Abstract
It is over 15 years since the identification through linkage, of “first wave” susceptibility genes for common cancers BRCA1, BRCA2, MLH1 and MSH2. These genes have strong frequency-penetrance profiles such that the associated clinical utility likely remains relevant regardless of the context of ascertainment. “Second wave” genes, not tractable by linkage, were subsequently identified by mutation screening of candidate genes (PALB2, ATM, CHEK2, BRIP1, RAD51C, and RAD51D). Their innately weaker frequency-penetrance profile has rendered delineation of cancer associations, risks and variant pathogenicity challenging, which has in turn compromised their clinical application. Early germline exome sequencing endeavours in common cancers did not yield the long anticipated slew of “next wave” genes, but instead infer a highly polygenic genomic architecture requiring much larger experiments to make any substantive inroads into gene discovery. As such, the ‘genetic economics’ of frequency-penetrance point firmly to focused identification of “wave 1” gene mutation carriers as most impactful for cancer control.
With screening, prevention and early detection at the forefront of the cancer agenda, we propose that the time is nigh for initiation of national population testing programmes to identify “first wave” gene mutation carriers. To deliver fully a precision prevention program, long-term large-scale studies of mutation carriers to capture longitudinal clinical data and serial biosamples are required.
Introduction
The last thirty years has witnessed significant improvements in the management of cancer, with striking successes in the treatment of testicular and paediatric cancers, and many haematological malignancies. Despite such advances, on account of the high proportion of patients presenting with late-stage disease, mortality rates have remained disappointingly poor for many common cancers, such as those of the colorectum and pancreas4. Initial euphoria that precision oncology would address the poor outcome for metastatic cancer has been tempered with realisation of the challenges associated with tumour heterogeneity, tumour evolution and emergence of resistance mutations5. Hence the precision oncology vision is unlikely to be a universal panacea in reducing the bulk cancer-related mortality associated with most common solid tumours10.
This recognition has in part driven renewed interest in exploring opportunities for optimising early detection of cancer through screening and prevention programmes. Any such strategy is likely a priori to have more impact if targeted to those at highest risk of developing cancer. This premise has been a central philosophy in the application of genetic testing for mutations in the cancer susceptibility genes (CSGs) that is now common place in family cancer clinics. Since most cancers have a significant heritable component14, there is now interest in the potential of genetic risk profiling to deliver personalised prevention programs to the wider population17. This prospect has been made possible as a result of the phenomenal progress in the identification of CSGs and risk variants.
Here we review our understanding of the genetic architecture of cancer susceptibility, current and future opportunities and consider the case for implementing large-scale genetic testing.
Identification of “first-wave” and “second-wave” cancer susceptibility genes
For most common cancers, risks for the same cancer in first-degree relatives of patients are increased two- to three-fold. Notable exceptions are chronic lymphocytic leukaemia, and thyroid and testicular cancers, for which risks are increased four- to eight-fold20. The genetic architecture underscoring these familial risks is now known to reflect a range of alleles with varying frequencies and effect sizes21.
Genetic linkage and positional cloning studies of multi-case families in the 1990s delivered the first tranche of CSGs for non-syndromic clusters of common cancers, most notably for breast and ovarian cancers (BRCA1 and BRCA2), colorectal cancer (mismatch repair (MMR) genes MLH1 and MSH2) and melanoma (CDKN2A)22–26. However, subsequent linkage analyses of the sizeable numbers of residual “unexplained” large pedigrees failed to yield ‘BRCA3’ or equivalent, leading to gradual acceptance that for these common cancers no further CSGs of “first wave” frequency-risk profile existed27,28. Alternative experimental approaches were required and focus moved to candidate gene experiments informed by the pathways revealed by the “first wave” CSGs. During the noughties, mutational screening of genes involved in DNA repair led to the identification of a second tranche of CSGs, including MuTYH, PALB2, CHEK2, BRIP1, ATM, RAD51C and RAD51D21,29–36. Given the simplistic biological rationale and limited size of experiment feasible with available low-throughput technologies, the yield of “second wave” CSGs from these early gene-screening endeavours was, in retrospect, surprisingly rich37.
