Abstract
Tapping into the metabolic cross-talk between a host and its virus can reveal unique strategies employed during infection. Viral infection is a dynamic process that generates an evolving metabolic landscape. Gaining a continuous view into the infection process is highly challenging and is limited by current metabolomics approaches, which typically measure the average of the entire population at various stages of infection. Here, we took an innovative approach to study the metabolic basis of host-virus interactions between the bloom-forming alga Emiliania huxleyi and its specific virus. We combined a classical method in virology, the plaque assay, with advanced mass spectrometry imaging (MSI), an approach we termed ‘in plaque-MSI’. Taking advantage of the spatial characteristics of the plaque, we mapped the metabolic landscape induced during infection in a high spatiotemporal resolution, unfolding the infection process in a continuous manner. Further unsupervised spatially-aware clustering, combined with known lipid biomarkers, revealed a systematic metabolic shift during infection towards lipids containing the odd-chain fatty acid pentadecanoic acid (C15:0). Applying ‘in plaque-MSI’ might facilitate the discovery of bioactive compounds that mediate the chemical arms race of host-virus interactions in diverse model systems.
Introduction
Microbial interactions, such as those between a host and its pathogen, are shaped by a dynamic metabolic cross-talk and chemical communication1,2. Metabolomics tools allow simultaneous identification of thousands of molecules in a given sample3, and have been used to monitor the metabolic fluctuations in diverse host-symbiont and host-pathogen model systems4,5.
Mapping the metabolic landscape of dynamic processes like viral infection requires high temporal resolution of the different stages of infection5,6. Most metabolic studies of viral infection are performed in bulk liquid cultures, measuring the average of the entire population at various time points during infection7,8. However, infection is not always a synchronous process, where all cells are infected at the same time. Asynchronous infection can lead to a mixed population of cells at different stages of infection. Consequently, bulk analysis might overlook rare subpopulations (e.g. resistant cells)9 and mask metabolic alterations that originate within them10.
Rewiring of host metabolic pathways by the virus during infection generates multiple metabolic states which may affect the infection outcome (i.e. cell death or resistance)9. Mapping these metabolic states into infection states might uncover unique strategies employed by the virus, which are essential for an optimal infection process1. Furthermore, occurrence of specialized metabolic capabilities encoded by viruses (i.e. auxiliary metabolic genes), common in the marine environment, provides an opportunity to reveal metabolic innovation during viral infection11. Recent advances in mass spectrometry imaging (MSI) technologies allow to localize specific metabolites of various dynamic processes and biotic interactions at the microscale level, and consequently to monitor metabolic changes in high spatiotemporal resolution12,13. MSI-based approaches were used to study the metabolic footprints of host-associated bacteria14, bacterial population dynamics15 and to detect human pathogens16.
An attractive host-virus model system is the cosmopolitan alga Emiliania huxleyi and its specific virus, E. huxleyi virus (EhV), which plays a key role in regulating the fate of carbon and sulfur in contemporary oceans17. E. huxleyi forms massive annual blooms that cover vast oceanic areas and are terminated following infection by EhV18. EhV infection causes profound remodeling of E. huxleyi’s transcriptome, which leads to massive changes in the metabolome and lipidome of infected cells. For example, changes in glycolysis, de novo fatty acid (FA) synthesis19, induction of triacylglycerols (TAGs) and rewiring of sphingolipid (SL) biosynthesis8,20,21. EhV is a large dsDNA virus with a genome harboring 472 predicted protein coding sequences, and it encodes almost a full biosynthetic pathway for SLs, a pathway that was not detected in any other viral genome to date22–24. Virus-derived glycosphingolipids (vGSLs), products of this virus-encoded pathway, were found to be central components of the EhV membranes and to trigger host programmed cell death (PCD)19,23,24. Hallmarks of PCD include the production of reactive oxygen species25, induction of caspase activity, metacaspase expression and compromised membrane integrity26,27. vGSLs are produced exclusively during viral infection and are used as an effective metabolic biomarker to detect viral infection during natural blooms of E. huxleyi in the ocean23,28.
In this study, we sought to gain higher spatiotemporal resolution of metabolites that are associated with viral infection, and in addition, to discover metabolites that are induced or reduced throughout this dynamic process. We combined a classical method in virology, the plaque assay29,30, with advanced MSI technologies, a unique approach we termed ‘in plaque-MSI’. This approach increased the spatiotemporal resolution by which we could study the metabolic profile of viral infection, taking advantage of the characteristics of the plaque. MSI data were used to visualize the changes of known lipid biomarkers for viral infection across the plaque. In parallel, spatially-aware unsupervised clustering and co-localization allowed for identification of lipids that were previously undescribed in this host-virus model system in addition to discovery of an unknown class of lipids. This approach uncovered a profound metabolic shift towards C15:0-based lipids during infection.
Results
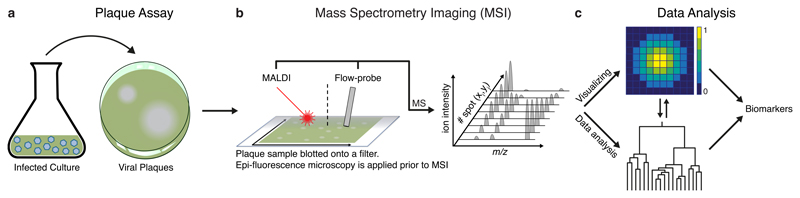
To gain high spatiotemporal resolution of the metabolic alterations during host-virus interactions, we mapped the metabolic profile of viral plaques (Fig. 1). As opposed to infection in liquid medium, a plaque originates in a single infected cell30. The plaque expands via concentric rings, and each ring corresponds to the plaque circumference at a different time point, keeping a metabolic record of the infection31. Thus, a snapshot of a plaque unfolds the infection process in a continuous manner, from the center of the plaque to the uninfected area in its periphery. We used the virus-susceptible E. huxleyi CCMP2090 strain and the lytic virus EhV201 to perform plaque assays (Fig. 1a). Accurate identification of a plaque and its center prior to MSI was based on the algal host chlorophyll autofluorescence, as detected by epi-fluorescence microscopy32. The microscopic image served as a reference for the following MSI analysis. Two MSI techniques were used to analyze the plaques: matrix assisted laser desorption ionization-MS (MALDI-MS, 100 µm resolution) and Flow-probe in situ micro-extraction33 (25 × 500 µm resolution on the x and y axes, respectively) (Fig. 1b). While MALDI-MS allows identification of spatial patterns of metabolites at a higher resolution, the Flow-probe-MS system has the advantage of faster sample analysis (~30 min vs. ~10 hours per sample) and a matrix-free preparation. The data collected by means of MSI was used to visualize the intensity of different metabolites along the plaque and to identify different spatial patterns (Fig. 1c). The spatial dimension of the plaque revealed the potential link between changes of specific metabolites and infection dynamics.
Fig. 1. Overview of the workflow of ‘in plaque-MSI’ analysis.
a, Viral plaques were prepared by mixing infectious EhV virions with E. huxleyi host cells in agarose and pouring the mixture into a petri dish, which was then incubated until the formation of plaques. b, A plaque sample was blotted onto a filter, which was imaged by epi-fluorescence microscopy prior to MSI in order to accurately identify the plaque morphology. Two MSI techniques were used to analyze plaque samples: MALDI-MS and Flow-probe-MS, each technique applied to a different biological sample. c, The collected spectra were used for visualization of specific metabolic markers for viral infection, as well as for untargeted data analysis using spatially-aware unsupervised clustering (Flow-probe-MS) and co-localization (MALDI-MS), aiming at discovering metabolites associated with viral infection.
Targeted MSI analysis of known lipid biomarkers across a viral plaque
We focused on known lipid biomarkers for viral infection, which were shown to be remodelled during infection in previous liquid chromatography-MS (LC-MS) based lipidomics experiments performed on liquid cultures8,20,21. These lipid biomarkers are usually classified based on their chemical category, such as: glycerolipids (GLs), glycerophospholipids (GPs) and sphingolipids (SLs). Since the viral genome also encodes the biosynthetic pathway for SLs22,23, the lipids can also be classified based on their biosynthetic origin (i.e. produced by alga- or virus-encoded enzymes). Additionally, these lipid biomarkers can be classified based on their relative abundance during infection (i.e. induction or reduction). Therefore, we grouped the lipids into three biological categories: virus-derived (encoded by the virus and induced during infection), host-induced and host-reduced (encoded by the host and induced or reduced during infection). vGSLs are the only virus-derived lipids known to date, with the majority of enzymes in their biosynthetic pathway encoded by the viral genome. Remodelled host lipids, on the other hand, include diverse classes: GLs such as BLL 36:6, TAG 48:1 and DGCC 40:7, and GPs such as PDPT 40:7 and PC 32:1, were all found to be induced during infection (i.e. ‘host-induced’), while others, such as BLL 38:6, were found to be reduced (i.e. ‘host-reduced’)8,20,21.
MS images of these lipid biomarkers across a plaque (Fig. 2) allowed us to follow their induction and reduction patterns during infection in a continuous manner based on a snapshot view of the plaque. Despite the differences in resolution and sample preparation, in most cases both MALDI-MSI (Fig. 2a) and Flow-probe-MSI (Fig. 2b) presented comparable spatial patterns (correlation > 0.8 for most lipids, see Supplementary Table 1) and highlighted the differences in the intensity of various lipids across the plaque. For example, vGSL (t17:0/h22:0) showed high relative intensity across the plaque area, while BLL 36:6 showed a different pattern, with lower intensity at the center of the plaque. Both lipids were detected only inside the plaque, whereas others, such as DGCC 40:7, PDPT 40:7 and TAG 48:1 were detected also outside the plaque area, albeit at a lower intensity, indicating a basal production in uninfected cells. Cross-section profiles (Fig. 2a and b) were used to present the relative abundance of each lipid along the plaque and revealed even finer differences, such as distance and degree of induction, between lipids with similar MS images. Interestingly, TAG 48:1 presented a unique ring-like pattern inside the plaque (visible only in the MALDI-MS, due to its higher spatial resolution), not identified in other known lipid species.
