Obesity results from a chronic imbalance between energy intake and energy output but remains difficult to prevent or treat in humans. AMP-activated protein kinase (AMPK) is an important regulator of energy homeostasis1–3, and is a molecular target of drugs used for the treatment of metabolic diseases, including obesity4,5. Here we show that mice expressing a gain-of-function AMPK mutant6 display a change in morphology of subcutaneous white adipocytes that is reminiscent of browning. However, despite a dramatic increase in mitochondrial content, Ucp1 expression is undetectable in these adipocytes. In response to a high fat diet, expression of skeletal muscle-associated genes is induced in subcutaneous white adipocytes from the gain-of-function AMPK mutant mice. Chronic genetic AMPK activation results in protection against diet-induced obesity due to an increase in whole-body energy expenditure most likely due to a substantial increase in the oxygen consumption rate of white adipose tissue. These results suggest that AMPK activation enriches, or leads to the emergence of, a population of subcutaneous white adipocytes that produce heat via Ucp1-independent uncoupling of ATP production on a high fat diet. Our findings indicate that AMPK activation specifically in adipose tissue could have therapeutic potential for the treatment of obesity.
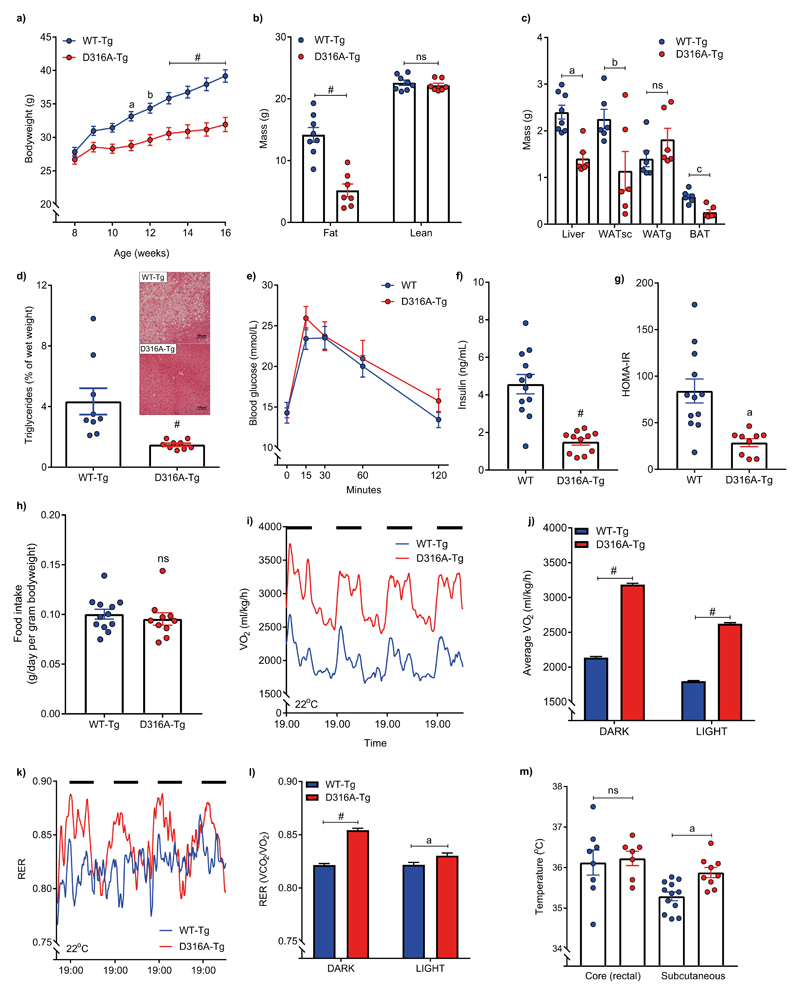
Previously, we reported on a mouse model expressing a gain-of-function mutation in the γ1 subunit of AMPK (mutation of aspartic acid residue 316 to alanine in mouse Prkag1; D316A) and showed that liver-specific activation of AMPK prevented steatosis on a high fructose diet6. However, AMPK activation in the liver had no detectable metabolic effect in mice fed either a standard chow diet or a high fat diet6. In order to determine the effect of more widespread AMPK activation, we crossed mice harbouring the gain-of-function AMPK γ1 transgene with mice expressing Cre-recombinase under the control of the β-actin promoter (β–actin-Cre), generating D316A-Tg mice. As a control, mice harbouring wild-type γ1 were crossed with β–actin-cre (hereafter referred to as WT-Tg). Both the WT-Tg and D316A-Tg mice were viable and transgene expression in a range of tissues was confirmed by blotting with an anti-Flag antibody (a Flag epitope was engineered at the C-terminus of the transgene; Supplementary Fig. S1a). In humans, gain-of-function mutations in AMPKγ2 lead to a cluster of severe cardiac abnormalities, including cardiac hypertrophy and ventricular pre-excitation (Wolff-Parkinson-White syndrome), as well as bradycardia1. There was a modest increase in heart weight but no change in PR interval, QRS complex duration or heart rate, in D316A-Tg mice compared to WT-Tg mice (Supplementary Table 1). Previous studies have indicated a role for AMPK in the regulation of feeding7,8, but there was no significant difference in bodyweight or food intake between WT-Tg and D316A-Tg mice maintained on a standard chow diet (Supplementary Fig. S1b,c). Similarly, no significant differences in oxygen consumption or body temperature were detected on a chow diet (Supplementary Fig. S1d,e). Strikingly, however, on a high fat diet (HFD), D316A-Tg mice gained much less weight than WT-Tg mice (Fig. 1a; Supplementary Fig. S2a). The reduction in bodyweight was accounted for by a decrease in fat mass, but not lean mass (Fig. 1b). Liver, subcutaneous white adipose tissue (WATsc) and brown adipose tissue (BAT) weights were all significantly reduced in the D316A-Tg mice, whereas gonadal WAT (WATg) weight was not reduced (Fig. 1c). Similar effects were seen in female mice (Supplementary Fig. S2b-d). Lipid accumulation in the liver was also significantly lower in the D316A-Tg compared to WT-Tg mice (Fig. 1d). There was no significant difference in glucose tolerance (Fig. 1e), but fasted plasma insulin levels were significantly lower in D316A-Tg mice (Fig. 1f), leading to a significant improvement in insulin sensitivity as determined by HOMA IR (Fig. 1g). Food intake was not significantly different on the HFD (Fig. 1h), but oxygen consumption in the D316A-Tg mice was significantly increased compared to WT-Tg mice (Fig. 1i,j), without any significant change in movement (Supplementary Fig. S2e). Increased oxygen consumption was still evident when calculated on a per mouse basis (Supplementary Fig. S2f). The respiratory exchange ratio (RER) was significantly increased in the D316A-Tg mice (Fig. 1k,l). Interestingly, although core body temperature was not altered, the surface temperature of the D316A-Tg mice was significantly raised (Fig. 1m), indicative of increased energy expenditure.
Figure 1. AMPK activation protects against diet-induced obesity by increasing energy expenditure.
