Abstract
The fetus is thought to be protected from exposure to foreign antigens, yet CD45RO+ T cells reside in the fetal intestine. Here we combined functional assays with mass cytometry, single-cell RNA-sequencing and high-throughput T cell antigen receptor (TCR) sequencing to characterize the CD4+ T cell compartment in the human fetal intestine. We identified 22 CD4+ T cell clusters, including naive-like, regulatory-like and memory-like subpopulations, which were confirmed and further characterized at the transcriptional level. Memory-like CD4+ T cells had high expression of Ki-67, indicative of cell division, and CD5, a surrogate marker of TCR avidity, and produced the cytokines IFN-γ and IL-2. Pathway analysis revealed a differentiation trajectory associated with cellular activation and proinflammatory effector functions, and TCR repertoire analysis indicated clonal expansions, distinct repertoire characteristics and interconnections between subpopulations of memory-like CD4+ T cells. Imaging-mass cytometry indicated that memory-like CD4+ T cells colocalized with antigen-presenting cells. Collectively, these results provide evidence for the generation of memory-like CD4+ T cells in the human fetal intestine that is consistent with exposure to foreign antigens.
Adaptive immunity is founded on the selection and expansion of antigen-specific T cells from a clonally diverse pool of naive precursors1. Naive T cells recirculate among lymph nodes to survey the array of peptide epitopes bound to major histocompatibility complex (MHC) proteins on the surface of antigen-presenting cells (APCs), and functional recognition of a given peptide-MHC molecule is governed by various danger signals and specific engagement via the clonotypically expressed T cell antigen receptor (TCR). This triggers a program of differentiation and proliferation that results in the generation of effector T cells, which home to the site of the primary infection and contribute to pathogen clearance, and memory T cells, which remain in the circulation and mediate anamnestic responses to secondary infection. In the last decade, it has also become clear that tissue-resident T cells are commonly present at barrier sites, including the intestine2.
Fundamental knowledge of adaptive immunity during early life remains sparse. The infantile intestine is known to harbor clonally expanded T cells3, which were also identified in the human fetal intestine, but rarely in fetal mesenteric lymph nodes, fetal thymus or fetal spleen, suggesting compartmentalization4. In addition, a rare population of CD4+ T cells displaying a memory and proinflammatory phenotype has been identified in umbilical cord blood5. Although the dogma of a sterile womb has been challenged by reports of bacteria colonization in the placenta6,7, amniotic fluid8,9 and meconium10, others have questioned these results11. Here we have combined functional studies with mass cytometry, RNA-sequencing (RNA-seq) and high-throughput TCR-sequencing to perform an in-depth analysis of the fetal intestinal CD4+ T cell compartment. Our results provide evidence for memory formation in the human fetal intestine, consistent with in utero exposure to foreign antigens.
Results
Human fetal intestinal CD4+ T cells are phenotypically diverse
To explore the CD4+ T cell compartment in the human fetal intestine, we applied a mass cytometry panel comprising 35 antibodies (Supplementary Table 1) that was designed to capture the heterogeneity of the immune system to seven lamina propria samples aged 14-21 gestational weeks12. After data acquisition, we selected CD45+ immune cells (Supplementary Fig. 1a) and mined the dataset via hierarchical stochastic neighbor embedding (HSNE)13. At the overview level, HSNE landmarks depicted the general composition of the immune system, with clear separation of the CD4+ T cell lineage (Supplementary Fig. 1b). We identified 110,332 CD4+ T cells, with an average of 15,761 events per fetal intestine, comprising 47.9% ± 9.6% of all immune cells. We then subjected HSNE-defined CD4+ T cells (Supplementary Fig. 1b) to t-distributed stochastic neighbor embedding (t-SNE)14 in Cytosplore15 to project their marker expression profiles onto a two-dimensional graph (Fig. 1a and Supplementary Fig. 1c). CD4+ T cells were characterized as CD45+CD3+CD4+CD7+ (Fig. 1a). Moreover, all CD4+ T cells were positive for the tissue-resident marker CD38 and approximately 50% of cells expressed CD161. 24.1% of the CD4+ T cell population co-expressed CD27, CD28, CD45RA and CCR7, indicative of a naive T cell (TN) phenotype, whereas 64.5% expressed CD45RO, indicative of a memory T cell (TM) phenotype (Fig. 1a,b). While all CD45RO+ TM cells were CD28+, differential expression of CD25, CD27, CD103, CD117, CD127, CCR6 and CCR7 was observed on these cells (Fig. 1a,b), reflecting substantial phenotypic diversity.
Fig. 1. Mass cytometric analysis of fetal intestinal CD4+ T cells.
a, t-SNE embedding of all CD4+ T cells (n = 110,332) derived from human fetal intestines (n = 7). Colors represent the ArcSinh5-transformed expression values of the indicated markers. b, t-SNE plot depicting the population cell border for TN cells (dashed yellow line), TM cells (dashed red line), and Treg cells (dashed green line). c, Density map describing the local probability density of cells, where black dots indicate the centroids of identified clusters using Gaussian mean-shift clustering. d, t-SNE plot showing cluster partitions in different colors. e, Heatmap showing median expression values and hierarchical clustering of markers for the identified subpopulations. f, Biaxial plots showing CD45RA and CCR7 expression on the indicated clusters analyzed by mass cytometry. The 22 clusters were merged into 6 phenotypic groups according to the heatmap shown in (e). g, Composition of the CD4+ T cell compartment in each fetal intestine represented by vertical bars, where the colored segment lengths represent the proportion of cells as a percentage of all CD4+ T cells in the sample. Colors as shown in (e).
We next applied Gaussian mean-shift clustering to the mass cytometry data using the t-SNE coordinates of the embedded CD4+ T cells (Fig. 1a). Based on cell density features (Fig. 1c), this identified 22 distinct CD4+ T cell clusters (Fig. 1d), each defined by a unique marker expression profile. Hierarchical clustering of the heatmap revealed eight major groups (CD161+CCR6–CD117– TM cells, CD161+CCR6+CD117+ TM cells, DN TM cells, Treg cells, CD161–CCR7+ TM cells, CD161– TN cells, CD161lo TN/TM cells and CD161–CCR7– TM cells) (Fig. 1e). High expression of CD25 and a lack of CD127 distinguished two regulatory T (Treg) cell clusters, with either a CD45RA+ TN or a CD45RO+ TM phenotype (Fig. 1a,b,e). CD161+CD4+ T cells branched into a CCR6–CD117–CD45RO+ TM and a CCR6+CD117+CD45RO+ TM cluster (Fig. 1e). Moreover, CD45RA+ TN and CD45RO+ TM cells were detected in both the CD161– and the CD161lo subpopulations. Additional diversity was observed for the expression of several activation markers, including CRTH2, HLA-DR, KLRG-1 and PD-1, the latter especially within the CD45RO+ TM cell clusters (Supplementary Fig. 1c). Of note, a small population of CD4–CD8a–TCRγδ– (DN) TM cells clustered among CD4+ T cells in both the HSNE and t-SNE plots. Biaxial plots confirmed coexpression of CD45RA and CCR7 on TN cells (Fig. 1f), whereas the CD161lo/–CD45RO+ TM subpopulation contained both CCR7+ central memory T (TCM) cells and CCR7– effector memory T (TEM) cells (Fig. 1f). All other CD45RO+ TM subpopulations harboured primarily TEM cells. Quantification of cellular frequencies for the CD4+ T cell clusters per fetal intestine revealed highly similar compositions with all CD45RO+ TM clusters detectable in all samples (Fig. 1g). In contrast, parallel analyses of CD4+ T cells isolated from three fetal livers and three fetal spleens from one shared and two additional fetuses aged 16-21 gestational weeks revealed a predominance of CD45RA+ TN cells (Supplementary Fig. 2a,b). These results delineated a phenotypically diverse array of human fetal intestinal CD4+ T cells, most of which displayed features associated with antigen exposure.
