Abstract
Conventional dendritic cells (cDCs) are found in all tissues and play a key role in immune surveillance. They comprise two major subsets, cDC1 and cDC2, both derived from circulating precursors of cDCs (pre-cDCs), which exited the bone marrow. We show that, in the steady state mouse, pre-cDCs entering tissues proliferate to give rise to differentiated cDCs, which themselves possess residual proliferative capacity. We use multi-colour fate mapping of cDC progenitors to show that this results in clones of sister cDCs, most of which comprise a single cDC1 or cDC2 subtype, suggestive of pre-cDC commitment. Upon infection, a surge in the influx of pre-cDCs into the affected tissue dilutes clones and increases cDC numbers. Our results indicate that tissue cDCs can be organized in a patchwork of closely positioned sister cells of the same subset whose co-existence is perturbed by local infection, when the bone marrow provides additional pre-cDCs to meet increased tissue demand.
Keywords: dendritic cells, fate mapping, clonal analysis, infection
Introduction
Conventional dendritic cells (cDCs) are leukocytes that play a key role in innate immunity as well as the initiation and regulation of T cell responses (1). They comprise two broad sub-types, cDC1s and cDC2s, that form a network of immune sentinel cells in most tissues of mice and humans (2). cDC1 and cDC2 originate from cDC-committed hematopoietic progenitors in bone marrow (BM) known as the common (but, more accurately, the conventional) DC progenitor (CDPs) (3–7). CDPs give rise to pre-cDCs that exit BM via the blood to seed lymphoid and non-lymphoid tissues(6–8). Individual pre-cDCs were originally envisaged to be bi-potential and generate both cDC1 or cDC2 (4, 9). More recently, cDC1/cDC2 subset specification was shown to be able to occur, at least under certain circumstances, during the CDP to pre-cDC transition to give rise to committed pre-cDC1 or pre-cDC2 (10–12). It remains unclear to what extent tissues are seeded by committed versus uncommitted pre-cDCs. Similarly, it is not known to which degree pre-cDCs arriving in non-lymphoid tissues have proliferative capacity (3) and can undergo local expansion to give rise to clones of differentiated cDC1 or cDC2 occupying a defined tissue territory.
The replacement of “old” tissue cDCs with “new” ones is thought to occur at a high rate as the half-life of cDCs in most tissues is 3-6 days (8). This steady-state cDC renewal is likely controlled at the level of the generation of pre-cDCs, their BM exit rate, their tissue seeding rate and the rate of pre-cDC proliferation and differentiation into cDC1 and cDC2. Importantly, there is indirect evidence to indicate that these parameters are not immutable and that the global generation of cDC (cDCpoiesis) can increase upon loss of cDCs in the periphery (13) or in response to infection or tissue injury (14–22). While all these studies suggest that cDC numbers in tissues can be regulated by local demand, it is unclear whether this requires changes in putative local proliferation of pre-cDCs or, alternatively, communication with the BM and recruitment of additional precursors.
Multicolor fate mapping of cell precursors allows for analysis of single-color cell clusters, which in turn informs on the clonal relationship of cells in tissues (23). Here, we use this approach to analyze the distribution and clonal architecture of cDCs in mice in which individual CDPs/pre-cDCs were fate-mapped with one of four possible fluorophores. We show that peripheral tissues such as lung and small intestine (SI) contain clones of sister cDCs that arise through local proliferation of immigrant pre-cDCs and their progeny. Most of these clones consist of a single cDC subtype, consistent with early fate specification at the level of the CDP to pre-cDC transition. Notably, upon lung infection with influenza A virus, cDC numbers increase through an acute influx of pre-cDCs from BM, which dilutes pre-existing cDC clones. Our findings offer deeper understanding of the organization and dynamics of cDCs in tissues and reveal an axis of emergency immune surveillance that could be manipulated to increase immunity in vaccination or immunotherapy.
Results
Pre-cDCs and their progeny retain proliferative capacity in peripheral tissues
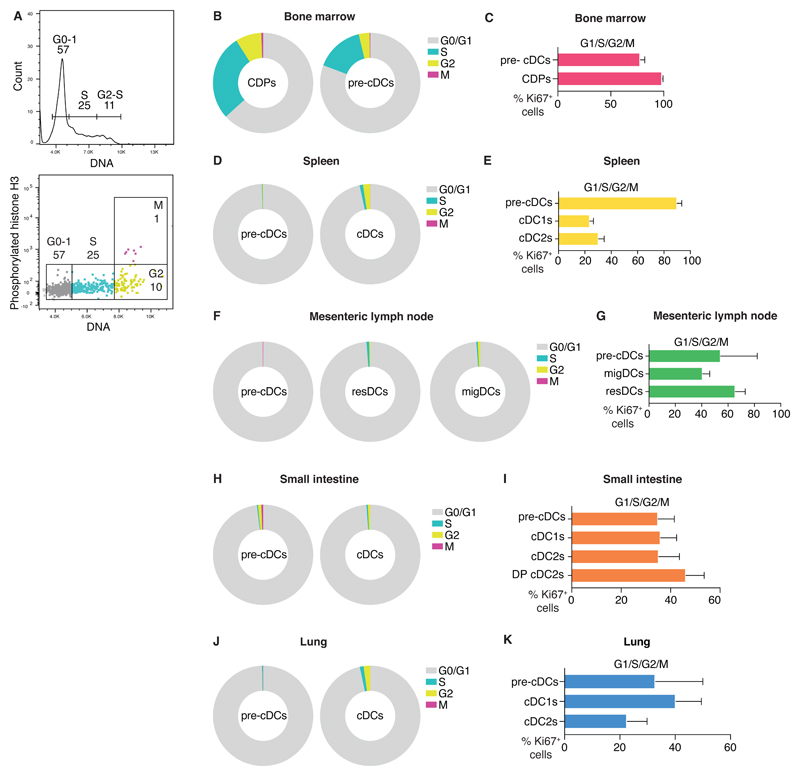
To analyze the cell cycle status of pre-cDCs and cDCs in tissues, we stained cells in bone marrow (BM), spleen, mesenteric lymph node (mesLN), small intestine (SI) and lung for both DNA content and phosphorylated histone H3 (phospho-H3). This technique allows us to identify cells in four different cell cycle phases, namely G0/G1, S, G2 and M (Fig. 1A). Consistent with their high turnover rate, a large fraction of BM CDPs and pre-cDCs was found in S/G2/M phases at any given time (Fig. 1B). That contrasted with pre-cDCs and fully differentiated cDCs in spleen, LN, lung and SI, in which the fraction of S/G2/M cells was much smaller, although not negligible, as previously observed (Fig. 1D, F, H, J, (8, 24, 25)). As the DNA/phospho H3 staining does not distinguish cells that are still cycling (G1) from resting cells that have exited the cell cycle (G0), we carried out a separate analysis for Ki67, which is expressed in the G1, S, G2 and M phases but not in G0. As expected, 100% of BM CDPs and 80% of BM pre-cDCs were Ki67+, consistent with cell cycle commitment (Fig. 1A, Fig. S1A). In spleen and mesenteric lymph nodes, 80% and 60% of pre-cDCs were Ki67+, while in lung and SI this figure was around 40% (Fig. 1E, G, I K). Differentiated cDCs in the spleen and mesenteric lymph nodes were also found to be Ki67+: 20% of splenic cDCs and 40-60% of lymph node cDCs. Similarly, we found that 30-40% of cDC1 and the two intestinal sub-types of cDC2 (CD103+ CD11b+ (double positive; DP) cDC2 and CD103- CD11b+ cDC2) were Ki67+ (Fig. 1I, Fig. S1B and (5, 26). In the lung, cDC1 and cDC2 stained for Ki67 at similar levels to cDCs in the SI (Fig. 1K, Fig. S1B). Overall, these data suggest that a substantial fraction of pre-cDCs that have entered peripheral tissues and of their cDC progeny remains in the G1 phase of the cell cycle and has not therefore become post-mitotic.
