Abstract
Purpose
Myeloma is a plasma cell malignancy characterized by the overproduction of immunoglobulin and is therefore susceptible to therapies targeting protein homeostasis. We hypothesized that heat shock factor 1 (HSF1) was an attractive therapeutic target for myeloma due to its direct regulation of transcriptional programs implicated in both protein homeostasis and the oncogenic phenotype. Here, we interrogate HSF1 as a therapeutic target in myeloma using bioinformatic, genetic and pharmacological means.
Experimental design and results
To assess the clinical relevance of this novel target, we analyzed publicly available gene expression datasets and found that expression of HSF1 and its target genes were associated with poorer myeloma patient survival. Sh-RNA-mediated knockdown or pharmacological inhibition of the HSF1 pathway with a novel chemical probe, CCT251236, or with KRIBB11, led to caspase-mediated cell death that was associated with an increase in EIF2α phosphorylation, CHOP expression and a decrease in overall protein synthesis. Importantly, both CCT251236 and KRIBB11 induced cytotoxicity in human myeloma cell lines and patient-derived primary myeloma cells with a therapeutic window over normal cells. Pharmacological inhibition induced tumor growth inhibition and was well-tolerated in a human myeloma xenograft murine model with evidence of pharmacodynamic biomarker modulation.
Conclusion
Taken together, our studies demonstrate the dependence of myeloma cells on HSF1 for survival and support the clinical evaluation of pharmacological inhibitors of the HSF1 pathway in myeloma.
Keywords: HSF1, multiple myeloma, ER stress
Introduction
Myeloma is a plasma cell neoplasm of the bone marrow that is characterized by the sustained secretion of large amounts of immunoglobulin (Ig). Myeloma cells therefore rely on intracellular protein homoeostasis mechanisms for survival [1]. This is highlighted by the clinical success of proteasome inhibitors that disable the ubiquitin-proteasome pathway [2]. Interestingly, one of the mechanisms by which bortezomib induces apoptosis is through the activation and overloading of a different protein homoeostasis pathway, the unfolded protein response (UPR) [3–5]. It can therefore be envisaged that myeloma would be susceptible to therapies targeting various protein handling mechanisms [6–7]. Indeed, there is current interest in the inhibition of these types of targets for myeloma, such as IRE1α in the UPR [8].
Another avenue for inducing proteotoxic stress is to target the heat shock response (HSR) and molecular chaperones. Although tanespimycin (17-AAG), a heat shock protein 90 (HSP90) inhibitor, showed convincing pre-clinical anti-myeloma efficacy, only modest responses were observed in clinical trials [9–10]. This is attributable to the upregulation of anti-apoptotic heat shock protein 70 protein 1 (HSP72) and other heat shock proteins (HSPs) following tanespimycin treatment by heat shock factor 1 (HSF1), the master transcription factor and regulator of the HSR [11–14]. A similar induction of HSPs and HSF1 activation is also observed following bortezomib treatment, suggesting that activation of the HSR limits the efficacy of a number of current myeloma drugs [6,15].
In addition to regulating HSP expression, HSF1 is indispensable for oncogenic transformation and cancer cell survival [16, 17]. Additional studies have implicated HSF1 in other aspects of tumor progression such as cell migration and angiogenesis [18–20]. Furthermore, higher levels of nuclear HSF1 are found in breast, kidney and oral cancers compared with normal tissues, with links to poor patient prognosis and increased metastasis [19, 21–22]. Extensive ChIP-seq experiments in breast cancer revealed that HSF1 promotes oncogenesis and cell survival through regulating a cancer-specific transcriptional program [23]. Not only is HSF1 strongly bound to its target genes in breast and colon tumors with high nuclear HSF1 levels, but the cancer-specific HSF1 gene expression profile is also a prognostic indicator for poor outcome [23].
With accumulating evidence for HSF1 and its transcriptional program in maintaining the malignant phenotype as well as its role in protein homeostasis, inhibition of HSF1 looks to be a promising therapeutic strategy [24]. Although several inhibitors of HSF1 have been reported, many of these have major limitations and none have progressed to the clinic [25]. Our own drug discovery efforts have resulted in a highly potent and selective inhibitor of the HSF1 stress pathway, CCT251236, which shows therapeutic activity at well-tolerated doses in human ovarian cancer xenografts [26]. In a similar fashion to our own phenotypic screen [26], Yoon et al. [27] have also identified an inhibitor of the HSF1 pathway, KRIBB11, which is well-tolerated in-vivo. Both these compounds facilitate the in-vitro and pre-clinical exploration of targeting HSF1-mediated transcription for myeloma therapy.
The role of the HSF1 pathway in hematological cancers is relatively unexplored compared with solid cancers. Given the evidence for HSF1 in mediating protein homeostasis and oncogenesis, we hypothesized it may be a good myeloma therapeutic target. Here, we describe the prognostic significance of HSF1 expression and demonstrate that shRNA-mediated knockdown of HSF1 in human myeloma cell lines (HMCLs) leads to a downregulation of global protein synthesis, activation of the UPR and caspase-mediated cell death. Utilizing CCT251236 and KRIBB11 as tool compounds, we show anti-myeloma activity in a human myeloma xenograft model and a potential therapeutic window for HSF1 pathway inhibition using primary patient-derived myeloma cells and peripheral blood mononuclear cells (PBMCs).
Materials and Methods
Expression and survival analysis
Expression data from CD138+ plasma cells (n=262), collected from relapsed patients enrolled in APEX, SUMMIT and CREST trials were examined (GEO accession GSE9782) [28]. Data for newly-diagnosed patients were obtained from the following clinical trials: Myeloma IX (n=258; GSE21349), Total Therapy 2/3 (n=559; GSE2658) and HOVON/GMMG-HD4 (n=320; GSE19784). Patients were separated into high and low HSF1 expression (Affymetrix probeset 202244_at) using the partitioning around medoids algorithm in R. Kaplan-Meier overall survival (OS) curves were generated and log-rank tests carried out using the R survival package. Hazard ratios (HR) and 95% confidence intervals (CI) were computed using a univariate Cox proportional hazards model in SPSS (IBM). To evaluate the impact of HSF1 target gene expression on OS, expression of genes in the cancer-specific HSF1 signature [23] was analyzed (456 genes; 793 probesets). Probesets with <5 samples with expression values >200 were removed. Data were min-max normalized and probeset intensities with low variance (<200) were discarded. Hierarchical clustering was performed on the remaining 359 probesets (Ward method) and heatmaps were generated using the R hclust and gplots packages. Further analysis was performed on RNA-seq data from CD138+ plasma cells from the MMRF CoMMpass trial (NCT01454297).
Cell lines
RPMI-8226, NCI-H929, U266, HEK293T/17 and HS-5 were purchased from ATCC. KMS-11 and MOLP-8 were a kind gift from Professor H. Johnsen (Aarhus University Hospital, Denmark). GFP-tagged bone marrow stromal cells, HS-5-GFP, were generated as previously described [29]. HEK293T/17 and HS-5-GFP cells were cultured in DMEM containing GlutaMAX™ and 10% FBS (Life Technologies). All other cells were cultured in RPMI-1640 containing GlutaMAX™ and 10% FBS. All cells tested negative for mycoplasma by PCR and were authenticated by STR analysis.
Primary cells
PBMCs were obtained from healthy donors. Patient primary myeloma cells were isolated from bone marrow aspirates by density gradient centrifugation using Ficoll-Paque Premium (GE Healthcare) according to manufacturer’s instructions. CD138+ cells were purified using CD138 Microbeads (Miltenyi Biotech) to a purity of >95%. All procedures were performed following informed consent. Approval for these studies was obtained from the Royal Marsden Hospital Review Board (CCR4238) and the Health Research Authority National Research Ethics Service Committee (14/YH/1317).
Compounds and plasmids
CCT251236 was synthesized as described [26]. Compounds were purchased as described: KRIBB11 (Merck Millipore), bortezomib (Cambridge Bioscience), Z-VAD-FMK and puromycin (InvivoGen), pactamycin and tunicamycin (Sigma). Plasmids were purchased or obtained as described: HSF1 shRNA pLKO.1 plasmids (Thermo Fisher; TRCN0000007480, TRCN0000007484), pLKO.1 empty vector, pLKO.1 GFP shRNA, pCMV-R8.72 lentiviral packaging and pCMV-VSV-G envelope plasmid (Addgene; plasmid ID 10878, 30323, 22036 and 8454).