A decade of clinical characterisation of cancer susceptibility genes
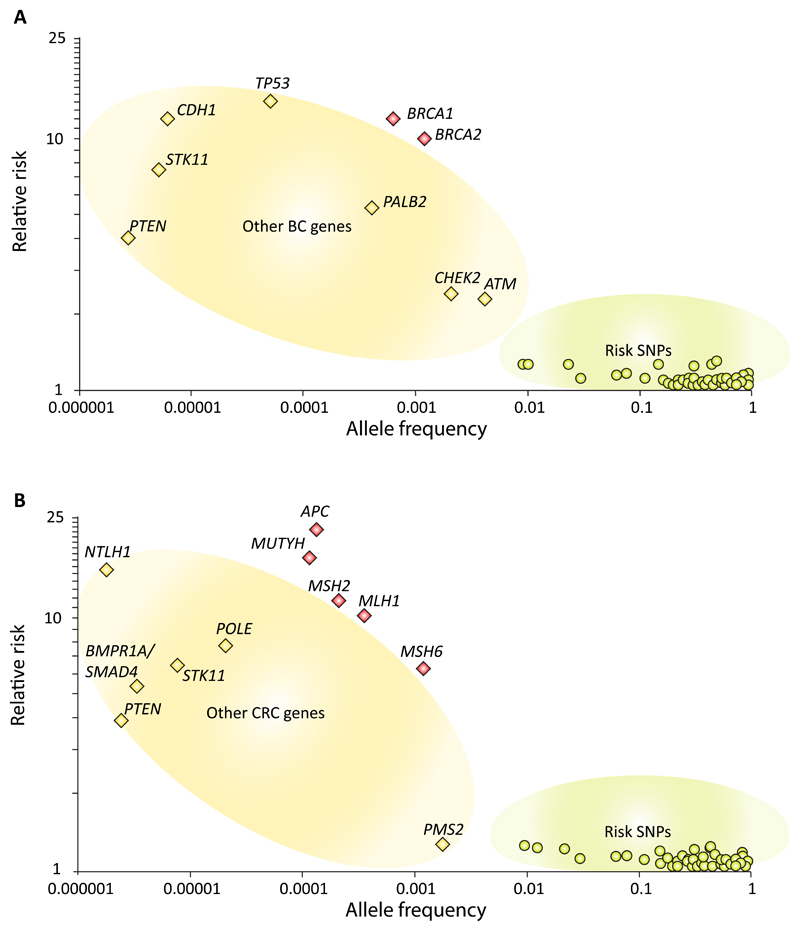
Despite more than 20 years of study (i) the estimates of cancer risk for the ‘first wave’ genes remain relatively imprecise with considerable variation between analyses and (ii) association of additional new cancers being regularly reported (and disputed). Plotting of frequency-risk profiles demonstrates neatly that both BRCA1 and BRCA2 lie well above the linear function obeyed by “second wave” and other breast cancer genetic susceptibility variants, with a similar pattern for MLH1 and MSH2 (Fig. 1). It is unsurprising therefore that characterisation of the “second wave” genes of much weaker frequency-risk profile has proved highly problematic, despite generation of high volumes of data over the last decade through both research and clinical high-throughput sequencing (HTS) of cancer gene panels.
Figure 1. Risk penetrance profile for genetic susceptibility factors for:
(a) Breast cancer; (b) Colorectal cancer
The first, seemingly basic, challenge has been establishing which cancers are truly associated with pathogenic mutations in these newer ‘second wave’ genes. Uncertainty persists as to whether reported ‘breast-cancer susceptibility genes’ such as BARD1, RAD50, NBS1 (NBN) and RECQL are actually associated with breast cancer risk38–42. BRIP1, originally reported as a CSG for breast cancer, has subsequently been shown through recent large-scale analyses to only influence ovarian cancer risk34,43–45. Likewise, there are multiple conflicting reports as to whether ovarian cancer susceptibility genes RAD51C and RAD51D also confer risk of breast cancer30,31,46–49. More tangentially, the purported association of mosaic mutations in PPM1D as a cause of susceptibility to breast and ovarian cancer seems instead simply to represent the confounding artefact of chemotherapy50–53.
The second challenge is establishing the magnitude of cancer risk (known also as penetrance) conferred by CSG mutations. Initial studies suggested PALB2 mutations conferred only a modest two-fold risk of breast cancer32. Subsequent (i) assembly of the world’s largest set of PALB2 mutation positive families with adjustment for ascertainment and (ii) huge case-control analyses of unselected breast cancer cases both support the true penetrance of PALB2 mutations for breast cancer as being of comparable magnitude to that of BRCA2. Disparity in risk estimates nevertheless caused ambivalence around implementation of clinical PALB2 testing 54,55. For ATM, epidemiological and pan-mutation analyses support intermediate penetrance with respect to breast cancer risk (Relative risks (RR) of 2 to 3); however again there are reports of specific missense ATM mutations having BRCA-equivalent risks33,54,56–58. For CHEK2, the breast cancer risks for the relatively frequent 1100delC mutation are well explored and reproducible (RR, 2-3) but for other mutations and cancer associations of CHEK2, the data are conflicting35,59–64. Such observations serve to illustrate that mutation penetrance can be different in heavily laden families as compared to the general population reflecting the influence of modifiers and environment. Only through very large unbiased studies of population-based data will the true associations and risks for variants in these “second wave” genes be ratified.
The third challenge lies in establishing which of the many variants in these genes are truly “pathogenic”. To date, there has been poor correlation with clinical pathogenicity for the majority of functional assays and in-silico predictions (largely derived from inter-species conservation and physio-chemical amino acid similarity)65. Accordingly, establishing pathogenicity for rare variants in “first wave” genes has been challenging and has largely relied on case-control comparison of variant frequencies incorporating tumour characteristics and family segregation. However, for “second wave” genes, except for founder mutations, such case-control comparisons are almost impossible due to very low frequencies of individual variants and/or modest risks. Accordingly, there have been minimal inroads in ascribing pathogenicity to anything other than nonsense and frameshift mutations in “second wave genes’, aside from occasional missense variants found in children with the respective rare bi-allelic phenotype (e.g. Ataxia Telangiectasia)33. Assessing pathogenicity for individual non-founder variants in “second wave” genes will likely only be feasible if robust functional assays can be developed (which correlate perfectly with clinical pathogenicity). Early data from CRISPR saturation editing of BRCA1 by multiplex homology-directed repair is heralding tentative enthusiasm of this long-sought nirvana19,66.