Fig. 1. Pre-cDCs and cDCs actively cycle in lymphoid and non-lymphoid organs.
(A) DNA content histogram (top) or dot plot with DNA content and phospho-H3 (bottom) of BM CDPs from one mouse as a representative example of cell cycle analysis. (B, D, F, H, J) Percentage of cells in G0/G1, S, G2 or M phases of the cell cycle determined as in (A) in the BM (B), spleen (D), mesenteric lymph node (F), SI (H) and lung (J). (C, E, G, I, K) Percentage of Ki67+ cells in in the BM (C), spleen (E), mesenteric lymph node (G), SI (I) and lung (K). Data in (B, D, F, H, J) are mean values from 6 C57BL/6 mice. Data in (C, E, G, I, K) are compiled from 12 C57BL/6 mice analyzed in 2 independent experiments. Error bars correspond to variation across mice using SD. Cells are gated as indicated in Materials and Methods.
Clec9aConfetti mice allow for multi-color labelling of cDCs in tissues
The above observations suggested the possibility that at least some tissue cDCs might be organized in patches of sister cells that arise through local cell division. To assess this, we resorted to multi-color fate mapping of cDC precursors. We crossed Clec9aCre mice (27) to Rosa26Confetti mice (23) (Fig. S2A; progeny henceforth called Clec9aConfetti). Both CDPs and pre-cDCs express DNGR-1, encoded by the Clec9a gene, and mice expressing Cre recombinase under the control of the Clec9a locus have been used to trace the cDC lineage in vivo (27). We expected that, in Clec9aConfetti mice, cDC precursors would become stochastically labelled with one of four fluorescent proteins (CFP, GFP, YFP or RFP) and transfer the fluorophore to daughter cells, allowing tracing of cDC clones. We validated Clec9aConfetti mice by flow cytometric spectral analysis (which allows separation of closely related fluorophores, including GFP and YFP) of spleen, mesLN, SI and lung cell suspensions. In all organs, we found labelled cDC1 and cDC2 (Fig. S2B-D). The percentage of labelled cells in Clec9aConfetti mice was lower than previously observed using Clec9aCre crossed to a Rosa26YFP single fluorophore fate reporter strain (27) (Fig. S2B), likely due to the complexity of the RosaConfetti locus, which reduces the efficiency of Cre-mediated recombination (28). This, together with the transience of Cre expression, leads to incomplete penetrance of the reporting event and causes labelling of only a fraction of cDCs, as previously reported (27). In contrast to cDCs, the frequency of labelled CD64+ cells, generally considered to correspond to monocytes and macrophages (MACROPHAGEs) (29, 30), was very low (Fig. S2B), as expected (27), although in lung, where CD64+ cells vastly outnumber CD64– cDCs, they contributed to a larger fraction of all labelled cells (Fig. S2C). As noted (23), the expression of the four fluorescent proteins in the Confetti reporter cassette was unequal, with clear underrepresentation of GFP+ cDCs (Fig. S2C, D). Importantly, cDC1s, but not cDC2s, are DNGR-1+ and hence express Cre recombinase after differentiation and become preferentially labelled in Clec9a-Cre-based reporter mice (27), including RosaConfetti (Fig. S2C, D). This also means that cDC1 can continue to recombine the inverted loxP sites of the RosaConfetti locus, switching back and forth between expression of GFP and YFP or of RFP and CFP. Consistent with that notion, GFP+YFP+ or CFP+RFP+ double positive cells were disproportionately more abundant among cDC1 than cDC2 subsets (Fig. S2C, D). Despite these limitations, the flow cytometric spectral analysis clearly demonstrates that CDP/pre-cDCs in Clec9aConfetti mice can be stochastically labelled with different fluorophores that are faithfully transmitted to daughter cells resident in tissues.
Imaging of Clec9aConfetti mice reveals cDC distribution in three dimensions
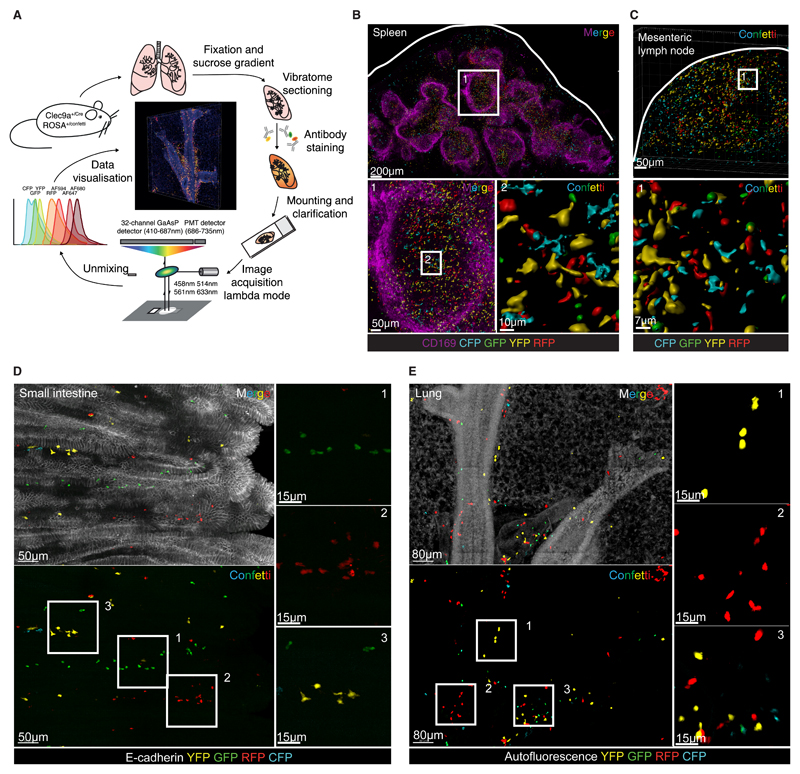
To visualize the spatial arrangement of labelled cDCs and assess possible clustering by fluorophore, we developed a protocol to fix and clarify tissues while preserving the Confetti fluorescent proteins (Fig. 2A). The method includes agarose embedding and vibratome sectioning, allowing cutting of 300µm sections, a thickness necessary to circumvent the scarcity of labelled cDCs in non-lymphoid tissues and to image enough cells for the analysis of clusters (Fig. 2A). It is also compatible with antibody staining of the sections (see below). Large tissue volumes (1020µm by 680µm in 100 z-steps of 3µm for SI, 1360µm by 1360µm in 40 z-steps of 5µm for lung) were then imaged by confocal microscopy using total spectrum acquisition (lambda mode scanning) followed by spectral unmixing to discriminate all fluorophores (Fig. 2A).
Fig 2. Spectral imaging of organs from Clec9aConfetti reveals the presence of single-color cDC clusters.
(A) Workflow of tissue processing, staining and imaging of Clec9aConfetti mouse organs. (B) 3D projection of a 300µm spleen section stained with anti-CD169. Confetti surfaces were generated with Imaris software to reduce autofluorescence. Zooming into T cell areas (1) was used to visualize Confetti+ cDCs (2). (C) 3D projection of a mesenteric lymph node with surfaces as in B. Square depicts selected zoom in area displayed at the bottom. (D) 3D projection of a 300µm vibratome section of the SI from a Clec9aConfetti mouse stained for E-cadherin to delineate the epithelium. (E) 3D projection of a 300µm vibratome section of the lung from a Clec9aConfetti mouse. Autofluorescence channel is displayed to visualize the lung structure.
