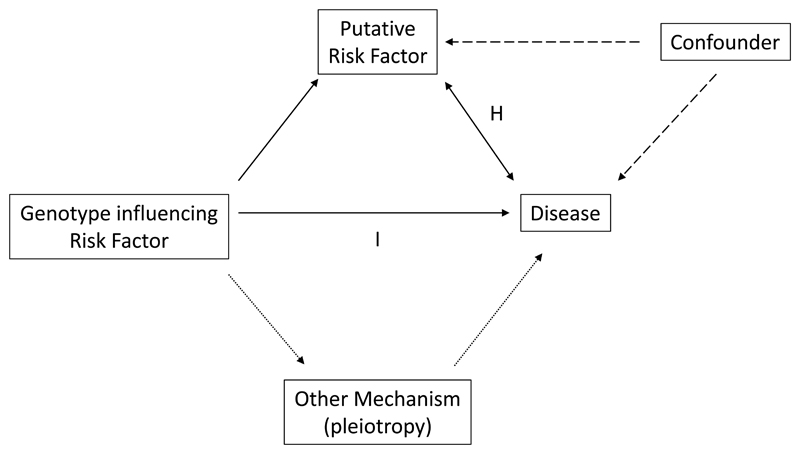
It is now nearly twenty years since my colleagues and I used the approach known as “Mendelian Randomisation” to investigate whether the association between plasma fibrinogen levels and myocardial infarction (MI), consistently documented in observational epidemiological studies, was causal, in a genetic study of around 5000 MI cases and 5000 controls.1 Since then, the approach has become ubiquitous in genetic epidemiology. The implementation of the method built upon papers by Katan, and by Gray and Wheatley, which originally highlighted the potential for exploiting the essentially unchanging nature through life of the individual’s genome, to overcome the potential effects of confounding and reverse causality in observational epidemiology.2, 3 Excellent reviews of the approach are available and increasingly sophisticated statistical approaches and study designs have been developed.4 At base, however, the concept is simple. If we can identify an allele which is associated with a change in a proposed disease risk factor, then provided the allele does not also affect disease risk in another way (termed pleiotropy), the association between the allele and the disease of interest should be predictable from the two composite associations: between allele and risk factor; and between risk factor and disease. The random segregation of alleles at meiosis mirrors a randomised controlled trial – individuals are “randomised” by genotype to different levels of the risk factor of interest, and hence, if the risk factor is causal, to congruent differences in disease risk. This method should be robust to confounding and able to distinguish true from reverse causality (Figure 1).
The relationships involved in a “Mendelian randomisation” study. The relationship of interest (arrow marked by H, the hypothesis) is between a putative risk factor and disease – or, as in the accompanying paper, between two disease phenotypes. This may be causal, reverse causal (indicated by bidirectional arrow), or confounded by one or more unmeasured factors (dashed arrows). In the absence of other genotype-mediated mechanisms, termed pleiotropy (dotted arrows), genotypes influencing levels of the putative risk factor should show a congruent association with disease if the factor-disease association is causal (arrow marked I, the relationship between the genetic “instrument” and disease).
“Mendelian randomisation” based investigations into certain putative risk factors for coronary artery disease have been revealing. In the case of fibrinogen and C-reactive protein, substantial causal associations have been essentially ruled out. By contrast, in the case of lipoprotein(a), a risk factor with complex epidemiology - exhibiting within-group association with coronary disease but higher levels in some lower-risk groups (eg those of Black ethnicity and women) - “Mendelian randomisation” provided key evidence in favour of a causal association.5 The approach has been extended to other uses – for example to genetically predict the side-effects of medications;6 and to investigate the effect on disease risk of environmental factors whose effect are mirrored by genotypes (for example folate deficiency and the MTHFR C677T genetic variant).7
The identification of hundreds of risk loci for cardiovascular and metabolic disease using genome-wide association study (GWAS) approaches has provided many genetic “instruments” which can be aggregated for use in “Mendelian randomisation” studies. In the accompanying paper, Sun and colleagues8 harness the power of the UK Biobank study to address a question of causality regarding hypertension and Type II diabetes. UK Biobank (www.ukbiobank.ac.uk) enrolled British volunteer participants aged 40-69 between the years 2006 and 2010. In addition to baseline phenotyping, volunteers agreed that details of their health records, held by the United Kingdom’s National Health Service, would be made available to UK Biobank for research purposes. In an audacious scientific move, UK Biobank carried out genome-wide genotyping for common genetic variants (single nucleotide polymorphisms, or SNPs) on all 500,000 of its volunteer participants. UK Biobank data is made available to the research community without preferential access, to any bona fide scientist from the academic or commercial sector, at minimal cost. Genetic studies in the UK Biobank resource have resulted, over the last couple of years, in the identification of many new genetic loci for coronary artery disease, hypertension, and obesity.9, 10
By contrast with early “Mendelian randomisation” studies, which used single or a small number of genetic polymorphisms, Sun et al. were able to deploy sets of 134 SNPs associated with Type II diabetes at conventionally accepted genome-wide levels of significance, and 233 SNPs similarly strongly associated with hypertension in previous genome-wide association studies, as their “genetic instruments”. They applied an approach termed “bidirectional Mendelian randomisation”. Here, the questions were (1) whether the SNPs robustly associated with Type II diabetes also predict hypertension (suggesting causality in one direction) and whether the SNPs associated with hypertension also predicted Type II diabetes (hence bidirectionality), among 318,664 UK Biobank participants of whom 13,931 had Type II diabetes and 172,344 had hypertension. Such a “bidirectional” approach has previously been used to investigate the direction of the “causal arrow” in a number of relationships, for example between adiposity and inflammation.11 A variety of sophisticated statistical approaches were adopted by Sun et al., in particular to attempt to detect and adjust for pleiotropy. Sun et al. conclude that participants at higher genetic risk of diabetes were also at higher risk of hypertension, though this increase in risk was of small magnitude, estimated at 7%. When considered quantitatively, the increase in blood pressure accompanying genetically instrumented diabetes was 0.67mmHg in systolic blood pressure, with no difference in diastolic blood pressure. In respect of the accuracy of the quantitative blood pressure analyses, it is worth noting that ~20% of participants were taking antihypertensive medication, and an across-the-board adjustment to their BP was made by adding 15mmHg to measured systolic and 10mm to measured diastolic values respectively. The effect of genetically instrumented Type II diabetes on hypertension was much smaller than the magnitude of the association between the phenotypes in observational data, both from UK Biobank and previous studies; among the possible explanations for this is that the “Mendelian randomisation” method was better able to avoid inflation of the estimated relationship due to confounding variables. By contrast, Sun et al. found no evidence in favour of the hypothesis that genetically instrumented hypertension caused diabetes; larger than 4% increases in risk were ruled out with 95% confidence.