Fetal CD4+ T cells display a memory gene expression profile
We next performed single-cell RNA-seq on flow-sorted fetal intestinal CD4+ cells from a lamina propria sample that was also included in the mass cytometry analysis. This yielded data for 1,804 CD4+ T cells, identifying cell-specific variable expression of 2,174 genes (Methods), which were further analyzed using the Seurat computational pipeline16. Unsupervised clustering revealed nine transcriptionally distinct subpopulations, seven of which corresponded to CD3+ T cell subsets while two displayed a gene expression profile matching CD86+HLA-DR+ APCs. The corresponding gene expression profiles of the seven T cell subsets were projected onto a single graph using t-SNE (Fig. 2a), and the top 20 upregulated genes were displayed in a heatmap (Fig. 2b). Five of the seven RNA-seq-identified CD4+ T cell subpopulations corresponded to the mass cytometry-defined CD4+ T cell major groups: CCR7+ TN with CD45RA+ TN, KLRB1lo/–SELL– TM with CD161lo/–CD45RO+ TM, KLRB1+CCR6–SELL– TM with CD161+CCR6–CD45RO+ TM, KLRB1+CCR6+SELL– TM with CD161+CCR6+CD45RO+ TM and FOXP3+ Treg cells with CD25+CD127lo Treg cells. The mass cytometry defined CD161– and CD161lo subpopulations (Fig. 1e) could not be discriminated in the RNA-seq dataset. One additional RNA-seq-identified subpopulation corresponded to proliferating cells, based on the expression of genes associated with cell division (CCNB2, CDK1 and MKI67) (Fig. 2a,b).
Fig. 2. Single-cell RNA-sequencing of fetal intestinal CD4+ T cells.
a, t-SNE embedding of fetal intestinal CD4+ T cells (n = 1,804) showing seven transcriptionally distinct clusters, including CCR7+ TN (n = 358), KLRB1lo/–SELL– TM (n = 237), KLRB1+CCR6–SELL– TM (n = 640), KLRB1+CCR6+SELL– TM (n = 336), undefined TM (n = 101), FOXP3+ Treg cells (n = 71), and proliferating cells (n =61). Colors indicate different cell clusters. b, Heatmap showing the normalized single-cell gene expression value (Z-Score, purple-to-yellow scale) for the top 20 differentially upregulated genes in each identified cluster. Colors as shown in (a). c–e, Expression of the indicated genes in each identified cluster at (c) the RNA level (log-normalized) and (d,e) the protein level analyzed by (d) mass cytometry (CyTOF, ArcSinh5-transformed) or (e) flow cytometry, presented as violin plots. Dashed lines indicate background levels. Colors as shown in (a).
As CD45RA and CD45RO were not detectable, we used other markers to distinguish TM from TN clusters. To compare gene or marker expression among cell clusters, we used violin plots, displaying the mode average as the thickest section (Fig. 2c-e). Consistent with the mass cytometry data, RNA-seq-defined TN cells were KLRB1–CCR7+SELL+/– (Fig. 2c,d), the latter confirmed by flow cytometry (Fig. 2e and Supplementary Fig. 3a). In the absence of TM-associated markers, SELL– TM cell populations were identified on the basis of differential expression of KLRB1 and CCR6 (Fig. 2c,d). Consistent with the mass cytometry data, expression of KIT (CD117) was restricted primarily to KLRB1+CCR6+SELL– TM cells (Supplementary Fig. 3b). Moreover, the gene expression profile of the IL2RA+IL7RloFOXP3+ Treg cell population (Fig. 2c) corresponded to the mass cytometry-defined CD25+CD127lo Treg cells (Fig. 2d). In addition, several RNA-transcripts including LAG3, TIGIT, CTLA4 and TNFRSF18 (GITR) ascertained the identity of FOXP3+ Treg cells (Fig. 2b). Finally, the RNA-seq data revealed an undefined TM cluster that was not identified by mass cytometry, but expressed genes similar to those detected in the KLRB1+CCR6+SELL– TM subpopulation, such as CD69, CCL5 and JAML. Cell population frequencies identified by mass cytometry and RNA-seq were comparable with the exception of mass cytometry-defined CD25+CD127lo Treg cells and RNA-seq-defined FOXP3+ Treg cells (Supplementary Fig. 3c).
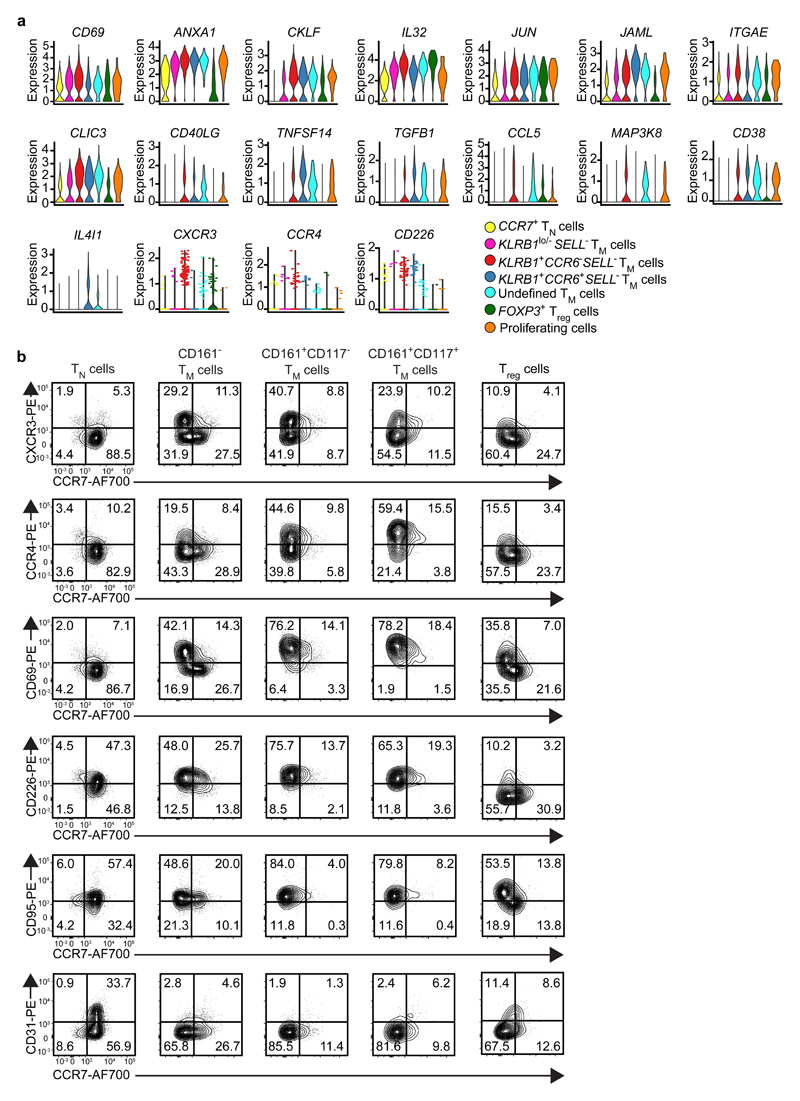
Compared with the CCR7+ TN population, KLRB1lo/–SELL– TM, KLRB1+CCR6–SELL– TM, KLRB1+CCR6+SELL– TM and undefined TM subpopulations had high expression of the tissue-resident and activation-associated gene CD69, the differentiation-promoting gene ANXA1 (Annexin A1), the chemokine-like factor CKLF, the cytokine IL32, the proliferation-associated gene JUN (C-Jun) and the adhesion molecule JAML (Fig. 3a). CD40LG (CD154), TNFSF14 and TGFB1 were specifically upregulated by KLRB1+CCR6–SELL– TM, KLRB1+CCR6+SELL– TM and undefined TM clusters, while CCL5 and MAP3K8 kinase upregulated by KLRB1+CCR6–SELL– TM and undefined TM subpopulations. Moreover, IL4I1 was specifically expressed by KLRB1+CCR6+SELL– TM and undefined TM cells. In addition, all fetal SELL– TM subpopulations had high expression of the tissue-resident genes ITGAE (CD103) and/or CD38 (Fig. 3a).