Validation of the system was initially carried out using lymphoid organs such as spleen and mesLN. It revealed an intricate network of Confetti+ cells predominantly localized to T cell areas, as expected (Fig. 2B, C). However, the large number and high density of labelled cells in these organs precluded analysis of clustering by fluorescent protein (Fig. 2B, C). We therefore focused on non-lymphoid tissues such as the SI and lung, where Confetti+ cells were easily detected but sufficiently sparse to allow cluster analysis (Fig. 2D, E). Strikingly, visual inspection of images from SI and lung of Clec9aConfetti mice revealed that Confetti+ cells were often found in discrete single-color clusters within individual villi or around airways, respectively (Fig. 2D, E and Videos S1, S2). Labelling was largely lost in Clec9aConfetti mice crossed to Flt3l-/- mice (Fig. S3A, B and Videos S3, S4), which lack cDCs but not MACROPHAGEs (31, 32), confirming that Confetti+ cells were bona fide cDCs. This was true even for the lungs, where a considerable number of Confetti+ cells was found to be CD64+ by flow cytometry (Fig S2D). To clarify the identity of the latter, we carried out a separate flow cytometric analysis of Clec9atdTomato mice deficient in Flt3L. As expected, the frequency of Tomato-labelled CD64– cells was reduced in Flt3l-/- mice (Fig. S3C), consistent with a decrease in total cDC numbers (Fig. S3D). In contrast, the frequency of CD64+ cells, irrespective of CD11c and CD11b expression, was not reduced in Flt3l-/- mice, which, again, is as expected (Fig S3D). However, the frequency of Tomato-labelled CD64+ cells was reduced in Flt3l-/- mice (Fig S3D), intimating that they represent atypical cDCs that express CD64, as previously suggested (Schraml et al., 2013). Nevertheless, to avoid ambiguity, CD64+ cells were excluded from further analysis.
ClusterQuant 3D analysis indicates single-color cDC clustering in Clec9aConfetti mice
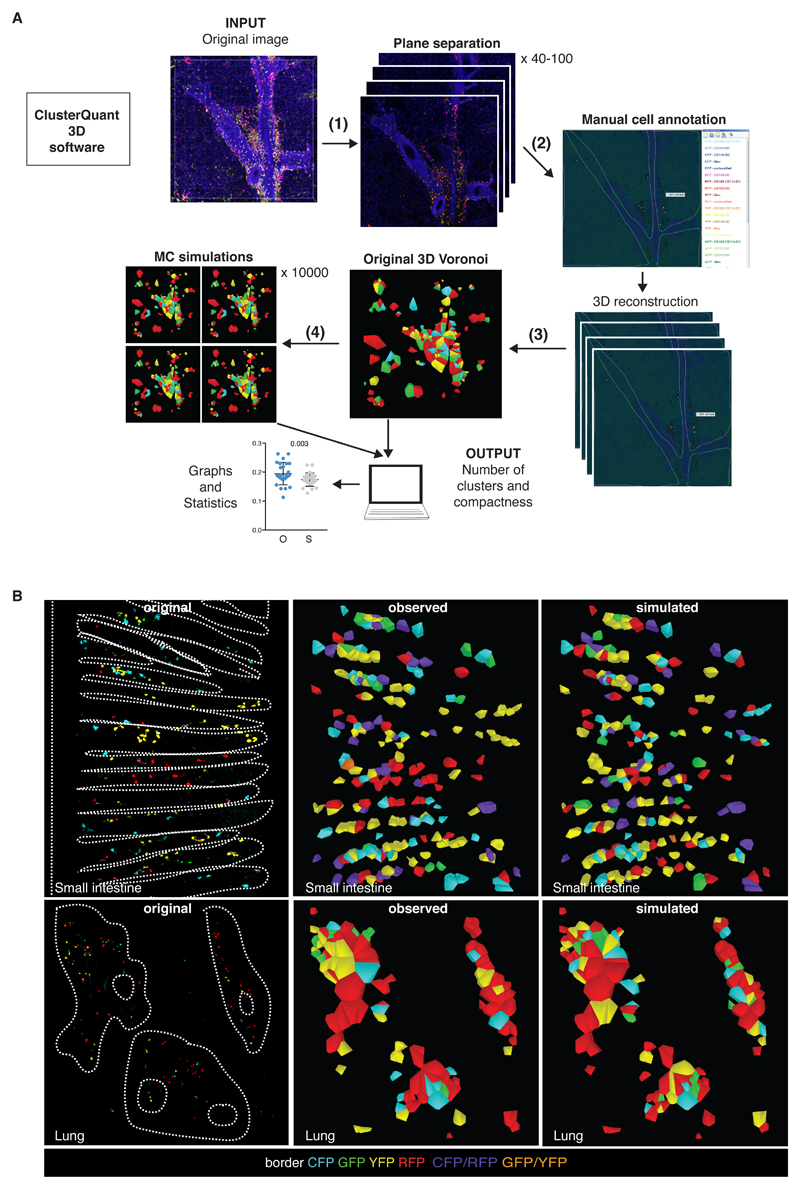
To quantify flurophore-based clustering of cDCs in SI and lung, we developed a 3D version of the ClusterQuant software previously used for analysis of cell clusters in single tissue planes (33). Briefly (Fig. 3A), the workflow involves different steps: (1) separation of 3D confocal images into individual z planes, corresponding to the optical slices used for image acquisition, (2) manual segmentation and annotation of cDCs in each plane, and (3) computation of 3D Voronoi polyhedrons using the x, y and z- cell coordinates. Each polyhedron contains all voxels closer to the centroid of that Confetti+ cell than to the centroids of all other cells and is used to compute neighbor and proximity relationships in subsequent analysis steps (Fig. 3B, C and Videos S5, S6). To aid tessellation and generate shapes that approximate cell volumes, in step (2) it is necessary to draw borders along anatomic barriers (crypts and airways). We also randomly inserted dummy cells (displayed as grey cells) into the spaces around Confetti+ cells, helping to remove artificially large Voronoi polyhedrons, avoiding false positive cluster formation and serving as a randomized control for the analysis (Fig. S4A, B, E – see below). Each Voronoi polyhedron was then assigned a color based on the fluorescent protein expressed by the annotated underlying cell (i.e. green Voronoi for a GFP cell). Double positive cells were assigned either purple (RFP+ CFP+) or orange (GFP+ YFP+). Adjacent Voronoi polyhedrons bearing the same color were considered a cluster (i.e., a group of 2 or more cells of the same color). The colors (but not the positions) of the Voronoi polyhedrons, representing individual cDCs, were then randomized in a step (4) through a Monte Carlo simulation with 10,000 possible realisations. Comparison of the original (O) vs. simulated (S) images was carried out by extracting relevant parameters such as number of clusters, cells per cluster and cluster compactness (Fig. 3A) to determine the probability that the observed clusters could have arisen by chance.
Fig 3. Analysis of single-color cDC clusters using ClusterQuant 3D.
(A) Workflow of plane separation (1), cell annotation (2), 3D Voronoi generation (3) randomisation (4) and analysis using ClusterQuant 3D. (B) Original images from the SI (top) and the lung (bottom) were annotated using ClusterQuant 3D software and converted to Voronoi polyhedrons (middle), which were then randomized through a Monte Carlo simulation (right). CD64 staining was used to exclude CD64+ cells from the analysis. Dashed lines indicate the structure of the SI (villi) or lung (airways), determined from CD11b staining or autofluorescence channel, respectively. Colors represent the different Confetti fluorescent proteins. Note the scarcity of double labelled cells, especially in lung.
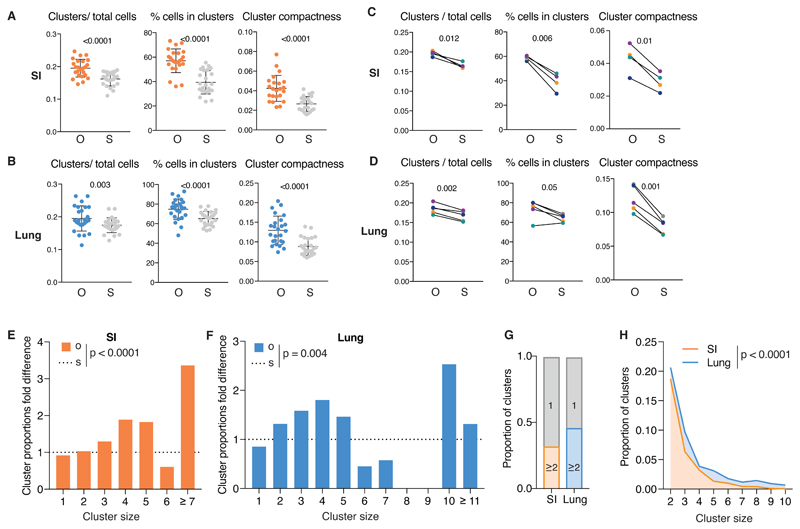
In all cases, the ClusterQuant 3D analysis clearly revealed a pattern of single-color cDC clustering in SI and lung that was not reproduced in randomized scenarios (Fig. 4). The difference between O and S images was statistically significant regardless of whether data were analyzed as number of clusters relative to total cell number, fraction of total cells in clusters (Fig. 4A, B) or number of clusters per unit volume of tissue (Fig. S4C, D). This was true for both SI and lung and was observed when images from all mice were pooled together (Fig. 4A, B) or when images were grouped per mouse (Fig. 4C, D). Confirming the validity of the 3D ClusterQuant algorithm, analysis of the randomly-placed dummy cells showed that they were not significantly clustered (Fig. S4E).
