Abstract
Background
Orolabial herpes simplex virus type 1 (HSV-1) infection has a wide spectrum of severity in immunocompetent persons. To study the role of viral genotype and host immunity, we characterized oral HSV-1 shedding rates and host cellular response, and genotyped viral strains, in monozygotic (MZ) and dizygotic (DZ) twins.
Methods
A total of 29 MZ and 22 DZ HSV-1–seropositive twin pairs were evaluated for oral HSV-1 shedding for 60 days. HSV-1 strains from twins were genotyped as identical or different. CD4+ T-cell responses to HSV-1 proteins were studied.
Results
The median per person oral HSV shedding rate was 9% of days that a swab was obtained (mean, 10.2% of days). A positive correlation between shedding rates was observed within all twin pairs, and in the MZ and DZ twins. In twin subsets with sufficient HSV-1 DNA to genotype, 15 had the same strain and 14 had different strains. Viral shedding rates were correlated for those with the same but not different strains. The median number of HSV-1 open reading frames recognized per person was 16. The agreement in the CD4+ T-cell response to specific HSV-1 open reading frames was greater between MZ twins than between unrelated persons (P = .002).
Conclusion
Viral strain characteristics likely contribute to oral HSV-1 shedding rates.
Keywords: Herpes simplex virus, herpes labialis, cold sores, twins, CD4 T cells
Oral herpes simplex virus type 1 (HSV-1) shedding rates, host cellular responses, and genotyped viral strains were characterized in monozygotic and dizygotic twins to determine whether the HSV-1 infection severity is influenced by host or viral genotype. Viral strain characteristics contribute to oral HSV-1 shedding rates.
Herpes simplex virus type 1 (HSV-1) is a ubiquitous pathogen that causes chronic infection [1]. Seroprevalence is high, with 67% of individuals aged 0–49 years infected globally and an estimated 3.7 billion individuals worldwide living with the infection, making it an important public health issue [2, 3]. Historically, HSV-1 is acquired during childhood through the oral route, and siblings can perhaps share viral strains [4]. Following acquisition, the virus undergoes retrograde axonal transport to establish latency in neural ganglia. Reactivation leads to viral shedding at epithelial surfaces, which can be asymptomatic or cause mucocutaneous disease. There is a remarkable spectrum of clinical severity among people with HSV-1 infection, ranging from asymptomatic disease to rare but major complications such as encephalitis or disseminated disease, even in immunocompetent persons [2, 5].
HSV-1 shedding frequency in the oral cavity varies widely in immunocompetent persons and the biological underpinnings for this variability are not understood. Twin studies have been used in pathogenesis studies to isolate the contribution of genetics to infectious diseases [6]. This is the first study to determine how HSV-1 infection severity, defined by oral shedding of HSV-1 DNA, is influenced by host or viral genotype and if host genetics influence CD4+ T-cell responses to HSV-1 proteins in twins.
METHODS
Study Population
Healthy, immunocompetent, human immunodeficiency virus type 1 (HIV-1)–seronegative, monozygotic (MZ) or dizygotic (DZ) twin pairs were recruited by the Virology Research Clinic from the community-based University of Washington Twin Registry [7]. Subjects were tested by Western blot for serum antibody to HSV-1 and HSV-2 [8]. HSV-1 antibody–positive persons were enrolled regardless of HSV-2 serostatus. Demographic and clinical information was obtained, and swab specimens were self-collected daily from the oral mucosa by participants, with the goal of collecting specimens on 60 consecutive days. Participants refrained from undergoing topical or systemic antiviral therapy during the study. Informed consent from participants and approval by the University of Washington Institutional Review Board and the Washington State Attorney General’s Office were obtained. Participants were asked to self-designate as a MZ or DZ twin [7]. Genomic zygosity testing was performed if a specimen was available. DNA was isolated from a blood specimen collected using the PAXgene Blood DNA system (Qiagen, Germantown, MD). The AmpFlSTR Identifiler (ThermoFisher, Waltham, MA) polymerase-chain reaction (PCR)–based test of 15 loci and a gender marker was used [9].
HSV Shedding
Swab specimens from the oral mucosa were obtained by participants and placed in 1 mL of 1× Proteinase K digestion buffer, as previously described [10]. Samples were evaluated for HSV DNA, using a real-time quantitative fluorescent probe PCR assay (Taqman; Applied Biosystems, Foster City, CA) [11]. Samples with >150 copies/mL were considered positive [12].
HSV-1 Genotyping
When samples were available from both twins, HSV-1 DNA was genotyped to determine if the twins were infected with the same or different strains. HSV-1 strains were considered the same or different on the basis of findings of Sanger sequencing and whole-genome sequencing (WGS). Five variable regions with informative single-nucleotide polymorphisms (SNPs) were PCR amplified (Supplementary Table 1) with methods similar to those previously reported [13, 14] and were deposited in GenBank (Supplementary Table 2) after low-quality termini were trimmed. Some regions were independently amplified and sequenced twice. The HSV-1 genome regions sequenced contained 1783–1933 base pairs, depending on which PCR primer yielded an amplicon (Supplementary Table 1). A review of GenBank in July 2012 showed these genomic areas had a substantial number of informative SNPs, insertions, or deletions. Strains were designated as presumptively the same if they had identical sequences of all available amplicons and as different if differences were observed at any loci or amplicon. For twins with 5 identical conventional HSV-1 amplicons, we confirmed identity of HSV-1 strains by WGS. We used HSV target capture and laboratory and analytical pipelines analogous to those reported for HSV-2 and human herpesvirus type 6 [15–18]. De novo–assembled contigs were mapped to the HSV-1 17+ reference genome (NC_001806) and deposited in GenBank (Supplementary Tables 2 and 3). Phylogenetic analysis was performed on concatenated unique long (UL)–unique short (US) sequences, using a randomized accelerated maximum likelihood [19] GTR+I+Γ model and 1000 bootstrap replicates and MAFFT alignment within Geneious v10 (Biomatters, Newark, NJ). Repeats flanking UL and US regions were excluded. Low-coverage regions within UL-US sequences were not manually removed, but the bioinformatic pipeline that constructs the consensus sequences assigns an “N” to bases with low coverage (<10×) [20]. This analysis was confirmed using a Bayesian approach, specifically MrBayes.