Clearly, for a given gene, analyses of (i) association with cancer, (ii) penetrance and (iii) variant “pathogenicity” are interdependent. Our “working” clinical estimates for these parameters have often been derived from overlapping analyses of the same underlying datasets, which are almost invariably distorted by ascertainment bias, limited by power and frequently subject to population stratification.
Clinical testing for cancer susceptibility: progress or misdirection?
Increasing affordability and throughput of HTS coupled with relaxation of gene patents has led to effective ‘deregulation’ of clinical testing of CSGs. For a given cancer type, the ‘clinical panel’ will typically include (i) relevant “first wave” and ‘second wave’ genes, (ii) selected genes only to date characterised within extremely rare, highly distinctive pleomorphic cancer syndromes, such as PTEN, TP53, CDH1 and STK11, and often (iii) additional genes with dubious or no evidence for association67.
For the ‘second wave genes’ of (seemingly) higher penetrance, mutations have proven strikingly infrequent, exemplified by the yield on testing of PALB2 in familial breast cancer or RAD51C/RAD51D in familial ovarian cancer 30,31,42,46,68. Furthermore, the conflicting published data around basic disease associations and penetrance have unsurprisingly resulted in disparity in clinical management for carriers of mutations in these genes (including individuals from the same family managed in different centres). For genes of intermediate penetrance such as CHEK2, effective management paradigms for families remain unclear69. Unlike for BRCA1, when a CHEK2 mutation is detected in a breast cancer proband, cascade testing in the family does not neatly place unaffected family members into dichotomised categories of high- and low-risk.
Furthermore, the complexity deepens as we extend genetic testing outside of the context of familial disease, into unselected incident cancer cases or indeed the general population. The inherently stronger frequency-penetrance function for “first wave genes”, has made possible (i) large studies of mutation positive families as well as (ii) analyses in large unselected cancer cohorts70–75. Triangulation of these data suggests a ‘true’ (or average) penetrance sufficiently high that interventions for screening and prevention likely remain relevant regardless of ascertainment73,74. For most ‘second wave’ genes, the ‘true’ penetrance is uncertain: ascertainment outside of the familial context thus further amplifies uncertainties around the efficacy of and justification for available clinical interventions.
The ‘rare syndromic’ genes such as TP53, STK11 and CDH1 also pose challenges when testing outside of a classical familial context. The case has been made for testing unselected cancer cases on account of concern that these conditions are underdiagnosed, especially because mutations can have pleomorphic effects, and can arise de novo. However, there are limited data on cancer risks outside of the families ascertained due to classic phenotype. Higher than predicted mutational frequencies in control data suggest the disease risks quoted in classic families grossly exceed those applicable to mutation carriers acertained from the general population76–80. Until such risks have been better established, extending testing for these genes beyond ‘classical’ or familial context may lead to interminable challenges in patient management. The risk-benefit trade-off for prophylactic gastrectomy would be highly uncertain if a pathogenic CDH1 variant were found in unaffected individual or isolated breast cancer case without relevant family history.
For many clinicians, initial enthusiasm for offering testing of a broader palette of genes has been tempered by such uncertainties as well as the low detection rates. Some would go so far as to argue that the ‘additional content’ on panels beyond “first wave” genes has only served to inflate costs of mainstreaming genetic testing whilst generating a spiralling industry in interpretation and over-management of variants of uncertain significance. Moreover, use of inflated estimates of cancer risk may unduly elevate anxiety and divert healthcare resource towards screening and preventative surgery of questionable benefit to the individual, let alone justification in the context of Public Health81.
Ten years on in genome-wide association studies
Early proponents of the common variant-common disease hypothesis had envisaged that genome-wide association studies (GWAS) might deliver a tractable ‘set’ of common variants for each tumour type, capturing a significant proportion of the heritable risk. The field of public health genomics arose in anticipation of implementing such information to stratify the population into neat tranches of risk for programmes of screening and prevention82 In 2007, the long-awaited first-wave results from GWAS for the common cancers were reported. What was striking were the modest effect sizes of the top associations, for example, the RR of 1.26 for the top breast cancer risk locus (intron 2 of FGFR2)83,84. Subsequent, larger GWAS identified respectable slews of additional hits but confirmed the portentous power calculations of the early GWAS, namely that each tumour type has an underlying genomic architecture comprising several hundred of loci of progressively more modest contribution82,85–88. Initiatives, such as the Breast Cancer Association Consortium (BCAC), have delivered experiments of increasing magnitude, each time adding to the proportion of familial relative risk (FRR) explained 89–94. The most recent Oncoarray experiment from BCAC, involving some 140,000 cases and a similar number of controls was sufficiently empowered to show 18% of the FRR for breast cancer being attributable to 140 risk SNPs. While statistical modelling indicates that ~40% of the FRR is likely to be enshrined in common variation, projections suggest that far larger sample sizes in excess of 300,000 will be required to explain 80% of this component of the heritable risk of breast and colorectal cancer91. Whilst large biobanks exist for some cancers, for many tumours is it likely prohibitive to assemble the magnitude of samples required to harvest a significant proportion of the FRR. Whilst issues of power may challenge clinical application of GWAS, this is not to detract from the wealth of insights GWAS has made to cancer biology which may in due course offer patient benefit95.