Fig. 2. Targeted MSI analysis of known lipid biomarkers reveals the metabolic landscape produced during viral infection.
Targeted analysis of two plaque samples at 5 dpi based on chlorophyll signal and seven lipid biomarkers for viral infection, using (a) MALDI-MS and (b) Flow-probe-MS. Chlorophyll autofluorescence (red pseudo-color) and MS images of specific lipids (false color representation of the relative intensity of detected ions, thresholding arrows were set to 0% and 100%) are presented. Cross-section graphs show the relative intensity across the plaque, from its periphery to the center, as depicted by the arrows (‘O’, out; ‘I’, in). vGSL, viral glycosphyingolipid (t17:0/h22:0) ([M+Na]+ = 826.6384 Da in MALDI-MS, [M+H]+ = 804.6565 Da in Flow-probe-MS); BLL 36:6, betaine-like lipid 36:6 ([M+H]+ = 802.5469 Da); BLL 38:6 ([M+H]+ = 830.5782 Da); TAG 48:1, triacylglycerol 48:1 ([M+Na]+ = 827.7105 Da in MALDI-MS, [M+H]+ = 805.7285 Da in Flow-probe-MS); DGCC 40:7, diacylglyceryl carboxyhydroxymethylcholine 40:7 ([M+H]+ = 826.6197 Da); PDPT 40:7, phosphatidyl-S,S-dimethylpropanethiol 40:7 ([M+H]+ = 849.5468 Da); PC 32:1, phosphatidylcholine 32:1 ([M+H]+ = 732.5543 Da)8,20,21. MS images were generated based on two samples (one for MALDI-MS and one for Flow-probe-MS). The analysis was performed on two additional samples, which presented similar intensity patterns (Supplementary Fig. 1). Scale bars: MALDI, 1 mm; Flow-probe, 2 mm.
Taken together, the targeted analysis indicates the existence of chemotypic heterogeneity during infection, as visible from the diverse intensity patterns of specific lipids across the plaque. This continuous metabolic view of the infection process would not have been possible in bulk LC-MS-based lipidomics.
Spatially-aware clustering of Flow-probe-MS data
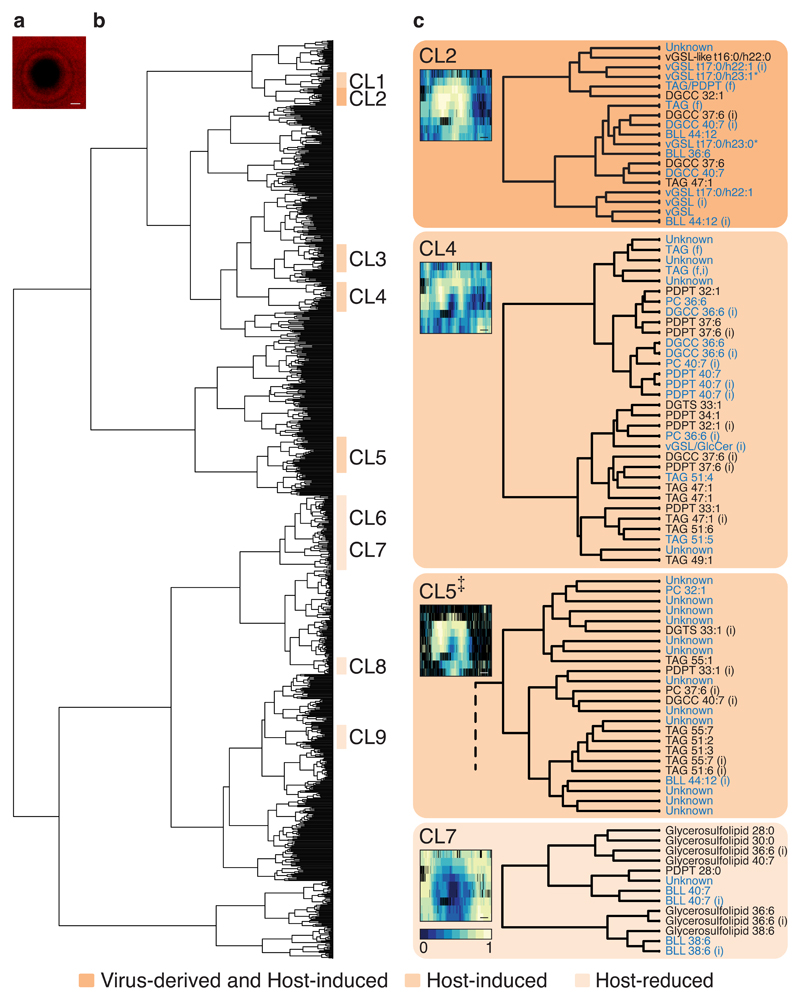
We applied an unsupervised spatially-aware clustering approach on Flow-probe-MS data generated from a plaque at 6 days post infection (dpi) in order to discover metabolites that are altered during infection34,35. First, m/z-images were generated for each m/z-value recorded, depicting the intensity of the m/z-values in each of the pixels analyzed in the sample. The m/z-images were clustered based on correlations between their spatial intensity profiles (Fig. 3a). A representative image was then generated for each cluster in the dendrogram, averaging the m/z-images of the cluster members. Based on these cluster-representative images, we selected clusters that presented distinct induction or reduction patterns across the plaque (Supplementary Fig. 2 and 3). This allowed us to focus on clusters with biological relevance to the infection dynamics. Next, we matched the m/z-values in each cluster with the known annotations of lipid biomarkers and used them as markers for the cluster category (i.e. virus-derived, host-induced or host-reduced). For example, cluster two (CL2) included several vGSL species (Supplementary Fig. 4) as well as other host-induced lipids, and therefore was classified as a ‘virus-derived and host-induced’ cluster. We then focused on four clusters which presented more pronounced changes across the plaque (Fig. 3b). Three clusters (CL2, 4 and 5) were induced in the plaque and one (CL7) was reduced. In total, the four clusters contained 129 mass features (see Methods section and Supplementary Table 2). Nineteen mass features were putatively annotated as lipids previously undescribed in this host-virus model system (eighteen induced and one reduced, see Table 1 and Supplementary Fig. 5), one of which was putatively identified as vGSL-like t16:0/h22:0 (as previously suggested by long-chain base analysis24, see Supplementary Fig. 6 and Supplementary Table 3). Five features that shared the same head group were not found in known repositories and were termed sulfonioglycerolipids (all decreased, see Table 1, Supplementary Fig. 5 and 7 and Supplementary Table 4).
Fig. 3. Unsupervised spatially-aware clustering of Flow-probe-MS data.
a, Chlorophyll autofluorescence (red pseudo-color) of the plaque sample at 6 dpi. b, Dendrogram clustering of all m/z-values based on their spatial Flow-probe-MSI distribution along the plaque (performed on a single experiment, as described in the Methods section). Nine clusters that presented distinct induction or reduction patterns across the plaque (as was visible by the representative average images of the clusters, see Supplementary Fig. 2 and 3) were classified and labelled. The category of each cluster was determined by the inclusion of known lipid biomarkers for viral infection. c, Four selected clusters are presented in detail, including their respective cluster-representative images and putatively annotated mass features. Mass features in blue indicate lipids that were previously described in the E. huxleyi-EhV model system; Mass features in black indicate either lipids previously undescribed in this model system or lipids of an unknown class, which were not found in known repositories (termed sulfonioglycerolipids, see Table 1). * Mass features of vGSL t17:0/h23:1 and vGSL t17:0/h23:0 were also putatively annotated as GlcCer t18:0/h22:1 and GlcCer t18:0/h22:0, respectively. ‡ Partial representation of CL5 (see Supplementary Fig. 3). A full m/z list of the four clusters can be found in Supplementary Table 2. (i), isotope; (f), fragment. Scale bar: 1 mm.
Table 1. Putative identification of previously undescribed lipids in the E. huxleyi-EhV model system.