Fig 4. SI and lung cDCs are organized in spatially-restricted clusters of sister cells.
(A) From left to right: number of clusters of 2 or more cells normalised to number of cells, percentage of number of cells in clusters and cluster compactness from 24 images of SI from 4 mice. Each point represents one image; observed (O), orange, compared to simulations (S), grey. (B) As in (A) from 25 lung images from 5 mice. (C) Data from (A) grouped per mouse. Lines link the observed and simulated scenarios associated with each mouse. Colors correspond to individual mice (observed (O) and simulation (S)). (D) Data from B grouped per mouse as for C. (E-F) Proportion of clusters of the indicated size in observed scenario (O, orange) normalised to the simulated scenario (S, dashed line). Data are from SI (E) or lung (F) of a representative mouse. Other mice are shown in Fig. S4. (G) Proportion of all clusters analysed of size 1 (grey) or ≥2 in SI (orange) or lung (blue).. (H) Comparison of proportion of clusters of size 2 to 10 in the SI (orange) and in the lung (blue). In all cases, CD64 staining was used to exclude CD64+ cells from the analysis.. Statistical analysis in (B-E) used a paired t-test and in (F-G) used a chi-squared test.
In additional analyses using statistical data binning with chi-squared testing, we examined the size distribution of clusters and normalised them to the value expected to be obtained by chance, which was calculated from the simulated scenarios. We found that there was a greater proportion of large cDC single color clusters than expected by chance alone (Fig. 4E, F, Fig. S4F, G). Indeed, in some cases, we observed very large clusters (> 15 cells), which are exceedingly improbable in a randomized scenario. However, cDCs were not always in observable clusters and in more than half of the cases appeared as isolated cells (Fig. 4G). Interestingly, we also found that clusters in the lung were significantly larger than in the SI, indicative of tissue differences (Fig. 4G, H). We conclude that at least some cDCs in SI and lung and, possibly, other tissues of Clec9aConfetti mice form single-color clusters, which represent clones of sister cells.
Single-color cDC clusters are predominantly composed of a single cDC subset
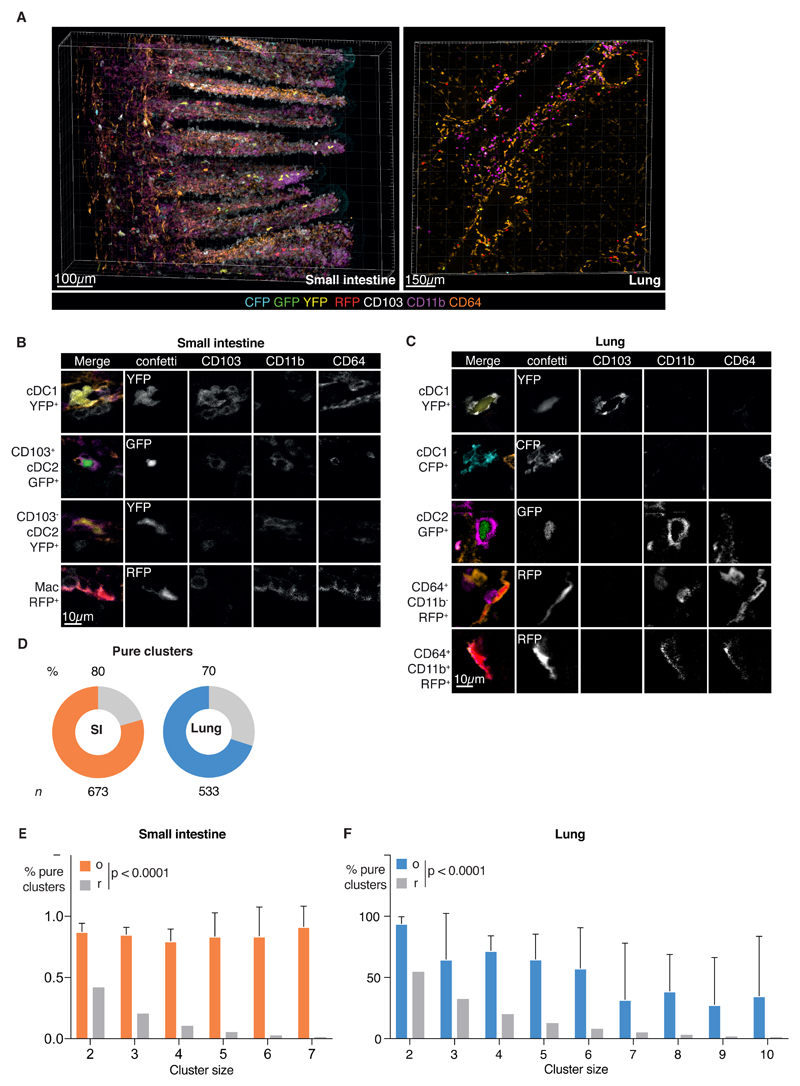
We next asked whether cDC clones comprise multiple subsets (i.e., arise from uncommitted pre-cDCs) or a single subset (i.e., originate from committed pre-cDC1s or pre-cDC2s or from dividing cDC1 and cDC2s) by analysing cluster composition. We used staining with Abs against CD11b and CD103 to allow discrimination between cDC1 and cDC2 and Ab against CD64 to exclude CD64+ cells (Fig. 5A-C and Videos S7, S8).
Fig 5. Single-color cDC clusters are predominantly composed of a single cDC subset.
(A) 3D projection of a Clec9Confetti image of SI (left) or lung (right) stained for CD103, CD11b and CD64. (B-C) Single z optical slices depicting individual cells from the SI (B) or lung (C) of Clec9Confetti mice; individual channels are shown on the right side of the merged image. (D) Pie charts representing percentage of pure clusters in SI (orange, 80%) or lung (blue, 70%) analyzed. Grey bar indicates mixed clusters. Data are pooled from all images and n indicates number of clusters analyzed. (E-F) Analysis of the proportion of pure clusters of cluster size 2 to 7 found in the SI (E, orange) or lung (F, blue) (o) vs the expected null distribution assuming random mixing (r, grey). Data are pooled from all images. Error bars correspond to variation across mice using SD. Statistical analysis was carried out using a Fishers exact test.
The distribution of fluorescent protein expression among cDC subsets in tissue sections of Clec9aConfetti mice as assessed by microscopy (Fig. S5A) was concordant with that observed in cell suspensions analyzed by spectral flow cytometry (Fig. S2C, D), cross-validating both experimental approaches. Cell composition analysis from ClusterQuant 3D analysis revealed that single-color clusters in both SI and lung often consisted of a single cDC subset (i.e., “pure”) although “mixed” clusters comprising cDC1 and cDC2 could also be observed (Fig. 5D, Fig. S5B). Pure clusters accounted for 80% of all clusters in the SI and 70% of clusters in the lung (Fig. 5D). The probability of a single-color cluster being pure by chance was calculated from the cluster size and the proportion of cDC1 and cDC2 cells in that tissue and revealed a far greater proportion of pure clusters than would be expected in a random scenario (Fig. 5E, F). Altogether, these data suggest that, during homeostasis, tissue cDC clones of sister cells are predominantly generated by local proliferation of incoming pre-committed cDCs and of their differentiated cDC progeny.