Rationalising strategies forwards informed by genomic architecture
As we reach the decade mark from rollout of both GWAS and HTS, it is imperative that we overlay our ‘real life genetic testing experiments’ from the clinic onto candid insights from our research endeavours.
Availability over the last few years of a ‘palette’ of long-awaited ‘second wave’ genes has impinged minimally on clinical care: a direct function of their risk-penetrance profile. Power analyses from initial exome sequencing endeavours in common cancers have made it likely that the ‘next wave’ susceptibility genes/rare alleles will be of equivalent or more modest risk-penetrance profile than the ‘second wave’ genes. Significant inroads towards identification of this ‘next wave’ of susceptibility genes/alleles will be achievable through exome/genome sequencing studies greater by scales of magnitude than those previously undertaken (likely >50,000 cases and 50,000 controls) 96,97. Furthermore, characterizing cancer association, penetrance and variant pathogenicity for any ‘next wave’ genes will be commensurately even more challenging than for ‘second wave’ genes.
Therefore, due to the immoveable truth of risk-penetrance function, clinical utility from germline genetic testing for cancer susceptibility has and will for the foreseeable future, be best effected through by identifying carriers of mutations in those stalwart ‘first wave’ genes, BRCA1, BRCA2, MLH1 and MSH2 (Fig 1). Hence, our clinical-research rhetoric and priorities should focus on leveraging full value from ‘first wave’ genes: (i) expanding identification of mutation carriers, (ii) improved statistical genetic epidemiologic studies of association, penetrance and variant pathogenicity and (iii) longitudinal biosampling to better understand cellular biology, pre-cancer states and tumourogenesis (Box 1).
Box 1. Priority research themes for BRCA and MMR: reaping new value from old hitters.
(i) A pre-cancer atlas to deliver insights into premalignant cell biology, develop of biomarkers for surveillance and evolve precision chemoprevention
Large-scale serial collection of multiple biosamples from mutation carriers, development of engineered models, application of single cell technologies, multi-omic analyses, studies of the microenvironment and immunological approaches will accelerate our understanding of pre-cancer biology and tumourogenesis, as well described by Lipman and colleagues1. This will in turn facilitate discovery of biomarkers and development of chemotherapeutic agents, chemopreventative agents and vaccines. Animal experiments of rank-ligand inhibition have shown potential for chemoprevention in BRCA-carriers: clinical trials are in early rollout2,3. Chemoprevention for colorectal cancer is an under-exploited area: exposition of the molecular basis for the protective effect of aspirin could yield additional benefit beyond MMR-carriers and beyond colorectal cancer. The predictable expressed epitopes of the MMR-deficient hypermutated tumours offer a tantalising model for immune modulation and cancer vaccines6. The distinctive hypermutated profile of MMR-deficient tumours, alongside their high a priori cancer risks, surely renders MMR-mutation carriers well-suited for early prospective studies of cDNA monitoring for CRC
(ii) Advanced clinical risk models and clinical interfaces for delivery of precision prediction
Early risk estimates derived from segregation analysis of early linkage families grouped together all MMR genes and BRCA1 with BRCA2. Through large consortia efforts (i) individual gene-level penetrance estimates (ii) variant-class and domain-specific cancer risks (iii) variant-specific cancer risks and (iv) risk-modifying common variants are being established. For example: (a) intriguing new profiles of disease association for the individual MMR genes reveal gene-specific indications for surgical management and screening7 (b) risk-modifying SNPs for BRCA1 and BRCA2 differentiate mutation carriers into clinically-meaningful different categories of risk8,9 (c) regions in both BRCA1 and BRCA2 have been delineated for which the comparative risks of breast and ovarian cancer differ significantly (BCCRs and OCCR) (d) For BRCA1, mutation-specific risks clearly distinct from the generally deployed ‘pan-mutation’ risks are emerging11–13. Larger datasets, controlled for ascertainment, fully typed for genetic factors and well characterised for non-genetic factors are urgently required to better model risk. These will also enable more rigorous testing for non-multiplicative interactions between risk factors. We also need clinician and patient decision-support tools, well designed to present and translate this complex data on clinical risk (along with its bounds of uncertainty).
(iii) Variant interpretation paradigms designed for cancer susceptibility genetics
Consistency in variant interpretation has been greatly advanced by (i) ClinVar emerging as the de facto centralised community variant classification repository (ii) ACMG standardisation of classification criteria15,16. However, the ACMG framework is best suited to rare dysmorphic syndromes: evolution of dedicated approaches for cancer susceptibility genes are in process. Each CSG is different both in terms of biology and mutational patterns. As currently being enacted by ENIGMA, InSIGHT and ClinGen expert groups, a combination of gene-specific expertise alongside universally consistent frameworks are required. Starting with wave 1 genes, such self-organising activities offer the prospect of consistent, systematic processing of genetic data, collaborative international deposition and comprehensive annotation. Furthermore, significant advances in the field are imminent from development of massively high-throughput functional assays, such as saturation editing of genomic BRCA1 regions by multiplex homology-directed repair and splicing assays such as MFASS (Multiplexed Functional Assay of Splicing using Sort-seq)18,19.