| CL | Putative annotation (lipid composition) |
Theoretical m/z | Chemical formula | Change |
|---|---|---|---|---|
| 2 | DGCC 32:1 (14:0/18:1) | 726.5884 | C42H77NO8 | Induced |
| 2 | DGCC 37:6 (15:0/22:6) | 786.5884 | C47H79NO8 | Induced |
| 2 | TAG 47:1 (14:0/15:0/18:1) | 791.7129 | C50H94O6 | Induced |
| 2 | vGSL-like (t16:0/h22:0) | 790.6408 | C44H87NO10 | Induced |
| 4 | DGTS 33:1 (15:0/18:1) | 724.6091 | C43H81NO7 | Induced |
| 4 | PDPT 32:1 (14:0/18:1) | 749.5155 | C40H77O8PS | Induced |
| 4 | PDPT 33:1 (15:0/18:1) | 763.5312 | C41H79O8PS | Induced |
| 4 | PDPT 34:1 (16:0/18:1) | 777.5468 | C42H81O8PS | Induced |
| 4 | PDPT 37:6 (15:0/22:6) | 809.5155 | C45H77O8PS | Induced |
| 4 | TAG 49:1 (15:0/16:0/18:1) | 819.7442 | C52H98O6 | Induced |
| 4 | TAG 51:6 (15:0/18:1/18:5) | 837.6972 | C54H92O6 | Induced |
| 5 | TAG 51:2 (15:0/18:1/18:1) | 845.7598 | C54H100O6 | Induced |
| 5 | TAG 51:3 (15:0/18:1/18:2) | 843.7442 | C54H98O6 | Induced |
| 5 | TAG 53:6 (15:0/16:0/22:0) | 865.7285 | C56H96O6 | Induced |
| 5 | TAG 55:1 (15:0/18:1/22:0) | 903.8381 | C58H110O6 | Induced |
| 5 | TAG 55:6 (15:0/18:0/22:6) | 893.7598 | C58H100O6 | Induced |
| 5 | TAG 55:7 (15:0/18:1/22:6) | 891.7442 | C58H98O6 | Induced |
| 5 | TAG 59:12 (15:0/22:6/22:6) | 937.7285 | C62H96O6 | Induced |
| 7 | PDPT 28:0 (14:0/14:0) | 695.4686 | C36H71O8PS | Reduced |
| 7 | Sulfonioglycerolipid 28:0 (14:0/14:0)* | 689.5026 | C38H72O8S | Reduced |
| 7 | Sulfonioglycerolipid 30:0 (14:0/16:0)* | 717.5339 | C40H76O8S | Reduced |
| 7 | Sulfonioglycerolipid 36:6 (14:0/22:6)* | 789.5339 | C46H76O8S | Reduced |
| 7 | Sulfonioglycerolipid 38:6 (16:0/22:6)* | 817.5652 | C48H80O8S | Reduced |
| 7 | Sulfonioglycerolipid 40:7 (18:1/22:6)* | 843.5797 | C50H82O8S | Reduced |
Nineteen lipids were previously undescribed in the E. huxleyi-EhV model system and five belong to an unknown class of lipids (termed sulfonioglycerolipids). The lipids were putatively annotated based on MS/MS spectra (Metabolomics Standards Initiative level 2 annotation73), except the sulfonioglycerolipids (marked with an asterisk), which belong to the same putatively characterized compound class based on MS/MS spectra (level 3 annotation). ‘CL’, cluster (Fig. 3). ‘Change’, as visible in the Flow-probe-MS images (Supplementary Fig. 5). [M+H]+ adduct is presented for all lipids. DGTS, Diacylglyceryl trimethylhomoserine.
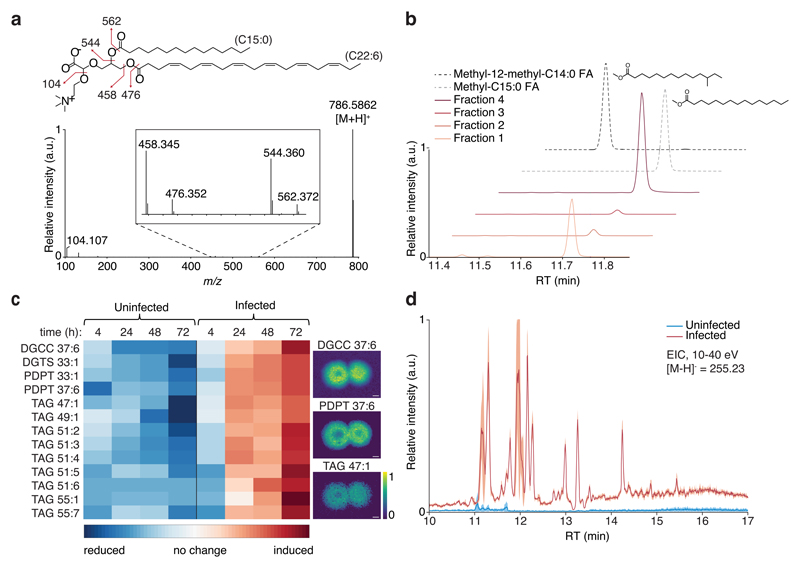
Interestingly, sixteen of the lipids that increased across the plaque were putatively identified as odd-chain fatty acid (OC-FA) lipids based on the Lipid Maps computationally-generated database of lipid classes and structure database (LMSD)36. Fourteen were previously undescribed in the E. huxleyi-EhV model system, while two were previously detected, however, their unique OC-FA characteristic was not identified21. MS/MS analyses of the OC-FA lipids using ‘in plaque’-Flow-probe revealed the existence of a C15:0 FA chain (pentadecanoic acid, Fig. 4a and Supplementary Data 1). The presence of C15:0 FA was further suggested by targeted LC-MS/MS analysis of infected cultures growing in liquid medium (Supplementary Fig. 8 and 9, Supplementary Tables 5 and 6 and Supplementary Data 2). The occurrence of C15:0 FA rather than a branched C14:0 FA (methyltetradecanoic acid) was confirmed by fatty acid methyl ester (FAME) analysis using gas chromatography-MS (GC-MS, see Fig. 4b and Supplementary Fig. 10).
Fig. 4. Induction of C15:0-based lipids during viral infection.
a, MS/MS-based putative identification of DGCC 37:6 with C15:0 FA (theoretical [M+H]+ = 786.5884 Da, observed = 786.5862 Da). The analysis was performed using both Flow-probe-MS/MS and LC-MS/MS based on two independent experiments in each technique, yielding similar results. Inset shows magnification of the mass range 450-570 Da. Double bonds in C22:6 FA were assigned based on the most common structure in the LMSD. b, GC-MS identification of C15:0 methyl ester in four different derivatized fractions collected from LC-MS runs of an infected culture at 48 hours post infection (hpi). Fraction 1 – DGCC 37:6 and PDPT 37:6; Fraction 2 – PDPT 33:1; Fraction 3 – DGTS 33:1, Fraction 4 – several TAGs. Methyl-C15:0 FA – Methyl pentadecanoate standard; Methyl-12-methyl-C14:0 FA – methyl 12-methylmyristate standard. FAME analysis was performed on two independent experiments, yielding similar results. c, Heatmap-representation of the alterations in C15:0-based lipids in uninfected and infected cells at different hpi (left) and MALDI-MS images of three representative lipids (right). Differences between control and infected samples were significant for all lipid species (2-way ANOVA, fdr-corrected, p < 0.02, n = 3). Scale bar: 2 mm. d, Systemic remodeling in C15:0-based lipids during viral infection presented by LC-MS extracted ion chromatogram (EIC) of C15:0 fragment ([M-H]- = 255.23 Da, negative ion mode, energy ramp of 10-40 eV) in uninfected and infected cells after 48 h. Values are presented as mean ± s.d. (light red or blue color), n = 3.
LC-MS-based analysis of these specific lipids during infection in liquid culture revealed a profound induction as compared to control uninfected cells (Fig. 4c and Supplementary Table 7). These OC-FA lipids were also detected in LC-MS-based analysis of the plaques, as well as in purified virions and in extracellular vesicles that originated from infected cultures (Supplementary Table 7). In order to explore the global distribution of C15:0-based lipids, we compared the abundance of C15:0 FA fragments in the lipidome of infected and uninfected cultures using LC-MS-based lipidomics of liquid cultures (Fig. 4d). This comparison revealed massive induction of C15:0-based lipids during lytic infection in diverse lipid classes, suggesting a systemic-level metabolic switch towards a C15:0-based lipidome. Following a targeted search, we were able to putatively identify seven more lipids containing a C15:0 FA (Supplementary Data 3). One of these lipids was TAG 48:1, which presented a unique ring-like intensity pattern across the plaque (Fig. 2) and has been shown to be highly induced during viral infection8, however, its FA composition was never explored. Our LC-MS/MS analyses indicated the existence of two structural isomers differing in their FA composition. While one isomer contained only even-chain FAs (C14:0, C16:0, C18:1) and was found both in uninfected and infected cells, the second isomer contained two OC-FAs (C15:0, C15:0, C18:1) and was found only in infected cells (Supplementary Fig. 11 and Supplementary Data 1).
A parallel analysis of MALDI-MS data by a computational co-localization approach was applied to three known lipid biomarkers: vGSL (t17:0/h22:0), BLL 36:6 and BLL 38:6 (Supplementary Tables 8-10). Most of the lipids co-localized with vGSL and BLL 36:6 were also found in the two clusters that originated from the Flow-probe-MS data (CL2 and CL4, see Fig. 3), including many of the C15:0-based lipids (Table 1). BLL 38:6, which is reduced during infection, was co-localized with all the sulfonioglycerolipids found in the host-reduced cluster in the Flow-probe-MS dataset (CL7, see Fig. 3 and Supplementary Table 10). Co-localization of TAG 48:1, which presented a unique ring-like intensity pattern, revealed seven additional mass features harboring a similar pattern, one of which was identified as TAG 46:1, and the remaining six features are currently unknown (Supplementary Fig. 12 and Supplementary Table 11).
Discussion
Tapping into the metabolic cross-talk between a host and its infecting virus can provide a unique way for the identification of metabolic pathways that are essential for viral infection or host defence strategies. E. huxleyi-EhV is an important host-pathogen model system with great ecological significance in the marine environment17,37. Viral infection leads to substantial rewiring of the lipidome of infected cells19, including a profound induction in saturated TAGs8 and specific vGSLs20,21,23. The production of both TAGs and vGSLs is essential for the assembly of virions8,24. TAGs are also enriched in extracellular vesicles, which are produced during infection and were shown to expedite infection38. A major obstacle in studying host-virus interactions is to track this highly dynamic process and to map the metabolic landscape produced to specific infection states. Gaining a continuous view into the infection process is limited by current metabolomics approaches (e.g. LC-MS), which typically measure the average of the entire population at various stages of infection. A viral plaque, on the other hand, captures the entire dynamics of the infection in one snapshot and resolves cells at different stages of infection in a spatiotemporal manner31. The spatial structure of the plaque might allow us to distinguish between the different metabolic states11 along the plaque and use them to predict the infection outcome (i.e. cell death or resistance)9. Segmentation of MALDI-MS data, having higher spatial resolution, could be instrumental to define these metabolic states that evolve during infection.