Fig. 3. Targeted analysis of memory-like fetal intestinal CD4+ T cells.
a, Expression (log-normalized) of the indicated genes in distinct CD4+ T cell clusters as determined by single-cell RNA-seq analysis, presented as violin plots. Colors indicate different cell clusters. b, Biaxial plots showing expression of CXCR3, CCR4, CD69, CD226, CD95, and CD31 vs. CCR7 on the indicated CD4+ T cell clusters analyzed by flow cytometry. Data represent two to three independent experiments.
In agreement with the RNA-seq data, flow cytometry indicated that the activation markers CXCR3, CCR4, CD69 and CD226 were highly expressed on CCR7– TEM cells (Fig. 3b). All CD4+ T cells expressed CD95, with the highest expression on CD161+ TEM cells (Fig. 3b). Expression of CD31, a marker associated with recent thymic emigrants17, was highest on CD45RA+ TN cells (Fig. 3b). Thus, RNA-seq confirmed the existence of distinct subpopulations of CD4+ T cells and indicate that many genes associated with inflammation and tissue residency were upregulated by fetal CD4+ TM cells, consistent with antigen-driven functionality and maturation.
Computational analysis reveals a differentiation pathway of CD4+ T cells
We next visualized the evolution of the t-SNE computation of the mass cytometry and RNA-seq data to reveal the ordering of single cells along putative differentiation trajectories12,15. At the onset of the mass cytometry data computation, where cells are grouped based on major shared features, CD25+CD127lo Treg cells clustered separate from the other cells, whereas the other cell clusters were ordered in a linear fashion with the CD45RA+ TN cells next to the CD161lo/–CD45RO+ TM cells, followed by the CD161+CCR6–CD45RO+ TM cells and the CD161+CCR6+CD45RO+ TM cells, consecutively (Fig. 4a). A similar phenotypic ordering was observed in parallel analyses of the RNA-seq data, although the KLRB1+CCR6+SELL– TM subpopulation aligned differently, but remained connected with the KLRB1+CCR6–SELL– TM cluster (Fig. 4b). Individual marker expression patterns at the middle of the t-SNE computation validated the ordering of the clusters and the comparability of the mass cytometry and RNA-seq data (Supplementary Fig. 4a,b). Similar patterns were identified using Diffusion map18, VorteX19 and principal component analysis (PCA)20 (Supplementary Fig. 4c–f). Thus, this analysis reveals a putative differentiation pathway leading to TM formation.
Fig. 4. Single-cell trajectory analysis of fetal intestinal CD4+ T cells.
a–b, t-SNE embeddings of all fetal intestinal CD4+ T cells analyzed by (a) mass cytometry (n = 10,436) and (b) single-cell RNA-seq (n = 1,743) at the onset and at the middle of the t-SNE computation. Colors indicate different cell clusters. c, A single-cell trajectory from the RNA-seq data (excluding Treg cells and proliferating cells) recovered by pseudotime analysis. Colors indicate different cell clusters as shown in (b). Grey arrows indicate three small branches. d, Three kinetic modules of pseudotime-dependent genes (n = 1,376) depicted in a log-variance-stabilized expression heatmap, indicating gene-enriched biological processes. Genes confirmed by mass cytometry and flow cytometry are denoted by black labels, and genes involved in TCR signalling are denoted by gray labels. The dashed grey box indicates the coordinated expression profile of TNF, FASL, and FYN. Euclidean distance values comparing gene expression profiles for each ordered pair of neighboring cells along the pseudotime trajectory are shown in the graph (right).
To extend our analysis of the gene expression profiles underlying this putative differentiation trajectory, we used the pseudotime algorithm in the Monocle toolkit21,22, which calculates the ordering of individual cells based on single-cell expression profiles. Based on this analysis, CCR7+ TN cells were separated from SELL– TM cells (Fig. 4c). When we clustered genes according to expression patterns along the pseudotime trajectory, cell-to-cell transitioning could be explained by the kinetics of 1,376 variable genes, which formed three large modules (Fig. 4d). The first module contained 540 genes associated with CCR7+ TN cells, including SELL, CCR7, CD27 and CD28 (Fig. 4d). The second module contained 453 genes, many of which were associated with an ongoing transcriptional program, such as RPL21, RPS2 and RPLP1. The highest activity of this transcriptional gene expression profile coincided with the transition of cells with a CCR7+ TN phenotype into cells with a SELL– TM phenotype (Fig. 4c,d). The third module contained 383 genes (Fig. 4d), 106 of which were associated with cellular activation and regulation of the immune system (Supplementary Fig. 5a), while 133 encoded proteins known to interact physically with each other (Supplementary Fig. 5b). In addition, 23 genes in module 3 could be assigned to cytokine or chemokine receptor pathways, including CCL20 and its receptor CCR6, the interferon receptor IFNGR1, TNF family members and IL-1 and IL-17 receptors (Supplementary Fig. 5b). Several signaling cascades were also represented in module 3, including the MAPK, TNF, IL-17 and TCR signaling pathways (FYN, LCP2, SOS1, MAP3K8 kinase, FASL and TNF) (Fig. 4d). The TH17-associated gene RORC was expressed in module 3 (Fig. 4d). In addition, the dynamic expression profiles of FYN, FASL and TNF clustered tightly with KLRB1 (CD161) (Fig. 4d) at the point in the pseudotime trajectory where CCR7+ TN cells were aligned next to SELL– TM cells (Fig. 4d). Finally, we quantified the smoothness of cell-to-cell transitioning based on gene expression changes along the trajectory, which showed that the pseudotime trajectory was most uncertain at the beginning and toward the end, but quite robust in the middle, where CCR7+ TN cells were aligned next to SELL– TM cells (Fig. 4d). In sum, these results identified temporal patterns of gene expression along the single-cell trajectory that is compatible with the transition of cells displaying a TN phenotype into cells with a TM phenotype.