Single-color cDC clusters are lost during infection
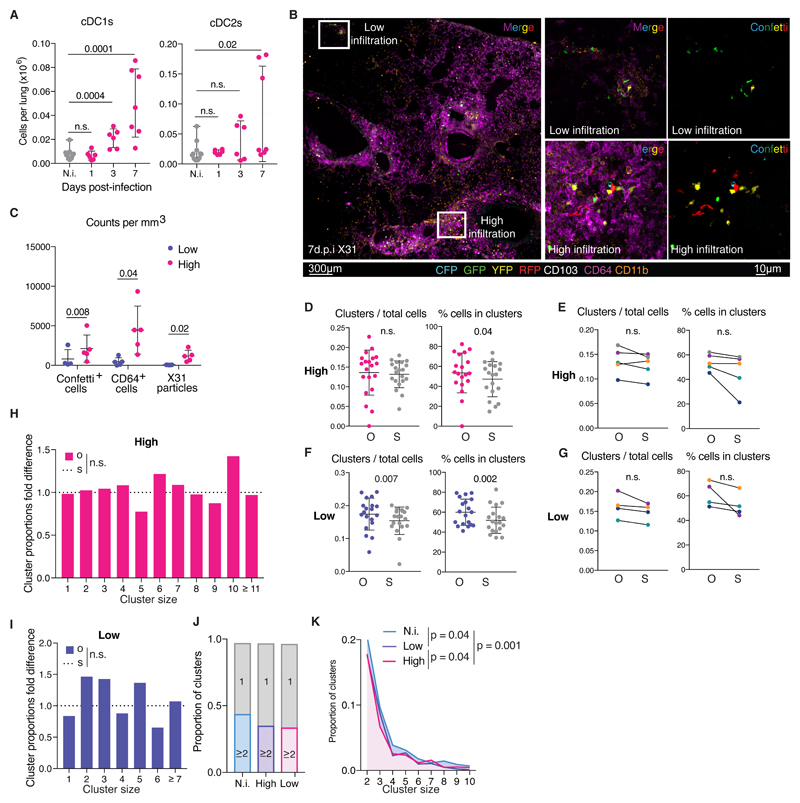
Next, we asked if cDC single-color clustering (and, inferentially, cDC clonality) in tissues holds true under inflammatory conditions when an increase in cDC numbers is often observed (16, 18–20). Clec9aConfetti mice were intranasally infected with influenza A virus (strain X31), which, over the course of a week, caused a large increase in lung cDC1 and cDC2 numbers, as assessed by flow cytometry (Fig. 6A). This was accompanied by large scale infiltration of innate immune cells into the lungs but did not affect the specificity of the genetic labelling of cDCs in Clec9aCre reporter mice (Fig. S6A-C). Images of infected lungs that were stained for viral proteins showed that the infection was patchy, with viral replication being confined to discrete foci (Fig. S6A and Video S9), as reported (34). At seven days post-infection, Confetti+ cells and CD64+ cells accumulated in much greater numbers around virus-containing foci than in uninfected regions (Fig. 6B, C). CD64+ cell infiltration was therefore used as a surrogate to discriminate high (infected) and low (uninfected) infiltrated areas (Fig. 6C). Notably, 3D ClusterQuant analysis revealed that the cDC single-color clustering that had been observed in steady state conditions was largely lost from highly infiltrated areas after infection, which no longer showed a statistically-significant difference between O and S scenarios irrespective of whether the data were pooled or analyzed by individual mouse (Fig. 6D, E, S7A and Video S10). Low infiltrated areas retained some clusters of single-color cDC in some of the mice but this did not reach statistical significance when all mice were considered together (Fig. 6F, G, Fig. S7B and Video S11).
Fig 6. Influenza A virus infection dilutes single-color cDC clustering.
(A) Number of cDC1s (left) or cDC2s (right) in lungs of Clec9aeYFP mice infected with influenza A virus (magenta) or non-infected (N.i., grey). (B) 3D projection of a Clec9Confetti lung section 7 dpi with Influenza A virus. (C) Quantification of Confetti+ cells, CD64+ cells and X31 particles from images with high (magenta) and low (purple) infiltration of cells. (D) Number of clusters of 2 or more cells normalised to number of cells and percentage of cells in clusters from 20 high infiltrated lung images from 5 mice. Each point represents one image; observed (O), magenta, compared to simulations (S), grey. (E) Data in D grouped per mouse. Lines link the observed and simulated scenarios associated with each mouse. Colors correspond to individual mice (observed (O) and simulations (S)). (F) Data as in D from 19 low infiltrated lung images from 4 mice. (G) Data in F grouped per mouse. (H-I) Proportion of clusters of the indicated size in original (O) normalised to the simulated (S, dashed line) scenario from the high (H) or low (I) infiltrated areas of a representative mouse. (J) Proportion of clusters of size 1 (grey) or ≥2 in non-infected lungs (N.i., blue) vs. high (magenta) and low (purple) infiltrated areas from lungs of infected mice. (K). Comparison of proportion of clusters of size 2-10 in non-infected lungs (N.i., blue, Fig.2), and high (magenta) and low (purple) infiltrated areas in the lungs of infected mice. Data correspond to the pool of all mice analyzed. Statistical analysis in (A) used an unpaired t-test in 2 independent experiments with 3mice/group, (C-G) paired t-test and in (H-K) used a chi-squared test.
We extended the analysis by comparing cluster size distribution between uninfected lungs and areas of low or high infiltration in infected mice (Fig. 6H-K). This confirmed that the data fitted a random scenario (Fig. 6H, I), although interestingly some low infiltrated areas in some mice still displayed statistically significant formation of clusters (Fig. S8). Despite the latter, overall there was a slight increase in the frequency of cDCs that were not in a cluster compared to the situation in uninfected mice (Fig. 6J). We found statistically significant differences at the level of larger clusters, which could be preferentially found in lungs from uninfected mice (Fig. 6K). Altogether, these results show that the single-color clustering of cDCs in lung is significantly reduced following influenza A virus challenge, predominantly in areas with active infection but also, to a lesser extent, in areas away from infectious foci.
Infection-driven loss of single-color cDC clustering correlates with influx of pre-cDCs
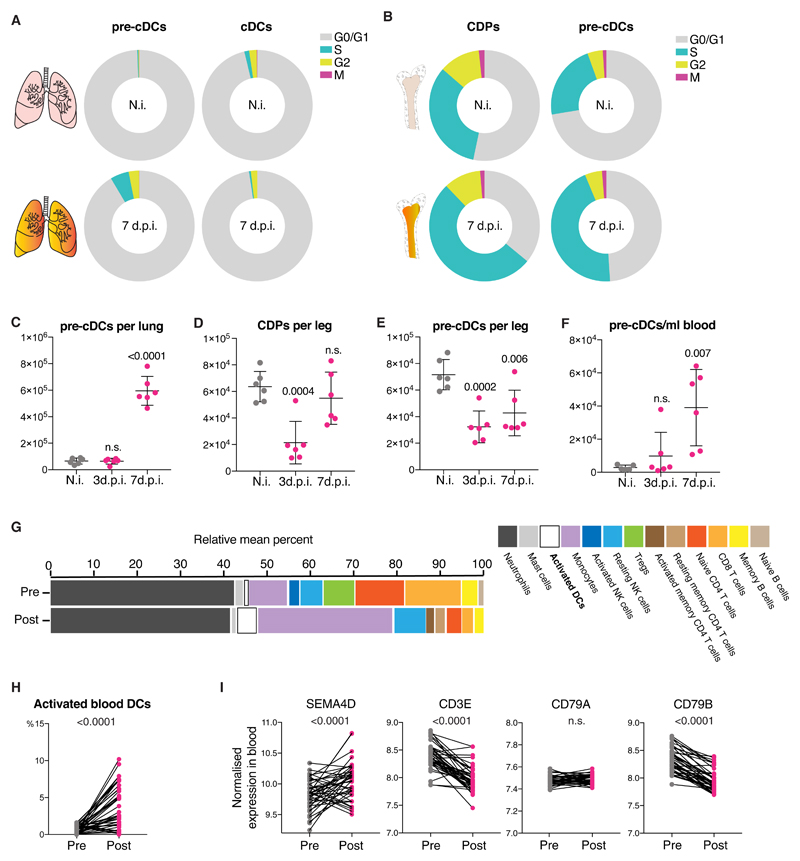
The above data suggested that infection-driven increases in local cDC numbers were not likely to be a result of increased local clonal expansion. Indeed, flow cytometric analysis of cell cycle status showed that infection led, if anything, to a decrease in the frequency of cDC1 and cDC2 in S/G2/M cell cycle phases and a reduction in % cells positive for Ki67 (Fig. 7A, Fig. S9A). Pre-cDC in lung and pre-cDC and CDP in BM did not show any major changes in Ki67 staining (Fig. S9A, B) although more of them were found to be in S phase at d7 post infection compared to uninfected mice (Fig. 7A, B). Therefore, the increase in cDC numbers in the lungs of mice post infection with influenza A virus (Fig. 6A) does not seem to be due to an increase in proliferation of cDCs or their immediate lung precursors.