By applying our MSI-based analyses to viral plaques, we were able to gain a high spatiotemporal resolution view of the metabolic continuum during infection. This approach enabled us to detect differences in the induction and reduction patterns of known lipid biomarkers of viral infection. These lipid biomarkers were shown to be remodelled during viral infection in previous LC-MS-based lipidomics experiments performed on liquid cultures and represent diverse lipid classes8,20,21. We could identify differences between lipids that previously presented similar trends during infection. vGSL and BLL 36:6, which are used as lipid biomarkers for E. huxleyi infection in the natural environment23,28,39, were both induced in the plaque. However, BLL 36:6 was reduced at the center of the plaque and was induced at a greater distance from the center than vGSL (Fig. 2). Lower intensity of BLL 36:6 at the center might suggest modification of this lipid or its consumption and recycling for the synthesis of other metabolites at later stages of infection. The different localization of induction might be explained by production of BLL 36:6 in cells at earlier stages of infection than vGSL, or even in uninfected cells due to a chemical signal diffusing from the infected cells. It is also possible that BLL diffuses at a faster rate than vGSL in the plaque. It is important to note that since the center of the plaque is consisting of mainly lysed cells32, the detected lipids in this area and their signal might originate not only from the lysed cells and virions, but also from different biodegradation processes that can occur in the agarose. The chemical stability of lipids, on the other hand, does not seem to play a significant role in the relative abundance measured across the plaque (Supplementary Fig. 13).
The structure of the plaque and the identification of distinct induction and reduction patterns across the plaque allowed us to apply unsupervised spatially-aware clustering on Flow-probe-MS data in order to discover metabolites that are induced or reduced during infection. The representative average image of each cluster was used to reduce the complexity of the untargeted data, allowing us to select clusters with biological relevance to the infection dynamics. A selected number of clusters, all of them with distinct patterns, enabled us to concentrate our identification efforts on a smaller number of infection-related mass features (Fig. 3). Thus, we were able to detect a group of functionally relevant metabolites from a plethora of unknown mass features based on their spatial intensity patterns. Such a dedicated identification might help to uncover metabolites that are masked in untargeted LC-MS-based metabolomics analyses. Applying ‘in plaque-MSI’ to other host-virus interactions might allow the discovery of bioactive molecules and potential anti-viral compounds.
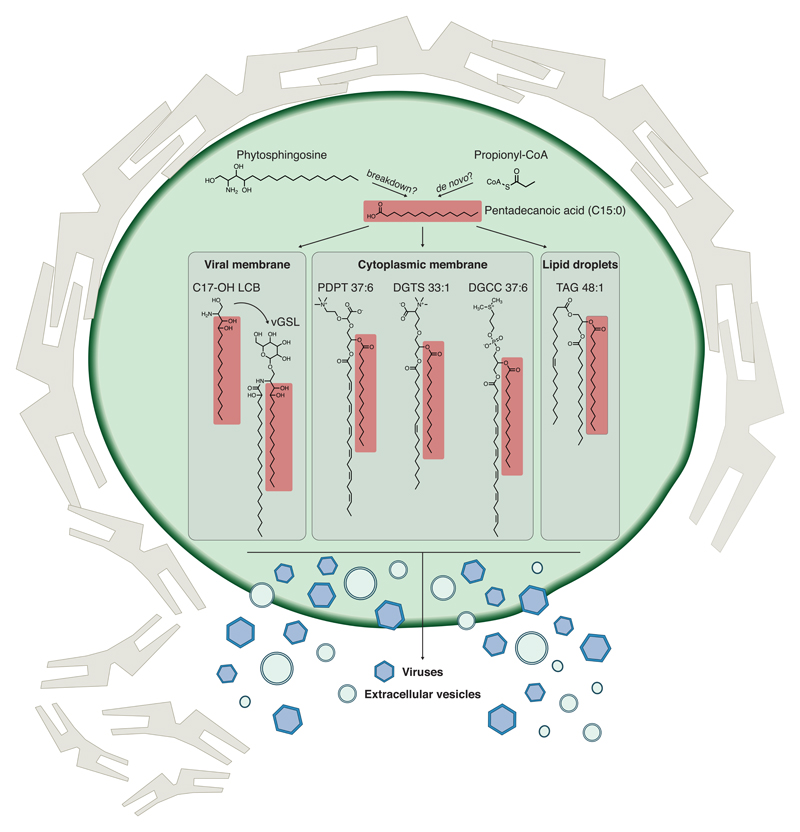
We putatively annotated nineteen lipid species that were previously undescribed in this host-virus model system and five of an unknown class of lipids, termed sulfonioglycerolipids. The sulfonioglycerolipids share the same head group and were highly reduced along the plaque and in infected liquid cultures (Supplementary Table 7). More than half of the lipids previously undescribed in this model system were putatively identified as C15:0-based lipids and were highly increased across the plaque and in infected liquid cultures (Fig. 4c). They were also detected in purified virions and in extracellular vesicles that originated from infected cultures (Supplementary Table 7) and represent a fraction of a systematic induction of C15:0-based lipids during infection (Fig. 4d). Interestingly, C15:0 FAs were incorporated in lipids usually found in the cytoplasmic membrane (e.g. SLs, DGCCs, DGTSs and PDPTs) or in lipid droplets (e.g. TAGs), but not in lipids found in the chloroplast (e.g. glycosyldiradylglycerols, see Fig. 5). While the biological function of OC-FA lipids is still unknown, C15:0 FA has been previously shown to be less preferred for β-oxidation than even-numbered FAs40. Thus, selective incorporation of OC-FA lipids in the viral membrane might increase the durability of the viral particles, and consequently attenuate their decay rate due to UV stress in the marine environment41. Induction of C15:0-based lipids during viral infection is also consistent with a previous report that indicated an increase in the levels of C15:0 FA during viral infection, although the specific lipids were not investigated42. Interestingly, C15:0 FA is also involved in the production of vGSLs during viral infection of E. huxleyi by EhV. vGSLs are products of a virus-encoded biosynthetic pathway and were found to be central components of the EhV membrane and to trigger E. huxleyi PCD19,23,24. EhV causes a shift in the substrate specificity of the virus-encoded enzyme pyridoxal 5′-phosphate (PLP)-dependent serine palmitoyl transferase (SPT; EC 2.3.1.50), the rate limiting enzyme of the entire SL biosynthetic pathway. In infected cells, the virus-derived SPT enzyme was shown to use C15:0-FA-CoA, while in uninfected cells the host-derived SPT typically utilizes the canonical C16:0-FA-CoA24. The C15:0-FA-CoA used by virus-derived SPT serves as a substrate for the unusual hydroxylated C17 long-chain base found in vGSLs24.
Fig. 5. Schematic model of the metabolic shift towards C15:0-based lipids in infected E. huxleyi cells.
Viral infection of E. huxleyi leads to a systematic induction of C15:0-based lipids. C15:0-based lipids are found in the cytoplasmic membrane and in lipid droplets and are also incorporated into virions and extracellular vesicles originating from the infected cell (Supplementary Table 7). The metabolic shift towards C15:0 FA42 during infection provides building blocks for C15:0-based lipids and is also required for the biosynthesis of vGSLs24, which were found to be central components of the EhV membranes and to trigger host PCD23. Several biosynthetic pathways have been proposed for C15:0 FA in other systems, among them de novo synthesis via propionyl-CoA, chain-shortening of longer FAs and degradation of phytosphingosine, a sphingoid base of GSLs47,50.
Recent years have witnessed a growing interest in lipids containing OC-FA and their role as markers for metabolic disorders in humans43. OC-FA lipids, including C15:0-based lipids, have been reported in humans, animals, microorganisms and plants44–46. Nevertheless, understanding of their biological function is still elusive. Several biosynthetic pathways have been proposed for OC-FAs, either de novo or by chain-shortening of longer FA chains (Fig. 5). De novo synthesis involves the conversion of propionic acid to propionyl-CoA, which, in turn, can replace acetyl-CoA in the initial step of fatty acid synthesis47,48. Excess of propionyl-CoA was reported to increase the levels of C15 and C17 FAs in humans43,49. In addition, phytosphingosine, a sphingoid base of GSLs, was previously reported as a source of C15:0 FAs in yeast. The odd FAs are later incorporated into GPs50. Future studies will enable elucidation of the biochemical origin and function of C15:0-based lipids during viral infection in the ocean.
Taken together, the ‘in plaque-MSI’ approach revealed a systematic metabolic shift towards C15:0-based lipids as a result of viral infection. Identification of this profound metabolic shift following an untargeted analysis might not have been possible in LC-MS-based analysis of bulk samples, since C15:0-based lipids are represented in diverse lipid classes. This shift might be part of the viral strategy to hijack host metabolism during infection, by inducing an orthogonal biosynthetic pathway for the synthesis of these lipids. Given their high and specific abundance during infection, C15:0-based lipids might serve as unique biomarkers for E. huxleyi-EhV interactions in the ocean.
Methods
Culture growth and viral infection dynamics
E. huxleyi strain CCMP2090 was used for this study. Cells were cultured in K/2 medium51 in artificial seawater (ASW)52 supplemented with ampicillin (100 µg mL-1) and kanamycin (50 µg mL-1), and incubated at 18 °C with a 16:8 h light:dark illumination cycle. A light intensity of 100 μmol photons m-2 s-1 was provided by cool white light-emitting diode lights. The virus used in this study was the lytic E. huxleyi virus EhV20153. Growing liquid cultures in the presence of antibiotics maintained a low basal abundance of bacteria throughout the experiments in both control and infected samples (Supplementary Fig. 14a). The lipid profile of the samples did not change significantly upon adding antibiotics, suggesting that the contribution of bacteria to the production of these specific lipids was insignificant (Supplementary Fig. 14b). The bacterial abundance was measured also in agarose samples, and no significant difference was detected between control and plaque samples (Supplementary Fig. 14c).
