1038/nrc.2017.102. [DOI] [PubMed] [Google Scholar]
- [13].Sussman DM, Schwarz JM, Marchetti MC, Manning ML. Phys Rev Lett. 2018;120:058001. doi: 10.1103/PhysRevLett.120.058001. [DOI] [PubMed] [Google Scholar]
- [14].Cayuso J, Xu Q, Wilkinson DG. Developmental Biology. 2015;401:122. doi: 10.1016/j.ydbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- [15].Bielmeier C, Alt S, Weichselberger V, La Fortezza M, Harz H, Jülicher F, Salbreux G, Classen A-K. Current Biology. 2016;26:563. doi: 10.1016/j.cub.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tracqui P. Reports on Progress in Physics. 2009;72:056701. [Google Scholar]
- [17].Cristini V, Frieboes HB, Gatenby R, Caserta S, Ferrari M, Sinek J. Clinical Cancer Research. 2005;11:6772. doi: 10.1158/1078-0432.CCR-05-0852. [DOI] [PubMed] [Google Scholar]
- [18].Poplawski NJ, Shirinifard A, Agero U, Gens JS, Swat M, Glazier JA. PLOS ONE. 2010;5:1. doi: 10.1371/journal.pone.0010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ciarletta P, Foret L, Ben Amar M. Journal of Royal Society Interface. 2011;8:345. doi: 10.1098/rsif.2010.0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ben Amar M, Chatelain C, Ciarletta P. Phys Rev Lett. 2011;106:148101. doi: 10.1103/PhysRevLett.106.148101. [DOI] [PubMed] [Google Scholar]
- [21].Lubkin SR, Murray JD. Journal of Mathematical Biology. 1995;34:77. doi: 10.1007/BF00180137. [DOI] [PubMed] [Google Scholar]
- [22].Miura T. The Journal of Biochemistry. 2015;157:121. doi: 10.1093/jb/mvu087. [DOI] [PubMed] [Google Scholar]
- [23].Bayly PV, Okamoto RJ, Xu G, Shi Y, Taber LA. Physical Biology. 2013;10:016005. doi: 10.1088/1478-3975/10/1/016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zimmermann J, Basan M, Levine H. The European Physical Journal Special Topics. 2014;223:1259. [Google Scholar]
- [25].Tarle V, Ravasio A, Hakim V, Gov NS. Integr Biol. 2015;7:1218. doi: 10.1039/c5ib00092k. [DOI] [PubMed] [Google Scholar]
- [26].Castro M, Molina-París C, Deisboeck TS. Phys Rev E. 2005;72:041907. doi: 10.1103/PhysRevE.72.041907. [DOI] [PubMed] [Google Scholar]
- [27].Mullins WW, Sekerka RF. Journal of Applied Physics. 1963;34:323. [Google Scholar]
- [28].Basan M, Joanny J-F, Prost J, Risler T. Phys Rev Lett. 2011;106:158101. doi: 10.1103/PhysRevLett.106.158101. [DOI] [PubMed] [Google Scholar]
- [29].Risler T, Basan M. New Journal of Physics. 2013;15:065011. [Google Scholar]
- [30].Drasdo D, Hoehme S. New Journal of Physics. 2012 [Google Scholar]
- [31].Lorenzi T, Lorz A, Perthame B. Kinetic and Related Models. 2017;10:299. [Google Scholar]
- [32].Risler T, Peilloux A. J Prost Phys Rev Lett. 2015;115:258104. doi: 10.1103/PhysRevLett.115.258104. [DOI] [PubMed] [Google Scholar]
- [33].Ninov N, Menezes-Cabral S, Prat-Rojo C, Manjón C, Weiss A, Pyrowolakis G, Affolter M, MartIn-Blanco E. Current Biology. 2010;20:513. doi: 10.1016/j.cub.2010.01.063. [DOI] [PubMed] [Google Scholar]
- [34].Bischoff M. Developmental Biology. 2012;363:179. doi: 10.1016/j.ydbio.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Montel F, Delarue M, Elgeti J, Malaquin L, Basan M, Risler T, Cabane B, Vignjevic D, Prost J, Cappello G, Joanny J-F. Phys Rev Lett. 2011;107:188102. doi: 10.1103/PhysRevLett.107.188102. [DOI] [PubMed] [Google Scholar]
- [36].Streichan SJ, Hoerner CR, Schneidt T, Holzer D, Hufnagel L. Proceedings of the National Academy of Sciences. 2014;111:5586. doi: 10.1073/pnas.1323016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Puliafito A, Hufnagel L, Neveu P, Streichan S, Sigal A, Fygenson DK, Shraiman BI. Proceedings of the National Academy of Sciences. 2012;109:739. doi: 10.1073/pnas.1007809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Benham-Pyle BW, Pruitt BL, Nelson WJ. Science. 2015;348:1024. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Mechanisms of Development. 2007;124:318. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- [40].Pan Y, Heemskerk I, Ibar C, Shraiman BI, Irvine KD. Proceedings of the National Academy of Sciences. 2016;113:E6974. doi: 10.1073/pnas.1615012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Levayer R, Dupont C, Moreno E. Current Biology. 2016;26:670. doi: 10.1016/j.cub.2015.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gudipaty SA, Lindblom J, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V, Rosenblatt J. Nature. 