Male mice expressing either wild-type γ1 (WT-Tg) or the D316A γ1 transgene (D316A-Tg) were fed a high fat diet (HFD) from 8 weeks of age. a, Bodyweight (n=20 for WT-Tg, and 17 for D316A-Tg). aP=0.002, bP=0.0006 and #P<0.0001 b, Total body fat and lean mass after 8 weeks on HFD (#P<0.0001) and (c) tissue weights after 16 weeks on HFD (n=8 per genotype; aP=0.0004, bP=0.0378, cP=0.0012, ns=not significant). d, Liver triglyceride levels in mice fed a HFD for 16 weeks (n=9 per genotype; #P<0.0001)). A representative image (from 9 independent mice per genotype) of H&E stained liver sections from HFD fed mice is shown as an inset. e, Oral glucose tolerance test and (f), fasted (6 h) serum insulin levels of mice fed a HFD for 12 weeks (n=12 for WT-Tg and 11 for D316A-Tg; #P<0.0001). g, HOMA IR calculated from 6 h fasted glucose and insulin levels (n=12 for WT-Tg and 9 for D316A-Tg, aP=0.0012). h, Food intake over a 5-day period (n=12 for WT-Tg and 10 for D316A-Tg, ns=not significant). i, Whole body oxygen consumption (VO2) monitored continuously over an 84-hour period, and (j) average VO2 during 12 h light and 12 h dark periods (dark periods represented by the solid black bars). Data points from individual mice were omitted in the graph to more clearly show mean values and error bar sizes. #P<0.0001. (k) Respiratory exchange ratio (RER) over 84 h and (l) average RER during light and dark periods. Data points from individual mice were omitted in the graph to more clearly show mean values and error bar sizes. aP=0.0086 #P<0.0001. For (i-l) mice were fed a HFD for 6 weeks (n=8 for WT-Tg and 6 for D316A-Tg mice). m, core (n=8 for WT-Tg and 7 for D316A-Tg mice) and subcutaneous (n=12 for WT-Tg and 9 for D316A-Tg mice) body temperature in mice fed a HFD for 12 weeks. aP=0.0022, ns=not significant). In all cases the results shown are the mean ±sem. Statistical analyses in panels a, b, e, j and l were performed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. Statistical analyses in panels c, f, g, h and m were performed by Student’s t-test, unpaired, 2-tailed, with Welch’s correlation applied to f and g. Data in panel d were analysed by Mann-Whitney test.
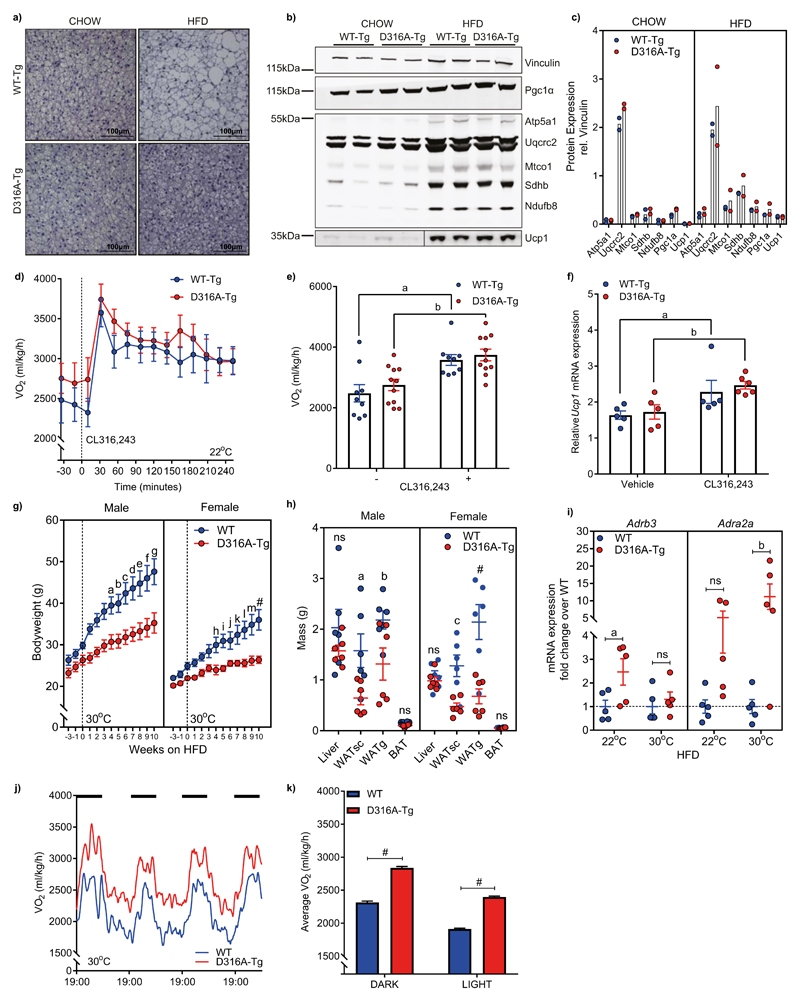
The findings described above suggest that the primary mechanism for decreased weight-gain and fat accumulation in the D316A-Tg mice is mediated by an increase in diet-induced thermogenesis, resulting in increased energy expenditure on a HFD. In mammals, BAT plays an important role in thermogenesis, contributing to increased energy expenditure. Numerous studies have revealed that proton leak across the inner mitochondrial membrane, mediated by the action of uncoupling protein 1 (Ucp1), generates heat rather than ATP production in BAT9,10. On a chow diet, there was no difference in BAT weight between WT-Tg and D316A-Tg mice and histological analysis revealed no obvious difference in BAT morphology. On a HFD adipocytes from BAT of D316A-Tg mice were smaller, and contained smaller lipid droplets, than cells isolated from WT-Tg mice (Fig. 2a). The transcriptional coactivator Pgc1α is a key regulator of BAT function and is upregulated in response to cold exposure, coordinating a number of changes including increased mitochondrial biogenesis. There was no difference in expression of Pgc1α or components of the mitochondrial electron transport chain between the two mouse lines in BAT, although there was increased expression of Pgc1α and mitochondrial proteins, including Ucp1, following a HFD (Fig. 2b,c), as has been reported previously11,12 In order to probe BAT function directly, we used the β3-adrenoreceptor (β3-AR) agonist CL316,243. Both WT-Tg and D316A-Tg mice showed a similar increase in oxygen consumption in response to acute treatment with CL316,243 (Fig. 2d, e) as well as a similar increase in mRNA expression for Ucp1 (Fig. 2f). These results suggest that AMPK activation does not have a significant effect on BAT function in vivo, and that BAT-mediated thermogenesis does not play a major role in the protection against DIO observed in our model. In order to confirm this, we housed mice at 30°C, which is within the thermoneutral zone for mice, where there is no requirement for thermogenesis to maintain body temperature13. Under thermoneutral conditions, both male and female mice expressing the D316A-Tg were protected against HFD-induced obesity (Fig. 2g) with significant reductions in white adipose tissue mass (Fig. 2h). As anticipated, expression of Adrb3 (encoding β3-AR) was significantly reduced in BAT from mice housed at 30°C relative to 22°C (Supplementary Fig. S2g). In WAT there was a modest increase in expression of Adrb3 in D316A-Tg relative to WT-Tg mice housed at 22°C, but this was ablated at 30°C (Fig. 2i). In contrast, expression of Adra2a (encoding α2a-AR) was significantly increased in D316A-Tg mice compared to WT-Tg at 30°C (Fig. 2i). Oxygen consumption in the D316A-Tg mice maintained at 30°C was significantly increased compared to WT-Tg mice (Fig. 2j,k). These results support the hypothesis that AMPK activation protects against DIO through increased energy expenditure mediated by BAT-independent mechanisms.