Plaque assay
150 mL of cells at 2-3×106 cells mL-1 were concentrated (2000 ×g, 5 min, 18 °C) to 2.7 mL. 300 μL of virions at a concentration of 104 virions mL-1 were added to the cells. After 2 h of incubation under normal growth conditions, the host-virus mixture was mixed with 9 mL of K/2 medium in ASW that contained 0.2% agarose (SeaKem LE agarose, Rockland, ME, USA) and then poured onto an agarose plate (12×12 cm) containing K/2 medium in ASW solidified by a 1.5%, supplemented with ampicillin (100 µg mL-1) and kanamycin (50 µg mL-1). Plaques were visible to the naked eye usually at 2 dpi. Plaque samples were subjected to MSI analysis at 5-6 dpi, when they were at a size large enough for spatial analysis (diameter of 4-8 mm) yet were still active and growing. Control samples were prepared following the same procedure, adding 300 µL ASW instead of virions. Blank samples were prepared following the same procedure, using K/2 medium only (i.e. without the host-virus mixture).
Enumeration of cells, viruses and bacteria
Cells were quantified using a Multisizer 4 Coulter Counter (Beckman Coulter, version 4.01) and an Eclipse (iCyt) flow cytometer (Sony Biotechnology, Champaign, IL, USA, using ec800 version 1.3.7 software), equipped with 405 and 488 nm solid state-air cooled lasers (both 25 mW on the flowCell) and standard filter setup. Algae were identified by plotting chlorophyll autofluorescence in the red channel (737 to 663 nm) versus green fluorescence (500 to 550 nm) or side scatter (Supplementary Fig. 15a). Extracellular viral particles were quantified as described previously (Supplementary Fig. 15b)24.
Quantification of E. huxleyi and bacterial cells from agarose samples was performed by suspending agarose samples in ASW. A sterile biopsy punch (8 mm diameter) was used for sampling. Each punched sample was added to 500 µL ASW, vortexed, pipetted and placed at 18 °C for 15 min to allow transfer of cells from the agarose to the ASW. Samples were then filtered through a 35 µm cell strainer. Plaques were smaller than 8 mm in diameter, and hence contained also uninfected areas. E. huxleyi cells were quantified as described above. Bacterial cells were quantified as described previously (Supplementary Fig. 15b)54.
Sample preparation for epi-fluorescence microscopy and MSI
Plaque samples were cut using a metal square punch and transferred to a glass slide. The upper layer of 0.2% agarose was blotted onto a 0.22 µm PVDF filter (Merck Millipore, Cork, Ireland). The filter containing the plaque was then dried for 10 min at 30 °C. The filter was immobilized on a microscope slide with double-sided adhesive tape for further analyses using epi-fluorescence microscopy and MSI.55–58
Epi-fluorescence microscopy
Microscopy images used for both Flow-probe-MS and MALDI-MS samples where obtained using Olympus IX81 motorized epi-fluorescence inverted optical microscope (Olympus, Tokyo, Japan) equipped with ×4 (numerical aperture (NA) 0.13, for MALDI-MS samples) and ×10 (NA 0.3, for Flow-probe-MS samples) objectives and a filter system for chlorophyll autofluorescence (Flow-probe-MS samples: ex: 470/40 nm, em: 632/60 nm; MALDI-MS samples: ex: 430/24 nm, em: 590 nm LP). Images were captured using ORCA-Flash 4.0 V2 (Hamamatsu Photonics KK, Hamamatsu City, Japan, for Flow-probe-MS samples) or Coolsnap HQ2 CCD (Photometrics, Tuscon, AZ, USA, for MALDI-MS samples) cameras and processed using MetaMorph (version 7.8.7, Molecular Devices) and CellSens Dimension (version 1.11, Olympus) software packages. Image analysis and processing was done using Fiji (Fiji Is Just ImageJ, version 1.51w). To correct for uneven illumination and remove background, a background image was generated by projecting the median of all frames and performing Gaussian blur (30 pixels diameter). Then, all frames were divided by the background image. Finally, the frames were stitched with 10% tile overlap. The resulting microscopy image allowed accurate identification of the plaque and its center prior to MSI. The center of the plaque can be detected by a reduced chlorophyll signal and is mainly consisting of lysed cells (Fig. 2)32,59. The infected area surrounding the center, on the other hand, is populated by living cells, as indicated by the chlorophyll signal.
Flow-probe-MS imaging
The analysis was performed at ambient conditions using a ‘Flow-probe’ in situ micro-extraction system33 (Prosolia, Indianapolis, IN, USA), connected to a Thermo Scientific ‘Q Exactive’ mass spectrometer. The ‘Flow-probe’ was operated using Flowprobe nMotion software (version 1.0.0.58, Prosolia). The instrument was operated in positive ion mode and the spray voltage set to 3 kV with a capillary temperature of 150 °C. A mixture of methanol:acetonitrile:toluene 50:35:15 + 0.1% TFA60, and a flow-rate of 10 μL min-1 was used. Samples were analyzed by measuring several parallel lines (pixel size: 25 × 500 µm, with spacing of one mm between lines) using a probe-speed of 100 μm sec-1. Chemical entities were monitored in a mass range between 125 and 1100 m/z at a resolution of 70,000 at 400 m/z for MS1 scans. Data-dependent fragmentation was performed on the ten most intense precursor ions per MS1 scan at a resolution of 17,500 at 400 m/z. Simultaneous Flow-probe imaging and data-dependent acquisitions were performed either using an inclusion list of known lipids, allowing ions not included in the inclusion list to be fragmented in the absence of listed ions, or without an inclusion list and using a dynamic exclusion window of three seconds. The full list of the lipids annotated using Flow-probe-MS/MS and their fragments can be found in Supplementary Data 1.
Processing of Flow-probe-MSI data
Raw Flow-probe-MSI data generated from a plaque at 6 dpi was converted from the vendor’s raw format to the open format mzML using the ‘msconvert’ software (part of ProteoWizard version 3.0)61 and from the mzML format to the MSI compatible data format imzML62 using the ‘imzMLConverter’ software (version 1.3)63. Next, imzML files corresponding with each experimental run were imported into the R open source programming environment (www.r-project.org) using the R package ‘MALDIquantForeign’64. Initially, 75 heading and tailing scans were removed from each horizontal acquisition line due to high levels of noise and measurement calibration issues, and in addition, the first horizontal acquisition line (out of eleven), which was detected in manual inspection of the raw data as an outlier, showing irregular visual pattern due to poor instrumental performance, was completely removed from analysis. This particular acquisition line was spatially located away from the center of infection, and we assume that no meaningful data was removed and that any subsequent data processing was not affected (Supplementary Fig. 16). Next, scans with a total ion current (TIC) lower than the 5% quantile of the overall per scan TIC (203 scans in total) were also removed immediately after raw data import and before any subsequent processing steps, to minimize unwanted shifts in mass and intensity measurement values (Supplementary Fig. 16a). Next, software parameters for mass feature detection, smoothing, alignment, binning and filtering were set according to the instrument’s mass measurement specifications and detailed manual inspection of known features in the raw data, as suggested by the software guidelines (Supplementary Table 12). The initial preprocessing step after the filtering procedures resulted in a matrix of 7035 mass features measured across 385.6 vertical scans per each of the ten horizontal acquisition lines (i.e. a matrix of 3856 scans by 7035 mass features). Next, matrix intensity values were log transformed and then scaled using a series of 25 specific mass features detected in a ‘blank’ sample (i.e. Flow-probe-MS analysis of a culture-free sample derived from K/2 medium in ASW containing 0.2% agarose). These 25 blank-specific features were used as reference values for the baseline analytical measurement variance, based on the assumption that they are independent of any biological factor. The median intensity value of these mass features was subtracted from the measured intensity values of all mass features across all scans, resulting in a feature matrix of log transformed and analytically-scaled intensity values in the arbitrary range of -6.46 to 4.91. The feature matrix was finally reduced column-wise by removing mass features which contained more than 99% missing values (corresponding with 5295 mass features or 75.26% of all mass features). The remaining matrix of 1740 mass features was then used to compute an intensity profile correlation matrix. Spearman's rho measure of association between all complete (i.e. non-NA) pairs of intensity values was calculated using the ‘cor.test’ function in R, resulting in a 1740 by 1740 correlation matrix. Agglomerative hierarchical clustering was then performed by transforming the correlation matrix into a distance matrix (i.e. scaling the correlation values to the range of zero to one) using the ‘agnes’ function in the R package ‘cluster’65. The ‘Ward’ clustering method was used after comparing the agglomerative coefficient (ac) scores between several methods (namely: ‘single’, ‘complete’, ‘average’ and ‘ward’) and using the method which resulted in the highest score (‘ac score’ = 0.98). Further cluster analysis was performed by converting the hierarchical cluster object into a dendrogram object and traversing the dendrogram structure to detect meaningful sub-clusters, focusing on sub-clusters containing mass features of previously identified, biologically meaningful lipids (i.e. ‘guilt-by-association’) or on clusters presenting meaningful spatial morphologies. Traversing the clustering tree and generating average spatial representations corresponding with different clusters was done using dedicated in-house R scripts based on the R function ‘dendrapply’. For plotting the average spatial representations, a simple moving average smoothing was applied using a rectangular box of 3 horizontal lines by 11 vertical scans. The detection of meaningful clusters was supported by tagging each cluster by its height in the dendrogram tree and the number of its members, using a minimum height of four and minimum member number of five (Supplementary Fig. 3). Plotting the average spatial representations was performed using the ‘YlGnBu’ in the R package ‘RColorBrewer’66. The uniqueness in each of the nine detected clusters was evaluated by generating a silhouette index as score of ‘goodness’ of clustering67,68, and are presented in Supplementary Fig. 17.