T-Cell Responses
Specificities of CD4+ T cells were determined as described elsewhere [21] for a subgroup of 10 MZ twin pairs who were randomly selected on the basis of criteria other than viral strain; 5 pairs were infected with the same HSV-1 strain, and 5 were infected with different HSV-1 strains. Briefly, cryopreserved, thawed peripheral blood mononuclear cells (PBMC) were stimulated with UV-treated lysate of HSV-1 17+ and live, CD3+CD4+CD137high cells were sorted and bulk-expanded [21]. To interrogate CD4 T-cell responses to HSV-1 proteins, we used an HSV-1 open reading frame (ORF) set [22]. Each HSV-1 protein was expressed, with overlapping, partial-length proteins observed in a few cases. CD4+ T-cell responses were assessed by determining the level of [3H] thymidine incorporated into the cells [22]. CD4+ T cells enriched for HSV-1 specificity were combined with irradiated autologous PBMCs and individual HSV-1 protein antigens. [3H] thymidine was added at 72 hours, cells were harvested, and counts per million were determined (TopCount NXT, Perkin-Elmer). Responses to each HSV-1 ORF were assigned as positive or negative, using a negative control antigen cutoff set from unrelated microbes [23] for a false-positivity rate of <1.0%. Control antigens were from Mycobacterium tuberculosis [24], Francisella tularensis [22], and Plasmodium falciparum [23]. No participant had a known infection history with these agents. Bulk expanded HSV-1–reactive CD4+ T cells (1 × 107) or ex vivo PBMCs (1.5 × 107) were submitted to Adaptive Biotechnologies (Seattle, WA) for TRB CDR3 survey sequencing [25].
Statistical Methods
Shedding rates were computed as the proportion of days with detectable virus, as described before [26, 27]. Spearman correlation was used to assess associations in shedding rates within twin pairs, by zygosity and HSV-1 strain. Regression lines were drawn using a linear regression. We used the bootstrap method to compare the percentage agreement in the distribution of the CD4+ T-cell responses to HSV-1–specific antigens in 20 unrelated persons within the twin cohort to that in the 10 MZ pairs. We created 5000 bootstrapped data sets containing 10 pairs of 20 randomly selected twins who were unrelated, and between-person agreement among the tested HSV-1 proteins was computed for each of these unrelated pairs. A percentage agreement was then computed for the 10 MZ pairs. The resulting distribution of agreement rates was used as the null distribution by which to compare the percentage agreement among twins. A similar bootstrap approach was used to assess whether the correlation in shedding rates between MZ twins was higher than expected under 5000 bootstrapped data sets, each containing 22 DZ pairs and sampled with replacement. We recorded the Pearson correlation coefficient each iteration.
RESULTS
Participants
Fifty-one twin pairs (102 participants) were enrolled. Twenty-two pairs identified themselves as DZ, and 29 pairs identified themselves as MZ (Table 1). Genomic or sex-based confirmation of zygosity was possible in 45 twinships (90 participants [90%]). Among the self-identified DZ participants, 18 twinships (36 participants) were of different sex or confirmed by genomic testing. One pair self-identified as DZ but was genomically MZ. Of the 29 pairs self-identified as MZ, 27 (54 participants) were confirmed as such by DNA testing. One twin pair self-identified as MZ but was DZ on DNA testing. Genomic zygosity results were used for our analyses when available. The final study population was 22 DZ twin pairs (43%) and 29 MZ twin pairs (57%). Eighty five percent of the subjects had a clinical history consistent with recurrent orolabial herpes, with similar proportions of MZ and DZ twins reporting the presence of cold sores.
Table 1.
Demographic, Clinical, and Viral Coinfection Characteristics of Study Participants
| Characteristic | Overall (n = 102) | MZ Twins (n = 58) | DZ Twins (n = 44) |
|---|---|---|---|
| Twin pairs, no. | 51 | 29 | 22 |
| Zygosity confirmed by sex difference or DNA testing | 90 (88) | 54 (93) | 36 (82) |
| Age, y | 37 (19–78) | 30 (19–78) | 54 (19–72) |
| Sex | |||
| Male | 32 (31) | 16 (28) | 16 (36) |
| Female | 70 (69) | 42 (72) | 28 (64) |
| HSV-2 seropositive | 13 (13) | 4 (7) | 9 (20) |
| History of symptomatic HSV-1 | 87 (85) | 50 (86) | 37 (84) |
| Time since first symptoms of HSV-1, ya | 21 (0.083–68) | 21(0.25–68) | 32 (0.083–60) |
| HSV shedding rate,b % | 9 (0–47.4) | 9.2 (0–38.2) | 9.3 (0–47.4) |
| Days with oral lesions, % | 0 (0–23.4) | 0 (0–23.4) | 0 (0–23.3) |
| Asymptomatic shedding rate,b % | 7.1 (0–47.4) | 6.8 (0–25.5) | 7.8 (0–47.4) |
Data are no. (%) of twins or median value (range), unless otherwise indicated.
aFor participants with data available.
bSee Methods for the definition of shedding rate.
A total of 5963 daily oral swab specimens were self-collected by participants on a median of 60 days/person (range, 39–69 days/person). Ninety-four percent of participants collected specimens on at least 50 of 60 anticipated swabbing days. HSV shedding on at least 1 day was detected in 77 participants (75%) and 45 twin pairs (88%). Of 5963 days, specimens collected on 607 (10.2%) were positive for HSV. Oral symptom diary data were available for a median of 62 days/person (range, 43–88 days/person). Thirty-one participants (30%) had lesions on at least 1 day during the study. Participants had lesions on 205 of 6386 days (3.2%), and HSV was detected on 123 of 199 days (61.8%) when oral lesions were present.
The median oral HSV shedding rate was 9.0% (range, 0%–47.4%), and the median percentage of days during which oral lesions were present was 0.0% (range, 0.0%–23.4%; Table 1). The median asymptomatic HSV shedding rate was 7.1% (range, 0.0%–47.4%).