Fig 7. Influenza A virus infection increases lung cDC numbers by recruiting BM progenitors.
(A-B) Percentage of lung (A) and BM (B) pre-cDCs and cDCs in G0/G1, S, G2 or M phases of the cell cycle determined as in Fig 1. Data are mean values from 6 non-infected (top) or 6 C57BL/6 mice 7 dpi with influenza virus (bottom). (C-F) Numbers of BM CDPs (D) or pre-cDCs in lung (C), BM (E) or blood (F) in non-infected (grey), or influenza A virus-infected C57BL/6 mice (magenta) at 3dpi or 7dpi. Each dot represents one mouse of 6 per group from one representative experiment. (G) Relative mean percentage of cells from peripheral blood of 41 patients pre and post natural infection with influenza A virus obtained from microarray data using CIBERSORT. (H) Percentage of activated blood DCs in individual patients from G. (I) Expression of SEMA4D, CD3E, CD79A and CD79B in peripheral blood from patients before (pre, grey) or in the first 48h of symptoms after natural infection with influenza A virus virus (post, magenta). Statistical analysis in C-F was based on an unpaired t test in a single experiment with 6 mice/group. Statistics in H, I employed a paired t-test.
To ask if it could be attributable to changes in the dynamics of lung seeding by pre-cDC, mice were examined at different time post-infection. At day 7 post-challenge, influenza virus infected mice had many more pre-cDCs in their lungs than uninfected controls (Fig. 7C). We employed markers used to distinguish pre-cDC1s and pre-cDC2s (10, 11) (with the caveat that such markers have not been validated in lung) and found that the infection-associated increase in lung pre-cDCs predominantly represented uncommitted cells (Fig. S9C), consistent with the fact that both cDC1 and cDC2 increased equally (Fig. 6A). In contrast to lung, there was loss of cDCs progenitors from BM, which reached their lowest numbers at day 3-4 post-infection and recovered by day 7 (Fig. 7D, E). As in lung, there was no major bias in terms of pre-cDC subset distribution in BM (Fig S9D).
These results suggested that high demand for cDCs induced by infection is met by rapid mobilization of cDC progenitors from the BM. Consistent with that notion, there was a clear increase in total number of pre-cDCs circulating in peripheral blood of mice at 7 days post-infection, when the increase in lung pre-cDCs and cDCs became apparent (Fig. 7F), with no differences in pre-cDC1/pre-cDC2 commitment (Fig. S9E). To extend these data to humans, we carried out an analysis of transcriptome datasets from peripheral blood of patients prior to or post infection with influenza A virus (35) using the CIBERSORT algorithm (36). This revealed a marked increase in the blood frequency of DCs, annotated in CIBERSORT as “activated DCs” (Fig. 7G, H), consistent with a demand-driven increase in DCpoiesis. To assess whether pre-cDC numbers were also increased in human blood, as in mice, we assessed levels of transcripts for SEMA4D, a recently identified marker for cDC progenitors (37). SEMA4D transcripts were increased post infection (Fig. 7I). Although SEMA4D can also be expressed by T and B cells, T and B cell markers (CD3E or CD79A/B) were either unchanged or decreased in the same datasets following influenza virus infection (Fig. 7I). Taken together, these data suggest that, in mice and, possibly, humans, influenza virus infection leads to local demand for cDCs that is met not by increased local clonal expansion of pre-cDC and their progeny but by communicating need to the BM, resulting in an efflux of pre-cDCs into blood and influx into lungs.
Discussion
The parameters underlying cDCpoiesis remain poorly understood. Here, we use a genetic model coupled to image analysis and three-dimensional cluster quantification to analyze how cDC precursors seed tissues at the single-cell level in the absence of cell transfer. We reveal that pre-cDCs in the steady-state enter peripheral tissues and can divide locally before differentiating into cDCs, which display residual proliferative capacity. This leads to formation of discrete clones of cDCs that remain in close proximity. Notably, we find that these sisters are predominantly composed of either cDC1 or cDC2, possibly providing in vivo corroboration for the notion that cDC subset commitment can occur at the pre-cDC level (10, 11). Finally, we demonstrate that cDC generation is an elastic process that responds to external tissue demand by exporting pre-cDCs from BM at times of need. The rapid influx of such pre-cDCs into tissues and reduction in local cell division likely leads to accelerated intermingling of clones and correlates with loss of single-color clustering in multi-color fate-mapping mice.
Quantification of images from tissues of Clec9aConfetti mice revealed that 45% of cDCs in lung and 35% of cDCs in SI are in single-color clusters of 2 or more cells. It is remarkable that single-color clusters can be detected at all. Because cDCs are motile, especially when activated, it is likely that clustering of clones dilutes over time. Motility will additionally cause cDC clones to intermingle and ongoing cDC death will lead to cluster dissolution. Additionally, the fact that cDC1 can switch fluorescent protein expression because of continued Cre expression can also break up single-color clusters (although in some case this can be accounted for in the analysis as the half-life of the proteins is sufficiently long to result in double labelling). Finally, the use of dummy cells to aid with segmentation in the analysis may inadvertently lead to artificial separation of single-color clusters. For all these reasons, the degree of cDC single-color clustering that we observe in tissues is likely to be an underestimate of the true extent of tissue cDC clonality and many of the isolated cells in tissues may therefore in fact have previously been part of a single-color cluster. Interestingly, large clones are sometimes observed. The issues discussed above (e.g., cDC migration) will disproportionately impact large clones, causing a bias towards detection of small single-colour clusters and isolated cells. The finding of some large clones suggests therefore the existence of tissue niches that allow for greater pre-cDC/cDC local proliferation and/or that prolong cDC half-life (25). Such niches may be more common in the lung than in SI as we noted a clear tendency for single-color cDC clusters in that organ to be larger. Why clonality does not result in observable patches of cDC1 vs cDC2 in tissues can be understood from the fact that large clones are relatively rare and that we visualize only a fraction of the tissue cDC network because of incomplete penetrance of the Cre-mediated recombination event. The superimposition of the mosaic pieces composed of cDC1 and cDC2 clones and single cells is expected to result in spatial mixing of the subsets, also explaining why we do not see obvious single color clusters in regions where cDCs are too abundant, such as T cell areas of lymph nodes and spleen.