Annotation of mass features to masses of known lipids was done using a dedicated in-house R script which performs mass-to-mass comparisons of observed mass features with the theoretical accurate mass of the protonated ([M+H]+), sodium ([M+Na]+) and ammonium ([M+NH4]+) ions of each known lipid, using a mass error tolerance of 6 parts per million (ppm). The annotation script further extracted the natural heavy isotope pattern of each putatively annotated parent ion and performed chemical formula decomposition using the ‘decomposeMass’ in the R package ‘Rdisop’69. Putative annotations of mass features to known lipids were then assigned using the combined criteria of: mass-to-mass, chemical formula decomposition and manual inspection of the corresponding mass peak in the raw data.
In total, the four clusters we focused on contained 129 mass features (Supplementary Table 2). Out of which, 94 mass features were putatively annotated as molecular ions, isotopes or fragments of lipids, corresponding with 41 m/z feature groups (of which, 33 were induced and 8 reduced, see Supplementary Data 1). Seventeen m/z feature groups were putatively annotated as lipids known from previous LC-MS-based studies (15 induced and 2 reduced)8,20,21.
MS images of specific m/z-values derived from the raw data (used in Fig. 2 and Supplementary Fig. 1, 4, 5, 6 and 16) were generated using the open-source software package MSiReader v1.0070,71, built on the platform of Matlab (Mathworks, Natick, MA, USA), allowing mass error of 5 ppm, using TIC normalization and a ‘Viridis’ colormap. Pixel smoothing was not applied to the images.
Matrix sublimation for MALDI-MSI
2,5-Dihydroxybenzoic acid (DHB) matrix was deposited using a sublimation apparatus (height × inner diameter, 250 mm × 152 mm, Sigma-Aldrich, St Louis, MO, USA). The sublimator was coupled to a rough pump and a digital thermocouple vacuum gauge controller and was placed on a sand bath heated by a hot plate. The temperature was monitored by a digital thermometer. 200 mg of DHB matrix were sublimated under a fixed pressure of 8×10-2 Torr at 140 °C for 4 min.
MALDI-MS imaging and data processing
MALDI imaging experiments were performed using a 7T Solarix FT-ICR (Fourier transform ion cyclotron resonance) mass spectrometer (Bruker Daltonics, Bremen, Germany). Datasets were recorded in positive ion mode using lock mass calibration (DHB matrix peak: [3DHB+H-3H2O]+, m/z 409.055408) at a frequency of 1 kHz and a laser power of 18%, with 100 laser shots per pixel and 100 µm pixel size. Each mass spectrum was recorded in the range of 150-3000 m/z in broadband mode with a Time Domain for Acquisition of 1M, providing an estimated resolving power of 115,000 at 400 m/z. The acquired spectra were processed using FlexImaging software (version 4.0, Bruker Daltonics, Bremen, Germany) and SCiLS Lab 2015b (SCiLS GmbH, Bremen, Germany). Data sets were normalized to Root Mean Square (RMS) intensity and MALDI images were plotted at the theoretical m/z ± 0.001% (FlexImaging) and m/z ± 5 ppm (SCiLS Lab), with pixel interpolation on. MS images were produced using SCiLS Lab 2015b using ‘Viridis’ colormap. Co-localization was carried out using SCiLS Lab based on Pearson’s correlation analysis (p = 0.05), using RMS normalization and an m/z interval of 5 ppm. Pixel smoothing was not applied to the images or to the co-localization analysis.
Cross-section graphs
Cross-section graphs were plotted by averaging the intensity of each lipid or chlorophyll signal across several lines from the outer part of the plaque to its center. The center was selected to be the minimum of chlorophyll signal. Ten lines were used in epi-fluorescence microscopy images and MALDI-MS and two in Flow-Probe-MS images. Intensity values for each line were extracted from the images using Fiji. Graphs were plotted using Matlab R2016b.
Chemical stability of lipid standards in MALDI-MS
A lipid standard solution containing 12.5 µM of Ceramide/Sphingoid internal standard mixture (Avanti Polar Lipids, Alabaster, AL, USA, LM6002) was mixed with freshly prepared DHB matrix solution (10 mg/mL in 100% MeOH and 0.1% TFA) in a 1:1 ratio (v/v). One µL of the mix was spotted onto a MALDI AnchorChip target (Bruker Daltonics) with six technical repeats per day, and the spotted target was allowed to air dry before MALDI-MS analysis. Each MALDI spot was analyzed with the same instrumental method as described for MALDI-MSI, and the spectrum was obtained from the sum of 5 scans in ‘smart walk’ mode with a 500 µm grid width and 10% grid increment.
Chemicals and internal standards for LC-MS analyses
Liquid chromatography-grade solvents were purchased from Merck (Darmstadt, Germany) and Bio-Lab (Jerusalem, Israel). Ammonium acetate was purchased from Sigma-Aldrich (St Louis, MO, USA). Internal standards for lipidomics analysis were purchased from Avanti Polar Lipids (Alabaster, AL, USA). The lipid internal standards were added to the extraction solution employed in the initial extraction step.
Lipidomics Analyses
Lipids were extracted from liquid cultures of E. huxleyi cells infected with EhV201 and from non-infected cells harvested at 4, 24, 48 and 72 hpi in three biological replicates. Infection was performed on exponential phase cultures (5×105 to 106 cells mL-1), which were infected 2-3 h after the onset of the light period, with a 1:100 volumetric ratio of viral lysate to culture (multiplicity of infection of 1:1 viral particles per cell). The samples (30-150 mL of each culture, equivalent to ~5×107 cells per sample) were collected on GF/C filters (pre-combusted at 400 °C for 8 h), plunged into liquid nitrogen, and stored at -80 °C until analysis23.
Lipid extraction and analysis were performed as previously described72 with some modifications: filters containing algae were placed in a 15 mL glass tube and extracted with 3 mL of a pre-cooled (-20 °C) homogenous methanol: methyl-tert-butyl-ether (MTBE) 1:3 (v/v) mixture containing 0.1 μg mL-1 of glucosylceramide (d18:1/12:0) which was used as an internal standard. The tubes were shaken for 30 min at 4 °C and then sonicated for 30 min. Ultra-performance liquid chromatography (UPLC) grade water:methanol (3:1, v/v) solution (1.5 mL) was added to the tubes followed by centrifugation. The upper organic phase (1.2 mL) was transferred into 2 mL Eppendorf tube. The polar phase containing algae debris and filter pieces was re-extracted with 0.5 mL of MTBE. Organic phases were combined and dried under N2 flow and then stored at -80 °C until analysis.
Extraction of lipids from plaque samples was performed by suspending the upper soft layer containing the plaque in the lipid extraction solution, following the same procedure described above (using third of the mentioned volumes). As a blank, soft layer without E. huxleyi cells (i.e. ASW + K/2 containing 0.2% agarose only) was extracted as well. Samples were extracted in three replicates. Isolation, concentration and lipid extraction of vesicles and virions was performed as previously described38. All dried lipid extracts were re-suspended in 300 μL buffer B (see below) and centrifuged again at 18,000 ×g and 4 °C for 5 min for LC-MS and LC-MS/MS analyses.
UPLC-q-TOF MS for lipidomics analysis
Post extraction, the supernatant was transferred to an autosampler vial and an aliquot of 1 µL was subjected to UPLC-MS analysis. Lipid extracts were analyzed using a Waters ACQUITY UPLC I-Class system coupled to a SYNAPT G2 HDMS mass spectrometer (Waters Corp., Milford, MA, USA). Chromatographic conditions were as described previously72. Briefly, the chromatographic separation was performed on an ACQUITY UPLC BEH C8 column (2.1×100 mm, i.d., 1.7 μm) (Waters Corp., Milford, MA, USA). The mobile phase consisted of water (UPLC grade) with 1% 1 M NH4Ac, 0.1% acetic acid (mobile phase A), and acetonitrile:isopropanol (7:3) with 1% 1 M NH4Ac, 0.1% acetic acid (mobile phase B). The column was maintained at 40 °C and the flow rate of the mobile phase was 0.4 mL min-1. The gradient separation was as follows: 1 min 45% A, 3 min linear gradient from 45% to 35% A, 8 min linear gradient from 35% to 11% A, 3 min linear gradient from 11% to 0% A. After washing the column for 4 min with 0% A, the buffer was set back to 45% A and the column was re-equilibrated for 4 min (22 min total run time). MS parameters were as follows: the source and de-solvation temperatures were maintained at 120 °C and 450 °C, respectively. The capillary voltage and cone voltage were set to 1.0 kV and 40 V, respectively. Nitrogen was used as de-solvation gas and cone gas at the flow rate of 800 L h-1 and 20 L h-1, respectively. The mass spectrometer was operated in full scan MSE positive resolution mode over a mass range of 50-1500 Da. For the high energy scan function, a collision energy ramp of 15-35 eV was applied. For the low energy scan function -4 eV was applied. Leucine-enkephalin was used as lock-mass reference standard. The major ions and specific fragment ions of the lipids were analyzed in positive ionization mode. For detection of C15:0 FA fragment ions of the lipids, samples were injected also in negative ionization mode. The MSE energies applied for the negative ionization were 15-40 eV. For MS/MS (in positive ionization mode), a collision energy ramp of 10-45 eV was applied.