Lentiviral production and transduction of HMCLs
Plasmids were propagated in bacterial cultures and purified using the PureLink HiPure Plasmid Filter Maxiprep Kit. 2x107 HEK293T/17 cells were transfected using the calcium phosphate method with a mixture of 16μg pCMV-R8.74, 5μg pCMV-VSV-G and 20μg of pLKO.1 empty vector, pLKO.1 GFP shRNA or HSF1 shRNA pLKO.1 and 125mM CaCl2 (Sigma) in 500μl nuclease-free water and 500μl HEPES-buffered saline pH 7.05. Conditioned medium was collected at 48-72 hours post-transduction and concentrated using the Lenti-X Concentrator Kit (Clonetech) according to manufacturer’s instructions. Up to 1ml of concentrated virus-containing media and 8μg/ml polybrene (Sigma) were used for the transduction of 5x106 cells.
Quantitative-PCR (q-PCR)
RNA was recovered from HMCLs using the RNeasy Mini Kit (Qiagen) and cDNA was synthesized from 50ng RNA using qSCRIPT cDNA Super Mix (Quanta Biosciences) according to manufacturer’s instructions. cDNA was diluted 1:10 in water in 25μl reaction volume with SYBR® Green Master Mix. The q-PCR cycling conditions were 15 sec at 95°C and 1 min at 60°C using the 7500 Fast Real-Time PCR System and analysis was carried out using the software (Applied Biosystems). The comparative CT method was used for the relative quantitation of cDNA, where GAPDH was used as the housekeeping gene. PCR primers used: HSP27 forward (300nM) 5’-CTGATGAAGGGGAAGCAGG-3’ and reverse (300nM) 5’GACGACTTTCTGTTGCTGGG-3’, CHOP forward (300nM) 5′-TGGAAATGAAGAGGAAGAATCAAAA-3′ and reverse (900nM) 5′-CAGCCAAGCCAGAGAAGCA-3′; GAPDH forward (500nM) 5’-GAAGGTGAAGGTCGGAGTC-3’ and reverse (500nM) 5’-GAAGATGGTGATGGGATTTC-3’. Positive controls for CHOP q-PCR were generated from cells treated with 10μg/ml tunicamycin for 4 hours.
Protein extraction
Proteins were harvested from cells with RIPA lysis buffer supplemented with protease inhibitor cocktail (Roche), PMSF and 1mM Na3VO4. Excised xenograft tumors were suspended in CHAPS lysis buffer in hard tissue grinding reinforced tubes with stainless steel beads (Precellys) and processed using a Precellys® 24 tissue homogenizer. Insoluble material and fatty tissue were removed by centrifugation at 16000 x g for 10 mins at 4°C, twice.
SDS-PAGE and Western blotting
Proteins were separated by SDS-PAGE on NuPAGE Bis-Tris gels (Life Technologies) and transferred onto PVDF membranes using the iBlot® Dry Blotting System (Life Technologies). Membranes were blocked with 5% milk or BSA in TBS with 0.1% Tween-20 and incubated with primary antibody overnight. Primary antibodies used were HSF1 (Cell Signaling, #4356), HSF1 phospho-Ser326 (Abcam, 76076), HSP72 (Enzo Life Science, ADI-SPA-810), HSP27 (Cell Signaling, #2402) PARP (Cell Signaling, #9542), eIF2α (Cell Signaling, #9722), eIF2α phospho-Ser51 (Cell Signaling, #9721) and GAPDH (Santa Cruz, 25778). Membranes were incubated with HRP-linked anti-rabbit IgG (Cell Signaling, #7072) or anti-mouse IgG (Cell Signaling, #7076) for 1 hour. ECL proteins were detected using Primer Western Blotting Detection Reagent (GE Healthcare) and visualized on Kodakκ® BioMax® XAR Film.
For HSF1 knockdown studies, proteins were collected 72 hours post-transduction. For CCT251236 and KRIBB11 treatment studies, proteins were collected as indicated. Quantitative densitometry analysis of protein bands was carried out using ImageJ software.
Light chain enzyme-linked immunosorbent assay (ELISA)
Ig light chain ELISA was performed as described previously [3] and quantified using the lambda (λ) or kappa (κ) ELISA Quantitation Kit (Bethyl Laboratories) according to manufacturer’s instructions. In-vitro secreted light chain was measured from conditioned media (1:100 dilution) collected from 3x106 cells following 24 hour culture. Intracellular light chain was measured from 500ng cell lysate protein collected. Plasma samples from mice bearing xenograft tumors and age-matched non-tumor bearing mice (as controls) were tested at 1:100 dilution. For HSF1 knockdown studies, cells were plated 72 hours post-transduction.
Puromycin protein synthesis assay
3x105 cells were plated following 72 hours transduction for 4 hours and treated with 10μM puromycin for 10 mins prior to whole cell lysis. Cells pre-treated with 10μM pactamycin, a ribosome inhibitor, were used as a control. SDS-PAGE and Western blotting was performed on lysates as described previously.
Cell proliferation and Caspase-Glo assay
The growth inhibitory response to CCT251236 and KRIBB11 were measured using WST-1 reagent (Roche) or CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS) (Promega), respectively. The Caspase-Glo 3/7 luminescent assay (Promega) was used to measure caspase activity. Absorbance was measured on an Epoch Microplate Spectrophotometer (Bio-Tek) and luminscence on a Mithras LB940 Microplate Reader (Berthold Technologies) according to manufacturer’s instructions. Cells were plated at 2x105 or 5x103 cells per well for 48-96 hours in 96 well-plates. Cells treated with 5nM bortezomib for 24 hours were used as positive controls for caspase activity.
Detection of cell viability by Annexin V/propidium iodide (PI) staining
Cells in Annexin V Binding Buffer (BD Biosciences) were stained with 2.5μg/ml PI Staining Solution and 5μl Annexin V-APC for 15 minutes and analyzed a BD LSR II™ flow cytometer. For HSF1 knockdown studies, cells were stained 72 hours following transduction. For rescue of cell death, cells were treated with 50μM Z-VAD-FMK for 24 hours prior to staining. In HSF1 pathway inhibitor studies, 3x105 cells were treated with 48 hour GI25 or GI50 concentrations of CCT251236 or KRIBB11. For patient cell studies, 5x105 CD138+ cells were co-cultured with 1x104 HS-5-GFP cells and treated with CCT251236 or KRIBB11 for 48 hours prior to staining with Annexin V-APC and DAPI (BD Biosciences). GFP-positive and negative cells were gated and analyzed separately to distinguish between stromal HS-5-GFP and myeloma cells.
Human myeloma xenograft model
Myeloma xenograft tumors were established subcutaneously (s.c.) in female NOD/SCIDγcnull mice (Charles River). 5x106 H929 cells in 100% Matrigel (BD Biosciences) were injected in a single flank. Mice with tumors that reached a mean diameter of 0.6-0.7 mm were randomly grouped for initiation of treatment (day 0) with daily intraperitoneal (i.p.) administration of KRIBB11 at 65mg/kg or vehicle (10% dimethylacetamide, 50% polyethylene glycol-300, 40% sterile water [27]), or CCT251236 at 20mg/kg orally (p.o.) or vehicle (10% DMSO, 90% 2-hydroxypropyl-β-cyclodextrin). Tumor volumes and mouse body weights were determined at regular intervals. At the end of treatment (day 18-19) 16 hours after the final dose, plasma samples were taken and tumors removed, weighed and extracted for proteins. The study was performed in accordance with UK Home Office regulations under the Animals Scientific Procedures Act 1986 and in accordance with UK National Cancer Research Institute guidelines [30].
Results
HSF1 and its transcriptional targets are associated with patient survival
HSF1 expression correlates with malignancy and poor prognosis in a number of solid tumors [18–19, 21–23]. To explore the pathobiological relevance of HSF1 in myeloma, we first assessed HSF1 expression levels in HMCLs. HSF1 expression by Western blot varied across the cell panel, with cell lines expressing higher HSF1 levels also generally showing higher expression of Ser326 HSF1 phosphorylation, an indicator of HSF1 transcriptional activity [31] (Supplementary Figure S1). Expression of HSP72 and HSP27 that are regulated by HSF1 showed considerable variation between HMCLs.