Experiments with cells in vitro, sorted ex vivo or transferred as bulk populations into mice have suggested that individual pre-cDCs can be either pre-committed to generate cDC1 or cDC2 or can be bi-potent and generate either cDC subset (10, 11). Here, we revisited this question in Clec9aConfetti mice where the composition of the Confetti+ clones is analyzed in an unperturbed state. We show that cDC clusters in lung and SI are predominantly pure cDC1 or cDC2 consistent with the possibility that they arise primarily from committed pre-cDC1s and pre-cDC2s. We also find that cDC1s and cDC2s retain residual capacity to proliferate after differentiation, which could contribute to the purity of the cDC clusters observed irrespective of pre-cDC commitment. The fact that cDC1s continue to express Cre could in theory introduce a bias towards detecting pure clusters of cDC1 expressing two fluorescent proteins surrounded by cDC2 expressing one of the two proteins. However, this was not observable in our images and the rarity of doubly labelled cDC1 events cannot explain the extremely high fraction of pure clusters detected. It should be noted that not all clusters are pure and that some mixed cDC ones are found in both SI and lung, possibly providing in vivo evidence for the existence of uncommitted pre-cDCs capable of seeding tissues and giving rise to sister cells of different fates.
Infection or inflammation are often accompanied by a local increase in macrophage and cDC numbers, which can be met through local proliferation or increased precursor recruitment. Tissue macrophages are self-renewing and can proliferate more rapidly in response to injury-induced signals and cytokines (38–40). Furthermore, blood monocytes can enter tissues upon demand and differentiate into cells that resemble tissue macrophages (41). Monocytes can also differentiate into cells that have cDC features but are distinct from those that arise from regular cDCpoiesis (42). Therefore, increased demand for cDCs in the periphery can only be met through increased generation of cDC precursors in BM or by greater mobilization and/or proliferation of pre-cDCs in tissues. We show that influenza A virus infection causes an increase in lung cDC numbers, which is not accompanied by increased local proliferation of pre-cDC or their differentiated progeny. Rather, it is accompanied by efflux of pre-cDCs from BM, which increase in number and frequency in peripheral blood and enter tissues, leading to increased seeding. In Clec9aConfetti mice, this acute influx of pre-cDCs into lungs results in dissolution of pre-existing single-color cDC clusters, a process that might be exacerbated by increased cDC mobility leading to increased intermingling of pre-existing clones and migration to mediastinal lymph nodes. Differently from a prior report (20), we did not observe a selective increase in cDC2 in influenza virus-infected lungs and, consistent with that observation, pre-cDC in BM or lung did not display any infection-induced changes in phenotype suggestive of a bias towards cDC1 or cDC2 commitment (10, 11). Whether this relates to the strain of influenza virus, the severity of the infection or the fact that the previous study (20) did not necessarily discriminate bona fide cDC2 from monocyte-derived cells remains to be established. Overall, our findings reveal that infected tissues can communicate to the BM their need for increased cDC, resulting in a release of pre-cDCs into blood and transient loss of cDC progenitors from their site of origin. By analogy with the increased release of granulocytes into the circulation during infection or inflammation (43, 44), the fact that cDC generation responds to demand might aptly be termed “emergency DCpoiesis”. The signals that regulate emergency DCpoiesis are likely to constitute useful targets for immunotherapeutic approaches to cancer, infectious disease and autoimmunity.
Materials and Methods
Mice
Clec9a-Cre, ROSA26-YFP (45), ROSA26-Confetti (23), Flt3l-/- (Taconic Biosciences) and C57BL/6J mice were bred at The Francis Crick Institute in specific pathogen-free conditions. All transgenic mouse lines were backcrossed to C57BL/6J. Six to twenty-week-old mice were used in all experiments unless otherwise specified. All animal experiments were performed in accordance with national and institutional guidelines for animal care.
Infection with influenza virus
Mice were anaesthetised via inhalation of isoflurane. Mice were infected intranasally with 35000 TCID50 of influenza A X31 (H3N2) in 30µl of PBS. Mice were monitored daily for weight loss and signs of infection and sacrificed at day 1, 3 and 7 post-infection.
Flow cytometry
Up to 4 million cells (see isolation protocol in Supplementary Methods) were pre-incubated with blocking anti-CD16/32 in FACS buffer for 10 min at 4°C and then stained for 20 min at 4°C with staining cocktail in FACS buffer in the presence of anti-CD16/32. DAPI was used to exclude dead cells, except for when cells were fixed. In the latter case, dead cells were excluded by live/dead fixable blue or aqua dye (Invitrogen). For Ki67, DNA content (FxCycle, Invitrogen) and phospho-H3 staining cells were fixed and permeabilised using Foxp3 Fix/Perm buffer set (eBioscience, 00-5523-00). For DNA content examination, samples were collected on a LSR Fortessa flow cytometer (BD Biosciences) or a SP6800 Spectral Analyzer (Sony) and analyzed using FlowJo 9 or 10 software (TreeStar Inc.). Gating strategies are shown in Fig. S10-12, including the discrimination of the four Confetti fluorophores by spectral analysis. Antibodies used for flow cytometry are listed in Table S2.
Microscopy
Mice were perfused with 20ml of PBS and 10ml of Antigenfix (Diapath). 1 ml of melted 2% low melting point agarose (Invitrogen) in PBS was inserted through the trachea. Lungs, spleen and lymph nodes were then removed and fixed overnight in Antigenfix at 4°C. The proximal SI (mostly duodenum) was also removed and contents were flushed with ice cold PBS and Antigenfix, and fixed for 2h at 4°C. Small intestines were subsequently cut longitudinally, rolled into a “Swiss roll” and fixed overnight in Antigenfix at 4°C. After fixation, tissues were transferred to 30% sucrose in PBS for 24h at 4C. Lungs, spleen and SI were embedded in 4% agarose in PBS and 300µm sections were cut using a Leica VT1200S vibratome and collected in PBS. Lymph nodes were unsectioned and imaged as whole mounts. Tissue sections were stained as explained in Supplementary Methods. Antibodies used in microscopy are listed in table S3. Sections were mounted on slides in the clearing solution Rapiclear 1.47 (SunJin Lab) for image acquisition while lymph nodes were clarified instead using uDISCO (46) and mounted on a metallic glass-bottom dish. Images were acquired on a LSM880 inverted confocal microscope as explained in supplementary information. Images and Videos were generated after adjusting channels using Imaris software.
Cluster analysis
To analyze the clustering of cells in 3 dimensions, we used a 3D version of ClusterQuant 2D (33) as described in detail in the Supplementary Methods. For cluster composition, we used_a probability-based method based on the ratio of cDC1:cDC2 in lung and SI as explained in supplementary methods and Fig.S5C, D.
Human influenza dataset analysis
Human microarray data from Influenza A virus infected cohort ((35), GEO GSE68310) was analysed using CIBERSORT(36). For gene expression data, samples were normalized and compared in a paired basis. Baseline was compared to first onset of symptoms.
Statistics
Statistical analyzes were performed using GraphPad Prism software (GraphPad), MATLAB (Mathworks) or RStudio. Statistical test used is specified in each figure legend.
Supplementary Material
One-sentence summary.
Many tissue cDCs are maintained as single subset clones, which disperse upon infection with acute recruitment of pre-cDCs.
Acknowledgements
This work was supported by The Francis Crick Institute, which receives core funding from Cancer Research UK (FC001136), the UK Medical Research Council (FC001136), and the Wellcome Trust (FC001136) and by ERC Advanced Investigator Grants (AdG 268670 and 786674), as well as, in part, by the Intramural Research Program of NIAID, NIH. M.C-C. and J.v.B. were supported by Boehringer Ingelheim Fonds. BS is member of the DFG funded Collaborative Research Center 914 (project A11). We thank members of the Immunobiology Laboratory for helpful discussions and suggestions. We thank the Crick Biological Resources, Flow Cytometry and Microscopy Facilities for assistance, as well as Maria Daly (Laboratory of Molecular Biology, Cambridge) and Roy Bongaerts (Sony) for facilitating access to spectral flow cytometry. We thank Andreas Wack for kindly sharing reagents and expertise.
Footnotes
Author contributions
M.C-C, J.v.B. and C.R.S. designed experiments, analyzed data and wrote the manuscript. M.C-C and J.v.B. conducted experiments with assistance from S.W., D.H., R.J., P.C., N.R., B.F., S.A., E.V., J.v.R., E.S. and F.K. for data analysis and study design. M.C-C, R.J. and P.C. did statistical analysis. H.C., M.B., M.G. and R.N.G. provided key reagents and advice. B.U.S. and N.R. helped with generating mice. C.R.S. supervised the project. All authors reviewed and edited the manuscript.