Implementation of large-scale population-level genetic testing: ‘primetime’ is now
The threshold for testing of BRCA1/BRCA2 and MMR genes based on family history have been reduced progressively over the past two decades and ascertainment of families through ‘mainstream’ testing at cancer diagnosis is becoming established. However, even in countries with well-developed genetics services, we have identified less than 10% of prevalent BRCA and MMR mutation carriers98. Even with expansion of testing in oncology, ascertainment of ‘the totality’ of prevalent BRCA1/BRCA2 and MMR mutations would take many decades under the current models98. Testing for BRCA1/BRCA2 founder mutations has been well demonstrated as economically and clinically effective: systematic rollout of founder testing in relevant subpopulations is long overdue98–103. Furthermore there is increasing clinical impetus and health economic evidence for offering testing to the general population for mutations in BRCA1/BRCA2 and MMR genes, even given the requirement for more expansive analysis of the genes and lower mutation detection than in founder mutation population programs99,104.
For each given cancer, the predictive value of the ‘testable SNP set’ feasibly tractable by GWAS is also now becoming clear105–113 [98-105] and we are reaching ‘jump-time’ on rollout-trials for population SNP cancer risk profiling. Whilst not quite the catholicon initially anticipated, in certain tumours SNP risk-profiling applied at population level can offer valuable risk discrimination, particular when combined with a predictive set of non-genetic risk factors114. Furthermore, intermediate penetrance susceptibility genes such as ATM, CHEK2 and BRIP1, whilst of equivocal use in the familial cancer clinic, although rare, add value in the context of population genetic risk stratification115,116.
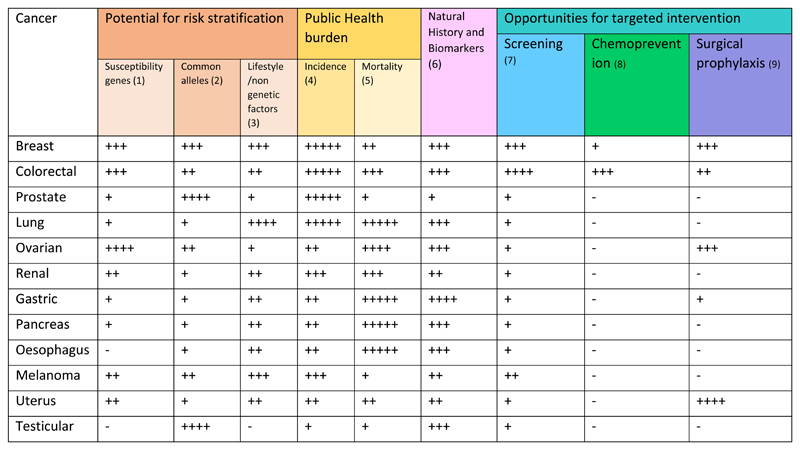
The value proposition of genomic risk profiling is not solely based on discriminatory performance of the ‘prediction tool’ but is heavily predicated on factors such as disease frequency, disease mortality, disease natural history, biomarkers of tumour behaviour and interventions available for screening and prevention (Fig 2). Breast (-ovarian) and colorectal cancers would seem to emerge as prime candidates for population risk profiling. Each cancer is common with high burden of mortality. For each there is a plausible tripartite ‘prediction tool’ (comprising common variants, non-genetic factors and high/intermediate penetrance susceptibility genes). For each there are effective options for intervention - screening, chemoprophylaxis and surgery. Modelling, health economic and pilot implementation studies for population genetic testing for these cancers are urgently required.
Figure 2. Attributes for a precision prevention programme, by cancer.
(1) Contribution by frequency-penetrance of high penetrance susceptibility genes
(2) Heritability and % excess familial risk explained by common (GWAS) alleles
(3) Receiver operator performance of totality of known lifestyle/non-genetic factors
(4) Incidence (Annual cases in UK: + <5,000; ++ 5,000-9,999; +++ 10,000-19,999; ++++ 20,000-39,999; +++++, ≥40,000)117
(5) Mortality (10 year survival + >80%; ++ 60-80%; +++ 40-60%; ++++: 20-40%; +++++ <20%)117
(6) Natural history of disease is well understood (eg robust biomarkers to predict poor prognosis disease)
(7) Effective and acceptable screening tool and confirmatory test consistent with delivery of national screening programme.
(8) Effective and acceptable chemoprophylaxis eg breast cancer (tamoxifen, AIs, SERMS) and colorectal cancer (aspirin)
(9) Elective and acceptable option for presymptomatic surgical removal of organ at risk.
Conclusion
Emergence of transformative new technologies has unsurprisingly fostered great expectation for gene discovery and delivery of new paradigms in genomic risk prediction for cancer. HTS offered both clinicians and researchers the promise of new clinic-ready CSGs with which to ‘diagnose’ our outstanding breast and colorectal cancer families. Instead, our HTS experiments to date have instead shown the genomic architecture of these common cancers to be much more complex than originally anticipated.
Testing dozens of genes of vanishingly low mutational frequency, poorly characterised risk and/or questionable association with disease has delivered limited gains. Instead guided by basic frequency-penetrance ‘economics’, we should re-embrace our ‘first wave’ genes and focus our efforts on identifying as many mutation carriers as possible. We should denounce the procrastination of awaiting additional future ‘new genes’ to ‘add value’ to the population screening proposition. The time is ripe for large-scale implementation studies of population BRCA and MMR gene testing (potentially combined ‘tripartite’ with risk profiling using common genetic and non-genetic factors). In parallel, we need to shift now to delivery-mode on proper ‘individualised’ precision prediction and management for mutation carriers. To deliver individualised risk and precision prevention, we require urgently clinical tools which integrate existing data on gene-, location-, and individual mutation-specific risk, common genetic and non-genetic modifiers, family history and context of ascertainment7,9,113.