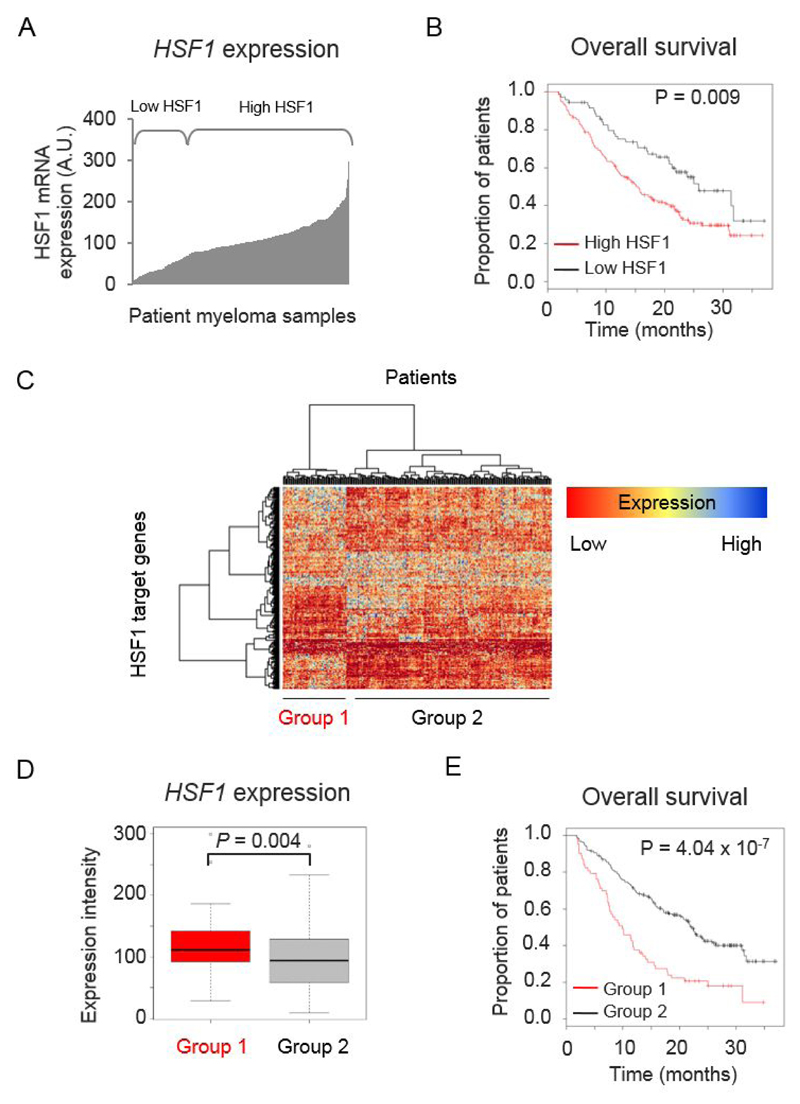
To determine whether HSF1 is a clinically relevant prognostic factor in myeloma, we looked for the impact of HSF1 expression on patient survival using publicly available gene expression profiling (GEP) datasets. HSF1 mRNA levels were assessed in CD138+ plasma cells isolated from newly-diagnosed (Supplementary Figure S2) and relapsed patients (Figure 1A, 1B). In the relapsed dataset [28], high HSF1 expression correlated with poorer OS (Figure 1B) (log rank P=0.009; Cox regression P=0.014, HR=1.749, CI=1.098-2.785). Although this was not observed in the GEP of newly-diagnosed patient datasets (Supplementary Figure S2A-C), we found that high HSF1 expression was significantly associated with poorer OS in an independent dataset derived from RNA-sequencing of newly-diagnosed patients (Supplementary Figure S2D). These findings suggest that HSF1 stress response proteins have an important role in myeloma.
Figure 1. HSF1 expression has prognostic significance in relapsed myeloma.
(A) Distribution of HSF1 mRNA expression across CD138+ plasma cell samples from relapsed patients (n=264) in Mulligan et al. dataset (GSE9782). (B) Kaplan-Meier plot of patient overall survival (OS) (median survival: high HSF1 = 15.7 months (n=71), low HSF1 = 25.8 months (n=191)). (C) Heatmap and dendrogram representing hierarchical clustering of patient samples based on the expression of HSF1 Ca-Sig genes. (D) Boxplot representing relative HSF1 expression between group 1 and 2. P value was determined using the Mann-Whitney U test. (E) Kaplan-Meier plot of patient OS of Group 1 (n=63) and Group 2 (n=201) patients separated by clustering (median survival: Group 1 = 9.9 months, Group 2 = 22.2 months).
As HSF1 regulates oncogenic processes by driving a specific transcriptional program, we used the HSF1 cancer signature (HSF1 Ca-Sig) of 456 genes identified by Mendillo et al. [23] to determine whether differential expression of these HSF1 target genes can also be an indicator for patient prognosis. Hierarchical clustering of relapsed patient samples, based on expression of the HSF1 Ca-Sig, separated the patients into two groups with distinct patterns of HSF1 target gene expression (Figure 1C). Not only did group 1 patients have a higher expression of HSF1 Ca-Sig genes than group 2 (Figure 1D), but they also performed worse in terms of OS compared with group 2 (Figure 1E; log rank P=4.04x10-7). These results suggest HSF1 regulates the expression of a subset of HSF1 Ca-Sig genes that impact the clinical outcome of myeloma patients.
HSF1 knockdown leads to myeloma cell death through caspase-mediated apoptosis
In order to study the cellular function of HSF1 in myeloma, we established stable HSF1 knockdown models using RPMI-8226 and KMS-11 HMCLs. We achieved 80-90% depletion of HSF1 expression using two independent HSF1-targeted shRNAs (sh1 and sh2) in both HMCLs following 72 hours transduction as compared with empty vector and GFP-targeted shRNA transduced (shGFP) controls (Figure 2A). The downregulation of HSF1 phospho-Ser326, HSP72 and HSP27 was also observed, giving confidence that this model is useful for assessing both the functionality of HSF1 and its downstream transcriptional targets.
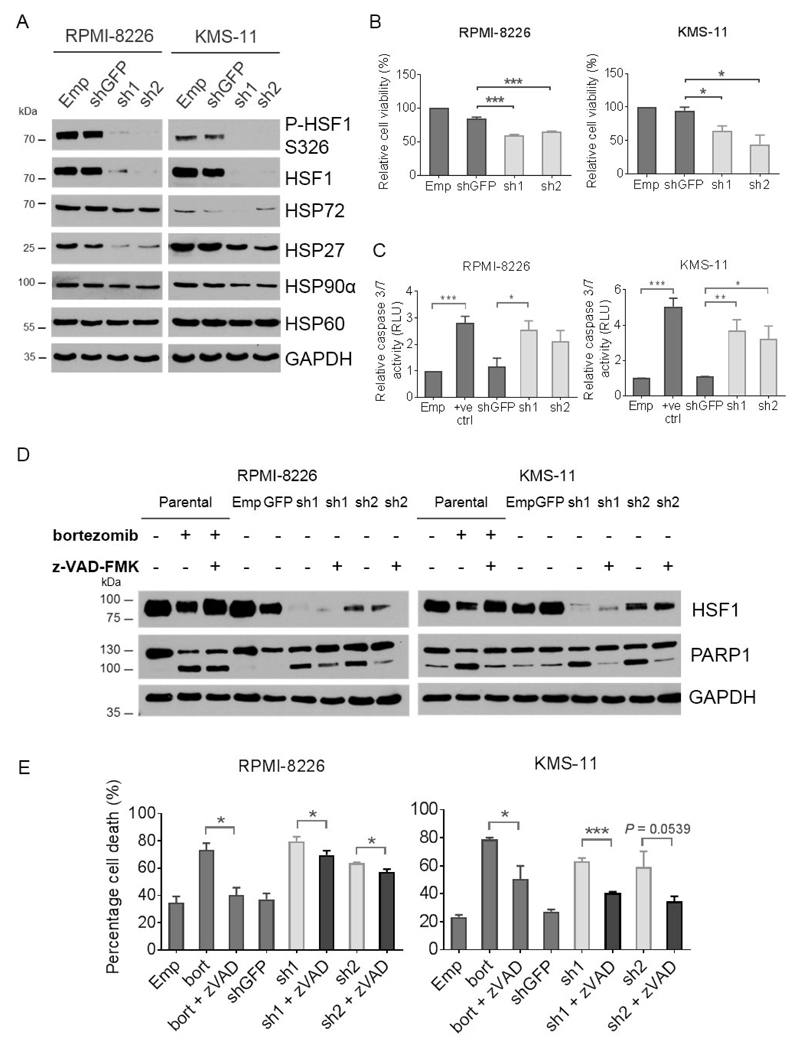
Figure 2. HSF1 silencing leads to caspase-mediated cell death.