The authors declare that they have no competing interests.
References
- 1.Merad M, Sathe P, Helft J, Miller J, Mortha A. The Dendritic Cell Lineage: Ontogeny and Function of Dendritic Cells and Their Subsets in the Steady State and the Inflamed Setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naik SH, Metcalf D, van Nieuwenhuijze A, Wicks I, Wu L, O'Keeffe M, Shortman K. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7:663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 4.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 5.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, et al. The origin and development of nonlymphoid tissue CD103+ DCs. Journal of Experimental Medicine. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt W-D, Shakhar G, Jung S. Intestinal Lamina Propria Dendritic Cell Subsets Have Different Origin and Functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, et al. Origin of the Lamina Propria Dendritic Cell Network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu F-F, Randolph GJ, Rudensky AY, Nussenzweig M. In Vivo Analysis of Dendritic Cell Development and Homeostasis. Science. 2009;324:392–396. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naik SH, Sathe P, Park H-Y, Metcalf D, Proietto AI, Dakic A, Carotta S, O'Keeffe M, Bahlo M, Papenfuss A, Kwak J-Y, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 10.Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HRB, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, Renia L, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 11.Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, KC W, Kretzer NM, Briseño CG, Durai V, Bagadia P, Haldar M, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8[alpha]+ conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.See P, Dutertre C-A, Chen J, Günther P, McGovern N, Irac SE, Gunawan M, Beyer M, Händler K, Duan K, Sumatoh HRB, et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;66:eaag3009–15. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochweller K, Miloud T, Striegler J, Naik S, Hämmerling GJ, Garbi N. Homeostasis of dendritic cells in lymphoid organs is controlled by regulation of their precursors via a feedback loop. Blood. 2009:1–12. doi: 10.1182/blood-2008-11-188045. [DOI] [PubMed] [Google Scholar]
- 14.Sevilla N, McGavern DB, Teng C, Kunz S, Oldstone MBA. Viral targeting of hematopoietic progenitors and inhibition of DC maturation as a dual strategy for immune subversion. J Clin Invest. 2004;113:737–745. doi: 10.1172/JCI20243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuniga EI, McGavern DB, Pruneda-Paz JL, Teng C, Oldstone MBA. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat Immunol. 2004;5:1227–1234. doi: 10.1038/ni1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson C, Ingman M, Jo Wick M. Elevated neutrophil, macrophage and dendritic cell numbers characterize immune cell populations in mice chronically infected with Salmonella. Microbial Pathogenesis. 2006;41:49–58. doi: 10.1016/j.micpath.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Singh P, Yao Y, Weliver A, Broxmeyer HE, Hong S-C, Chang C-H. Vaccinia Virus Infection Modulates the Hematopoietic Cell Compartments in the Bone Marrow. Stem Cells. 2008;26:1009–1016. doi: 10.1634/stemcells.2007-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welner RS, Pelayo R, Nagai Y, Garrett KP, Wuest TR, Carr DJ, Borghesi LA, Farrar MA, Kincade PW. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–3761. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GeurtsvanKessel CH, Willart MAM, van Rijt LS, Muskens F, Kool M, Baas C, Thielemans K, Bennett C, Clausen BE, Hoogsteden HC, Osterhaus ADME, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. Journal of Experimental Medicine. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballesteros-Tato A, León B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat Immunol. 2010;11:216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farache J, Koren I, Milo I, Gurevich I, Kim K-W, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal Bacteria Recruit CD103+ Dendritic Cells into the Intestinal Epithelium to Sample Bacterial Antigens for Presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano H, Lyons-Cohen MR, Whitehead GS, Nakano K, Cook DN. Distinct functions of CXCR4, CCR2, and CX3CR1 direct dendritic cell precursors from the bone marrow to the lung. Journal of Leukocyte Biology. 2017 doi: 10.1189/jlb.1A0616-285R. jlb.1A0616–285R–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Kabashima K, Banks TA, Ansel KM, Lu TT, Ware CF, Cyster JG. Intrinsic Lymphotoxin-β Receptor Requirement for Homeostasis of Lymphoid Tissue Dendritic Cells. Immunity. 2005;22:439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 26.Scott CL. Characterisation of dendritic cells in the intestine. PhD thesis. Glagow Thesis Service; 2014. pp. 96–134. http://theses.gla.ac.uk/4829/ [Google Scholar]
- 27.Schraml BU, van Blijswijk J, Zelenay S, Whitney PG, Filby A, Acton SE, Rogers NC, Moncaut N, Carvajal JJ, Reis e Sousa C. Genetic Tracing via DNGR-1 Expression History Defines Dendritic Cells as a Hematopoietic Lineage. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 29.Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D, Bain CC, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. European Journal of Immunology. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 30.Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, Gregoire C, Malissen B, Guilliams M. CD64 Expression Distinguishes Monocyte-Derived and Conventional Dendritic Cells and Reveals Their Distinct Role during Intramuscular Immunization. J Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 31.McKenna HJ, peschon JJ, miller RE. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 32.Waskow C, Liu K, Darrasse-Jèze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghigo C, Mondor I, Jorquera A, Nowak J, Wienert S, Zahner SP, Clausen BE, Luche H, Malissen B, Klauschen F, Bajenoff M. Multicolor fate mapping of Langerhans cell homeostasis. Journal of Experimental Medicine. 2013;210:1657–1664. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuyama S, Katsura H, Zhao D, Ozawa M, Ando T, Shoemaker JE, Ishikawa I, Yamada S, Neumann G, Watanabe S, Kitano H, et al. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nature Communications. 2015;6:1–8. doi: 10.1038/ncomms7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, Shaw CA, et al. Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections - A Prospective Cohort Study. PLoS Pathog. 2015;11:e1004869–29. doi: 10.1371/journal.ppat.1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573–14. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieweke MH, Allen JE. Beyond Stem Cells: Self-Renewal of Differentiated Macrophages. Science. 2013;342 doi: 10.1126/science.1242974. 1242974–1242974. [DOI] [PubMed] [Google Scholar]
- 39.Minutti CM, Jackson-Jones LH, Garcia-Fojeda B, Knipper JA, SUtherland TE, Logan N, Ringqvist E, Guillamat-Prats Raque, Ferenbach DA, Artigas A, Stamme C, et al. Local amplifiers of IL-4Ra–mediated macrophage activation promote repair in lung and liver. 2017:1–7. doi: 10.1126/science.aaj2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Y, Weinstein JS, Licona-Limon P, Schmid ET, Pelorosso F, Gagliani N, et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. 2017;356:1072–1076. doi: 10.1126/science.aai8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varol C, Mildner A, Jung S. Macrophages: Development and Tissue Specialization. Annu Rev Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- 42.Briseño CG, Haldar M, Kretzer NM, Wu X, Theisen DJ, KC W, Durai V, Grajales-Reyes GE, Iwata A, Bagadia P, Murphy TL, et al. Distinct Transcriptional Programs Control Cross-Priming in Classical and Monocyte-Derived Dendritic Cells. CellReports. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boettcher S, Manz MG. Regulation of Inflammation- and Infection-Driven Hematopoiesis. Trends in Immunology. 2017;38:1–13. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:1–13. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 45.Srinivas S, Watanabe T, Lin C-S, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001:1–8. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, Ertürk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nature Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- 47.Vray B, Plasman N. Separation of murine peritoneal macrophages using percoll density gradients. J Immunol Methods. 1994:53–59. doi: 10.1016/0022-1759(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 48.Wienert S, Heim D, Kotani M, Lindequist BR, Stenzinger A, Ishii M, Hufnagl P, Beil M, Dietel M, Denkert C, Klauschen F. CognitionMaster: an object-based image analysis framework. Diagnostic Pathology. 2013;8:1–1. doi: 10.1186/1746-1596-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.