TCR analysis reveals clonal expansion of fetal CD4+ T cells
Surface expression of CD5 correlates with TCR avidity23–26. Because CD5 gene expression was upregulated in TM cells compared to TN cells, we quantified CD5 expression on all identified fetal intestinal CD4+ T cell subsets using flow cytometry and observed that all the CD4+ T cell subsets expressed CD5 (Fig. 5a), but that the median fluorescence intensity (MFI) was higher in CD161– TM, CD161+CD117– TM and CD161+CD117+ TM cells and lower in CD25+CD127lo Treg cells and CD45RA+ TN cells (Fig. 5a–c), suggesting that cells with a TM phenotype express TCR with a higher avidity compared to TN cells.
Fig. 5. CD5 expression analysis and high-throughput TCR-sequencing of fetal intestinal CD4+ T cells.
a–b, CD5 expression on the indicated CD4+ T cell clusters. (a) The biaxial plots depict one representative experiment, and (b) the bar graphs depict the median fluorescence intensity (MFI) of CD5 expression for each cluster relative to the corresponding CD161+CD117– TM subpopulation in each fetal intestine (n = 7). Data represent six independent experiments. Error bars indicate mean ± SEM. *p < 0.05, Two-tailed Wilcoxon matched-pairs signed-ranks test. c, Expression (log-normalized) of CD5 gene transcripts in the indicated cell clusters, presented as violin plots. d, Dot plots showing the percentage of TCR cDNA molecules per unique TCRβ sequence in each cluster from each fetal intestine. Data are from two independent samples. A single duplicate is shown for samples with technical replicates. e, Dot plots showing the normalized Shannon-Wiener index for TCRα (TRA) and TCRβ (TRB) sequences in each cluster from each fetal intestine. Data are from two independent samples. f, Dot plots showing averaged TCR repertoire characteristics weighted per clonotype for each cluster. Data are from two independent samples. g, Dendrogram showing weighted clonal overlaps for TCRβ nucleotide sequences among clusters, analyzed using the F2 similarity metric in VDJtools. Colors indicate different fetal intestines.
Next, we evaluated the TCR clonotypic architecture of flow-sorted fetal intestinal CD45RA+ TN, CD45RO+ TM and CD25+CD127lo Treg subpopulations. Analysis of the TCRβ rearrangements in a single fetal intestine indicated limited overlap among the distinct subpopulations, most of which were highly polyclonal (not shown). Distinct clonotypes were expanded among CD45RO+ TM cells compared to CD45RA+ TN cells (Supplementary Fig. 6a). We then used a quantitative high-throughput approach for deep sequencing of TCRα and TCRβ rearrangements in all identified fetal intestinal CD4+ T cell subsets isolated from two additional fetal intestines (Supplementary Table 2). Post-analysis of the obtained repertoires was conducted using VDJtools27. As expected, all TM subpopulations showed greater clonality compared to the TN subpopulation (Fig. 5d,e). The averaged characteristics of CDR3 length, added N-nucleotides and physicochemical characteristics of the 5 amino acid residues located in the middle of the CDR3 loop, which are most likely to contact the peptide-MHC complex28, also differed among all subpopulations (Fig. 5f). The latter analysis included the averaged statistical potential of the CDR3 loop with respect to epitope interactions, comprising the estimated “energy” of the interaction with a random epitope29, the “strength” of the interaction (derivative of “energy”, VDJtools27), hydrophobicity (Kidera factor 4)30,31 and “volume” (values from http://www.imgt.org/IMGTeducation/Aide-memoire/_UK/aminoacids/IMGTclasses.html). These analyses provided no evidence for clonal expansion of CD4+ T cells as a function of intrinsically high TCR-avidities for self-derived peptide-MHC complexes (Fig. d-f), suggesting indirectly that antigen-specific interactions triggered clonal selection of CD4+ T cells from the TN cell pool. Analysis of V-J segment use (Jensen-Shannon divergence; Supplementary Fig. 6b) and overlaps among repertoires in terms of the weighted proportion of shared TCRβ clonotypes revealed tightly clustered technical replicates and clearly distinguished all subpopulations of CD4+ T cells (Fig. 5g). At the same time, the CD161– TM, CD161+CD117– TM and CD161+CD117+ TM cells clustered similarly in each fetus, with minimum overlap with the CD45RA+ TN and CD25+CD127lo Treg cells (Fig. 5g). Analysis of the clonal overlap of amino acid CDR3 repertoires between the same populations in the two fetal intestines revealed that the CD161– TM, CD161+CD117– TM and CD161+CD117+ TM populations displayed much stronger overlap than the CD45RA+ TN and CD25+CD127lo Treg CD4+ T cells (Supplementary Fig. 6c), which could be explained by TCR selection due to exposure to similar foreign antigens. Finally, although the majority of the TCR repertoire was specific for each population, up to 20% of the T cell clones were shared between the CD45RA+ TN and the three CD45RO+ TM cell populations (Supplementary Fig. 6d), suggesting a potential clonal relationship between CD45RA+ TN and CD45RO+ TM cells. These results indicate that avidity-based, clonotype-specific expansion of the TN pool was associated with TM formation and confirmed the close relationship between CD161– TM, CD161+CD117– TM and CD161+CD117+ TM cells.
Fetal CD4+ TM cells secrete proinflammatory cytokines
To determine the functional profiles of fetal intestinal CD4+ T cells, we flow-sorted CD3+CD4+ T cells and measured expression of TNF, IL-2, IFN-γ, IL-4, granzyme B and IL-17A in CD45RA+ TN cells and CD117– and CD117+ TM cells after cross-linking CD3 and CD28. The activation marker CD154 (CD40L) was upregulated on all cells analyzed (Fig. 6a,b), indicating efficient stimulation. All three subpopulations secreted large amounts of TNF (Fig. 6a,b), but CD117– TM cells and CD117+ TM cells displayed the highest MFIs (Supplementary Fig. 7a). Moreover, IL-2, IFN-γ, IL-4 and granzyme B were more commonly expressed in CD117– TM and CD117+ TM cells relative to CD45RA+ TN cells (Fig. 6a,b). The majority of cytokine-producing CD4+ T cells did not express Ki-67 (Supplementary Fig. 7b). Importantly, higher frequencies of IL-2+IFN-γ+ cells were detected in the CD117– TM and CD117+ TM cells compared with the CD45RA+ TN population (Supplementary Fig. 7c), suggesting greater polyfunctionality. Although the TH17-associated RORC gene was expressed by 1.3% of KLRB1+CCR6+SELL– TM cells (Fig. 4d), IL-17A production was undetectable in all TM cells. Thus, fetal intestinal CD117– TM and CD117+ TM cells deployed multiple effector functions reminiscent of classical CD4+ TM cells in response to TCR-mediated signal transduction and costimulation via CD28.