(A) Western blot of whole cell lysates from human RPMI-8226 and KMS-11 myeloma cells following 72 hours transduction with HSF1 shRNA (sh1 and sh2) for the depletion of HSF1 expression, empty pLKO.1 vector (Emp) or GFP shRNA (shGFP) as controls. Membranes were probed with antibodies against HSF1, phospho-HSF1 (P-HSF1) at Ser236 (S326), HSP72, HSP27, and GAPDH as a loading control. (B) Viability of human HSF1 shRNA transduced cells. Cells were analyzed by flow cytometry following staining with Annexin V-APC and PI. Graphs represent Annexin V-APC and PI negative cells as a percentage of the Emp control. (C) Caspase 3/7 activity of HSF1 shRNA transduced cells was detected by the Caspase-Glo® assay. Graphs represent RLU relative to the Emp control. Empty vector transduced cells treated with 5 nM bortezomib for 24 hours were used as a positive control (+ve ctrl). (D) Western blot of whole cell lysates from HSF1 shRNA transduced cells for PARP cleavage following treatment with and without 50μM z-VAD-FMK for 24 hours. Parental cells treated with 5 nM bortezomib with and without z-VAD-FMK treatment were used as positive controls for PARP cleavage and inhibition of caspase activity, respectively. Western blotting was independently repeated twice. (D) Viability of HSF1 shRNA transduced cells with or without z-VAD-FMK treatment were analyzed by flow cytometry following Annexin V-APC and PI staining. Empty vector transduced cells treated with bortezomib and z-VAD-FMK were used as positive controls for the rescue of cell death (bort and bort + zVAD). Graphs represent Annexin V-APC and PI negative cells as a percentage of the Emp control. Data are shown as means ±S.E.M. of three independent experiments. Significant differences were calculated by Student’s t-tests and P-values are indicated where * ≤ 0.05, ** ≤ 0.01, and *** ≤ 0.001.
Knockdown of HSF1 has previously been shown to reduce the viability of various cancer cell lines [16, 32]. It was, therefore, of interest to assess myeloma cell survival following HSF1 silencing. Knockdown of HSF1 led to a marked reduction in cell viability (>40% decrease) compared with shGFP controls (Figure 2B). This was accompanied by an increase in caspase 3/7 activity (Figure 2C) and the cleavage of poly-ADP ribose polymerase (PARP) (Figure 2D), which is a downstream substrate of caspase 3. Concurrent treatment with a pan-caspase inhibitor (z-VAD-FMK) partially rescued PARP cleavage (Figure 2E) and decreased the population of dead cells showing Annexin V and PI-positive staining (Figure 2E). These data demonstrate that the depletion of HSF1 expression is associated with caspase-mediated apoptosis, indicating a dependence of myeloma cells on HSF1 for survival.
Pharmacological inhibitors of the HSF1 pathway have in-vitro activity on HMCLs
To evaluate the feasibility of HSF1 as a therapeutic target, two chemically distinct inhibitors of the HSF1 pathway were employed: CCT251236, our novel potent and selective small molecule inhibitor of the HSF1 pathway [26], and KRIBB11, a pharmacologically distinct inhibitor of the HSF1 pathway previously described [27]. Both CCT251236 and KRIBB11 led to a growth inhibitory effect in HMCLs (Supplementary Figure S3A). Using concentrations of CCT251236 and KRIBB11 corresponding to the 48 hour GI25 and GI50 (Supplementary Figure S3B), pharmacological inhibition of the HSF1 pathway led to a concentration and time-dependent increase in caspase 3/7 activity (Figure 3A) that was accompanied by a decrease in cell viability (Figure 3B).
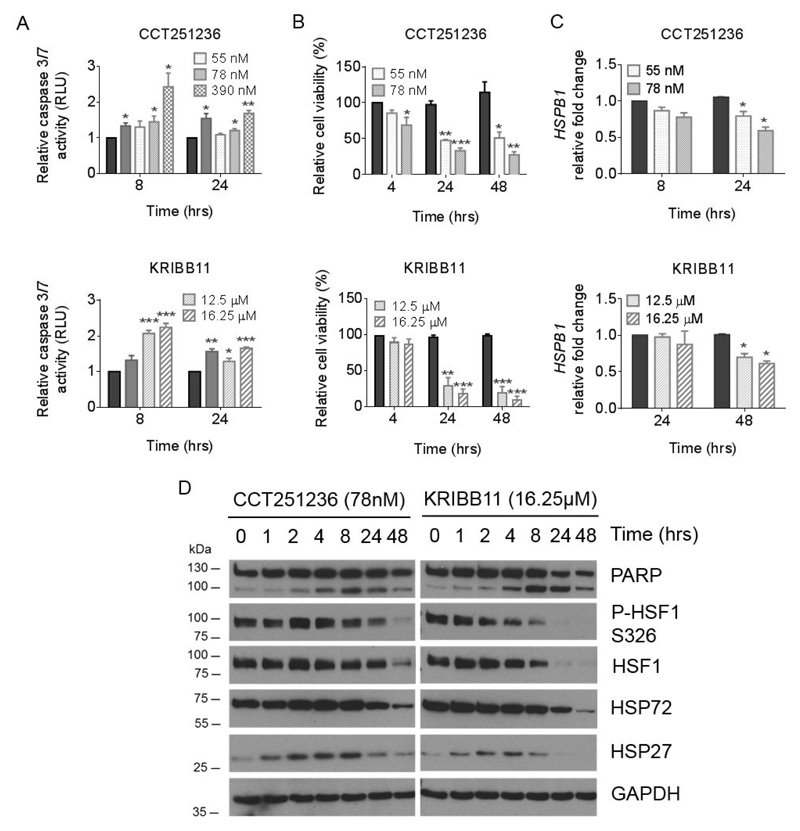
Figure 3. CCT251236 and KRIBB11 treatment induces caspase-mediated cell death concomitant with the downregulation of HSF1 target genes.
Human RPMI-8226 myeloma cells were treated with CCT251236 or KRIBB11 at concentrations corresponding to the 48 hour GI25 and GI50 values. (A) Caspase 3/7 activity was detected by the Caspase-Glo® assay. Graphs represent RLU relative to the vehicle-treated control. Cells treated with 5 nM bortezomib for 24 hours (solid grey bars) were used as positive controls for caspase activity. (B) Cells were analyzed by flow cytometry following staining with Annexin V-APC and PI over a 48 hour time course. Graphs represent Annexin V-APC and PI negative cells as a percentage of the vehicle-treated control. (C) HSPB1 mRNA expression normalized to GAPDH mRNA presented relative to vehicle-treated controls. For all bar graphs, solid black bars represent vehicle-treated controls at indicated time points. Data are shown as means ±S.E.M. of three independent experiments. Significant differences were calculated by Student’s t-tests and P-values are indicated where * ≤ 0.05, ** ≤ 0.01, and *** ≤ 0.001 compared with vehicle-treated control at the same time point. (D) Western blot of whole cell lysates from RPMI-8226 cells treated with KRIBB11 or CCT251236 over a 48 hour time course. Membranes were probed with antibodies against PARP, HSF1, phospho-HSF1 (P-HSF1) at Ser236 (S326), HSP72, HSP27, with GAPDH as a loading control.
To demonstrate on-pathway activity, levels of HSP27 (HSPB1) mRNA expression were assessed by qPCR. A time-dependent decrease in HSPB1 mRNA expression was observed following treatment (Figure 3C) that corresponded to the downregulation of HSP27 protein expression (Figure 3D). Downregulation of HSP72 expression and an increase in PARP cleavage were also observed in a time-dependent fashion (Figure 3D). Similar results were obtained in human KMS-11 and H929 cells in response to pharmacological inhibition of the HSF1 pathway (Supplementary Figure S4).