Figure 2. AMPK activation protects against diet-induced obesity through a brown adipose tissue-independent mechanism.
a, Representative images (from 6-7 mice per genotype from a single experimental cohort) of haematoxylin stained BAT from 16 week old mice fed chow or 12 week old mice fed for 4 weeks on a HFD (scale bar = 100 μm). b, Western blot analysis and (c) quantification, of mitochondrial electron transport chain proteins, Pgc1α and Ucp1 in BAT. In each case, samples from two mice are shown and vinculin is used as a loading control. d, e, Whole body oxygen consumption (VO2) under basal conditions and following injection with the β3-adrenoreceptor agonist, CL316,243 (n=9 for WT-Tg, and 11 for D316A-Tg; aP=0.0081, bP=0.0084). f, Ucp1 mRNA expression in BAT harvested 6 h post injection with CL316,243 (n-5 for WT-Tg and 6 for D316A-Tg mice) or vehicle control (n=5 for WT-Tg and D316A-Tg mice). aP=0.0159, bP=0.0087. g, Bodyweight of mice transferred to 30°C (represented by the dashed line) and fed a HFD for 10 weeks (n=8 for male and 6 for female WT, and 6 for male and female D316A-Tg mice). aP=0.0433, bP=0.0304, cP=0.0056, dP=0.0031, eP=0.0017, fP=0.001, gP=0.0006, hP=0.02, iP=0.0068, jP=0.0477, kP=0.0044, lP=0.0005, mP=0.0001, #P<0.0001. h, Tissue weights from mice housed at 30°C and fed a HFD for 11 weeks (n=6 mice per group). aP=0.0216, bP=0.0368, cP=0.0044, #P<0.0001. i, mRNA expression of Adrb3 and Adra2a in WATsc from mice housed at 22°C or 30°C and fed a HFD for 11 weeks (n=5 mice per group). aP=0.0297, bP=0.0063, ns=not significant. j, Whole body oxygen consumption (VO2) over an 84-hour period, and (k) average VO2 during 12 h light and 12 h dark periods (dark periods represented by the solid black bars) in mice housed at 30°C fed a HFD for 11 weeks (n=6 per genotype, #P<0.0001). Data points from individual mice were omitted in the graph to more clearly show mean values and error bar sizes. In all cases, results shown are the mean ±sem. Statistical analyses in panels e, g, h, I and k were performed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. Statistical analyses in panel f was performed by Mann-Whitney test.
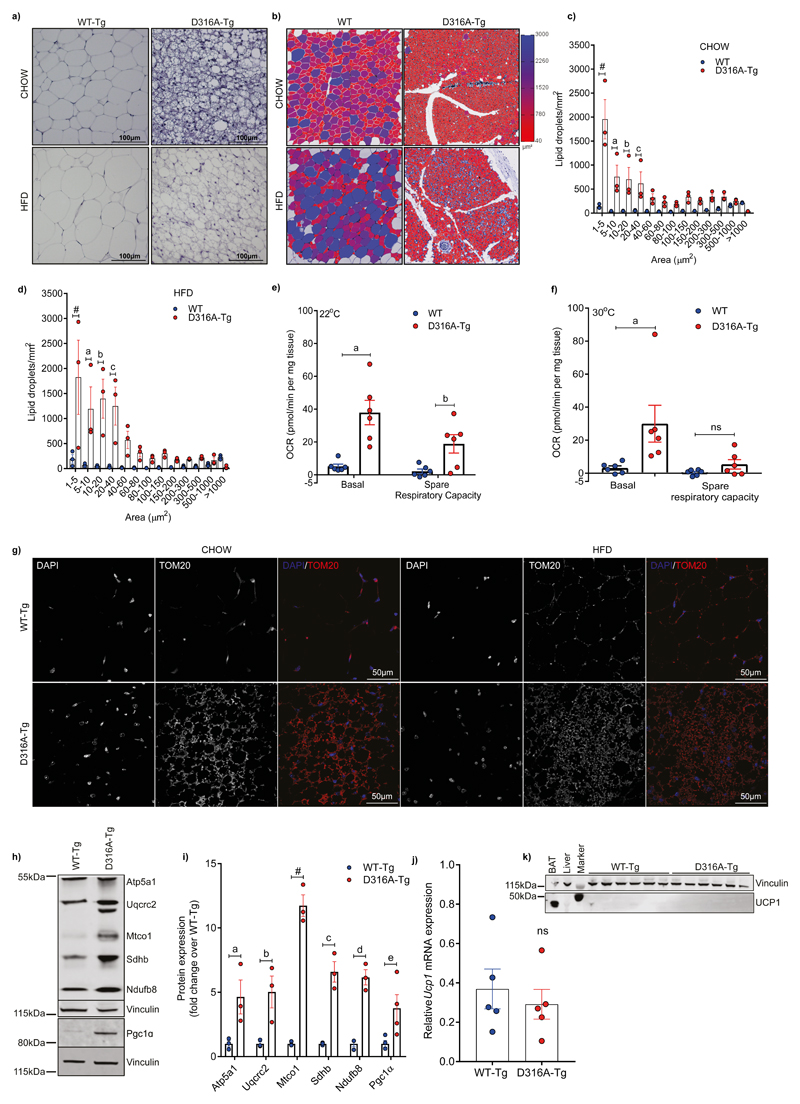
In contrast to BAT, most adipocytes in WAT do not express Ucp1. Adaptation to cold exposure significantly increases the number of cells expressing Ucp1, a process that has been termed “browning”. These beige, or brite (brown-like in white), adipocytes have a similar overall morphology to brown adipocytes, including increased numbers of mitochondria and multilocular fat droplets14,15, and express genes (including Ucp1) that are usually associated with brown adipocytes16. We observed dramatic changes in the morphology of WATsc (Fig. 3a), but not the gonadal WAT (Supplementary Fig. S3a), between WT-Tg and D316A-Tg mice maintained either on a chow diet or HFD. There was a striking increase in the number of cells containing multilocular lipid droplets in the D316A-Tg mice on both chow and HFD. Quantification revealed a marked shift towards smaller lipid droplets in adipocytes from D316A-Tg mice (Fig. 3b-d). We measured oxygen consumption rate (OCR) in WATsc explants and found that there was a significant increase in basal OCR, as well as an increase in spare respiratory capacity, in tissue explants from D316A-Tg mice (Fig. 3e). Importantly, these changes were maintained in explants isolated from D316A-Tg mice housed at 30°C (Fig. 3f). Consistent with increased OCR, there was a marked increase in mitochondrial content in WATsc isolated from D316A-Tg mice on both chow and HFD, as determined by immunofluorescence using an antibody against Tom-20, a mitochondrial outer membrane protein (Fig. 3g). Similar to classical browning, this effect was spread throughout the WATsc depot but with a heterogeneous distribution. On a HFD there was a marked increase in Pgc1α protein expression, together with increased expression of components of the mitochondrial electron transport chain (Fig. 3h,i). More modest changes in protein expression of Pgc1α and electron transport chain proteins were also seen in mice maintained on a chow diet (Supplementary Fig. S3b,c). These changes are consistent with the process of browning typically seen in WAT in response to cold adaptation17. Importantly, however, there was no difference in Ucp1 mRNA expression in WATsc from D316A-Tg mice versus WT-Tg fed a HFD (Fig. 3j), and Ucp1 protein was not detectable (Fig. 3k). Although the role of Ucp1 is generally accepted as being essential for dissipation of the mitochondrial proton gradient and thermogenesis in brown fat, it has been reported that Ucp1-independent mechanisms exist that contribute to heat generation in beige adipocytes18–20. The results of the current study are consistent with a Ucp1-independent phenomenon. Crucially, in our current study, we observe increased thermogenesis in response to high fat feeding, rather than cold adaptation or β3-adrenergic stimulation as has been reported previously.