Whilst we have already made great progress on characterisation and clinical applications of these genes, to deliver fully their impact for cancer prevention, significant and sustained investment in research platforms will be required to deliver the necessary long-term epidemiological, biological and clinical studies (Box 2).
Box 2. Research ‘Platforms’ for studying BRCA and MMR genes.
To establish and expand biological, epidemiological and clinical trials ‘platforms’ will require (i) coordinated, systematic assembly of large cohorts of mutation carriers, (ii) flexible and considered structures of consent and (iii) long-term, sustainable funding. These include:
-
(i)
Prospective cohorts to advance studies of penetrance and association (observational epidemiology), including longitudinal data linkage of germline genetic status to cancer occurrence, treatment and outcomes, with information on screening and preventative surgery and details of ascertainment.
-
(ii)
Longitudinal acquisition of biological samples (‘pre-cancer’ and cancer atlases): Biomarker discovery and insights into pre-cancer biology will be accelerated through largescale serial collection from mutation carriers of multiple biosamples (e.g. blood, urine, saliva, normal colonic tissue, exhaled gases, lavage from breast ducts, peritoneal lavage, polyps, CIS and tumour tissue).
-
(iii)
National infrastructure for clinical trials in CSG mutation carriers: comprehensive national networks of traceable, contactable mutation carriers, with which to deliver rapid enactment of well-powered trials of screening or chemoprophylaxis.
Acknowledgements
We thank our many colleagues for decades of interesting discussions around these themes, but in particular the invaluable and wise thoughts of Will Foulkes, Marc Tischkowitz, Helen Hanson, Amy Taylor, Katie Snape and Anju Kulkani. We should also like to thank Doug Easton and Peter Devilee for providing the data used to generate Figure 1a. R.S.H. is supported by Cancer Research UK (C1298/A8362 Bobby Moore Fund for Cancer Research UK). C.T. is supported by the Movember Foundation.
Footnotes
Competing Interests
The authors declare no competing financial or non-financial interests as defined by Nature Research.
Author Contributions
C.T, A.S and R.S.H researched, reviewed, drafted and edited the manuscript. A.S and C.T generated the images.
References
- 1.Yurgelun MB, Chenevix-Trench G, Lippman SM. Translating Germline Cancer Risk into Precision Prevention. Cell. 2017;168:566–570. doi: 10.1016/j.cell.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, et al. Leveraging premalignant biology for immune-based cancer prevention. Proceedings of the National Academy of Sciences. 2016;113:10750–10758. doi: 10.1073/pnas.1608077113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nolan E, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med. 2016;22:933–9. doi: 10.1038/nm.4118. [DOI] [PubMed] [Google Scholar]
- 4.Cancer incidence statistics. Cancer Research UK; 2017. [Google Scholar]
- 5.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095–111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloor M, et al. Vaccination of MSI-H colorectal cancer patients with frameshift peptide antigens: A phase I/IIa clinical trial. Journal of Clinical Oncology. 2015;33:3020–3020. [Google Scholar]
- 7.Møller P, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne RL, Antoniou AC. Genetic modifiers of cancer risk for BRCA1 and BRCA2 mutation carriers. Ann Oncol. 2011;22(Suppl 1):i11–7. doi: 10.1093/annonc/mdq660. [DOI] [PubMed] [Google Scholar]
- 9.Milne RL, Antoniou AC. Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers. Endocr Relat Cancer. 2016;23:T69–84. doi: 10.1530/ERC-16-0277. [DOI] [PubMed] [Google Scholar]
- 10.Prasad V. Perspective: The precision-oncology illusion. Nature. 2016;537:S63. doi: 10.1038/537S63a. [DOI] [PubMed] [Google Scholar]
- 11.Spurdle AB, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012;49:525–32. doi: 10.1136/jmedgenet-2012-101037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moghadasi S, et al. The BRCA1 c. 5096G>A p.Arg1699Gln (R1699Q) intermediate risk variant: breast and ovarian cancer risk estimation and recommendations for clinical management from the ENIGMA consortium. J Med Genet. 2017 doi: 10.1136/jmedgenet-2017-104560. [DOI] [PubMed] [Google Scholar]
- 13.Rebbeck TR, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. Jama. 2015;313:1347–61. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 15.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanock S. Charting a Course toward Precision Cancer Prevention. 2016 [Google Scholar]
- 18.Cheung R, et al. Large-scale screening of rare genetic variants in humans reveals frequent splicing disruptions. bioRxiv. 2017 [Google Scholar]
- 19.Findlay GM, et al. Accurate functional classification of thousands of BRCA1 variants with saturation genome editing. bioRxiv. 2018 doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houlston R, Peto J. Genetics and the common cancers. In: Eeles R, Ponder BA, Easton D, Horwich A, editors. Genetic Predisposition to Cancer. SpringerLink; pp. 208–226. [Google Scholar]
- 21.Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer. 2010;10:353–61. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- 22.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 23.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulos N, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–9. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 25.Leach FS, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 26.Hussussian CJ, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 27.Elston RC, Cordell HJ. Overview of model-free methods for linkage analysis. Adv Genet. 2001;42:135–50. doi: 10.1016/s0065-2660(01)42020-7. [DOI] [PubMed] [Google Scholar]
- 28.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–41. [PubMed] [Google Scholar]
- 29.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 30.Loveday C, et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet. 2012;44:475–6. doi: 10.1038/ng.2224. author reply 476. [DOI] [PubMed] [Google Scholar]