LC-MS and LC-MS/MS data analysis, lipid identification and quantification
LC-MS data were analyzed and processed with MassLynx and QuanLynx (version 4.1, Waters Corp., Milford, MA, USA). The putative identification of the various lipid species was performed according to the lipid accurate mass and LC-MS/MS fragmentation pattern, as described previously20,21,24. The putative identification of lipid species that were previously undescribed in this host-virus model system (listed in Table 1) was based on the Lipid Maps computationally-generated database of lipid classes and structure database (LMSD)36 (and carried out following the Metabolomics Standards Initiative level 2 annotation73. See Fig. 4a and Supplementary Fig. 8, 9 and 11 for MS/MS spectra and the list of fragments of representative species of each class). Five unidentified mass features, which we termed sulfonioglycerolipids (listed in Table 1), did not exist in our in-house repository or in public repositories such as Lipid Maps36 and METLIN74. Putative structures of these sulfonioglycerolipids, which belong to the same putatively characterized compound class, was based on accurate mass and LC-MS/MS fragmentation pattern (corresponding with level 3 annotation), as elaborated in Supplementary Fig. 7 and Supplementary Table 4. A full list of the all lipids annotated using LC-MS/MS and their detected fragments can be found in Supplementary Data 1-3. Relative levels of lipids extracted from liquid cultures were normalized to the internal standard and the number of algal cells used for analysis. Heatmap was generated using Matlab R2016b. Mean intensity values (normalized peak intensity per cell, n = 3) were log10 transformed and standardized per lipid (row-wise). Zero values were replaced with 0.5 of the minimal value prior to log10 transformation.
Fatty acid composition
Four fractions enriched with potential C15:0-based lipids were collected manually from LC-MS runs of lipidomics extracts from infected cells at 48 hpi. Fractions were collected at the following RT ranges: 4.3-5.3 min (fraction 1, containing DGCC 37:6 and PDPT 37:6); 6.6-7.4 min (fraction 2, containing PDPT 33:1); 7.5-8.4 min (fraction 3, containing DGTS 33:1); 16.1-18.3 min (fraction 4, containing several TAGs). The presence of the above-mentioned lipids in the collected fractions was verified by re-injecting the collected fractions to the LC-MS, using the same conditions for lipidomics analysis, as described above. The collected samples were dried under N2 flow. As a negative control, 4 mL of mobile phase A and 4 mL of mobile phase B were dried as well. As a positive control, 6 mL of the extraction solution for lipids containing isotopically labelled TAG 48:1 (C15:0/C18:1/C15:0) standard (Avanti Polar Lipids, Alabaster, AL, USA) were dried as well.
The FA composition of lipids was analyzed using derivatized fatty acid methyl esters (FAMEs). Derivatization by trans-esterification was performed as follows: the samples were dissolved in 500 µL 1.25 M methanolic hydrochloride (MeOH/HCl, Sigma-Aldrich, St. Louis, MO, USA), incubated for 1 h at 80 °C while shaking and then dried under N2 flow. The resulting FAMEs were extracted twice with 250 µL hexane:chloroform 1:1 and once with 250 µL chloroform, each extraction for 30 min at room temperature while shaking. The extracts were dried under N2 flow. The dried FAME extracts were re-suspended in 100 µL hexane and centrifuged at 18,000 ×g and 4 °C for 5 min for GC-MS analysis.
GC-MS for FAME analysis
Post extraction, the supernatant was transferred to an autosampler vial and an aliquot of 1 µL was subjected to GC-MS analysis. The GC-MS system comprised of an Agilent 7890A gas chromatograph equipped with split/splitless injector and a LECO Pegasus HT Time-of-Flight Mass Spectrometer (TOF-MS, LECO Corp., St Joseph, MI, USA). GC was performed on a 30 m × 0.25 mm × 0.25 μm Rxi-5Sil MS column (Restek). Samples were analyzed in the splitless mode; injector temperature was set at 280 °C. Analytes by 1 μL injected were separated using the following chromatographic conditions: helium was used as carrier gas at a flow rate of 1.0 mL min-1. The thermal gradient started at 80 °C and was held at this temperature for 2 min, ramped to 330 °C at 15 °C min-1 and then held at 330 °C for 6.0 min. Eluents were fragmented in the electron impact mode with an ionization voltage of 70 eV. The MS mass range was 45-800 m/z with an acquisition rate of 20 spectra per second. The ion source chamber was set to 250 °C and the transfer line to 250 °C. LECO ChromaTOF (version 4.50.8.0, LECO Corp.) was used for acquisition control and data processing. FAMEs were identified by comparison of their mass spectra and retention times to the corresponding analytical standards, injected in the same GC-MS conditions. Two standards were used: methyl pentadecanoate (methyl-C15:0 FA, Sigma, St. Louis, MO, USA) and methyl 12-methylmyristate (methyl-12-methyl-C14:0 FA, Santa Cruz Biotechnology, CA, USA), at a concentration of 10 µg mL-1 in hexane (Fig. 4b and Supplementary Fig. 10).
Data availability
Data supporting the findings of this study are available within the paper (and supplementary information files). Raw data generated or analyzed in this study have been deposited to the EMBL-EBI MetaboLights repository75 with the identifiers MTBLS767 (Including LC-MS of plaque samples, LC-MS/MS of specific lipids and GC-MS) and MTBLS769 (including MALDI-MSI and Flow-probe-MSI). The data can be accessed with the following links: http://www.ebi.ac.uk/metabolights/MTBLS767, http://www.ebi.ac.uk/metabolights/MTBLS769.
Supplementary Material
Acknowledgements
We thank Constanze Kuhlisch from the Vardi lab for her assistance with GC-MS analyses and fruitful discussions, Shiri Graff van Creveld from the Vardi lab for her assistance in designing the figures for this manuscript, and Avia Mizrachi from the Vardi lab for her assistance with image analysis and processing. We also thank Alexander Brandis from the Targeted Metabolomics Unit at the Life Sciences Core Facilities, Weizmann Institute of Science, for his assistance in FAME derivatization, Ron Rotkopf from the Bioinformatics Unit, Department of Biological Services, Weizmann Institute of Science, for his assistance with the statistical analysis, Sung Sik Lee from the scientific Center for Optical and Electron Microscopy (ScopeM), ETH Zürich for his assistance with epi-fluorescence microscopy, and Tal Luzzatto-Knaan from the Department of Marine Biology, University of Haifa, for her useful comments on the manuscript. This research was supported by the European Research Council CoG (VIROCELLSPHERE grant no. 681715) awarded to A.V and by EMBO Short Term Fellowship (ASTF 601 - 2015) awarded to G.S.
Footnotes
Author contributions
G.S. and A.V. conceptualized the project and conceived and designed the experiments, G.S. and A.V. wrote the manuscript; G.S. performed all experiments; N.S. developed computational analysis of MS-data.; C.Z. conducted lipid extractions and LC-MS experiments; R.A.M and E.J.H.E conducted Flow-probe-MS experiments; Y.D. conducted MALDI-MS experiments; I.R. conducted GC-MS experiments; D.S. isolated vesicles and virions for lipidomics analysis; all authors provided useful feedback on the experimental design and comments on the manuscript.
Competing interests
The authors declare no competing financial interests.