Figure 3. AMPK activation increases subcutaneous white adipose mitochondrial content and Ucp1-independent respiration.
a, Representative image (from 6-7 mice per group from a single experimental cohort) of haematoxylin stained sections of WATsc from mice maintained on a chow or fed a HFD for 4 weeks (scale bar = 100 μm). b, Examples of 1 mm2 region-of-interest (ROI) selected for quantification. 3 ROIs per mouse were quantified and this was repeated for 3 mice per group. Lipid droplet size is represented using a heat-scale shown alongside. c, Quantification of lipid droplet size on chow diet. aP=0.001, bP=0.0038, cP=0.0212, #P<0.0001. d, Quantification of lipid droplet size on HFD. aP=0.004, bP=0.0003, cP=0.0015, #P<0.0001. e, Basal oxygen consumption rate (OCR) and spare respiratory capacity measured in adipose tissue explants from WT and D316A-Tg mice (n=6 per genotype) housed at 22°C and fed a HFD for 8 weeks. aP=0.0001, bP=0.0269. f, OCR and spare respiratory capacity in adipose tissue explants from mice housed at 30°C and fed a HFD for 11 weeks. aP=0.0076, ns=not significant. g, Representative images (3 images per section from 6-7 mice per group from a single experimental cohort) of immunofluorescent staining of mitochondrial outer membrane protein Tom20 (shown red in the merged images) counterstained with DAPI to stain nuclei (shown in blue in the merged images) in WATsc (scale bar = 50 μm). Similar results were obtained from two independent cohorts. h, Representative western blot analysis of mitochondrial electron transport chain proteins and Pgc1α in WATsc from mice fed a HFD and (i) quantification of protein expression (n=3-4 mice per genotype). aP=0.0137, bP=0.0055, cP=0.0001, dP=0.0003, eP=0.0397, #P<0.0001. j, mRNA expression and (k) Western blot of Ucp1 in WATsc from mice fed a HFD for 4 weeks. A sample of BAT is included as a positive control and vinculin is used as a loading control. In all cases, results shown are the mean ±sem. Statistical analyses in panels c, d, e, f, and i, were performed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. Statistical analyses in panel j was performed by Student’s t-test, unpaired, 2-tailed.
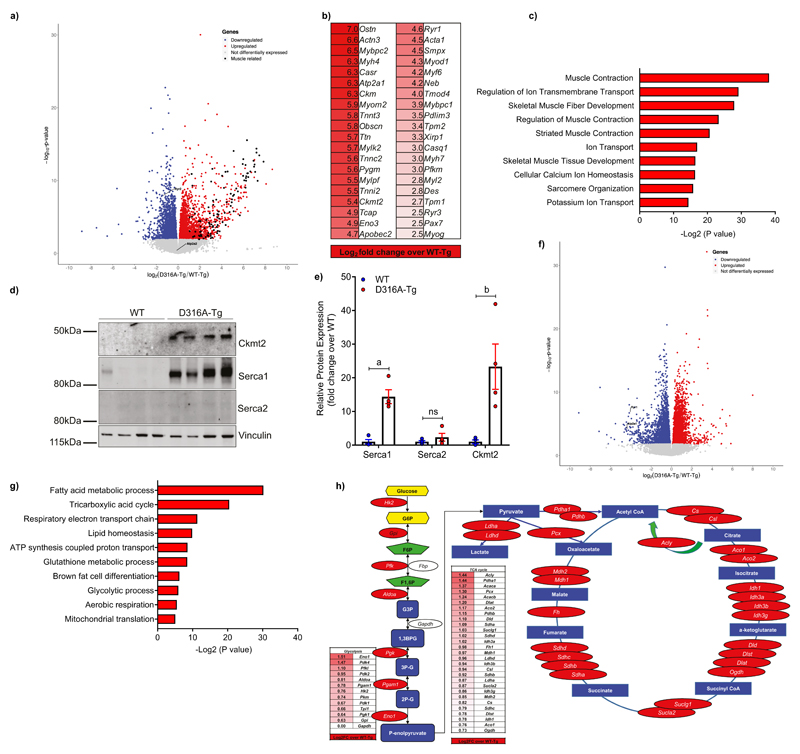
In order to investigate further the mechanism underlying the Ucp1-independent thermogenesis in the D316A-Tg model we performed a global transcriptomic analysis from WATsc isolated from mice fed a HFD. An obvious feature to emerge was the marked increase in expression of skeletal muscle-selective genes in tissue from the D316A-Tg mice (Fig. 4a-c). Of the 100 most differentially expressed genes, 71 were identified as being associated with skeletal muscle function and/or development. Notably, genes encoding proteins involved in Ucp1-independent thermogenic pathways are included in these most highly up-regulated transcripts. Ckmt2, which encodes the sarcomeric form of mitochondrial creatine kinase, was recently identified as an important component of a creatine-dependent ADP/ATP substrate cycling mechanism that is activated in inguinal beige adipocytes in response to cold-exposure20. On a HFD, Ckmt2 mRNA was increased over 30-fold in WATsc from D316A-Tg mice compared to WT-Tg, and consistent with this, Ckmt2 protein was also significantly increased (Fig. 4d,e). In a previous study20, β-guanidinopropionic acid (β-GPA), a creatine analogue that inhibits creatine transport21, was shown to antagonize creatine-driven thermogenesis. However, we were unable to detect any significant effect of β-GPA on bodyweight or WATsc mass (Supplementary Fig. S4) in either WT-Tg or D316A-Tg mice fed a HFD. These results suggest that creatine-futile cycling is not essential for the effects on energy expenditure seen in our model. Studies in fish have revealed that a specialized type of muscle, called heater organs, have evolved that use Ca2+ transport to produce heat without muscle contraction22. In this mechanism, a naturally leaky ryanodine receptor increases cytosolic Ca2+ activating Ca2+ transport via Ca2+-ATPases. Some of the energy released from ATP hydrolysis by the Ca2+-ATPase is released in the form of heat23. Two of the key components required for Ca2+-cycling mediated thermogenesis are sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 1 (Serca1; gene name Atp2a1) and ryanodine receptor 1 (Ryr1) were also significantly up-regulated in D316A-Tg mice (Fig. 4b), and Western blotting revealed a significant increase in Serca1 protein expression (Fig. 4d,e). A recent study reported that Serca2b and Ryr2 were involved in Ca2+-cycling dependent thermogenesis in beige adipose tissue in response to β3-AR stimulation24. Interestingly, neither Atp2a2 (encoding Serca2) or Ryr2 mRNA expression was increased in our model (Fig. 4a), and Serca2 protein expression was not detected in WATsc (Fig. 4d). These findings suggest that different signaling pathways could lead to isoform-selective Ca2+-cycling in WAT. Further studies are required to elucidate whether the thermogenic response stimulated by the combined effect of HFD feeding and AMPK activation utilises a Ca2+-cycling mechanism involving Serca1 and Ryr1, analogous to the mechanism reported in beige adipose tissue24. It is possible that multiple mechanisms e.g. creatine cycling and Ca2+-cycling, contribute to the thermogenic phenotype and/or that other novel mechanisms are involved.