Twin Zygosity and HSV-1 Shedding Rate
The median overall HSV-1 shedding rate was 9.3% (range, 0.0%–47.4%) in DZ twins and 9.2% (range, 0.0%–38.2%) in MZ twins (Table 1). The median asymptomatic shedding rate was 7.8% (range, 0.0%–47.4%) in DZ twins and 6.8% (range 0.0–25.5%) in MZ twins.
There was a positive correlation in shedding rates within all twin pairs (r = 0.34; P = .015), as well as a similar magnitude of correlation between shedding rates within MZ (r = 0.37; P = .047) and DZ (r = 0.31; P = .16) twin pairs. Among bootstrapped data sets, the mean correlation was 0.30 (5th and 95th percentiles, −0.18 and 0.72, respectively) over all bootstrapped data sets . The observed correlation in MZ twin pairs was 0.37, which is at the 60th percentile of the bootstrapped DZ correlations, for a 2-sided P value of .81.
Viral Strain and HSV-1 Shedding Rate
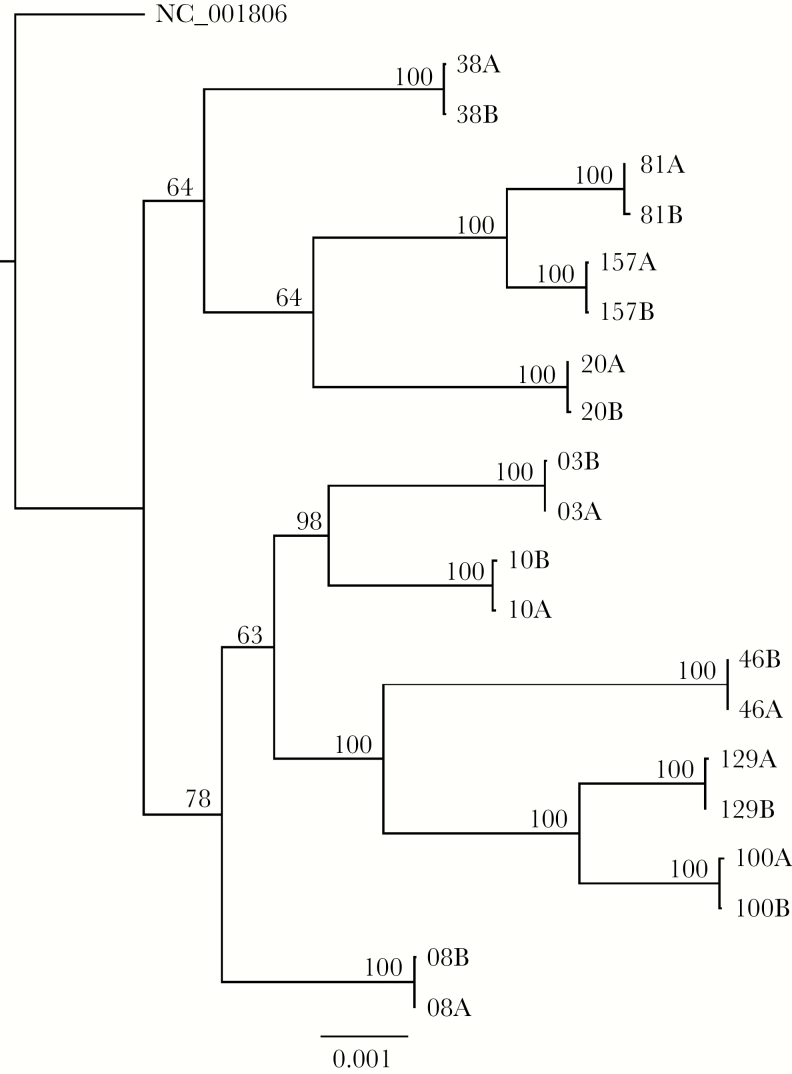
Twenty-two pairs had low-level HSV-1 shedding and insufficient HSV-1 DNA levels to classify viral strains as the same or different. Sanger sequencing data showed that 15 twin pairs (30 participants) likely had the same strain and that 14 twin pairs (28 participants) had different strains (Table 2). We performed WGS on specimens from the 15 twin pairs identified with the same strain by Sanger sequencing. Data from samples from 10 twins pairs met WGS acceptance criteria of >30000 reads aligning to the HSV-1 reference genome and >80% of HSV-1 loci sequenced for both persons. WGS reads included aligned read number, depth, and genome coverage (Supplementary Table 3). Phylogenetic analyses using maximum-likelihood (Figure 1) and Bayesian (Supplementary Figure 2) approaches showed that samples from each twin pair were highly related. Since Sanger sequencing correctly detected same-strain HSV-1 infection in the 10 pairs tested by WGS, we retained the Sanger sequence–based strain identity assignments for all twin pairs.
Table 2.
Herpes Simplex Virus Type 1 (HSV-1) Genotypes and Oral HSV Shedding Rates Among Twin Subjects
| Zygosity, Variable | HSV-1 Genotype | Total | ||
|---|---|---|---|---|
| Same | Different | No Dataa | ||
| Monozygotic | ||||
| Twins, no. | 9 | 6 | 14 | 29 |
| Shedding rateb | 16.1 (167/1035) | 13.6 (98/719) | 3.5 (60/1700) | 9.4% (325/3454) |
| Dizygotic | ||||
| Twins, no. | 6 | 8 | 8 | 22 |
| Shedding rateb | 14.8 (105/708) | 14.6 (129/886) | 5.2 (48/915) | 11.2% (282/2509) |
| Overall | ||||
| Twins, no. | 15 | 14 | 22 | 51 |
| Shedding rateb | 15.6 (272/1743) | 14.1 (227/1605) | 4.1 (108/2615) | 10.2% (607/5963) |
aInsufficient HSV-1 DNA from twins from a given twinship to classify viral strains as the same or different within that twinship.
bData are percentage of oral swabs positive for HSV DNA (no. of swabs positive/no. of swabs tested).
Figure 1.
Phylogenetic tree of near-complete HSV-1 sequences, determined by whole-genome sequencing, from 10 twin pairs with identical amplicon sequencing supports accurate diagnosis of same-strain infection. “A” and “B” refer to cotwins from numbered twin pairs. Numbers at branch points are bootstrap support values. The reference genome for laboratory strain 17+ was used as the outgroup.