HSF1 knockdown or HSF1 pathway inhibition modulates protein homeostasis
Given that HSF1 and the HSPs play a pivotal role in protein folding, the consequence of HSF1 depletion on the Ig production capacity of myeloma cells was assessed using an Ig light chain ELISA. The ELISA was performed on cell supernatants and whole cell lysates to detect secreted and intracellular levels of Ig light chains from RPMI-8226 and KMS-11 cells. Loss of HSF1 was associated with a significant downregulation of secreted Ig light chains compared with shGFP controls (Figure 4A, Supplementary Figure S5). Interestingly, no significant differences were observed in the intracellular levels of Ig light chains. We had previously observed that silencing of HSC70 and HSP72 reduced the secretion but increased the retention of Ig light chains in HMCLs [33]. The lack of intracellular Ig accumulation in this model suggested that knocking down HSF1, which in turn alters the expression of several hundred HSF1-regulated genes, reduces overall Ig light chain synthesis.
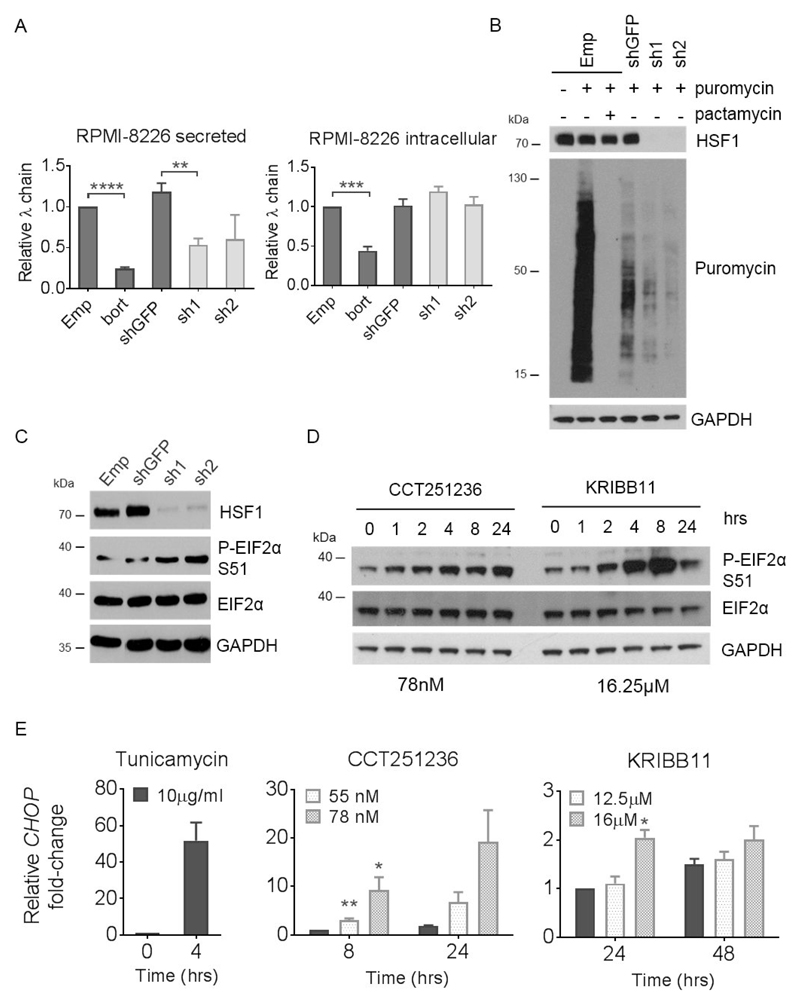
Figure 4. HSF1 silencing and HSF1 pathway inhibition leads to a downregulation of global protein synthesis concomitant with eIF2α phosphorylation and CHOP expression.
Human RPMI-8226 myeloma cells were transduced with HSF1 shRNA (sh1 and sh2) and empty pLKO.1 vector (Emp) or GFP shRNA (shGFP) as controls for 72 hours. (A) Relative levels of secreted and intracellular λ Ig light chains produced over a 24 hour period following HSF1 silencing were analysed by ELISA. shGFP cells treated with bortezomib (bort) for 24 hours were used as a positive control for a reduction in secreted and intracellular light chains. (B) Relative protein synthesis was determined by Western blot of cells treated with 4 μg/ml puromycin for 10 mins following HSF1 silencing. Cells pre-treated with 10 μM pactamycin, a ribosome inhibitor, for 15 minutes were used as controls for reduced protein synthesis. Membranes were probed with antibodies against puromycin and HSF1. (C) Levels of phospho-EIF2α (P-EIF2α) at Ser 51 (S51) from RPMI-8226 whole cell lysates following HSF1 silencing were also determined by Western blot. (D) Levels of P-EIF2α were also analysed by Western blotting of whole cell lysates extracted from RPMI-8226 cells treated with CCT251236 or KRIBB11 over a 48 hour timecourse. GAPDH was used as a loading control. (E) CHOP mRNA expression quantified by q-PCR from RPMI-8226 following 48 hours CCT251236 or KRIBB11 treatment. Cells treated with 10μg/ml tunicamycin for 4 hours were used as positive controls. Graphs represent CHOP expression fold-change relative to vehicle-treated controls. Data are shown as means ±S.E.M. of three independent experiments. Significant differences were calculated by Student’s t-tests and P-values are indicated where * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001 and **** ≤ 0.0001.
To assess whether protein synthesis is impaired by HSF1 knockdown, a protein translation assay based on puromycin incorporation into elongating polypeptide chains at the ribosome was used. Knockdown of HSF1 decreased the relative level of puromycin incorporation compared with shGFP transduced cells (Figure 4B, Supplementary Figure S5B). The level of puromycin incorporation observed was comparable to the positive control treated with pactamycin, a ribosome inhibitor. Interestingly, one of the signaling cascades that can block protein translation is activation of the PERK arm of the UPR. Specifically, eukaryotic initiation factor 2α (EIF2α) is phosphorylated at Ser 51 by PERK [34]. Consistent with this, knockdown of HSF1 (Figure 4C) as well as pharmacological inhibition of the HSF1 pathway using CCT251236 or KRIBB11 (Figure 4D, Supplementary Figure S5C) led to a marked increase in phospho-EIF2α. The transcript of CHOP, a downstream transcriptional target of PERK activation, was also upregulated by both agents (Figure 4E, Supplementary Figure S5D). These data suggest an additional downstream effect of HSF1 knockdown on other cellular protein homeostasis pathways.
Pharmacological inhibitors of HSF1 decrease viability of primary patient-derived myeloma cells while sparing PBMCs
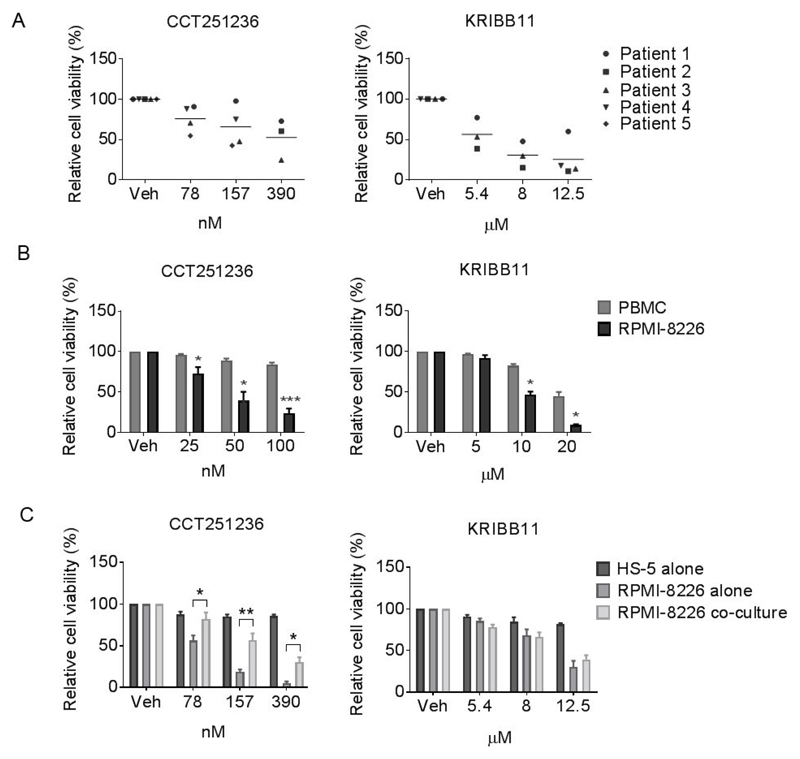
To gain insight into the potential therapeutic index of agents targeting HSF1, the anti-myeloma activity of CCT251236 and KRIBB11 was further assessed in-vitro using CD138+ cells isolated from myeloma patient bone marrows. A concentration-dependent decrease in cell viability was observed in these primary cells following 48 hour treatment (Figure 5A). We also examined the cytotoxicity of both compounds in PBMC and the bone marrow stromal cell line (BMSC), HS-5. Both CCT251236 and KRIBB11 decreased PBMC and HS-5 BMSC viability in a concentration-dependent fashion, but this was seen to a much lesser extent than in RPMI-8226 myeloma cells (Figure 5B, 5C). Importantly, when tested under conditions of the co-culture of RPMI-8226 with HS-5 BMSCs, there was still significant anti-myeloma activity of CCT251236. Although as seen with other effective myeloma therapies, the bone marrow microenvironment offers some protection (Figure 5C).