Figure 4. AMPK activation induces a skeletal muscle-like gene signatures in WATsc in mice fed a HFD.
RNAseq analysis was used to determine changes in gene expression in WATsc isolated from WT-Tg and D316A-Tg mice fed a HFD for 16 weeks (n=6 mice per genotype). a, Volcano plot with down-regulated genes shown in blue, up-regulated in red, and unchanged in grey. 1353 genes were upregulated and 1102 genes downregulated (fold change >1.5). Differentially expressed genes (DEGs) were identified (P<0.05) using DESeq2 (Wald test) in R. DEGs were expressed as Log2 fold change over WT-Tg with an adjusted P value for each gene. Skeletal muscle-associated genes are shown in black. Cardiac isoforms of Serca (Atp2a2) and ryanodine receptor (Ryr2) are also highlighted. b, The 40 most highly up-regulated skeletal muscle-associated genes and (c) the most significantly enriched gene ontology (GO) annotations for the 100 most highly up-regulated genes are shown. Association P values determined by DAVID analysis (Fisher Exact P-value). d, Western blot and (e) quantification of Ckmt2, Serca1 and Serca2 in WATsc isolated from mice fed a HFD for 16 weeks (n=4 per genotype from two independent experimental cohorts). Protein expression was normalized to vinculin and expression is shown as fold change relative to WT. Significant differences from WT are shown as aP=0.0026 and bP=0.0494; ns=not significant. f, Volcano plot as in (a) from WATsc from mice fed a chow diet (n=6 mice per genotype). 1361 genes were upregulated and 1808 genes downregulated. Atp2a1 and Ryr1 are highlighted on the plot. g, The most significantly enriched gene ontology (GO) annotations for significantly up-regulated genes are shown. h, A schematic diagram showing up-regulated genes (highlighted in red) in glycolysis and the TCA cycle in D316A WATsc in mice fed a chow diet. Results shown in panel e are the mean ±sem and statistical analysis performed by multiple t-test adjusted for multiple comparisons.
In contrast to the changes in gene expression observed on a HFD, transcriptomic analysis of WATsc from mice fed a chow diet revealed a strikingly different pattern of expression. Over 3000 genes were significantly changed (fold change>1.5, P<0.05) between WT-Tg and D316A-Tg mice (Fig. 4f). Genes in pathways involved in fatty acid metabolism, TCA cycle, mitochondrial ATP production and glycolysis were all highly enriched in WATsc from D316A-Tg mice (Fig. 4g,h), whereas skeletal muscle-associated genes, including Atp2a1 and Ryr1, were down-regulated (Fig. 4f). Taken together, these results show that on a HFD, but not on a chow diet, AMPK activation induces a switch in WATsc towards a skeletal muscle-like phenotype. Conversely, on a chow diet, AMPK activation leads to changes in expression of genes involved in catabolic pathways, including glycolysis and the TCA cycle.
Our findings suggest that activating AMPK increases the proportion of brown-like adipocytes within the WATsc depot, but these cells differ from conventional beige adipocytes since they do not express Ucp1. Switching mice from a chow to high fat diet results in a marked change in transcriptional response in these brown-like cells leading to increased expression of skeletal muscle associated genes. We propose that these adipocytes, referred to as skeletal muscle-like AMPK reprogrammed thermogenic (SMART) adipocytes, might represent a previously unrecognized cell type or a phenotype that ordinary subcutaneous white adipocytes can attain during sustained AMPK activation. Elegant studies using lineage tracing techniques have revealed that different types of adipocytes can develop from the same pool of progenitor cells that give rise to skeletal muscle cells16,25,26. Current evidence suggests that myocytes and brown adipocytes arise from a Myf5+-precursor, whereas white adipocytes descend from a different precursor. Most studies indicate that beige adipocytes derive from a white adipocyte precursor (Myf5-) stemming mainly from the WATsc depot, and their development is increased in response to cold-adaptation16,25,26.
In order to study further the origin of these SMART adipocytes, we crossed the γ1 transgenic mice with mice expressing Cre-recombinase under the control of the adiponectin-promoter (Adipoq-Cre) which drives expression in mature white and brown adipocytes27, but not adipocyte precursor cells28, to generate WT-TgAdipoq-Cre and D316A-TgAdipoq-Cre mice. In contrast to the results obtained in the D316A-Tg mice crossed with β–actin-Cre, there was no change in bodyweight, fat mass, or liver, WATsc or BAT histology between the two genotypes on a HFD (supplementary Fig. S5). These results demonstrate that AMPK activation in mature white or brown adipocytes is not sufficient to recapitulate the metabolic phenotype observed in the β–actin-Cre line. This suggests that AMPK activation in a different population of white adipocytes is responsible for the metabolic phenotype, distinguishing this model form many previous models showing changes in thermogenesis mediated by expression in mature adipocytes. Next, we crossed the γ1 transgenic lines with mice expressing Cre-recombinase under the control of the platelet-derived growth factor receptor (Pdgfr) α-promoter to generate WT-TgPdgfrα–Cre and D316A-TgPdgfrα–Cre mice. Previous studies have shown that this Cre-line drives expression in most adipocyte precursors present in WAT28,29. Similar to the results with Adipoq-Cre, there were no significant phenotypic changes between the genotypes (Supplementary Fig. S5). This finding indicates that the precursor cell leading to the newly defined SMART adipocytes is distinct from Pdgfrα-expressing preadipocytes. Although unlikely, it remained possible that activation of AMPK in skeletal muscle could drive the phenotype seen in the global mouse model. We therefore crossed the γ1 transgenic lines with mice expressing Cre-recombinase under the control of the Mef2c promoter to drive expression in skeletal muscle. Again, there were no significant phenotypic changes between the genotypes on a HFD (Supplementary Fig. S5), ruling out skeletal muscle as the primary tissue driving protection against DIO.
The studies described above utilize a gain-of-function AMPK model that is present from birth, which would not be a desirable therapeutic strategy. To better reflect a therapeutic approach, we used an inducible β-actin-Cre line (CAGGCre-ER™) to determine the effect of AMPK activation following the onset of diet-induced obesity. Male mice aged 8 weeks were switched to a HFD, and 4 weeks later transgene expression was induced by tamoxifen injection. Mice expressing either the floxed γ1 allele or the CAGGCre-ER™ alone were injected with tamoxifen and used as controls (Control). Tamoxifen caused a drop in bodyweight in both Control and D316A-Tg mice, but the D316A-Tg mice gained significantly less weight over the next 7 weeks on HFD (Supplementary Fig. S6). At this point, 7 weeks after administering tamoxifen, the Control mice had gained more than three-fold the weight of the D316A-Tg mice (5.6 ± 1.3 g vs 1.8 ± 1.4 g (n=9 per group), compared to their bodyweight immediately prior to tamoxifen injection). This was reflected by a significantly lower total fat mass in the D316A-Tg mice (Supplementary Fig. S6). Immunohistochemical analysis revealed that adipocytes isolated from both BAT and WATsc of D316A-Tg mice had smaller lipid droplets compared to Control mice and liver triglyceride content was markedly reduced in the D316A-Tg mice (Supplementary Fig. S6). Consistent with the findings in the constitutive β-actin-cre model, western blotting showed that components of the mitochondrial electron transport chain were increased in WATsc of D316A-Tg mice, with significant increases in Atp5a1 and Sdhb (Supplementary Fig. S6). Taken together, these results demonstrate that the effects of AMPK activation are not developmental in origin, adding considerable strength to the translational potential of AMPK activation in protecting against diet-induced obesity.