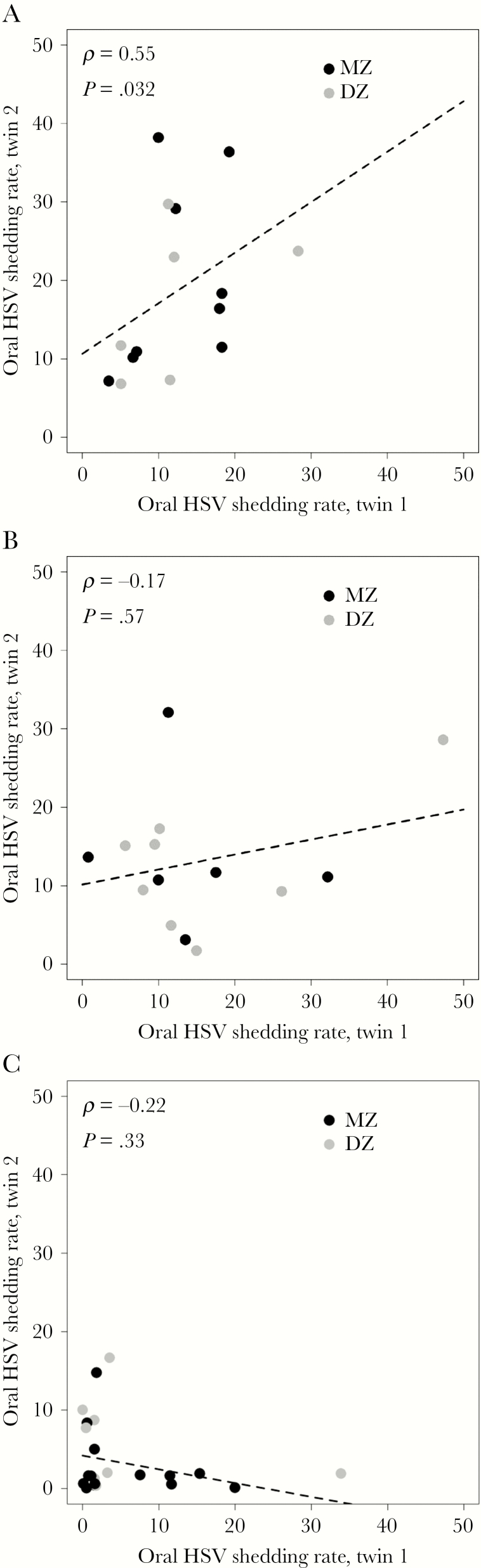
Nine MZ and 6 DZ twin pairs were infected with the same strain, and 6 MZ and 8 DZ twin pairs were infected with different HSV strains. Twenty-nine twin pairs had a known viral strain status for both persons. We found a positive correlation in shedding rates in twin pairs with the same viral strain (r = 0.55; P = .032; Figure 2A) but not among pairs with different strains (r = −0.17; P = .57; Figure 2B). Figure 2C depicts correlation in HSV shedding rates between twins when the HSV-1 strain could not be analyzed because of low-level or absent shedding. Within half of the cohort (approximately 25 twin pairs), we would have 80% power to detect a shedding correlation of >0.54, which was observed between twins infected with the same strain (Figure 2A).
Figure 2.
HSV-1–Specific CD4+ T-Cell Responses
The CD4+ T-cell response among PBMCs was studied in a subgroup of 10 MZ twin pairs, of which 5 had the same HSV-1 strain and 5 had different strains. The median age was 30 years (range, 19–67 years). The median shedding rate was 12.7% (range, 0%–38.2%).
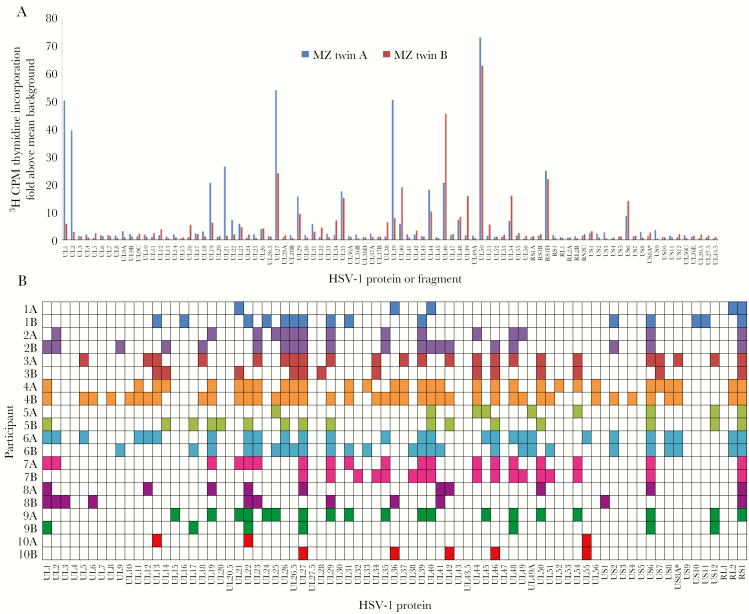
For each person, CD4+ T cells among PBMCs were enriched for HSV-1–reactive cells and expanded polyclonally [22]. These bulk cultures had low proliferative response to a panel of negative control antigens (Supplementary Figure 1). The median breadth of the CD4+ T-cell response was 16 HSV-1 antigens/person (range, 3–41 antigens/person; Figure 3B). The HSV-1 proteins eliciting prevalent CD4+ T-cell responses included ICP4 (gene RS1), in 17 persons (85%); glycoprotein B (gene UL27), in 16 persons (80%); and glycoprotein D (gene US6), in 15 persons (75%); the proteins encoded by UL39 and UL46 that we previously reported [22] as population prevalent for CD8+ T-cell responses to HSV-1 were recognized by 11 persons (55%) each ( Figure 3B). Overall, 64 of 77 HSV-1 proteins (83%) were recognized by CD4+ T cells from at least 1 subject.
Figure 3.