Fig. 6. Functional profiling of fetal intestinal CD4+ T cells.
a–b, Purified fetal intestinal CD4+ T cells were treated with a control antibody or stimulated with anti-CD3 and anti-CD28 for 4 h. Intracellular expression of TNF, IL-2, IFN-γ, IL-4, granzyme B, and CD154 was determined for each subpopulation by flow cytometry. (a) The biaxial plots show data from one representative experiment after stimulation with anti-CD3 and anti-CD28, and (b) the bar charts show data for each subpopulation from each fetal intestine (TNF: n = 4 samples in two independent experiments; IL-2 and granzyme B: n = 3 samples in two independent experiments; IFN-γ, IL-4 and CD154: n = 7 samples in four independent experiments). Error bars indicate mean ± SEM. *p < 0.05, **p <0.01, Kruskal-Wallis test with Dunn’s test for multiple comparisons.
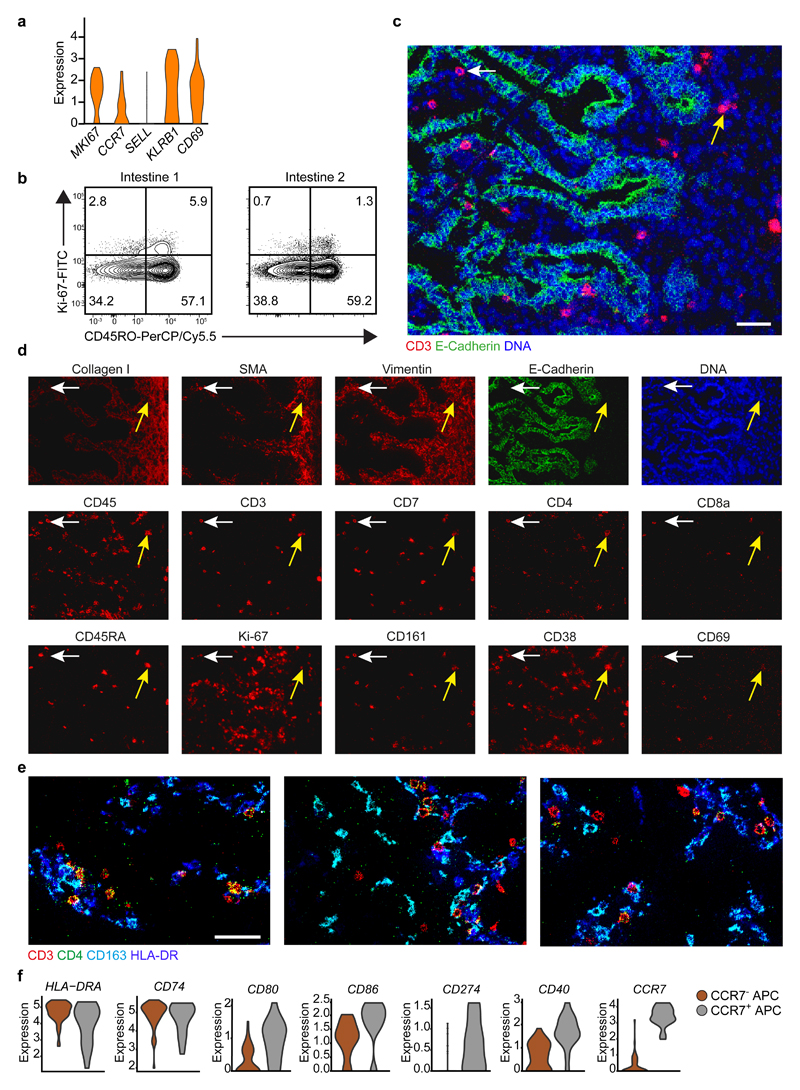
Fetal CD4+ T cells are co-localized with antigen presenting cells
The single-cell RNA-seq analysis revealed a MKI67+ cluster of proliferating cells, together with high expression of the TM cell-associated markers KLRB1 (CD161) and CD69 and low expression of the TN cell-associated markers CCR7 and SELL (CD62L) (Fig. 7a). Flow cytometry of fetal intestinal CD4+ T cells indicated the presence of Ki-67+ cells, predominantly within the CD45RO+ compartment (Fig. 7b). To assess the spatial distribution of CD4+ T cells in situ, we employed a panel of 15 antibodies (Supplementary Table 3) combined with a DNA stain to perform imaging-mass cytometry on tissue sections of four human fetal intestinal samples. Stains for collagen I and smooth muscle actin were used to visualize the extracellular matrix of the basement membrane, and the epithelium and lamina propria were distinguished as vimentin–E-cadherin+ and vimentin+E-cadherin–, respectively (Fig. 7c,d). Most CD4+ T cells localized to the lamina propria (Fig. 7c,d). Differential expression of CD45RA further allowed discrimination of CD45RA+ TN (Fig. 7c,d) from CD45RA– TM cells in the lamina propria (Fig. 7c,d). In addition, all CD4+ T cells expressed CD38, whereas only some CD4+ T cells expressed CD69 (Fig. 7d). Using a second panel comprising 10 antibodies (Supplementary Table 3), we found that CD4+ T cells frequently colocalized with CD163+HLA-DR+ APCs (Fig. 7e). Moreover, the single-cell RNA-seq analysis of fetal intestinal cells revealed two cluster of cells displaying high expression of gene transcripts encoding HLA-DR, CD74 (HLA-class II invariant chain), inhibitory molecule PD-L1 (CD274), CD80 and CD86, typically found in APCs. Moreover, these APCs expressed gene transcripts encoding CD40, consistent with an activated phenotype (Fig. 7f), whereas stimulated fetal intestinal CD4+ T cells expressed CD40L (CD154) (Fig. 6b). In addition, 25.8% of APCs had high expression of CCR7, potentially enabling migration to the mesenteric lymph nodes (Fig. 7f). Collectively, these results indicated the existence of CD4+ TM cells in the fetal intestine, many of which colocalized in the lamina propria with activated CD163+HLA-DR+ APCs.
Fig. 7. Characterization and spatial localization of fetal intestinal CD4+ T cells and APCs.
a, Expression (log-normalized) of the indicated genes in proliferating fetal intestinal CD4+ T cells, presented as violin plot. b, Biaxial plots showing expression of Ki-67 vs. CD45RO in the fetal intestinal CD4+ T cell compartment analyzed by flow cytometry. Data represent two independent experiments. c, Representative mass cytometry image of a fetal intestine showing the overlay of CD3 (red), Ecadherin (green), and DNA (blue). Scale bar, 100 μm. d, Representative mass cytometry images of fetal intestines showing expression of the indicated stromal markers, immune markers, Ki-67 and DNA by the cells identified in (c). Yellow arrows: CD4+CD45RA+ TN cells; white arrows: CD4+CD45RA– TM cells. e, Representative mass cytometry images of a fetal intestine showing the overlay of CD3 (red), CD4(green), CD163 (cyan), and HLA-DR (blue). Scale bar, 50 μm. Colors and scale bars are similar in all three panels. Data in (b-d) represent four independent samples in four independent experiments. f, Expression (log-normalized) of the indicated genes in two clusters of APCs, presented as violin plots. CCR7– APCs (n = 49), CCR7+ APCs (n = 17).