- 31.Loveday C, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renwick A, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 34.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 35.Meijers-Heijboer H, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 36.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 37.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–45. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 38.Cybulski C, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015;47:643–6. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 39.Heikkinen K, et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27:1593–9. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tommiska J, et al. Evaluation of RAD50 in familial breast cancer predisposition. Int J Cancer. 2006;118:2911–6. doi: 10.1002/ijc.21738. [DOI] [PubMed] [Google Scholar]
- 41.Slavin TP, et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer. 2017;3:22. doi: 10.1038/s41523-017-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buys SS, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 43.Easton DF, et al. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J Med Genet. 2016;53:298–309. doi: 10.1136/jmedgenet-2015-103529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sopik V, Foulkes WD. Risky business: getting a grip on BRIP. J Med Genet. 2016;53:296–7. doi: 10.1136/jmedgenet-2015-103648. [DOI] [PubMed] [Google Scholar]
- 45.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–7. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 46.Pelttari LM, et al. RAD51C is a susceptibility gene for ovarian cancer. Human Molecular Genetics. 2011;20:3278–3288. doi: 10.1093/hmg/ddr229. [DOI] [PubMed] [Google Scholar]
- 47.Couch FJ, et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osorio A, et al. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet. 2012;21:2889–98. doi: 10.1093/hmg/dds115. [DOI] [PubMed] [Google Scholar]
- 49.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–4. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 50.Pharoah PDP, et al. PPM1D Mosaic Truncating Variants in Ovarian Cancer Cases May Be Treatment-Related Somatic Mutations. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swisher EM, et al. Somatic Mosaic Mutations in PPM1D and TP53 in the Blood of Women With Ovarian Carcinoma. JAMA Oncol. 2016;2:370–2. doi: 10.1001/jamaoncol.2015.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zajkowicz A, et al. Truncating mutations of PPM1D are found in blood DNA samples of lung cancer patients. Br J Cancer. 2015;112:1114–20. doi: 10.1038/bjc.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruark E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–10. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Southey MC, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J Med Genet. 2016;53:800–811. doi: 10.1136/jmedgenet-2016-103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antoniou AC, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Concannon P. ATM heterozygosity and cancer risk. Nat Genet. 2002;32:89–90. doi: 10.1038/ng0902-89. [DOI] [PubMed] [Google Scholar]
- 57.Gatti RA, Tward A, Concannon P. Cancer risk in ATM heterozygotes: a model of phenotypic and mechanistic differences between missense and truncating mutations. Mol Genet Metab. 1999;68:419–23. doi: 10.1006/mgme.1999.2942. [DOI] [PubMed] [Google Scholar]
- 58.Swift M, Reitnauer PJ, Morrell D, Chase CL. Breast and other cancers in families with ataxia-telangiectasia. N Engl J Med. 1987;316:1289–94. doi: 10.1056/NEJM198705213162101. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt MK, et al. Age- and Tumor Subtype-Specific Breast Cancer Risk Estimates for CHEK2*1100delC Carriers. J Clin Oncol. 2016;34:2750–60. doi: 10.1200/JCO.2016.66.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hale V, Weischer M, Park JY. CHEK2 (*) 1100delC Mutation and Risk of Prostate Cancer. Prostate Cancer. 2014;2014 doi: 10.1155/2014/294575. 294575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han FF, Guo CL, Liu LH. The effect of CHEK2 variant I157T on cancer susceptibility: evidence from a meta-analysis. DNA Cell Biol. 2013;32:329–35. doi: 10.1089/dna.2013.1970. [DOI] [PubMed] [Google Scholar]
- 62.Liu C, Wang QS, Wang YJ. The CHEK2 I157T variant and colorectal cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13:2051–5. doi: 10.7314/apjcp.2012.13.5.2051. [DOI] [PubMed] [Google Scholar]
- 63.Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26:542–8. doi: 10.1200/JCO.2007.12.5922. [DOI] [PubMed] [Google Scholar]
- 64.Schutte M, et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet. 2003;72:1023–8. doi: 10.1086/373965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Human Molecular Genetics. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Starita LM, et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics. 2015;200:413–22. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Easton DF, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–57. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osher DJ, et al. Mutation analysis of RAD51D in non-BRCA1/2 ovarian and breast cancer families. Br J Cancer. 2012;106:1460–3. doi: 10.1038/bjc.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tung N, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13:581–8. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeRycke MS, et al. Targeted sequencing of 36 known or putative colorectal cancer susceptibility genes. Mol Genet Genomic Med. 2017;5:553–569. doi: 10.1002/mgg3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hampel H, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 72.Hampel H, et al. Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology. 2005;129:415–21. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 73.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–90. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antoniou AC, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rich T, Lotito M, Kidd J, Saam J, Lancaster J. Abstract PD7-03: Characterization of Li-Fraumeni syndrome diagnosed using a 25-gene hereditary cancer panel. Cancer Research. 2016;76 PD7-03-PD7-03. [Google Scholar]