References
- 1.Olive AJ, Sassetti CM. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat Rev Microbiol. 2016;14:221–234. doi: 10.1038/nrmicro.2016.12. [DOI] [PubMed] [Google Scholar]
- 2.Magnúsdóttir S, Thiele I. Modeling metabolism of the human gut microbiome. Curr Opin Biotechnol. 2018;51:90–96. doi: 10.1016/j.copbio.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Aldridge BB, Rhee KY. Microbial metabolomics: innovation, application, insight. Curr Opin Microbiol. 2014;19:90–96. doi: 10.1016/j.mib.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Ankrah NYD, et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014;8:1089–100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smet J, et al. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016;10:1823–1835. doi: 10.1038/ismej.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods. 2015;12:1091–1097. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 7.Roe B, Kensicki E, Mohney R, Hall WW. Metabolomic profile of hepatitis C virus-infected hepatocytes. PLoS One. 2011;6:e23641. doi: 10.1371/journal.pone.0023641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malitsky S, et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol. 2016;210:88–96. doi: 10.1111/nph.13852. [DOI] [PubMed] [Google Scholar]
- 9.Frada MJ, et al. Morphological switch to a resistant subpopulation in response to viral infection in the bloom-forming coccolithophore Emiliania huxleyi. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 11.Rosenwasser S, Ziv C, van Creveld SG, Vardi A. Virocell metabolism: Metabolic innovations during host-virus interactions in the ocean. Trends Microbiol. 2016;24:821–832. doi: 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Petras D, Jarmusch AK, Dorrestein PC. From single cells to our planet—recent advances in using mass spectrometry for spatially resolved metabolomics. Curr Opin Chem Biol. 2017;36:24–31. doi: 10.1016/j.cbpa.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Dong Y, Li B, Aharoni A. More than pictures: when MS imaging meets histology. Trends Plant Sci. 2016;21:686–698. doi: 10.1016/j.tplants.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Ryffel F, et al. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 2015 doi: 10.1038/ismej.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watrous JD, et al. Microbial metabolic exchange in 3D. ISME J. 2013;7:770–780. doi: 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasch P, et al. Identification of highly pathogenic microorganisms by matrix-assisted laser desorption ionization-time of flight mass spectrometry: results of an interlaboratory ring trial. J Clin Microbiol. 2015;53:2632–2640. doi: 10.1128/JCM.00813-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simó R. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol. 2001;16:287–294. doi: 10.1016/s0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- 18.Lehahn Y, et al. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr Biol. 2014;24:2041–2046. doi: 10.1016/j.cub.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 19.Rosenwasser S, et al. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26:2689–2707. doi: 10.1105/tpc.114.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulton JM, et al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ Microbiol. 2014;16:1137–1149. doi: 10.1111/1462-2920.12358. [DOI] [PubMed] [Google Scholar]
- 21.Hunter JE, Frada MJ, Fredricks HF, Vardi A, Van Mooy BAS. Targeted and untargeted lipidomics of Emiliania huxleyi viral infection and life cycle phases highlights molecular biomarkers of infection, susceptibility, and ploidy. Front Mar Sci. 2015;2:81. [Google Scholar]
- 22.Wilson WH, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 23.Vardi A, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326:861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 24.Ziv C, et al. Viral serine palmitoyltransferase induces metabolic switch in sphingolipid biosynthesis and is required for infection of a marine alga. Proc Natl Acad Sci U S A. 2016;113:E1907–1916. doi: 10.1073/pnas.1523168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheyn U, et al. Expression profiling of host and virus during a coccolithophore bloom provides insights into the role of viral infection in promoting carbon export. ISME J. 2018;12:704–713. doi: 10.1038/s41396-017-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bidle KD, Haramaty L, Barcelos e Ramos J, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci U S A. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 2016 doi: 10.1038/ismej.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laber CP, et al. Coccolithovirus stimulation of carbon export in the North Atlantic. Nat Microbiol. 2018;3467:1–26. doi: 10.1038/s41564-018-0128-4. [DOI] [PubMed] [Google Scholar]
- 29.Ellis EL, Delbrück M. The growth of bacteriophage. J Gen Physiol. 1939;22:365–384. doi: 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper PD. The plaque assay of animal viruses. Adv Virus Res. 1961;8:319–378. doi: 10.1016/s0065-3527(08)60689-2. [DOI] [PubMed] [Google Scholar]
- 31.Yin J. Evolution of bacteriophage T7 in a growing plaque. J Bacteriol. 1993;175:1272–1277. doi: 10.1128/jb.175.5.1272-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llewellyn CA, et al. The response of carotenoids and chlorophylls during virus infection of Emiliania huxleyi (Prymnesiophyceae) J Exp Mar Bio Ecol. 2007;344:101–112. [Google Scholar]
- 33.Hsu C-C, et al. Real-time metabolomics on living microorganisms using ambient electrospray ionization flow-probe. Anal Chem. 2013;85:7014–7018. doi: 10.1021/ac401613x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deininger SO, Ebert MP, Fütterer A, Gerhard M, Röcken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Proteome Res. 2008;7:5230–5236. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 35.Alexandrov T, Chernyavsky I, Becker M, von Eggeling F, Nikolenko S. Analysis and interpretation of imaging mass spectrometry data by clustering mass-to-charge images according to their spatial similarity. Anal Chem. 2013;85:11189–11195. doi: 10.1021/ac401420z. [DOI] [PubMed] [Google Scholar]
- 36.Sud M, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcolombri U, et al. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348:1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 38.Schatz D, et al. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat Microbiol. 2017;2:1485–1492. doi: 10.1038/s41564-017-0024-3. [DOI] [PubMed] [Google Scholar]
- 39.Vardi A, et al. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc Natl Acad Sci U S A. 2012;109:19327–19332. doi: 10.1073/pnas.1208895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotoh N, et al. Metabolism of odd-numbered fatty acids and even-numbered fatty acids in mouse. J Oleo Sci. 2008;57:293–299. doi: 10.5650/jos.57.293. [DOI] [PubMed] [Google Scholar]
- 41.Long AM, Short SM. Seasonal determinations of algal virus decay rates reveal overwintering in a temperate freshwater pond. ISME J. 2016;10:1602–1612. doi: 10.1038/ismej.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans C, Pond DW, Wilson WH. Changes in Emiliania huxleyi fatty acid profiles during infection with E. huxleyi virus 86: physiological and ecological implications. Aquat Microb Ecol. 2009;55:219–228. [Google Scholar]
- 43.Sperl W, et al. Odd-numbered long-chain fatty acids in propionic acidaemia. Eur J Pediatr. 2000;159:54–58. doi: 10.1007/s004310050010. [DOI] [PubMed] [Google Scholar]
- 44.Řezanka T, Vítová M, Nováková A, Sigler K. Separation and identification of odd chain triacylglycerols of the protozoan Khawkinea quartana and the mold Mortierella alpina using LC–MS. Lipids. 2015;50:811–820. doi: 10.1007/s11745-015-4042-8. [DOI] [PubMed] [Google Scholar]
- 45.Böer M, Graeve M, Kattner G. Impact of feeding and starvation on the lipid metabolism of the Arctic pteropod Clione limacina. J Exp Mar Bio Ecol. 2006;328:98–112. [Google Scholar]
- 46.Narayanan S, Tamura PJ, Roth MR, Prasad PVV, Welti R. Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant Cell Environ. 2016;39:787–803. doi: 10.1111/pce.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingram LO, Chevalier LS, Gabbay EJ, Winters K. Priopionate-induced synthesis of odd-chain-length fatty acids by Escherichia coli. J Bacteriol. 1977;131:1023–1025. doi: 10.1128/jb.131.3.1023-1025.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oizumi J, Giudici TA, Ng WG, Shaw KNF, Donnell GN. Propionate metabolism by cultured skin fibroblasts from normal individuals and patients with methylmalonicaciduria and propionicacidemia. Biochem Med. 1981;26:28–40. doi: 10.1016/0006-2944(81)90027-2. [DOI] [PubMed] [Google Scholar]
- 49.Wendel U. Abnormality of odd-numbered long-chain fatty acids in erythrocyte membrane lipids from patients with disorders of propionate metabolism. Pediatr Res. 1989;25:147–150. doi: 10.1203/00006450-198902000-00014. [DOI] [PubMed] [Google Scholar]
- 50.Kondo N, et al. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat Commun. 2014;5:5338. doi: 10.1038/ncomms6338. [DOI] [PubMed] [Google Scholar]
- 51.Keller MD, Selvin RC, Claus W, Guillard RRL. Media for the culture of oceanic ultraphytoplankton. J Phycol. 2007;23:633–638. [Google Scholar]
- 52.Goyet C, Poisson A. New determination of carbonic acid dissociation constants in seawater as a function of temperature and salinity. Deep Sea Res Part A Oceanogr Res Pap. 1989;36:1635–1654. [Google Scholar]
- 53.Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch Virol. 2002;147:1685–98. doi: 10.1007/s00705-002-0841-3. [DOI] [PubMed] [Google Scholar]
- 54.Barak-Gavish N, et al. Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci Adv. 2018;4 doi: 10.1126/sciadv.aau5716. eaau5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–94. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann T, Dorrestein PC. Homogeneous matrix deposition on dried agar for MALDI imaging mass spectrometry of microbial cultures. J Am Soc Mass Spectrom. 2015;26:1959–1962. doi: 10.1007/s13361-015-1241-8. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y-L, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–7. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu W-T, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci U S A. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin J. Spatially resolved evolution of viruses. Ann N Y Acad Sci. 1994;745:399–408. doi: 10.1111/j.1749-6632.1994.tb44392.x. [DOI] [PubMed] [Google Scholar]
- 60.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adusumilli R, Mallick P. Data conversion with ProteoWizard msConvert. In: C L, K J, M P, editors. Methods in Molecular Biology. Vol. 1550. Humana Press; New York, NY: 2017. pp. 339–368. [DOI] [PubMed] [Google Scholar]
- 62.Schramm T, et al. imzML — A common data format for the flexible exchange and processing of mass spectrometry imaging data. J Proteomics. 2012;75:5106–5110. doi: 10.1016/j.jprot.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 63.Race AM, Styles IB, Bunch J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J Proteomics. 2012;75:5111–5112. doi: 10.1016/j.jprot.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 64.Gibb S, Strimmer K. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics. 2012;28:2270–2271. doi: 10.1093/bioinformatics/bts447. [DOI] [PubMed] [Google Scholar]
- 65.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. cluster: Cluster Analysis Basics and Extensions. R package version 2.0.5. 2016 [Google Scholar]
- 66.Neuwirth E. RColorBrewer: ColorBrewer Palettes. R package version 1.1-2. 2014 [Google Scholar]
- 67.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 68.Wehrens R. Chemometrics with R: Multivariate Data Analysis in the Natural Sciences and Life Sciences. Springer; Berlin Heidelberg: 2011. [Google Scholar]
- 69.Böcker S, Letzel MC, Lipták Z, Pervukhin A. SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics. 2009;25:218–224. doi: 10.1093/bioinformatics/btn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on matlab platform. J Am Soc Mass Spectrom. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bokhart MT, Nazari M, Garrard KP, Muddiman DC. MSiReader v1.0: evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J Am Soc Mass Spectrom. 2018;29:8–16. doi: 10.1007/s13361-017-1809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hummel J, et al. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci. 2011;2:1–17. doi: 10.3389/fpls.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guijas C, et al. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haug K, et al. MetaboLights - An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013;41:D781–D786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the paper (and supplementary information files). Raw data generated or analyzed in this study have been deposited to the EMBL-EBI MetaboLights repository75 with the identifiers MTBLS767 (Including LC-MS of plaque samples, LC-MS/MS of specific lipids and GC-MS) and MTBLS769 (including MALDI-MSI and Flow-probe-MSI). The data can be accessed with the following links: http://www.ebi.ac.uk/metabolights/MTBLS767, http://www.ebi.ac.uk/metabolights/MTBLS769.