CD4+ T-cell herpes simplex virus type 1 (HSV-1) protein-level specificity patterns are similar between monozygotic (MZ) twins. A, Representative HSV-1 proteome-wide reactivity to 90 HSV-1 proteins or protein fragments for a MZ twin pair. Responses to each HSV-1 open reading frame, measured in counts per million (CPM), were designated as positive or negative by using a cutoff set with negative control antigens from unrelated microbes. Data are from twin pair 7 in panel B. B, Summarized peripheral blood mononuclear cell (PBMC) CD4+ T-cell reactivity to 77 HSV-1 proteins (fragment responses condensed) in 10 MZ twin pairs. Twin pair participants are listed on the y-axis. Responses are designated as positive (darker shading or colored) or negative per the cutoff described in Methods. MZ twins in adjacent rows have the same shading/color.
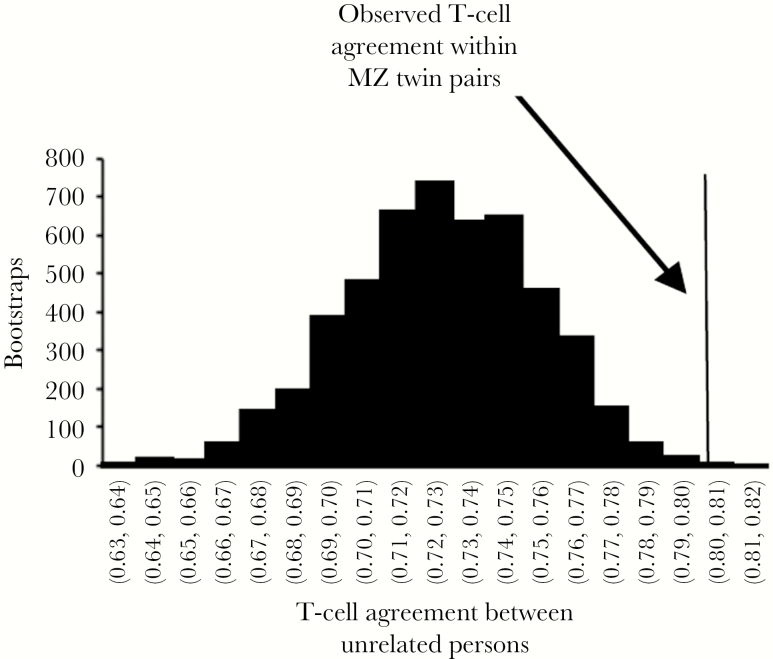
CD4+ T-cell responses to individual HSV-1 antigens showed concordance between MZ twins when analyzed visually (Figure 3A and 3B).We hypothesized that the pattern of reactivity to the HSV-1 proteins would be more similar between MZ twins than between unrelated persons, owing to shared genetic identity between HLA and, possibly, TRA/TRB allelic variants. Unrelated persons had a mean percentage agreement of 73% (2.5th and 97.5th percentiles, 67% and 78%, respectively) among CD4+ T-cell responses to HSV-1 proteins (Figure 4). In contrast, the percentage agreement of HSV-1 protein–level reactivity in MZ twin pairs was 80% (608 of 760 protein-level positive responses). This is the 99.9th percentile of agreement rates for unrelated individuals in a bootstrap analysis (2-sided P = .002), indicating a low probability that the higher within-twinship agreement for MZ twins arose by chance.
Figure 4.
Protein-wise peripheral blood mononuclear cell (PBMC) CD4+ T-cell responses to herpes simplex virus type 1 (HSV-1) in monozygotic (MZ) twins agree more than expected by chance. Data for unrelated persons were analyzed 5000 times by bootstrap analysis, with the percentage agreement shown as a histogram, with bins labeled on the x-axis. The observed agreement for presence or absence of protein-level CD4+ T-cell responses for the 10 MZ twin pairs was 80%.
The specificity of T cells is determined by the amino acids encoded by TCR TRA/TRB CDR3 regions. To assess this complexity, TRB CDR3 genomic DNA was sequenced in a platform [28] in which read and cell numbers correlate. We compared HSV-1–reactive CD4+ T-cell lines to whole PBMCs from 2 MZ twin pairs (twins 3A and 3B and twins 4A and 4B; Figure 3B). Bulk HSV-1–reactive CD4+ T-cell lines had higher clonality indices than PBMCs (Table 3), as expected for memory cells recognizing a limited number of epitopes. We compared identity in TRB CDR3 amino acid–level sequences between persons to examine the hypothesis that MZ twins would share CDR3 use more than nontwins. Within MZ twin pair 3, shared CDR3 sequences comprised 1.15% and 1.04% of the HSV-1–reactive CD4+ T-cell line TRB CDR3 reads in twins 3A and 3B, respectively. For twin pair 4, shared sequences comprised 2.72% and 1.25% of total reads in twins 4A and 4B, respectively. These abundances were generally higher than the sharing rates between unrelated persons (Supplementary Table 4).
Table 3.
TRB CDR3 Analyses of Peripheral Blood Mononuclear Cells (PBMCs) and Herpes Simplex Virus Type 1–Specific CD4+ T-Cell Lines From Monozygotic Twin Pairs
| Participant, Specimen | Productive Template | Unique CDR3a | Clonalityb | Maximum Frequency, %c |
|---|---|---|---|---|
| Twin 3A | ||||
| PBMCs | 403678 | 327307 | 0.05 | 0.52 |
| CD4+ T-cell line | 305871 | 7976 | 0.40 | 6.51 |
| Twin 3B | ||||
| PBMCs | 397131 | 339611 | 0.03 | 0.06 |
| CD4+ T-cell line | 227141 | 6918 | 0.38 | 4.95 |
| Twin 4A | ||||
| PBMCs | 327237 | 267244 | 0.04 | 0.36 |
| CD4+ T-cell line | 269787 | 7092 | 0.39 | 4.70 |
| Twin 4B | ||||
| PBMCs | 484997 | 392072 | 0.04 | 0.12 |
| CD4+ T-cell line | 242678 | 7323 | 0.39 | 7.31 |
aData are no. of unique productive TRB CDR3 nucleotide sequences detected.
bData are clonality of TRB CDR3 nucleotide reads calculated per the Adaptive Biotechnologies algorithm, ranging from 0, for a population with no shared sequences, to 1.0, for a monoclonal population.
cData are the abundance of the most frequently observed TRB CDR3 clonotype in the population.