Figure 5. HSF1 pathway inhibition decreases viability of patient-derived myeloma cells whilst sparing human PBMCs and a bone marrow stromal cell line.
Cell viability of Annexin V-APC and PI stained (A) CD138+ primary myeloma cells from 5 patients, (B) human RPMI-8226 cell line and PBMCs from healthy donor, (C) RPMI-8226 and the bone marrow stromal cell line (HS-5) were assessed by flow cytometry following 48 hour treatment with CCT251236 or KRIBB11. Graphs represent the population of Annexin V-APC negative and PI negative cells as a percentage of vehicle-treated controls (Veh). Data are shown as means ±S.E.M. of three independent experiments. Significant differences were calculated by Student’s t-tests and P-values are indicated where * ≤ 0.05, ** ≤ 0.01 and *** ≤ 0.001 comparing RPMI-8226 with PBMC or HS-5 cell viability at the same compound concentration.
These results demonstrate in-vitro anti-myeloma activity of HSF1 pathway inhibitors on patient primary myeloma cells and strongly suggest a potential therapeutic window for a clinical candidate HSF1 inhibitor. Next, as single agent activity was observed with HSF1 pathway inhibitors in in-vitro models, we tested the activity in-vitro of CCT251236 or KRIBB11 combined with bortezomib, a proteasome inhibitor that is a current myeloma front-line therapy. CCT251236 and KRIBB11 at sub-toxic concentrations potentiated the anti-myeloma effect of bortezomib in HMCLs (Supplementary Figure S6), supporting the further study of HSF1 pathway inhibitors with proteasome inhibitors.
KRIBB11 and CCT251236 have anti-myeloma efficacy in a human xenograft model
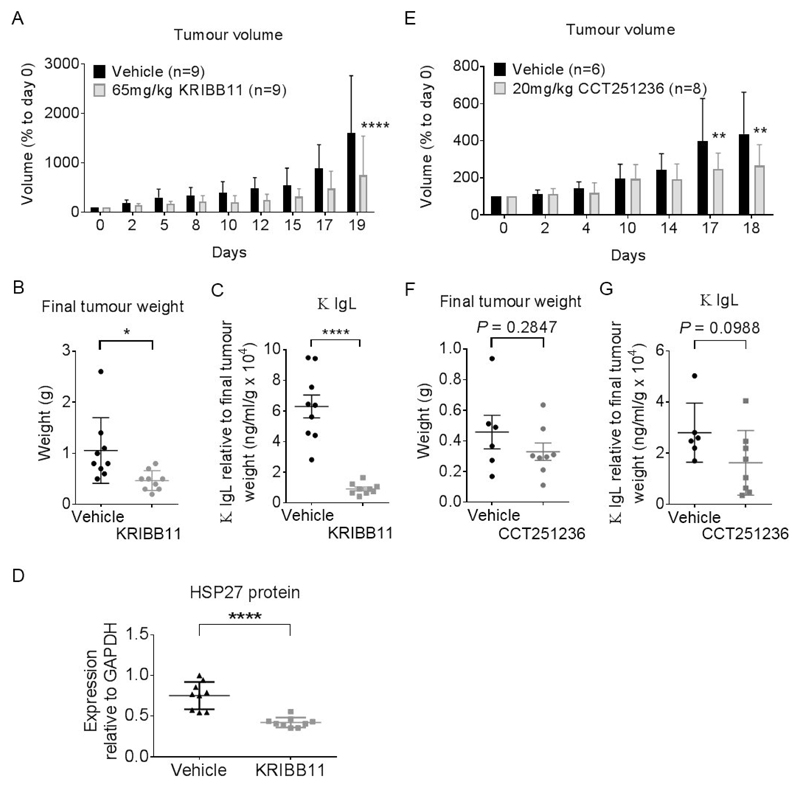
Yoon et al. [27] previously demonstrated that KRIBB11 administered at 50 mg/kg i.p. daily was well-tolerated by mice and inhibited the growth of HCT-116 human colon cancer xenograft tumors. Based on this work, a pilot study of KRIBB11 at 50 mg/kg i.p. daily was similarly well-tolerated in mice carrying s.c. H929 human myeloma xenografts, but the effects on tumor growth were marginal (data not shown). We therefore increased the dose to 65 mg/kg KRIBB11 i.p. daily. At this slightly higher dose, we observed a significant decrease in tumor volume and tumor weight in KRIBB11-treated mice compared with vehicle treated controls at end of therapy (day 19) (Figure 6A, 6B). The dose schedule was well-tolerated with no loss in body weight over the treatment period (Supplementary Figure S7A) indicating a therapeutic index. The therapeutic benefit was associated with a significant decrease in serum κ Ig light chain as well as a significant reduction in HSP27 protein expression in treated tumors compared with controls (Figure 6C, 6D, Supplementary Figure S7B). Similar results were observed with s.c H929 human myeloma xenografts with treated with 20 mg/kg p.o. CCT251236 (Figure 6E, 6F, Supplementary Figure S7C, S7D). These data indicate that inhibition of the HSF1 pathway has anti-myeloma efficacy in a human myeloma xenograft model and that HSP27 expression may be a suitable pharmacodynamic (PD) biomarker of efficacy.
Figure 6. KRIBB11 and CCT251236 are efficacious in a subcutaneous H929 human myeloma xenograft model.
Athymic mice bearing well-established tumors were dosed daily with 65 mg/kg KRIBB11 i.p. or 20 mg/kg CCT251236 p.o. or vehicle over 19 and 18 days, respectively. At the end of the study, 16 hours after the final dose, plasma was collected and tumors were harvested and weighed. (A) Mean tumor volume as a percentage of day 0 (start of treatment). Statistical differences were calculated by two-way ANOVA followed by Bonferroni’s multiple comparisons test with FWER ≤ 0.05. (B) Mean final tumor weights. (C) Mean κ Ig light chains detected in plasma samples. Values are normalised to final tumor weight. (D) Mean HSP27 protein expression as determined by densitometry analysis of Western blot bands relative to GAPDH loading control presented in Supplementary Figure S7B. Data are shown as means ±S.D. Statistical differences were calculated by Student’s t-tests. P-values are indicated where * ≤ 0.05, ** ≤ 0.01 and **** ≤ 0.0001.
Discussion
Our study provides both in-vitro and in-vivo evidence for the pro-oncogenic role of HSF1 in myeloma and the tractability of the HSF1 pathway as a therapeutic target. This is the first report of a significant association between HSF1 expression and the survival of myeloma patients, which adds to the array of studies in solid tumors showing the prognostic impact of HSF1 expression and clinical outcome [19, 21–22, 35]. Furthermore, the difference in survival of patients exhibiting differential expression of a subset HSF1 target genes that was previously reported as prognostic for other cancers [23] supports the notion that HSF1 drives a transcriptional program in maintaining the cancer phenotype with effects on patient outcome.
Clearly, the above prognostic ability of HSF1 and its target genes is insufficient evidence to infer essentiality in myeloma or to conclude that the HSF1 pathway is a good target for therapeutic intervention in this disease. However, we also show here, employing both shRNA knockdown and the use of two chemically distinct HSF1 pathway inhibitors, that HSF1 is indeed required for myeloma cell survival: since either genetic or pharmacological perturbation induces myeloma cell death. Such complementary, orthogonal use of two technically distinct perturbation approaches is accepted as an important means of derisking target validation, building confidence in the target-of-interest and the potential for small molecule tractability [36].