One of the major pharmacological approaches to target obesity is activation of pathways that increase energy expenditure in order to reduce positive energy balance. Here we show that genetic activation of AMPK increases energy expenditure in mice fed a HFD through Ucp1-independent thermogenesis in WATsc. Previous studies using AMPK deletion models reported effects of AMPK on UCP1-dependent thermogenesis in both BAT and WAT30–32. Pharmacological activation of AMPK was reported to increase BAT mass in the offspring of obese mice30, and to increase UCP1 expression in WAT31. In our study, UCP1 expression was not affected by AMPK activation using either global or adipose tissue-specific models. Taken together, these studies indicate that AMPK plays an important role in development of both BAT and WAT. However, further studies are required to elucidate the precise role of AMPK activation in regulating the thermogenic program in adipose tissue. Importantly, our findings strengthen the idea that AMPK activators have considerable potential in treating metabolic diseases, including obesity. In this study we show that AMPK activation increases the population of a novel type of adipocyte within the subcutaneous white adipose depot that resemble brown adipocytes, but lack Ucp1 expression. To our knowledge, this is the first identification of these cells, although at present their origin remains unknown and further characterization of these cells is a major challenge for future studies. Additionally, the mechanism leading to thermogenesis in these cells remains to be established. Nonetheless, induction of these precursor cells provides an exciting novel pharmacological target for exploiting as a therapeutic strategy in treating obesity.
Methods
Animals
All in vivo studies were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act (1986) and approved by the Animal Welfare and Ethical Review Board at Imperial College London. The Rosa26 gene targeting vector was prepared from a mouse C57BL/6 bacterial artificial chromosome with homology arms 5.6 kb and 1.7 kb flanking the XbaI site in the Rosa26 gene and a sequence encoding the Flag epitope (DYKDDDDK) at the C-terminus was engineered into the constructs to allow recognition by an anti-Flag antibody. Targeted ES cells were injected into BalbC/cANnCrl (Charles River, Germany) blastocysts and embryos were implanted into pseudo pregnant C57Bl6NCrl female mice. The resulting chimeric animals were mated with C57BL/6N mice to produce agouti heterozygous animals (F1). To generate animals without the Neo cassette, F1 mice were bred with CAG-FlpO. These mice were crossed with mice expressing Cre-recombinase under the control of the β-actin promoter (Tmeme163Tg(ACTB-cre)2Mrt (stock number 003376; Jackson Laboratories, Maine, USA) to generate mice with ubiquitous expression of the γ1 transgene (referred to as WT-Tg and D316A-Tg mice). Adipose-specific expression was achieved by crossing the γ1 floxed mice with mice expressing Cre-recombinase under the control of the adiponectin promoter (Adipoq-Cre; B6.FVB-Tg(Adipoq-cre)1Evdr/J; stock number 028020; Jackson Laboratories, Maine, USA) or the Pdgfrα promoter (C57BL/6-Tg(Pdgfra-cre)1Clc/J; stock number 013148; Jackson Laboratories, Maine, USA). Muscle-specific expression was achieved by crossing the γ1 floxed mice with mice expressing Cre-recombinase under the control of the Mef2c promoter (C57BL/6-Tg(Mef2c-cre)2Blk; a generous gift from Brian L. Black (University of California San Francisco)). For inducible expression, mice were crossed with mice expressing Cre-recombinase under the control of a tamoxifen-inducible chicken β-actin promoter (CAGGCre-ERTM; B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J; stock number 004682; Jackson Laboratories, Maine, USA). Induction was achieved by i.p injection of mice with 3 mg tamoxifen in 0.15 ml corn oil (4 daily injections). Unless stated otherwise, male mice were used for all studies and were maintained on a 12 hour light/dark cycle at 22°C with free access to food and water, group-housed in specific-pathogen free barrier facilities. Chow-standard breeding diet number 3 was from Special Diets Services and high fat diet (45% energy from fat) was obtained from TestDiet St. Louis, USA. Unless otherwise stated, for high fat diet feeding, diet was switched at 8 weeks of age. At the end of the procedure, animals were killed by cervical dislocation and organs harvested rapidly, weighed and either frozen in liquid nitrogen for further analysis or placed into 4% paraformaldehyde for subsequent histological analysis.
Metabolic Phenotyping
Whole-animal oxygen consumption rate (VO2) was measured using a Columbus Instruments Comprehensive Laboratory Animal Monitoring System. Prior to these studies, mice were singly housed for 1 week to acclimatise to housing conditions. Animals were fed ad libitum in all studies using this system. Animals were weighed prior to being placed into designated cages, with even distribution of genotypes between each rack. VO2 consumption was calculated by built in software (Oxymax) and normalised to bodyweight. Movement was measured by counting horizontal beam breaks. Recordings were taken at regular intervals throughout the duration of the experiment. In some cases, mice were housed at 30°C (within their thermoneutral zone). For the β3-agonist study, mice were removed from the cages, injected i.p with CL316, 243 (1 mg/kg) or vehicle control, before returning to the cage for monitoring of oxygen consumption. In some cases, VO2 time series data were smoothed by using a running average method, with GraphPad Prism software, and average VO2 values calculated. Food intake on chow was measured manually by recording the daily mass of diet used over a 3-week period. Food intake on the HFD was measured using BioDAQ food intake monitoring cages (Research Diets Inc.), measuring the ad libitum feeding activity over a 5-day period. Mice were fed from gated hoppers mounted outside of the cage to reduce variability. Data was recorded using the BioDAQ DataViewer. Total body composition was measured using an EchoMRI body composition analyser. Core body temperature was measured using a rectal thermal probe and subcutaneous temperature measured using IPTT300 BMDS transponders, inserted subcutaneously into the left or right flank of the mouse. Liver triglyceride content was measured using Triglyceride liquid (Sentinel Diagnostics). Glucose tolerance tests were performed on mice after a 6-h overnight fast. Animals were given an oral bolus of glucose (2 g/kg lean body mass) and blood glucose levels determined by a glucometer at the indicated time points. Serum insulin was measured in mice either fasted for 6h or fasted/re-fed for 2 h using an Ultra-Sensitive Mouse Insulin ELISA kit (CrystalChem). ECGs from conscious, unrestrained mice were recorded using the ECGenie System (Mouse Specifics Inc.).
Treatment with β-guanidinopropionic acid
In some cases, mice were transferred to a HFD and after 1 week were given access to water containing 0.5% β-guanidinopropionic acid and 0.13 % saccharin or water containing only 0.13% saccharin. Water, food intake and bodyweight were monitored over the next 3 weeks.