DISCUSSION
In our study, we found that the median oral HSV shedding rate was 9%. There was a positive correlation in shedding rates in twin pairs with the same viral strain, regardless of zygosity. We also found a positive correlation in HSV-1 shedding frequencies within all twin pairs, but we could not detect a stronger correlation between MZ twins than between DZ twins. Thus, viral genetic factors appear to correlate with oral HSV-1 shedding rates, while the data do not allow us to conclude that host genetics are a factor. As hypothesized, CD4+ T-cell responses were more similar between MZ twins than between DZ twins.
While the mechanisms influencing HSV-1 shedding frequency remain unknown, prior infections with cross-reactive alphaherpesviruses [25] or HIV-1 [29], the size of the inoculum [30], and environmental factors might influence HSV-1 severity. Host genetic factors influencing severe HSV-1 infection include rare defects in innate immunity, specifically central nervous system–intrinsic viral sensing and the type I interferon pathway [31]. A family cohort study found a chromosome 21 locus linked with the presence or absence of orolabial lesions [32]. Stress can influence HSV-specific acquired immunity and molecular correlates of reactivation in animal models [33], although in the human host, Strachan et al [34] found that stress did not influence viral shedding rates. Last, HSV inoculum size influences the number of latently infected neurons [30], which might affect the frequency of shedding and lesions. Together, it appears that a variety of factors can influence HSV severity, but the exact reasons for heterogeneity in HSV-1 shedding largely remain unknown. In this limited sample size, we did not demonstrate that HSV-1 shedding is associated with host genetics; while shedding rates correlated between twins, the correlation was similar between MZ and DZ twins, and this correlation could be an effect of viral strain. Larger cohorts might be able to establish a separate host genetic effect. Limited studies are present in the literature that demonstrate an influence of HSV-1 strains on the pattern of disease, and many studies are restricted to animal models [35, 36]. Different HSV-1 strains circulate globally [37], but their impact on clinical symptoms in the human host remains unclear.
CD4+ T-cell responses to HSV-1 within MZ twin pairs agreed more closely than expected by chance. Our findings are consistent with known biological mechanisms, as MZ twins have identical HLA haplotypes. HLA class II allelic variation controls CD4+ T-cell specificity, and HLA genotype influences the shedding of genital HSV-2 [38]. T cells can be cross-reactive between HSV-1 and HSV-2 even if the HSV-1 and HSV-2 peptide homologs are nonidentical [22]. Our cohort had a low HSV-2 infection rate (Table 1). Therefore, HSV-2 coinfection was unlikely to confound our findings concerning the similarity of T-cell responses between twins. CD4+ T-cell responses were analyzed in MZ twins with the same and those with different strain HSV-1 infections. However, the protein coding sequence variation between epidemiologically unrelated HSV-1 strains is very low, at <1% of amino acids [37], and we combined immunology data for the set of MZ twin pairs regardless of HSV-1 genotypes.
By pooling both MZ and DZ twin pairs, we detected a contribution of viral genotype to HSV-1 shedding rates. While in vitro studies of strains recovered from persons with different severity show no consistent differences, HSV-1 strains vary at coding and noncoding loci and show variable virulence in animal models [39–42]. Owing to a lack of sample availability, we were unable to determine the viral strain status within many twinships. It is possible that attenuating variations are present in some strains infecting the persons in whom we did not detect shedding. We did not correlate specific viral polymorphisms or haplotypes with shedding rates.
Strengths of this study include the coordinated evaluation of both host and viral genotype, high adherence of study participants to the intensive swabbing regimen over a long period, and the use of an objective virologic outcome as our primary measure. Studies evaluating HSV-1 shedding in the oral cavity in asymptomatic persons by using highly sensitive PCR-based assays are limited. Many reports obtain samples by culture-based assay at 1 time point or irregular intervals and likely underestimate HSV-1 shedding rates [43, 44]. With oral sampling, HSV-1 shedding detected by PCR is observed on 9%–26% of days [45, 46]. To our knowledge, this is the largest cohort of individuals evaluated for oral HSV-1 shedding by using daily sampling [45–48]. In this cohort we found many twins were infected with different viral strains. This could occur if twins were infected at a young age by different persons or there was acquisition of HSV-1 later in life. A recently published article by McQuillan et al [49] found that only 27% of individuals in a US cohort were seropositive for HSV-1 at ages 14–19 years during 2015–2016, with the seroprevalence increasing with age. This suggests that, in recent years, children are less often infected with HSV-1 and that acquisition of virus might occur following sexual debut [50–52]. Although possible, it is unlikely that the HSV-1 genome evolved into that of a different strain over time in an individual [53].
Our study was limited by a small sample size, and it was only powered to detect large effects of either host genetics or viral strain on HSV-1 shedding rates. An additional limitation was the approximation of the severity of HSV-1 infection provided by daily samples over the defined 60-day period. It is possible that the 10 MZ twin pairs randomly selected to have their CD4+ T-cell responses evaluated were not representative of all twin pairs. We were unable to compare CD4+ T-cell responses to HSV-1 proteins in unrelated persons with the same HLA haplotype. We were able to definitively assign zygosity for most twinships and used self-reported zygosity for the remainder. We did detect a low rate of subject misidentification of zygosity at 4% (2 of 45) when evaluated by genomic testing. This rate is in agreement with the literature, which generally finds self-reported accuracy rates near 95% [54, 55]. It is possible that the association between zygosity and shedding rate would weaken if the error rate in subject self-reported zygosity in the 6 twinships we could not verify was atypically high, but we have no reason to suspect this. Of note, 85% of our study population had a history of orolabial herpes, which is higher than that of the general population of HSV-1–infected persons [3], reflecting partly our identification of potentially eligible twins by the history of oral herpes.