The caspase-mediated cell death observed following HSF1 knockdown in RPMI-8226 and KMS-11 HMCLs clearly suggests that HSF1 and the genes in the HSF1 transcriptional program have a pro-survival and/or anti-apoptotic function in myeloma. Our findings are supported by observations by Heimberger et al. [37] where 72 hour HSF1 shRNA-mediated silencing in two other HMCLs (MM1.s, INA-6) also led to loss in cell viability. Shah et al. [15] however, did not observe a decrease in cell viability to the same extent following 48 hour HSF1 siRNA-mediated knockdown, suggesting that a sustained and prolonged loss of HSF1 is required for an anti-myeloma effect. Myeloma is therefore another example where HSF1 enables cell proliferation or suppresses cell death in the oncogenic context, as shown using Hsf1-/- MEFs transformed with Pdgf-b or C-myc [16]. The sensitivity of HMCLs, patient primary cells, as well as a tumour xenograft, to two chemically distinct small molecule compounds that inhibit the HSF1 stress pathway, CCT251236 and KRIBB11, is an encouraging sign for the potential efficacy of inhibitors of the HSF1 pathway for myeloma patients.
Given the challenge of therapeutically targeting transcription factors such as HSF1 with small molecules [25], other more chemically tractable targets within the HSR previously have been investigated in myeloma, namely HSP90 and HSP70. A number of HSP90 inhibitors have been tested both pre-clinically and/or clinically including tanespimycin [38]. However, the induction of anti-apoptotic HSP72 expression following HSP90 inhibition in myeloma and other solid tumors has been reported to limit their effects [10, 38–39]. The cytoprotective action of HSP70 family proteins are illustrated by our findings where HSP72 siRNA knockdown enhanced sensitivity to tanespimycin [14]. Targeting one HSP70 isoform alone however is insufficient, as we showed that silencing the constitutively expressed HSP70 isoform (HSC70) led to the upregulation of HSP72 and that dual knockdown of both HSP70 isoforms induced heightened caspase-mediated cell death compared with silencing either isoform alone [13, 33]. Indeed, we and others have reported that small molecule HSP70 inhibitors that target both isoforms, such as VER155008 and MAL3-101, increase the anti-myeloma effect of HSP90 inhibition [33, 40]. Besides HSP70 family proteins, the compensatory upregulation of anti-apoptotic HSP27 has also been reported following tanespimycin treatment in HMCLs [41]. Therefore, we propose that targeting the broad downstream HSF1 transcriptional program may be a more effective therapeutic strategy than inhibiting HSPs individually.
Additional compounds, notably triptolide and KNK437, have been described as inhibitors of HSF1-mediated HSP72 expression and used to demonstrate the anti-myeloma efficacy of pharmacological HSF1 pathway blockade [37, 42]. However, as these compounds are reported to have general transcription inhibitor characteristics, we utilized two novel HSF1 pathway inhibitors as tools to explore the response of myeloma models to pharmacological inhibition of the HSF1 pathway.
CCT251236, a bisamide compound, was derived from an initial hit compound that suppressed the induction of HSF1-mediated HSP72 expression in a cell-based phenotypic screen [26]. We showed by extensive profiling that CCT251236 possesses no detectable off-target kinase activity, making it superior to compounds from another chemical series, 4,6-disubstituted pyrimidines, which were also discovered in the same screen [43]. KRIBB11 was a hit compound from a cell-based screen designed to identify molecules that attenuated heat shock element (HSE)-driven luciferase reporter signals [27]. Although the off-target effects of KRIBB11 are unknown, Yoon et al. [27] demonstrated that KRIBB11 directly interacts with HSF1 and does not attenuate NF-kB promoter activity. Furthermore, the similar phenotypic changes observed following KRIBB11 treatment and HSF1 knockdown eases potential concerns over KRIBB11 off-target activity at concentrations used in our present work. Indeed, the growth inhibitory activity of KRIBB11 on HMCLs has also been demonstrated in MM1.s cells [44]. The more potent growth inhibitory activity of CCT251236 compared with KRIBB11 can be attributed to the much lower cellular potency of KRIBB11 in the inhibition of HSF-driven luciferase activity (EC50=1.2 μM) compared with that of CCT251236 in the cellular inhibition of HSP72 expression (EC50=19 nM) [26–27].
Given the challenges in discovering selective inhibitors of HSF1 [25, 45], compounds such as CCT251236 or ones recently described by Vilaboa et al. [46] are steps in the right direction towards developing a potent and selective HSF1 pathway inhibitor for clinical use. Of note, one of the compounds reported, IHSF058 [46], causes depletion of HSF1 protein – as we also observe here with CCT251236 and KRIBB11, albeit at 24–48 hours when cell death is underway. Understanding the mechanism and significance of this HSF1 depletion requires further study.
Importantly, the therapeutic activity of CCT261236 and KRIBB11 in our xenograft models as well as the in-vitro cell killing effects in HMCLs observed that was concomitant with a decrease HSP72 and HSP27 expression, provides additional evidence that HSF1 pathway inhibition is a viable means to suppress the cytoprotective and pro-survival actions of HSF1, the HSPs and also other HSF1-regulated genes. The further development of compounds with improved potency and pharmacokinetic properties, as well as additional work identifying robust biomarkers that correlate with response will be very important. Furthermore, the use of additional pre-clinical myeloma models that more closely recapitulate human disease, together with refinement of optimally efficacious doses and schedules, will be essential in determining whether HSF1 pathway inhibitors merit progression into the clinic.
Another important question that remains to be answered is whether targeting the HSF1 pathway would be a universal approach for myeloma patients, as with proteasome inhibition, or whether particular genetically or clinically-defined subgroup of patients would benefit more from such therapy. Additional studies are required to examine whether factors – such as expression of HSF1, the transcriptional program it regulates (including the HSF1 Ca-Sig derived from other cancers [23] or a signature that could be determined specifically in myeloma) or indeed Ig production – are able to predict sensitivity to HSF1 pathway inhibition. These studies could potentially provide insights into myeloma subtypes that might best respond to HSF1-targeted therapy. Combining HSF1 pathway inhibitors with other modulators of protein handling pathways should also be explored further, in line with the idea of using combinatorial proteotoxic stress targeted therapy for myeloma [1, 6]. Indeed, initial evidence reported here showing the combinatorial benefit with bortezomib supports this approach. Furthermore, the induction of CHOP expression and EIF2α phosphorylation following HSF1 knockdown or pharmacological HSF1 pathway inhibition suggests that PERK inhibitors may also potentiate HSF1 inhibitors.
Despite the clinical progress made with novel agents for myeloma in the past decade, patients continue to relapse [47]. There is therefore an unmet need for new classes of myeloma therapeutics. With increasing evidence for intraclonal genetic heterogeneity in myeloma [48], targeting fundamental cellular stress support pathways may be an alternative and potentially be more effective than targeting specific genetic lesions or oncogenic pathways. Thus, inhibiting one target with multiple downstream effects is an attractive proposition.
In summary, our work demonstrates that HSF1 pathway activity is essential for myeloma cell survival. More importantly, our findings provide proof-of-concept evidence to support the further development of highly potent and selective HSF1 pathway inhibitors as an additional and promising therapeutic option for myeloma.
Statement of Translational Relevence.
Despite the advent of newly approved treatments and improved clinical outcome for myeloma patients, this disease has a high rate of relapse. In light of increasing evidence for intraclonal heterogeneity, targeting fundamental cellular stress pathways, such as the HSF1 pathway, may provide an alternative therapeutic approach. Here, we not only highlight the prognostic significance of HSF1 expression in relapsed myeloma patients, but more importantly demonstrate for the first time the anti-myeloma efficacy of a novel and highly selective inhibitor of the HSF1 pathway, CCT251236, in human myeloma cell lines, primary patient myeloma cells and a human myeloma xenograft model. Taken together, this work provides proof-of-concept evidence to support inhibition of the HSF1 pathway and the clinical development of HSF1 inhibitors as an anti-myeloma strategy to add to the therapeutic armamentarium for a disease greatly in need of novel agents.
Financial Support
This work was supported by Cancer Research UK (grant number C20826/A12103). J.H.L.F. was funded by a studentship from Cancer Research UK. F.E.D. was a Cancer Research UK Senior Cancer Research Fellow. P.W. is a Cancer Research UK Life Fellow. We acknowledge NHS funding to the NIHR Biomedical Research Centre at the Royal Marsden Hospital and the Institute of Cancer Research, Cancer Research UK funding to the Cancer Research UK Cancer Therapeutics Unit and funding from Cancer Research Technology Pioneer Fund and the Battle Against Cancer Investment Trust for the project leading to the discovery of CCT251236 and other HSF1 pathway inhibitors.