Discussion
Here we used mass cytometry and single-cell RNA-seq to characterize CD4+ T cells in the human fetal intestine. Mass cytometry revealed three major populations of fetal intestinal CD4+ T cells (TN, TM and Treg cells), that could be further distinguished into eight distinct cells clusters that displayed additional heterogeneity. These cell clusters were present in seven human fetal intestines, suggesting a physiologically robust immune composition. Single-cell RNA-seq revealed the presence of seven CD4+ T cell subpopulations, five of which displayed phenotypic overlap with the mass cytometry-defined CD4+ T cell subpopulations. We used computational tools to construct putative CD4+ T cell differentiation trajectories. Using adapted t-SNE32, we obtained remarkably similar trajectories for the mass cytometry and RNA-seq data. We identified three distinct gene expression modules along the differentiation trajectory that correspond to an increase in gene translation and subsequent activation of immune related genes. In addition, high-throughput TCR sequencing indicated clonal expansions within the CD4+ TM cell pool, consistent with the evidence for cell proliferation within the CD45RO+ TM pool that was obtained at both the mRNA and protein level. Moreover, CD4+ TM cells secreted higher amounts of pro-inflammatory cytokines upon TCR triggering compared to CD4+ TN cells. Finally, fetal intestinal CD4+ TM cells displayed a tissue-resident profile and were frequently found to colocalize with APCs in the lamina propria. Together, this suggested that clonotype-specific transcriptional programs regulated by antigen encounter underpinned the formation of CD4+ TM cells in the fetal intestine.
T cells in umbilical cord and peripheral blood obtained of infants aged 2 months were reported to display a typical CD45RA+ TN phenotype3. The observation herein that a large pool of CD45RO+ cells with a tissue-resident profile populated the fetal intestine suggests the compartmentalization of the immune system early in life. In conjunction with the earlier finding that clonally expanded T cells were present in the fetal intestine, but virtually absent in other fetal organs4, our results further suggest that memory formation was driven by local exposure to foreign antigens. The observation that there is a substantial overlap in the amino acid CDR3 repertoires of the memory CD4+ T cells compartment in the two fetuses analyzed may indicate exposure to similar foreign antigens.
Approximately 50% of all fetal intestinal CD4+ T cells were CD161+ and transcriptionally distinct from their CD161– counterparts, consistent with a recent study33. The kinetics of KLRB1 (CD161) expression was preceded by increased expression CD5 and coincided with increased expression of several TCR signaling genes, including FYN, FASL and TNF, suggesting a coordinated program of transcription. Of note, CD161 was identified as a costimulatory molecule in the context of TCR stimulation33.
Although the mass cytometry and RNA-seq data were largely compatible, there were exceptions. For example, coexpression of CCR6 and KIT among KLRB1+CCR6+SELL– TM cells was not reflected in the gene expression profiles. Conversely, expression of ITGAE (CD103) mRNA was not reflected by protein expression. These anomalies were likely attributable to discordant gene transcription and protein expression34 and may also relate to differences in sensitivity of the employed techniques.
The presence of a large population of TN cells in the fetal intestine is in stark contrast to the predominace of TM cells in the adult intestine. As the TN cells expressed relatively high amounts of CD31, which demarcates recent thymic emigrants, our results indicate direct migration of recent thymic emigrants into the intestine35,36. We propose that antigen-specific priming of TN cells takes place in the mesenteric lymph nodes followed by migration of the resulting TM cells to the lamina propria leading to a progressive loss of TN cells. Similarly memory formation is taking place in the CD8+ T cell compartment (not shown).
Distinct subpopulations of fetal intestinal Treg cells were distinguished by several markers, including high expression of CD25 and Foxp3, and a lack of CD127. In line with previous results37, approximately 50% of these cells expressed CD45RO, while the remainder expressed CD45RA. The CD45RA+ Treg cells expressed TCRs with longer CDR3β loops, higher numbers of added N-nucleotides and distinct physicochemical characteristics, suggesting higher affinities for self-antigens compared to CD45RO+ Treg cells38. The presence of oligoclonal T cell expansions in fetuses with autoimmune conditions associated with a genetic absence of Treg cells indicate a key role for these cells in immune suppression in utero39.
In conclusion, our study revealed a putative differentiation trajectory in the fetal intestinal CD4+ T cell compartment, consistent with the formation of TM cells in utero, presumably as a consequence of exposure to foreign antigens. These could include non-inherited maternal HLA-molecules40 and pathogen-derived ligands, which could be derived from amniotic fluid8,9. We propose that immune priming in the fetal intestine prepares the infant for the massive influx of bacteria that occurs immediately after birth, with anamnestic responses in situ facilitated by the colocalization of CD4+ TM cells with APCs.
Methods
Sample processing and cell isolation
Fetal tissues were obtained from elective abortions with informed consent. The gestational age ranged from 14 to 22 weeks. Intestines were separated from mesentery, cut into small pieces, embedded in optimal cutting temperature compound, and snap-frozen in isopentane. The remaining intestines were used for single-cell isolation as described previously12. Briefly, fetal intestines were cleared of meconium, cut into fine pieces, treated with 1 mM dithiothreitol (Fluka) for 2 x 10 min (replacing buffer) at room temperature (rT), and then incubated with 1 mM ethylenediaminetetraacetic acid (Merck) for 2 x 1 h (replacing buffer) at 37 °C under rotation to separate the epithelium from the lamina propria. To obtain single-cell suspensions from the lamina propria, the intestines were rinsed with Hank’s balanced salt solution (Thermo Fisher Scientific), incubated with 10 U/mL collagenase IV (Worthington) and 200 µg/mL DNAse I grade II (Roche Diagnostics) overnight at 37 °C, and filtered through a 70 µm nylon mesh. Isolated cells were then further purified with a Percoll gradient (GE Healthcare). Fetal liver and spleen tissues were cut into small pieces and filtered through a 70 µm nylon cell strainer and the immune cells were isolated with Ficoll-Paque™ density gradient (provided by apothecary LUMC). All the isolated cells were stored in liquid nitrogen. Study approval was granted by the Medical Ethics Commission of Leiden University Medical Centre (protocol P08.087). All experiments were conducted in accordance with local ethical guidelines and the principles of the Declaration of Helsinki.
Cell suspension-mass cytometry
Antibodies used for cell suspension-mass cytometry are listed in Supplementary Table 1. Purified antibodies lacking carrier protein were conjugated with metal reporters by using a MaxPar X8 Antibody Labeling Kit (Fluidigm). Procedures for antibody staining and data acquisition were described previously41. Briefly, cells from fetal intestines were incubated with 5 µM Cell-ID Intercalator-103Rh (Fluidigm) for 15 min at rT and then stained with a cocktail of metal-conjugated antibodies for 45 min at rT. After washing, cells were incubated with 125 nM Cell-ID Intercalator-Ir (Fluidigm) in MaxPar Fix and Perm Buffer (Fluidigm) overnight at 4 °C. Data were acquired using a CyTOF 2™ mass cytometer (Fluidigm) and normalized using EQ Four Element Calibration Beads with the reference EQ Passport P13H2302 (Fluidigm).