In conclusion, this study of healthy adult twins revealed that viral genetics contributes to oral HSV-1 shedding rates. Persons with the same HSV-1 strain have greater agreement in viral shedding than persons with different strains. Oral HSV-1 shedding rates are correlated between twins, but we were unable to establish a separate host effect. Multilocus sequencing appears to be accurate for diagnosing HSV-1 genotype, although the decreased costs of near full-length WGS and the richness of the data set favor the newer technology. Virus-specific T-cell responses are highly complex at the clonotype level but are generally more similar between MZ twins than between unrelated persons. Further investigation will be required as to how specific viral and host loci, together with environmental factors, contribute to the spectrum of recurrent HSV-1 severity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the research participants and staff of the University of Washington Virology Research Clinic, the University of Washington Molecular Virology Laboratory, the Twin Registry at the University of Washington, and the participating twins.
Financial support. This work was supported by the National Institutes of Health (grants T32AI007044-38 [to M. S. R.], R01AI094019 [to D. M. K.], P01AI030731 [to A. W., A. S. M., and D. M. K.], and R21AI081347-01 and RC2HL103416 [to E. S.]).
Potential conflicts of interest. D. M. K. and A. W. are coinventors listed on patents owned by the University of Washington that involve herpes simplex vaccines. D. M. K. has been a consultant for or conducted research sponsored by Glaxo SmithKline, Merck, Admedus, Immune Design, Vical, Biomedical Research Models, and Sanofi Pasteur. A. W. has conducted research sponsored by Genocea and Vical and is a consultant for Aicuris. A. S. M. has been a consultant for Immune Design and AiCuris. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: IDWeek 2014, Philadelphia, Pennsylvania, 8–12 October 2014. Abstract 1154.
References
- 1. Baringer JR, Swoveland P. Recovery of herpes-simplex virus from human trigeminal ganglions. N Engl J Med 1973; 288:648–50. [DOI] [PubMed] [Google Scholar]
- 2. Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet 2001; 357:1513–8. [DOI] [PubMed] [Google Scholar]
- 3. Looker KJ, Magaret AS, May MT, et al. Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012. PLoS One 2015; 10:e0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tunbäck P, Bergström T, Claesson BA, Carlsson RM, Löwhagen GB. Early acquisition of herpes simplex virus type 1 antibodies in children–a longitudinal serological study. J Clin Virol 2007; 40:26–30. [DOI] [PubMed] [Google Scholar]
- 5. Whitley RJ. Herpes simplex encephalitis: adolescents and adults. Antiviral Res 2006; 71:141–8. [DOI] [PubMed] [Google Scholar]
- 6. Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet 2002; 3:872–82. [DOI] [PubMed] [Google Scholar]
- 7. Strachan E, Hunt C, Afari N, et al. University of Washington Twin Registry: poised for the next generation of twin research. Twin Res Hum Genet 2013; 16:455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ashley RL, Militoni J, Lee F, Nahmias A, Corey L. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol 1988; 26:662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang MJ, Tzeng CH, Tseng JY, Huang CY. Determination of twin zygosity using a commercially available STR analysis of 15 unlinked loci and the gender-determining marker amelogenin–a preliminary report. Hum Reprod 2006; 21:2175–9. [DOI] [PubMed] [Google Scholar]
- 10. Jerome KR, Huang ML, Wald A, Selke S, Corey L. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J Clin Microbiol 2002; 40:2609–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wald A, Huang ML, Carrell D, Selke S, Corey L. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J Infect Dis 2003; 188:1345–51. [DOI] [PubMed] [Google Scholar]
- 12. Magaret AS, Wald A, Huang ML, Selke S, Corey L. Optimizing PCR positivity criterion for detection of herpes simplex virus DNA on skin and mucosa. J Clin Microbiol 2007; 45:1618–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin ET, Koelle DM, Byrd B, et al. Sequence-based methods for identifying epidemiologically linked herpes simplex virus type 2 strains. J Clin Microbiol 2006; 44:2541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muller WJ, Dong L, Vilalta A, et al. Herpes simplex virus type 2 tegument proteins contain subdominant T-cell epitopes detectable in BALB/c mice after DNA immunization and infection. J Gen Virol 2009; 90:1153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koelle DM, Norberg P, Fitzgibbon MP, et al. Worldwide circulation of HSV-2×HSV-1 recombinant strains. Sci Rep 2017; 7:44084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnston C, Magaret A, Roychoudhury P, et al. Highly conserved intragenic HSV-2 sequences: Results from next-generation sequencing of HSV-2 UL and US regions from genital swabs collected from 3 continents. Virology 2017; 510:90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnston C, Magaret A, Roychoudhury P, et al. Dual-strain genital herpes simplex virus type 2 (HSV-2) infection in the US, Peru, and 8 countries in sub-Saharan Africa: A nested cross-sectional viral genotyping study. PLoS Med 2017; 14:e1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greninger AL, Roychoudhury P, Makhsous N, Hanson D, Chase J, Krueger G, et al. Copy number heterogeneity, large origin tandem repeats, and interspecies recombination in HHV-6A and HHV-6B reference strains. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stamatakis A. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics 2015; 51:6.14.1–14. [DOI] [PubMed] [Google Scholar]
- 20. Greninger AL, Roychoudhury P, Xie H, Casto A, Cent A, Pepper G, et al. Ultrasensitive capture of human herpes simplex virus genomes directly from clinical samples reveals extraordinarily limited evolution in cell culture. mSphere 2018, 3. doi: 10.1128/mSphereDirect.00283-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jing L, Haas J, Chong TM, et al. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin Invest 2012; 122:654–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jing L, Haas J, Chong TM, et al. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin Invest 2012; 122:654–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laing KJ, Russell RM, Dong L, et al. Zoster Vaccination Increases the Breadth of CD4+ T Cells Responsive to Varicella Zoster Virus. J Infect Dis 2015; 212:1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nayak K, Jing L, Russell RM, et al. Identification of novel Mycobacterium tuberculosis CD4 T-cell antigens via high throughput proteome screening. Tuberculosis (Edinb) 2015; 95:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jing L, Laing KJ, Dong L, et al. Extensive CD4 and CD8 T Cell Cross-Reactivity between Alphaherpesviruses. J Immunol 2016; 196:2205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wald A, Zeh J, Selke S, Ashley RL, Corey L. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med 1995; 333:770–5. [DOI] [PubMed] [Google Scholar]