This paper contributes large-scale genetic data to the ongoing debate as to whether diabetes causes hypertension, or vice versa, which has not been entirely resolved by observational epidemiology. Regarding pathophysiological mechanisms, Sun et al. speculate that the association of diabetes genotypes with systolic rather than diastolic blood pressure may result from effects of the pro-diabetes alleles on arterial stiffness. This hypothesis could also be tested in UK Biobank, as participants had arterial stiffness measured. Regarding the population health implications of their work, Sun et al. state that their findings illustrate the importance of an optimal population glycemic profile, and of blood pressure monitoring in patients with type II diabetes. This view, entirely correct in my opinion, is very much in line with current recommendations. It perhaps highlights a gap that might be perceived between the scientific elegance of some “Mendelian randomisation” studies and their capacity, thus far, to deliver results that change clinical practice.
Finally, what can be said about this study from the point of view of reproducibility and data-sharing? One of the stipulations of using UK Biobank data is that results, derived data fields, and analytical code used to generate published results are to be returned to UK Biobank within six months of publication. Thus, in due course readers of Circulation Research will be empowered to check the working of Sun et al. for themselves, and confirm, refute or expand their conclusions – the ultimate guarantee.
Sources of Funding Statement
BK is supported by a British Heart Foundation Personal Chair
Footnotes
Conflict of Interest Statement:
I have had a long-term involvement in the UK Biobank study and am currently Chair of the International Scientific Advisory Board. I have no financial conflict of interest in respect of this or any other activity.
References
- 1.Youngman LD, Keavney BD, Palmer A, Parish S, Clark S, Danesh J, Delepine M, Lathrop M, Peto R, Collins R. Plasma fibrinogen and fibrinogen genotypes in 4685 cases of myocardial infarction and 6002 controls: Test of causality by “Mendelian randomisation”. Circulation. 2000;102:31–32. [Google Scholar]
- 2.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet (London, England) 1986;1:507–8. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 3.Gray R, Wheatley K. How to avoid bias when comparing bone marrow transplantation with chemotherapy. Bone marrow transplantation. 1991;7(Suppl 3):9–12. [PubMed] [Google Scholar]
- 4.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human molecular genetics. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. The New England journal of medicine. 2009;361:2518–28. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 6.Martin RI, Pogoryelova O, Koref MS, Bourke JP, Teare MD, Keavney BD. Atrial fibrillation associated with ivabradine treatment: meta-analysis of randomised controlled trials. Heart (British Cardiac Society) 2014;100:1506–10. doi: 10.1136/heartjnl-2014-305482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamasoula C, Prentice RR, Pierscionek T, Pangilinan F, Mills JL, Druschel C, Pass K, Russell MW, Hall D, Topf A, Brown DL, et al. Association between C677T polymorphism of methylene tetrahydrofolate reductase and congenital heart disease: meta-analysis of 7697 cases and 13,125 controls. Circulation Cardiovascular genetics. 2013;6:347–53. doi: 10.1161/CIRCGENETICS.113.000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, Zhou T, Heianza Y, Li X, Fan M, Fonseca VA, Qi L. Type 2 Diabetes and Hypertension: A Study on Bidirectional Causality. Circulation Research. 2019;124 doi: 10.1161/CIRCRESAHA.118.314487. xxx-xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 11.Welsh P, Polisecki E, Robertson M, Jahn S, Buckley BM, de Craen AJ, Ford I, Jukema JW, Macfarlane PW, Packard CJ, Stott DJ, et al. Unraveling the Directional Link between Adiposity and Inflammation: A Bidirectional Mendelian Randomization Approach. Endocr Rev. 2009;30:927–928. doi: 10.1210/edrv.30.7.9996. [DOI] [PubMed] [Google Scholar]