- 77.Susswein LR, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genetics in Medicine. 2016;18:823–832. doi: 10.1038/gim.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Andrade KC, et al. Higher-than-expected population prevalence of potentially pathogenic germline TP53 variants in individuals unselected for cancer history. Hum Mutat. 2017;38:1723–1730. doi: 10.1002/humu.23320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jung K, Amanda F, P SKA, H D Ashley, R SD. The prevalence of DICER1 pathogenic variation in population databases. International Journal of Cancer. 2017;141:2030–2036. doi: 10.1002/ijc.30907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loveday C, et al. p.Val804Met, the most frequent pathogenic mutation in RET, confers a very low lifetime risk of medullary thyroid cancer. J Clin Endocrinol Metab. 2018 doi: 10.1210/jc.2017-02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Narod SA. The tip of the iceberg: A Countercurrents Series. Current Oncology. 2012;19:129–130. doi: 10.3747/co.19.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 83.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 86.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 87.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 88.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 89.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. 361e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017 doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amos CI, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiology Biomarkers & Prevention. 2017;26:126. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al Olama AA, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakoda LC, Jorgenson E, Witte JS. Turning of COGS moves forward findings for hormonally mediated cancers. Nat Genet. 2013;45:345–348. doi: 10.1038/ng.2587. [DOI] [PubMed] [Google Scholar]
- 95.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017 doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 96.Guo Michael H, et al. Determinants of Power in Gene-Based Burden Testing for Monogenic Disorders. The American Journal of Human Genetics. 2016;99:527–539. doi: 10.1016/j.ajhg.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chubb D, et al. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat Commun. 2016;7 doi: 10.1038/ncomms11883. 11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Manchanda R, et al. Current detection rates and time-to-detection of all identifiable BRCA carriers in the Greater London population. J Med Genet. 2018 doi: 10.1136/jmedgenet-2017-105195. [DOI] [PubMed] [Google Scholar]
- 99.Manchanda R, et al. Cost-effectiveness of Population-Based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 Mutation Testing in Unselected General Population Women. J Natl Cancer Inst. 2018 doi: 10.1093/jnci/djx265. [DOI] [PubMed] [Google Scholar]
- 100.Manchanda R, et al. Cost-effectiveness of population based BRCA testing with varying Ashkenazi Jewish ancestry. Am J Obstet Gynecol. 2017;217:578.e1–578.e12. doi: 10.1016/j.ajog.2017.06.038. [DOI] [PubMed] [Google Scholar]
- 101.Manchanda R, et al. Cost-effectiveness of population screening for BRCA mutations in Ashkenazi jewish women compared with family history-based testing. J Natl Cancer Inst. 2015;107:380. doi: 10.1093/jnci/dju380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Manchanda R, et al. Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: a randomized controlled trial. J Natl Cancer Inst. 2015;107:379. doi: 10.1093/jnci/dju379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lieberman S, et al. Population screening for BRCA1/BRCA2 founder mutations in Ashkenazi Jews: proactive recruitment compared with self-referral. Genet Med. 2017;19:754–762. doi: 10.1038/gim.2016.182. [DOI] [PubMed] [Google Scholar]
- 104.Levy-Lahad E, Lahad A, King M-C. Precision Medicine Meets Public Health: Population Screening for BRCA1 and BRCA2. JNCI: Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/dju420. dju420-dju420. [DOI] [PubMed] [Google Scholar]
- 105.Pashayan N, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104:1656–63. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garcia-Closas M, Gunsoy NB, Chatterjee N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mavaddat N, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frampton MJ, et al. Implications of polygenic risk for personalised colorectal cancer screening. Ann Oncol. 2016;27:429–34. doi: 10.1093/annonc/mdv540. [DOI] [PubMed] [Google Scholar]
- 109.Litchfield K, et al. Polygenic susceptibility to testicular cancer: implications for personalised health care. Br J Cancer. 2016;114:e22. doi: 10.1038/bjc.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kong SW, et al. Summarizing polygenic risks for complex diseases in a clinical whole-genome report. Genet Med. 2015;17:536–44. doi: 10.1038/gim.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genet Med. 2017;19:30–35. doi: 10.1038/gim.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gray E, et al. Evaluation of a Stratified National Breast Screening Program in the United Kingdom: An Early Model-Based Cost-Effectiveness Analysis. Value Health. 2017;20:1100–1109. doi: 10.1016/j.jval.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 113.Evans DG, et al. The impact of a panel of 18 SNPs on breast cancer risk in women attending a UK familial screening clinic: a case-control study. J Med Genet. 2017;54:111–113. doi: 10.1136/jmedgenet-2016-104125. [DOI] [PubMed] [Google Scholar]
- 114.Jervis S, et al. A risk prediction algorithm for ovarian cancer incorporating BRCA1, BRCA2, common alleles and other familial effects. J Med Genet. 2015;52:465–75. doi: 10.1136/jmedgenet-2015-103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee AJ, et al. Incorporating truncating variants in PALB2, CHEK2, and ATM into the BOADICEA breast cancer risk model. Genet Med. 2016;18:1190–1198. doi: 10.1038/gim.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jones MR, Kamara D, Karlan BY, Pharoah PDP, Gayther SA. Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecol Oncol. 2017 doi: 10.1016/j.ygyno.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 117.UK Cancer Incidence Statistics. Cancer Research UK; 2018. [Google Scholar]