Footnotes
Disclosure of Conflicts of Interest:
The authors are employees of The Institute of Cancer Research, which has a commercial interest in the development of chaperone and stress pathway inhibitors. Work on molecular chaperones and stress pathway signaling at the Cancer Research UK Cancer Therapeutics Unit has been funded by Vernalis and AstraZeneca and HSP90 inhibitors have been licensed to Vernalis and Novartis. Research on HSF1 inhibitors has been funded by the Battle Against Cancer Investment Trust and the Cancer Research Technology Pioneer Fund. P.W. has ownership interest in Chroma Therapeutics and is or has been an advisor to Vernalis, Astex Therapeutics, Nuevolution and Chroma Therapeutics.
Author Contributions:
JHLF, KJ, GJM, PW, SAE, FED conceived the study, performed experiments, analyzed data and wrote the manuscript.
SH, LZ, LIA, FM, CP, JJK, MDB, CPW, EDB, CSR, NEAC performed experiments and analyzed data.
All authors reviewed and approved the manuscript.
References
- 1.Aronson LI, Davies FE. DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica. 2012;97:1119–30. doi: 10.3324/haematol.2012.064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson KC. Progress and Paradigms in Multiple Myeloma. Clin Cancer Res. 2016;22(22):5419–5427. doi: 10.1158/1078-0432.CCR-16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obeng EA, et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meister S, et al. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer research. 2007;67:1783–92. doi: 10.1158/0008-5472.CAN-06-2258. [DOI] [PubMed] [Google Scholar]
- 5.Davenport EL, et al. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–9. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- 6.Neznanov N, et al. Proteotoxic stress targeted therapy (PSTT): induction of protein misfolding enhances the antitumor effect of the proteasome inhibitor bortezomib. Oncotarget. 2011;2:209–21. doi: 10.18632/oncotarget.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Workman P, Davies FE. A stressful life (or death): combinatorial proteotoxic approaches to cancer-selective therapeutic vulnerability. Oncotarget. 2011;2(4):277–80. doi: 10.18632/oncotarget.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auner HW, Cenci S. Recent advances and future directions in targeting the secretory apparatus in multiple myeloma. Br J Haematol. 2015;168(1):14–25. doi: 10.1111/bjh.13172. [DOI] [PubMed] [Google Scholar]
- 9.Mitsiades CS, et al. Antimyeloma activity of heat shock protein-90 inhibition. Blood. 2006;107:1092–100. doi: 10.1182/blood-2005-03-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson PG, et al. Tanespimycin and bortezomib combination treatment in patients with relapsed or relapsed and refractory multiple myeloma: results of a phase 1/2 study. British journal of haematology. 2011;153:729–40. doi: 10.1111/j.1365-2141.2011.08664.x. [DOI] [PubMed] [Google Scholar]
- 11.Bagatell R, et al. Induction of a Heat Shock Factor 1-dependent Stress Response Alters the Cytotoxic Activity of Hsp90-binding Agents. Clin Cancer Res. 2000;6:3312–3318. [PubMed] [Google Scholar]
- 12.Maloney A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67(7):3239–53. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 13.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell. 2008;14(3):250–62. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Davenport EL, et al. Leukemia : official journal of the Leukemia Society of America. Vol. 24. Leukemia Research Fund; U.K: 2010. Targeting heat shock protein 72 enhances Hsp90 inhibitor-induced apoptosis in myeloma; pp. 1804–7. [DOI] [PubMed] [Google Scholar]
- 15.Shah SP, et al. Bortezomib-induced heat shock response protects multiple myeloma cells and is activated by heat shock factor 1 serine 326 phosphorylation. Oncotarget. 2016;7(37):59727–59741. doi: 10.18632/oncotarget.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai C, et al. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J-N, et al. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–97. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 18.Hoang AT, et al. A novel association between the human heat shock transcription factor 1 (HSF1) and prostate adenocarcinoma. The American journal of pathology. 2000;156:857–64. doi: 10.1016/S0002-9440(10)64954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang F, Chang R, Yang L. Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer. 2011 doi: 10.1002/cncr.26482. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 20.Gabai VL, et al. Heat Shock Transcription Factor Hsf1 is Involved in Tumor Progression via Regulation of HIF-1 and RNA-binding Protein HuR. Molecular and cellular biology. 2012;32:929–940. doi: 10.1128/MCB.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiwata J, et al. State of heat shock factor 1 expression as a putative diagnostic marker for oral squamous cell carcinoma. International Journal of Oncology. 2012;40:47–52. doi: 10.3892/ijo.2011.1178. [DOI] [PubMed] [Google Scholar]
- 22.Santagata S, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18378–83. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendillo Marc L, et al. HSF1 Drives a Transcriptional Program Distinct from Heat Shock to Support Highly Malignant Human Cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13(4):469–78. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 25.de Billy E, Powers Marissa V, Smith Jennifer R, Workman Paul. Drugging the heat shock factor 1 pathway: Exploitation of the critical cancer cell dependence on the guardian of the proteome. Cell Cycle. 2009;8:3806–3808. doi: 10.4161/cc.8.23.10423. [DOI] [PubMed] [Google Scholar]
- 26.Cheeseman MD, et al. Discovery of a Chemical Probe Bisamide (CCT251236): An Orally Bioavailable Efficacious Pirin Ligand from a Heat Shock Transcription Factor 1 (HSF1) Phenotypic Screen. J Med Chem. 2017;60(1):180–201. doi: 10.1021/acs.jmedchem.6b01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon YJ, et al. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286(3):1737–47. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan G, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–88. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 29.Pawlyn C, et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer J. 2017;7(3):e549. doi: 10.1038/bcj.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Workman P, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102(11):1555–77. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guettouche T, et al. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC biochemistry. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204–13. doi: 10.1038/onc.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, et al. Hsp70 inhibition induces myeloma cell death via the intracellular accumulation of immunoglobulin and the generation of proteotoxic stress. Cancer Lett. 2013;339(1):49–59. doi: 10.1016/j.canlet.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 35.Engerud H, et al. High level of HSF1 associates with aggressive endometrial carcinoma and suggests potential for HSP90 inhibitors. British journal of cancer. 2014;111:78–84. doi: 10.1038/bjc.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blagg J, Workman P. Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell. 2017;32(1):9–25. doi: 10.1016/j.ccell.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heimberger T, et al. The heat shock transcription factor 1 as a potential new therapeutic target in multiple myeloma. British journal of haematology. 2012 doi: 10.1111/bjh.12164. [DOI] [PubMed] [Google Scholar]
- 38.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18(1):64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson PG, et al. Tanespimycin monotherapy in relapsed multiple myeloma: results of a phase 1 dose-escalation study. British journal of haematology. 2010;150:438–45. doi: 10.1111/j.1365-2141.2010.08265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braunstein MJ, et al. Antimyeloma Effects of the Heat Shock Protein 70 Molecular Chaperone Inhibitor MAL3-101. J Oncol. 2011;2011 doi: 10.1155/2011/232037. 232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasui H, et al. BIRB 796 enhances cytotoxicity triggered by bortezomib, heat shock protein (Hsp) 90 inhibitor, and dexamethasone via inhibition of p38 mitogen-activated protein kinase/Hsp27 pathway in multiple myeloma cell lines and inhibits paracrine tumour growth. British journal of haematology. 2007;136:414–23. doi: 10.1111/j.1365-2141.2006.06443.x. [DOI] [PubMed] [Google Scholar]
- 42.Bustany S, et al. Heat shock factor 1 is a potent therapeutic target for enhancing the efficacy of treatments for multiple myeloma with adverse prognosis. Journal of hematology & oncology. 2015;8:40. doi: 10.1186/s13045-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rye CS, et al. Discovery of 4,6-disubstituted pyrimidines as potent inhibitors of the heat shock factor 1 (HSF1) stress pathway and CDK9. Medchemcomm. 2016;7(8):1580–1586. doi: 10.1039/c6md00159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiita AP, et al. Global cellular response to chemotherapy-induced apoptosis. eLife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santagata S, et al. Tight Coordination of Protein Translation and HSF1 Activation Supports the Anabolic Malignant State. Science. 2013;341:1238303–1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilaboa N, et al. New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival. Nucleic Acids Res. 2017;45(10):5797–5817. doi: 10.1093/nar/gkx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlowski RZ, Lonial S. Integration of Novel Agents into the Care of Patients with Multiple Myeloma. Clin Cancer Res. 2016;22(22):5443–5452. doi: 10.1158/1078-0432.CCR-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker BA, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–90. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]