- 27. Wald A, Corey L, Cone R, Hobson A, Davis G, Zeh J. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J Clin Invest 1997; 99:1092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeWitt WS, Emerson RO, Lindau P, et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol 2015; 89:4517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Velzen M, Ouwendijk WJ, Selke S, et al. Longitudinal study on oral shedding of herpes simplex virus 1 and varicella-zoster virus in individuals infected with HIV. J Med Virol 2013; 85:1669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lekstrom-Himes JA, Pesnicak L, Straus SE. The quantity of latent viral DNA correlates with the relative rates at which herpes simplex virus types 1 and 2 cause recurrent genital herpes outbreaks. J Virol 1998; 72:2760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A 2015; 112:E7128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kriesel JD, Bhatia A, Thomas A. Cold sore susceptibility gene-1 genotypes affect the expression of herpes labialis in unrelated human subjects. Hum Genome Var 2014; 1:14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freeman ML, Sheridan BS, Bonneau RH, Hendricks RL. Psychological stress compromises CD8+ T cell control of latent herpes simplex virus type 1 infections. J Immunol 2007; 179:322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strachan E, Saracino M, Selke S, Magaret A, Buchwald D, Wald A. The effects of daily distress and personality on genital HSV shedding and lesions in a randomized, double-blind, placebo-controlled, crossover trial of acyclovir in HSV-2 seropositive women. Brain Behav Immun 2011; 25:1475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H, Davido DJ, Morrison LA. HSV-1 strain McKrae is more neuroinvasive than HSV-1 KOS after corneal or vaginal inoculation in mice. Virus Res 2013; 173:436–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim JK, Kim YK, Hong J, et al. Isolation of the enhanced neurovirulent HSV-1 strains from Korean patients. Virus Genes 2003; 26:115–8. [DOI] [PubMed] [Google Scholar]
- 37. Szpara ML, Gatherer D, Ochoa A, et al. Evolution and diversity in human herpes simplex virus genomes. J Virol 2014; 88:1209–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Magaret A, Dong L, John M, et al. HLA Class I and II alleles, heterozygosity and HLA-KIR interactions are associated with rates of genital HSV shedding and lesions. Genes Immun 2016; 17:412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spruance SL, Chow FS. Pathogenesis of herpes simplex labialis. I. Replication of herpes simplex virus in cultures of epidermal cells from subjects with frequent recurrences. J Infect Dis 1980; 142:671–5. [DOI] [PubMed] [Google Scholar]
- 40. Cavallero S, Huot N, Francelle L, Lomonte P, Naas T, Labetoulle M. Biological features of herpes simplex virus type 1 latency in mice according to experimental conditions and type of neurones. Invest Ophthalmol Vis Sci 2014; 55:7761–74. [DOI] [PubMed] [Google Scholar]
- 41. Kolb AW, Lee K, Larsen I, Craven M, Brandt CR. Quantitative Trait Locus Based Virulence Determinant Mapping of the HSV-1 Genome in Murine Ocular Infection: Genes Involved in Viral Regulatory and Innate Immune Networks Contribute to Virulence. PLoS Pathog 2016; 12:e1005499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brandt CR, Grau DR. Mixed infection with herpes simplex virus type 1 generates recombinants with increased ocular and neurovirulence. Invest Ophthalmol Vis Sci 1990; 31:2214–23. [PubMed] [Google Scholar]
- 43. Spruance SL. Pathogenesis of herpes simplex labialis: excretion of virus in the oral cavity. J Clin Microbiol 1984; 19:675–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kameyama T, Sujaku C, Yamamoto S, Hwang CB, Shillitoe EJ. Shedding of herpes simplex virus type 1 into saliva. J Oral Pathol 1988; 17:478–81. [DOI] [PubMed] [Google Scholar]
- 45. Mark KE, Wald A, Magaret AS, et al. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J Infect Dis 2008; 198:1141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramchandani M, Kong M, Tronstein E, et al. Herpes simplex virus Type 1 shedding in tears and nasal and oral mucosa of healthy adults. Sex Transm Dis 2016; 43:756–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gilbert SC. Oral shedding of herpes simplex virus type 1 in immunocompetent persons. J Oral Pathol Med 2006; 35:548–53. [DOI] [PubMed] [Google Scholar]
- 48. Liljeqvist JA, Tunbäck P, Norberg P. Asymptomatically shed recombinant herpes simplex virus type 1 strains detected in saliva. J Gen Virol 2009; 90:559–66. [DOI] [PubMed] [Google Scholar]
- 49. McQuillan G, Kruszon-Moran D, Flagg EW, Paulose-Ram R. Prevalence of Herpes simplex virus Type 1 and Type 2 in persons aged 14–49: United States, 2015–2016. NCHS Data Brief 2018:1–8. [PubMed] [Google Scholar]
- 50. Woestenberg PJ, Tjhie JH, de Melker HE, et al. Herpes simplex virus type 1 and type 2 in the Netherlands: seroprevalence, risk factors and changes during a 12-year period. BMC Infect Dis 2016; 16:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bradley H, Markowitz LE, Gibson T, McQuillan GM. Seroprevalence of herpes simplex virus types 1 and 2–United States, 1999-2010. J Infect Dis 2014; 209:325–33. [DOI] [PubMed] [Google Scholar]
- 52. Korr G, Thamm M, Czogiel I, Poethko-Mueller C, Bremer V, Jansen K. Decreasing seroprevalence of herpes simplex virus type 1 and type 2 in Germany leaves many people susceptible to genital infection: time to raise awareness and enhance control. BMC Infect Dis 2017; 17:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Minaya MA, Jensen TL, Goll JB, Korom M, Datla SH, Belshe RB, et al. Molecular evolution of Herpes simplex virus 2 complete genomes: comparison between primary and recurrent infections. J Virol 2017, 91. doi: 10.1128/JVI.00942-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen WJ, Chang HW, Wu MZ, et al. Diagnosis of zygosity by questionnaire and polymarker polymerase chain reaction in young twins. Behav Genet 1999; 29:115–23. [DOI] [PubMed] [Google Scholar]
- 55. Reed T, Plassman BL, Tanner CM, Dick DM, Rinehart SA, Nichols WC. Verification of self-report of zygosity determined via DNA testing in a subset of the NAS-NRC twin registry 40 years later. Twin Res Hum Genet 2005; 8:362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.