2017 doi: 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fletcher GC, Diaz-de-la Loza M-d-C, BorregueroMuñoz N, Holder M, Aguilar-Aragon M, Thompson BJ. Development. 2018 doi: 10.1242/dev.159467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].LeGoff L, Lecuit T. Cold Spring Harbor perspectives in biology. 2016;8:a019232. doi: 10.1101/cshperspect.a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Basan M, Risler T, Joanny J-F, Sastre-Garau X, Prost J. HFSP Journal. 2009;3:265. doi: 10.2976/1.3086732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ranft J, Aliee M, Prost J, Jülicher F, Joanny J-F. New Journal of Physics. 2014:1. [Google Scholar]
- [47].Recho P, Ranft J, Marcq P. Soft Matter. 2016;12:2381. doi: 10.1039/c5sm02857d. [DOI] [PubMed] [Google Scholar]
- [48].Cochet-Escartin O, Ranft J, Silberzan P, Marcq P. Biophys J. 2014;106:65. doi: 10.1016/j.bpj.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Proceedings of the National Academy of Sciences. 2007;104:15988. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].See Supplemental Material for further theoretical calculations, estimates of parameters, and Refs. [63–71]
- [51].Farooqui R, Fenteany G. J Cell Sci. 2004;118:51. doi: 10.1242/jcs.01577. [DOI] [PubMed] [Google Scholar]
- [52].Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Nature Physics. 2009;5:426. [Google Scholar]
- [53].Scarpa E, Mayor R. The Journal of Cell Biology. 2016;212:143. doi: 10.1083/jcb.201508047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ranft J, Basan M, Elgeti J, Joanny J-F, Prost J, Jülicher F. Proc Natl Acad Sci. 2010;107:20863. doi: 10.1073/pnas.1011086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Harris AR, Peter L, Bellis J, Baum B, Kabla AJ, Charras GT. PNAS. 2012;109:16449. doi: 10.1073/pnas.1213301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hayes P, Solon J. Mechanisms of Development. 2017;144:2. doi: 10.1016/j.mod.2016.12.005. [DOI] [PubMed] [Google Scholar]
- [57].Cross MC, Hohenberg PC. Rev Mod Phys. 1993;65:851. [Google Scholar]
- [58].Mark S, Shlomovitz R, Gov NS, Poujade M, Grasland-Mongrain E, Silberzan P. Biophysical Journal. 2010;98:361. doi: 10.1016/j.bpj.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Antal T, Rácz Z. Physical Review E (Statistical Physics. 1996;54:2256. doi: 10.1103/physreve.54.2256. [DOI] [PubMed] [Google Scholar]
- [60].Taylor HB, Khuong A, Wu Z, Xu Q, Morley R, Gregory L, Poliakov A, Taylor WR, Wilkinson DG. Journal of The Royal Society Interface. 2017;14 doi: 10.1098/rsif.2017.0338. 20170338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rodríguez-Franco P, Brugues A, Marín-Llauradó A, Conte V, Solanas G, Batlle E, Fredberg JJ, Roca-Cusachs P, Sunyer R, Trepat X. Nature Materials. 2017;16:1029. doi: 10.1038/nmat4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Frieboes HB. Cancer Research. 2006;66:1597. doi: 10.1158/0008-5472.CAN-05-3166. [DOI] [PubMed] [Google Scholar]
- [63].Saffman PG, Taylor G. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 1958;245:312. [Google Scholar]
- [64].Edwards SF, Wilkinson DR. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 1982;381:17. [Google Scholar]
- [65].Blanch-Mercader C, Vincent R, Bazellières E, Serra-Picamal X, Trepat X, Casademunt J. Soft Matter. 2017;13:1235. doi: 10.1039/c6sm02188c. [DOI] [PubMed] [Google Scholar]
- [66].Bonnet I, Marcq P, Bosveld F, Fetler L, Bellaiche Y, Graner F. Journal of The Royal Society Interface. 2012;9:2614. doi: 10.1098/rsif.2012.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bambardekar K, Clément R, Blanc O, Chards C, Lenne PF. Proceedings of the National Academy of Sciences. 2015;112:1416. doi: 10.1073/pnas.1418732112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sepúlveda N, Petitjean L, Cochet O, Grasland-Mongrain E, Silberzan P, Hakim V. PLoS Computational Biology. 2013;9:e1002944. doi: 10.1371/journal.pcbi.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Curtain R, Morris K. Automatica. 2009;45:1101. [Google Scholar]
- [70].Trächtler A. arXiv:1603.01059 [math.DS] 2016 ArXiv e-prints. [Google Scholar]
- [71].A. Trächtler, private communication.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.