Measurement of respiration in white adipose tissue explants
Subcutaneous white adipose tissue (1.5-2 mg) was obtained using a Biopsy Puncher (2 mm) and placed into XF24 Islet Capture Microplates and incubated in assay medium (substrate-free DMEM (Sigma D5030), 30 mM NaCl, pH7.4, 25 mM glucose, 0.5 mM sodium pyruvate). Oxygen consumption rate was measured on a Sehaorse XF24 Flux analyser following sequential addition of oligomycin (5 μM), carbonyl cyanide 3-chlorophenyl hydrazone (CCCP, 5 μM) and antimycin (5 μM).
Histological analysis
Tissues were fixed in 4% paraformaldehyde overnight and dehydrated by incubation in 50%, then 70%, ethanol. Samples were wax embedded in paraffin and sectioned to a thickness of 4 microns. Tissues were deparaffinised and rehydrated using Acquaclear, 100% then 70% ethanol and boiled in sodium citrate antigen retrieval solution for 5 minutes in a pressure cooker. Sections were stained with haematoxylin and eosin, or with an anti-glycogen phosphorylase antibody on a Discovery Ultra (Ventana Medical Systems). The secondary antibody used was Discovery OmniMap anti-rabbit HRP (RUO) and detected with ChromoMap DAB Kit (RUO). Stained sections were visualised under bright-field microscopy. For immunofluorescent staining, slides were washed in TBS and blocked by incubation with 0.2% (w/v) fish skin gelatin (FSG) for 1 hour at room temperature. Sections were then incubated with primary antibody in 0.2% FSG overnight at 4°C. Slides were washed in 10 mM Tris-HCl pH 7.4, 150 mM NaCl (TBS) containing 0.1% Tween-20 and incubated with an Alexa Fluor conjugated secondary antibody (488 green or 633 red) for 1 hour at room temperature. After subsequent washes with TBS, sections were incubated with DAPI to stain nuclei for 5 minutes, rinsed under running water and mounted using VectaShield immunofluorescent mounting medium. Immunofluorescence-stained sections were imaged using a Leica TCS SP5 confocal microscope at 200Hz, with with either a Leica 20x/0.7NA Plan-Apochromat (PL-APO), 40x/1.25NA PL-APO and 63x/1.4NA PL-APO objective lens and analysed using LASAF software (Leica). A custom ImageJ macro script was developed to automate lipid droplet identification and area quantification from 1 mm2 regions-of-interest. Briefly, images of tissue sections were acquired on a Zeiss AxioScan.Z1 digital slide scanner with a 20X/0.8NA PL APO objective lens. Images were converted to grayscale and filtered to enhance the stained boundaries. Following this, images were thresholded and binarised for detection of the lipid droplet boundaries, the resultant binary mask inverted, and a watershed operation applied to segment individual adipocytes. Further refinement of individual adipocyte and lipid droplet identification was achieved by filtering based on morphological measurements, specifically circularity and aspect ratio, in addition to manual editing.
Western blot analysis
Tissues were homogenised using an Ultra-Turrax homogeniser in 10x w/v ice cold homogenisation buffer containing 50 mM Tris, 50 mM NaF, 5 mM Napyrophosphate, 1 mM EDTA, 0.25 M mannitol, 1 mM dithiothreitol, 157 μg/ml benzamidine, 4 μg/ml trypsin inhibitor and 0.1 mM phenylmethylsulphonyl fluoride. Homogenates were centrifuged at 13,000 x g for 15 minutes to remove insoluble material. Protein content of the soluble fraction was quantified using a BCA assay kit (ThermoScientific). Proteins (50 μg total) were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore Immobilon-FL) at 100 V for 90 minutes. Membranes were stained with PonceauS to check protein transfer and blocked in 4% (w/v) bovine serum albumin (BSA) for 1 hour at room temperature. Unless stated otherwise, primary antibodies were diluted 1:1000 in TBS containing 4 % BSA and 0.1% Tween-20, and incubated with the membrane for 4 hours at room temperature or overnight at 4°C. Membranes were washed extensively with TBS containing 0.1% Tween-20 before incubation with an appropriate IRDye secondary antibody (LI-COR Biosciences) in TBS for 1 hour at room temperature. Blots were visualised using the Odyssey Imaging System (LI-COR Biosciences) and quantified using ImageStudio 4.0.
Antibodies
The following antibodies were used in this study: total OXPHOS antibody cocktail (Abcam, ab110413); Ckmt2 (Abcam, ab55963); Flag (Cell Signaling, 14793); Pgc1-α (Abcam, ab54481); Tom-20 (SantaCruz, sc-11415); Serca1 (Abcam, ab109899); Serca2 (Invitrogen, MA3-919); Ucp1 (Abcam, ab 10983); vinculin (Sigma, V9131).
RNA isolation
Total RNA was isolated from snap-frozen tissue by homogenisation in 1 ml TRIzol (Life Technologies) per 100 mg tissue on ice. Samples were stored overnight at -20°C, centrifuged at 10,000 x g for 15 minutes and the homogenate removed to a fresh tube. Chloroform (400 ml per ml) was added and the mixture centrifuged at 10,000 x g for 15 minutes at room temperature. The aqueous phase was transferred to an RNAse-free Eppendorf and absolute ethanol (0.53 x volume) added. RNA was purified using RNeasy Mini spin columns (Qiagen). RNA was eluted in 50 μl RNase free H2O and quantified using a NanoDrop spectrophotometer. Samples were frozen at -80°C until required.
RT-qPCR
3 μg RNA was incubated with 1 μl random hexamers (50 ng/μl), 1 μl 10 mM dNTP mix and DEPC H2O to 10 μl. Samples were incubated at 65°C for 5 minutes on a thermal cycler. 10 μl cDNA synthesis mix (containing 2 μl 10 x RT buffer, 4 μl 25 mM MgCl2, 2 μl 0.1 M DTT, 1 μl RNaseOut and 1 μl Superscript II (Qiagen)) was added to each sample and cycled as follows: 25°C (10 min), 85°C (50 min) and the tubes transferred onto ice until cool. 1 μl RnaseH was added to each sample and incubated at 370C for 10 min. To determine primer linearity, 5 μl cDNA from each sample were pooled and serially diluted to give a standard curve. For each PCR reaction, 5 μl cDNA was added to 10 μl 2x SYBR-HiROX (Bioline), 1.6 μl forward (AGCCATCTGCATGGGATCAAA) and reverse (GGGTCGTCCCTTTCCAAAGTG) primer mix and ddH2O to a total reaction volume of 20 μl. The qPCR plate was analysed using an Opticon thermal cycler with Opticon monitor software to generate c(t) values for each reaction.
To quantify gene expression, c(t) value replicates were checked for primer efficacy (%) and consistency. Average c(t) values were then calculated for each sample, and quantified using a linear equation (y=mx+c) previously determined from the standard curve, corresponding to the gene of interest. The equation was solved for x, with y as c(t) value. This value was then transformed using 10x, as the standard curve was generated on a logarithmic scale. This process was repeated for all genes, including a designated housekeeping gene. Unless otherwise stated, all experiments were normalised to Polr2a, an RNA polymerase which has been shown to remain stable across dietary interventions.
Gene expression was then presented as both a ratio to the expression of the housekeeping gene for the experiment, and as fold change over the designated control.