Imaging-mass cytometry
Antibodies used for imaging-mass cytometry are listed in Supplementary Table 3. Purified antibodies lacking carrier protein were conjugated with metal reporters by using a MaxPar X8 Antibody Labeling Kit (Fluidigm). Snap-frozen human fetal intestinal biopsies were sectioned at a thickness of 5 μm and fixed by incubating with 1% paraformaldehyde for 5 min at rT followed by 100% methanol for 5 min at –20 °C. After fixation, tissue sections were washed in Dulbecco’s phosphate-buffered saline (Thermo Fisher Scientific) containing 0.5% bovine serum albumin and 0.05% Tween, rehydrated in additive-free Dulbecco’s phosphate-buffered saline, washed again, and blocked with Superblock Solution (Thermo Fisher Scientific). Tissue sections were then stained with a cocktail of metal-conjugated antibodies overnight at 4 °C, washed, and incubated with 125 nM Cell-ID Intercalator-Ir for 30 min at rT. After a further wash, tissue sections were dipped in Milli-Q water (Merck Millipore) for 1 min and dried for 20 min at rT. Data were acquired using a Hyperion™ imaging-mass cytometer (Fluidigm) at a resolution of 1 µm, with settings aligned to company guidelines. The ablation frequency was 200 Hz, and the energy was 6 dB. Regions of interest were acquired at a size of 1 by 1 mm2. All data were stored as MCD files and txt files.
Single-cell RNA-sequencing
Single, live, CD8a–TCRγδ–CD4+ cells from the intestines of fetus #6 were sorted using a FACSAria III flow cytometer (BD Biosciences). Post-sort purity was 96.5%. Single-cell RNA-sequencing was performed as described previously42. Briefly, cells combined with oil, reagents, and beads were loaded on a Chromium Single Cell Controller (10x Genomics). Lysis and barcoded reverse transcription of polyadenylated mRNA from single cells were performed inside each gel bead emulsion. Next-generation sequencing libraries were prepared in a single bulk reaction, and transcripts were sequenced using a HiSeq4000 System (Illumina).
Integrated data analysis
For cell suspension-mass cytometry, data from single, live, CD45+ cells, gated individually using Cytobank as shown in Supplementary Fig. 1a, were sample-tagged and hyperbolic-arcsinh-transformed with a cofactor of 5 using Cytosplore+HSNE software13. The major immune lineages shown in Supplementary Fig. 1b were then identified at the overview level by performing a 3-level HSNE analysis carried out with default parameters (perplexity: 30; iterations: 1,000). All t-SNE plots and Gaussian Mean-Shift clustering-derived cell clusters were generated in Cytosplore15. Hierarchical clustering of the phenotype heatmap was created with Euclidean correction and average linkage clustering in Cytosplore+HSNE. Violin plots for cytometry data were generated in R. Diffusion map plots for mass cytometry data were generated using the “density” package in R. Single-cell force-directed layouts for mass cytometry data were generated using “VorteX” software19. For imaging-mass cytometry, all images were generated using MCD Viewer software v1.0.560 (Fluidigm). For single-cell RNA-seq, single-cell transcriptome sequencing data were processed using the single-cell RNA-seq package “Seurat” in R16. The Seurat object was generated by following the criteria that each gene was expressed by at least 3 cells and that at least 200 genes were expressed per cell. Data were further filtered based on the parameters: (i) unique gene count per cell >200 and <2,000; and (ii) mitochondrial percentage of all genes <0.05. After log-normalization, a PCA-reduction analysis (pcs.compute = 20) was performed based on the 2,174 variable genes across single cells. Next, graph-based clustering detection and a t-SNE algorithm were applied to the top 13 PCA-dimensions. The resolution for cluster detection was 0.8. Heatmaps, PCA plots, diffusion map plots, and violin plots of the RNA-seq data were generated using the “Seurat” package. The t-SNE plots for RNA-seq data shown in Fig. 4b were generated in Cytosplore+HSNE. Only genes with local standardization (>0.5) across all cells were taken into account. Bar graphs and dot plots (showing mean and SD) were generated in Prism (GraphPad). The pseudotime analysis shown in Fig. 4c,d was performed using the Monocle 2 toolkit in R as described previously22, excluding unrelated Treg cells. Briefly, the single-cell trajectory was inferred using the dpFeature unsupervised procedure to identify variable genes, and the dimensions were reduced using t-SNE on the top high-loading principal components. The top 1,000 significant genes were selected as the ordering genes and reduced with the DDRTree method for the single-cell graph shown in Fig. 4c. Variable genes were selected at a significant false discovery rate of <10%, clustered by pseudo-temporal expression patterns, and visualized in a heatmap in Fig. 4d. Gene list enrichment analysis was performed using ToppGene43, gene interaction network analysis was performed using the BioGrid interaction database44, and gene pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes45.
Flow cytometry
For surface staining, cells were incubated with fluorochrome-conjugated antibodies and human Fc block (BioLegend) for 30–45 min at 4 °C. For intracellular cytokine/CD154 staining, cells were stimulated with CD3/CD28-specific (2.5 µg/mL each, BioLegend) or control antibodies (5 µg/mL, BioLegend) for 4 h at 37 °C. Brefeldin A (10 µg/mL, Sigma) was added for the final 3 h. Cells were then fixed/permeabilized using Fixation Buffer and Intracellular Staining Perm Wash Buffer (BioLegend). For intracellular Foxp3/Ki-67 staining, cells were prepared using a Foxp3 Staining Buffer Set (eBioscience). Electronic compensation was performed using individually stained CompBeads (BD Biosciences). Cells were acquired using an LSR II cytometer (BD Biosciences) or sorted using a FACSAria III flow cytometer (BD Biosciences) as shown in Supplementary Fig. 3a. Data were analyzed with FlowJo software v10 (Tree Star Inc.). The antibodies used in this study are listed in Supplementary Table 4.
TCR repertoire analysis
CD4+ T cell subsets were sorted according to the gating strategy shown in Supplementary Fig. 3a. For conventional sequencing, a total of 5,000 cells per subset was sorted directly into RNAlater (Applied Biosystems) using a FACSAria III flow cytometer (BD Biosciences). All expressed TCRβ rearrangements were amplified using a template-switch anchored RT-PCR, sequenced, and analyzed as described previously46. Gene use was determined according to the ImMunoGeneTics (IMGT) nomenclature47.
For high-throughput sequencing, an average of 6,700 ± 2,000 cells per subset was sorted directly into RLT buffer (Qiagen) using a FACSAria III flow cytometer (BD Biosciences). Four volumes of TRIzol (Invitrogen) were then added to the RLT cell lysate. RNA was extracted according to the TRIzol Reagent User Guide. Unique molecular identifier (UMI)-labelled 5’RACE TCRα and TCRβ cDNA libraries were prepared using a Human TCR Profiling Kit (MiLaboratory LLC). All extracted RNA was used for cDNA synthesis, and all synthesized cDNA was used for PCR amplification. Libraries were prepared in parallel using the same number of PCR cycles and sequenced in parallel using a 150 + 150 bp MiSeq System (Illumina). This approach generated a total of 11,310,000 TCRα and TCRβ sequencing reads (250,000 ± 150,000 reads per library), from which 625,000 unique unique UMI-labelled TCR cDNA molecules (13,500 ± 7,000 molecules per library) were extracted using MIGEC48 and MiXCR49 software with a threshold of at least 2 sequencing reads per UMI. Each library contained an average of 3,500 ± 1,300 functional (in-frame, without stop-codons) CDR3 nucleotide sequences. Averaged TCR repertoire characteristics weighted by clonotype size were analyzed using VDJtools software27. Gene use was determined according to the ImMunoGeneTics (IMGT) nomenclature47.