RNA-seq analysis
RNA was extracted from subcutaneous white adipose (6 mice per genotype fed either chow or a HFD for 16 weeks, processed individually). For each sample, 2 μg RNA in 60 μl RNase-free H2O was quality assessment using a BioAnalyzer RNA kit (Agilent CA, US). An RNA Integrity Number (RIN) score of >7 was required for further analysis. The RNA libraries were prepared using a TruSeq Stranded mRNA Library Prep Kit (Illumina) and standard Illumina protocol. Libraries were quantified with Qubit HS (ThermoFisher) and Agilent BioAnalyzer adjusted to the appropriate concentration for sequencing. Indexed libraries were pooled and sequenced at a final concentration of 1.6 pM on an Illumina NextSeq 500 high-output run using paired-end chemistry with 75 bp read length. The sequencing data was demultiplexed using Illumina bcl2fastq2-v2.16. The quality of the reads was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were processed and mapped to the mouse genome mm10 using the Bcbio-nextgen framework version 0.9.0 (https://github.com/chapmanb/bcbio-nextgen). The aligner used was STAR 2.4.1d and alignment quality was assessed with QualiMap v.2.1.1.
Identification of differentially expressed genes was performed using DESeq2 in R. A principle component analysis was generated as a quality control step, to assess the clustering of samples in terms of gene expression. A list of differentially expressed genes (DEGs) was generated, expressed as Log2 fold change over the control sample (WT-Tg), with an adjusted P value for each gene.
Analysis of differentially expressed genes (DEGs)
Using original DESeq2 output Excel files, genes were listed in order of Log2 fold change, with an adjusted P value threshold of 0.05 and a biological threshold of ± 1.5 fold change relative to WT-Tg. Ingenuity Pathway Analysis (IPA) (Qiagen Bioinformatics, US) was used to analyse the lists of differentially expressed genes defined according to P<0.05. Gene ontology (GO) enrichment analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to find Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched within differentially expressed genes. For the HFD analysis, an input list consisting of top 100 most upregulated genes (by fold change) was used. The GEO accession number for the RNA sequencing data is GSE120429.
Statistical Analysis
Unless stated otherwise, data presented are shown of mean ±sem. Graphpad Prism software (v7) was used for graphing and statistical analysis. For comparison between two groups, datasets were analysed by a Student’s t tests, with statistical significance defined as a P value of <0.05. To compare three or more data sets, a one-way analysis of variance (one-way-ANOVA) was used, followed by Bonferroni’s range test to measure significance between means. Multiple comparisons were analysed by two-way ANOVA), followed by a Bonferroni’s Multiple Comparisons Test to determine statistical significance between groups based on one variable. For RNA sequencing analyses, an adjusted P value threshold was determined at 0.05.
Supplementary Material
Acknowledgements
This work was funded by the Medical Research Council UK (grant MC_US_A654_0003 to DC). AEP was funded by a BBSRC-CASE Studentship Award (BB/L502662/1). LW was funded by a British Heart Foundation Studentship Award. We would like to thank members of the Whole Animal Physiology Team (MRC LMS) for their assistance.
Footnotes
Data Availability
The datasets that support the findings of this study are available from the corresponding author upon request. RNA-sequence datasets used in this study are available from Gene Expression Omnibus (GEO) with the accession number GSE120429.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Author Contributions
AEP, LM, PJM, TA, MB, LW and RF carried out the in vivo and in vitro studies. SK, AB and MC carried out the RNAseq and analysis. CW and DS helped with immunohistochemistry analysis. AS and MBY helped with the metabolic phenotyping. LP and AW helped in the phenotypic analysis of the mice. AEP, LM, WK and MRD carried out the cellular respiration studies. AEP, LM, AW, DMS, JAR, MAS and DC designed and planned the study. All authors contributed to the preparation of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Carling D. AMPK signalling in health and disease. Curr Opin Cell Biol. 2017;45:31–37. doi: 10.1016/j.ceb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Hardie DG. AMP-activated protein kinase: maintaining energy homeostasis at the cellular and whole-body levels. Annu Rev Nutr. 2014;34:31–55. doi: 10.1146/annurev-nutr-071812-161148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 4.Cool B, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Xiao B, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4 doi: 10.1038/ncomms4017. 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods A, et al. Liver-specific activation of AMPK prevents steatosis on a high fructose diet. Cell Reports. 2017;18:3043–3051. doi: 10.1016/j.celrep.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson U, et al. AMP-activated protein kinase plays a role in the control of food intake. J Biol Chem. 2004;279:12005–12008. doi: 10.1074/jbc.C300557200. [DOI] [PubMed] [Google Scholar]
- 8.Minokoshi Y, et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428:569–574. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 12.García-Ruiz E, et al. The intake of high-fat diets induces the acquisition of brown adipocyte gene expression features in white adipose tissue. Int J Obesity. 2015;39:1619–1629. doi: 10.1038/ijo.2015.112. [DOI] [PubMed] [Google Scholar]
- 13.Gordon CJ. Thermal physiology of laboratory mice: defining thermoneutrality. J Therm Biol. 2012;37:654–685. [Google Scholar]
- 14.Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harms M, Seale P. Brown and beige fat: development, function and therapeurtic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granneman JG, Burnazi M, Zhu Z, Schwamb LA. White adipose tissue contributes to UCP1-independent thermogenesis. Am J Physiol Endocrinol Metab. 2003;285:E1230–E1236. doi: 10.1152/ajpendo.00197.2003. [DOI] [PubMed] [Google Scholar]
- 19.Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, Kozak LP. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1-/- mice. J Biol Chem. 2006;281:31894–31908. doi: 10.1074/jbc.M606114200. [DOI] [PubMed] [Google Scholar]
- 20.Kazak L, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. 2015;163:643–655. doi: 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitch CD, Chevli R. Inhibition of creatine and phosphocreatine accumulation in skeletal muscle and heart. Metabolism. 1980;29:686–690. doi: 10.1016/0026-0495(80)90115-8. [DOI] [PubMed] [Google Scholar]
- 22.Block BA, O’Brien J, Meissner G. Characterization of the sarcoplasmic reticulum proteins in the thermogenic muscles of fish. J Cell Biol. 1994;127:1275–1287. doi: 10.1083/jcb.127.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Costa DC, Landeira-Fernandez AM. Thermogenic activity of the Ca2+-ATPase from blue marlin heater organ: regulation by KCl and temperature. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1460–R1468. doi: 10.1152/ajpregu.90993.2008. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23:1454–1465. doi: 10.1038/nm.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2106;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Gurmaches J, Hung CM, Guertin DA. Emerging Complexities in Adipocyte Origins and Identity. Trends Cell Biol. 2016;26:313–326. doi: 10.1016/j.tcb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eguchi J, et al. Transcriptional Control of Adipose Lipid Handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffery E, et al. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, et al. AMPK/α-Ketoglutarate Axis Dynamically Mediates DNA Demethylation in the Prdm16 Promoter and Brown Adipogenesis. Cell Metab. 2016;24:542–554. doi: 10.1016/j.cmet.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, et al. AMP-activated protein kinase (AMPK) regulates energy metabolism through modulating thermogenesis in adipose tissue. Front Physiol. 2018;9:122. doi: 10.3389/fphys.2018.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mottillo EP, et al. Lack of Adipocyte AMPK Exacerbates Insulin Resistance and Hepatic Steatosis through Brown and Beige Adipose Tissue Function. Cell Metab. 2016;24:118–129. doi: 10.1016/j.cmet.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.