Statistics
Results are shown as mean ± SEM. The statistics test used were two-tailed Wilcoxon matched-pairs signed-ranks test and Kruskal-Wallis test with Dunn’s test for multiple group comparisons, as appropriate (after normality test). P ≤ 0.05 was considered to be statistically significant. All statistics were analysed using GraphPad Prism 7 software.
Supplementary Material
Acknowledgments
We thank the Center for Contraception, Abortion and Sexuality (Leiden and The Hague) for collection and provision of fetal material, K. Lodder, T. van Herwaarden, M. Bialecka and F. Wang for dissection of fetal tissues, and S. L. Kloet for assistance with single-cell RNA sequencing. This research was supported by Leiden University Medical Center (N.L., V.v.U., N.G., F.K.), The Netherlands Organization for Scientific Research (NWO Applied Technical Sciences grant 12721 and ZonMW grant 91112008) (T.A., T.H., J.E., B.P.F.L.), the Russian Science Foundation (grant 16-15-00149), (S.A.K., E.S.E., M.I., O.V.B, D.M.C.), the Wellcome Trust (grant 100326/Z/12/Z) (K.L., J.E.M., D.A.P), China Scholarship Council (N.L. and N.G.) and the European Commission under a MSCA-ITN award (grant number 675743/ISPIC) (T.A.).
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Life Sciences Reporting Summary linked to this article.
Data availability
Mass cytometry data are available via Flow Repository (https://flowrepository.org/id/FR-FCM-ZYRD). scRNA-seq data are available via Gene Expression Omnibus accession code GSE122846. The remaining data that support the findings of this study are available from the corresponding author upon reasonable request.
ORCID: Frits Koning: 0000-0002-4007-5715
Author Contributions
N.L., V.v.U. and F.K. conceived the study and wrote the manuscript. N.L. performed most of the experiments with help from V.v.U. and N.G.. N.L. performed most of the data analyses with help from V.v.U., T.A. and B.P.F.L.. S.A.K., K.L., J.E.M., E.S.E., M.I., D.M.C., O.V.B. and D.A.P. performed TCR repertoire analyses. D.A.P. revised the manuscript. N.F.C.C.d.M. helped with imaging-mass cytometry experiments. T.H., V.v.U., J.E., and B.P.F.L. developed Cytosplore. S.M.C.d.S.L. provided human fetal tissues. All authors discussed the results and helped prepare the final manuscript.
Competing Interests
The authors declare no competing financial interests.
References
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Fan X, Rudensky AY. Hallmarks of Tissue-Resident Lymphocytes. Cell. 2016;164:1198–1211. doi: 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thome JJC, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat Med. 2015;22:72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunders MJ, et al. Memory CD4(+)CCR5(+) T cells are abundantly present in the gut of newborn infants to facilitate mother-to-child transmission of HIV-1. Blood. 2012;120:4383–4390. doi: 10.1182/blood-2012-06-437566. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci Transl Med. 2014;6:238ra72. doi: 10.1126/scitranslmed.3008748. [DOI] [PubMed] [Google Scholar]
- 6.Stout MJ, et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208:226.e1–7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aagaard K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiulio DB. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med. 2012;17:2–11. doi: 10.1016/j.siny.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Ardissone AN, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornef M, Penders J. Does a prenatal bacterial microbiota exist? Mucosal Immunol. 2017;10:598–601. doi: 10.1038/mi.2016.141. [DOI] [PubMed] [Google Scholar]
- 12.Li N, et al. Mass cytometry reveals innate lymphoid cell differentiation pathways in the human fetal intestine. J Exp Med. 2018;215:1383–1396. doi: 10.1084/jem.20171934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Unen V, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun. 2017;8:1740. doi: 10.1038/s41467-017-01689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 15.Höllt T, et al. Cytosplore: Interactive Immune Cell Phenotyping for Large Single-Cell Datasets. Comput Graph Forum. 2016;35:171–180. [Google Scholar]
- 16.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimmig S, et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J Exp Med. 2002;195:789–794. doi: 10.1084/jem.20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angerer P, et al. destiny – diffusion maps for large-scale single-cell data in R. Bioinformatics. 2015;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 19.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nat Methods. 2016;13:493–496. doi: 10.1038/nmeth.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolliffe I. International Encyclopedia of Statistical Science. 2011. Principal Component Analysis; pp. 1094–1096. [Google Scholar]
- 21.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzam HS, et al. CD5 Expression Is Developmentally Regulated By T Cell Receptor (TCR) Signals and TCR Avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T Cell-Positive Selection Uses Self-Ligand Binding Strength to Optimize Repertoire Recognition of Foreign Antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persaud SP, Parker CR, Lo W-L, Scott Weber K, Allen PM. Intrinsic CD4 T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol. 2014;15:266–274. doi: 10.1038/ni.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulton RB, et al. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol. 2015;16:107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shugay M, et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egorov ES, et al. The Changing Landscape of Naive T Cell Receptor Repertoire With Human Aging. Front Immunol. 2018;9:1618. doi: 10.3389/fimmu.2018.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 30.Kidera A, Konishi Y, Oka M, Ooi T, Scheraga HA. Statistical analysis of the physical properties of the 20 naturally occurring amino acids. J Protein Chem. 1985;4:23–55. [Google Scholar]
- 31.Rackovsky S. Global characteristics of protein sequences and their implications. Proc Natl Acad Sci U S A. 2010;107:8623–8626. doi: 10.1073/pnas.1001299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezzotti N, et al. Approximated and User Steerable tSNE for Progressive Visual Analytics. IEEE Trans Vis Comput Graph. 2017;23:1739–1752. doi: 10.1109/TVCG.2016.2570755. [DOI] [PubMed] [Google Scholar]
- 33.Fergusson JR, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014;9:1075–1088. doi: 10.1016/j.celrep.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edfors F, et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol. 2016;12:883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McFarland RD, Douek DC, Koup RA, Picker LJ. Identification of a human recent thymic emigrant phenotype. Proc Natl Acad Sci U S A. 2000;97:4215–4220. doi: 10.1073/pnas.070061597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staton TL, et al. CD8+ recent thymic emigrants home to and efficiently repopulate the small intestine epithelium. Nat Immunol. 2006;7:482–488. doi: 10.1038/ni1319. [DOI] [PubMed] [Google Scholar]
- 37.Michaelsson J, Mold JE, McCune JM, Nixon DF. Regulation of T Cell Responses in the Developing Human Fetus. The Journal of Immunology. 2006;176:5741–5748. doi: 10.4049/jimmunol.176.10.5741. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y, et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature. 2015;528:132–136. doi: 10.1038/nature16141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allenspach EJ, et al. Absence of functional fetal regulatory T cells in humans causes in utero organ-specific autoimmunity. J Allergy Clin Immunol. 2017;140:616–619.e7. doi: 10.1016/j.jaci.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez de Agüero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 41.van Unen V, et al. Mass Cytometry of the Human Mucosal Immune System Identifies Tissue- and Disease-Associated Immune Subsets. Immunity. 2016;44:1227–1239. doi: 10.1016/j.immuni.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bean DM, et al. esyN: network building, sharing and publishing. PLoS One. 2014;9:e106035. doi: 10.1371/journal.pone.0106035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol. 2011;Chapter 10:Unit10.33. doi: 10.1002/0471142735.im1033s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefranc M-P, et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 49.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.