Abstract
Much less research on regulation and function of selenoproteins has been conducted in domestic pigs than in rodents or humans, although pigs are an excellent model of human nutrition and medicine and pork is a widely consumed meat in the world. Phylogenetically, the 25 identified porcine selenoproteins fell into two primitive groups, and might be further divided into three parallel branches. Despite a high similarity to that of humans and rodents, the porcine selenoproteome exhibited the closest evolutionary relationship with that of sheep and cattle among eight domestic species. Expression (mRNA, protein, and/or enzyme activity) of 2/3 of the 25 porcine selenoproteins in various tissues of pigs was affected by dietary Se intakes, and 14 of them showed responses to a high fat diet. When dietary Se deficiency mainly down-regulated the expression of selected selenoproteins, dietary Se excess exerted rather diverse effects on their expression. Overdosing pigs with dietary Se induced hyperinsulinemia, along with lipid accumulation and protein increase, in the liver and muscle by affecting key genes and(or) proteins involved in the metabolisms of glucose, lipid, and protein. In conclusion, expression of porcine selenoproteins was highly responsive to dietary Se and fat intakes, and was involved in body glucose, lipid, and protein metabolism as those of rodents and humans.
Keywords: Evolution, gene, pig, selenium, selenoprotein, tissue
Graphical Abstract
1. Introduction
Selenium (Se) was initially recognized as a toxic chemical to animals and humans [1]. Marco Polo described the hoof loss of horses as Se poison from his travels through Northwest China where soil contained high Se [2]. In 1934, Kurt Franke reported that alkali disease and blind staggers disease in grazing livestock in areas of the United States with Se-rich soil [3]. Acute Se poisoning in swine was characterized by an abnormal posture, unsteady gait, diarrhea, abdominal pain, increased pulse and respiratory rate, prostration, and death [4]. An endemic human disorder, characterized by loss of hair and nails, was observed in the 1960s in Hubei Province of China, and later was found to result from high intakes of Se from the local food, water, and air [5].
Because Se is an essential nutrient required by both humans and animals, Se deficiency has been a more practical problem than Se toxicity in many parts of the world. Dietary Se deficiency can cause various types of diseases including liver necrosis and mulberry heart disease in pigs [6]. It is well-accepted that metabolic functions of Se are mainly mediated by selenoproteins. Approximately 100 selenoprotein families have been identified including a set of 45 vertebrate selenoproteins [7]. While selenoproteomes of humans and rodents have been well-studied [8–11], there is relatively little research on the evolution and function of porcine selenoproteome. Because pork is one of the most widely consumed meats in the globe [12] and pigs are an excellent model of human medicine [13–15], our review is intended to help advance this particular area. We will explore the evolution of porcine selenogenome by comparing intraspecies and interspecies differences, and analyze expression, regulation, and function of the porcine selenogenome and (or) selenproteome under different nutritional conditions.
2. Evolution of porcine selenogenome
Previous analyses of selenogenomes have revealed differences in selenoprotein sets among various organisms [7, 16–18]. Some green algae and vertebrates have more than 20 selenoproteins, whereas red algae, insects, and nematodes have only less than 5 selenoproteins or higher plants and yeast do not have any selenoprotein [8]. Notably, aquatic organisms generally have larger numbers of selenoproteins than terrestrial organisms, and mammalian species show a trend toward a decreased use of these proteins [16, 17]. Selenoproteins were originated at the base of the eukaryotic domain and the environment has played an important role in their evolution [7, 8, 16, 17].
a). Role of natural selection
Through a very long natural selection, selenocysteine (Sec) has been retained and become utilizable by organisms [19]. The presence of Sec in more than 25 proteins, rather than in only one unique circumstance by chance, must bear irreplaceable roles [19]. Evolution is a population level process governed usually by four fundamental forces including natural selection, mutation, recombination, and random genetic drift [20]. The selective forces operating to maintain, lose, or acquire a given trait are considered to be important in the evolution of Se utilization traits [21]. Peng et al. [22] found new horizontal gene transfer events for components of different Se utilization traits by phylogenetic analyses.
b). Role of Se abundance in the environment
The variability of Se abundance in the environment is an important driving force in the evolution of Se utilization traits [21]. The organisms absolutely depending on Se can compromise their existence in Se deficiency as enzymes containing Sec in their catalytic residues could evolve into cysteine (Cys)-containing proteins or co-existing Sec- and Cys-containing forms [21,23]. A typical case is that the genome of M. maripaludis encodes several Sec-containing proteins and also homologs containing Cys in place of Sec, and synthesis of the latter is repressed when media contain adequate Se [23]. Furthermore, human migration around the world has changed soil Se levels of the habitation environments [24]. Localized adaptation to dietary differences has occurred in genes related to metabolism of macronutrients such as lactose and starch as well as micronutrients such as Se, iron and iodine [25]. Thus, the adequacy of Se supply can be a major factor to determine expression patterns of selenoprotein-related genes [25,26].
c). Intraspecies comparison of porcine selenoprotein genes
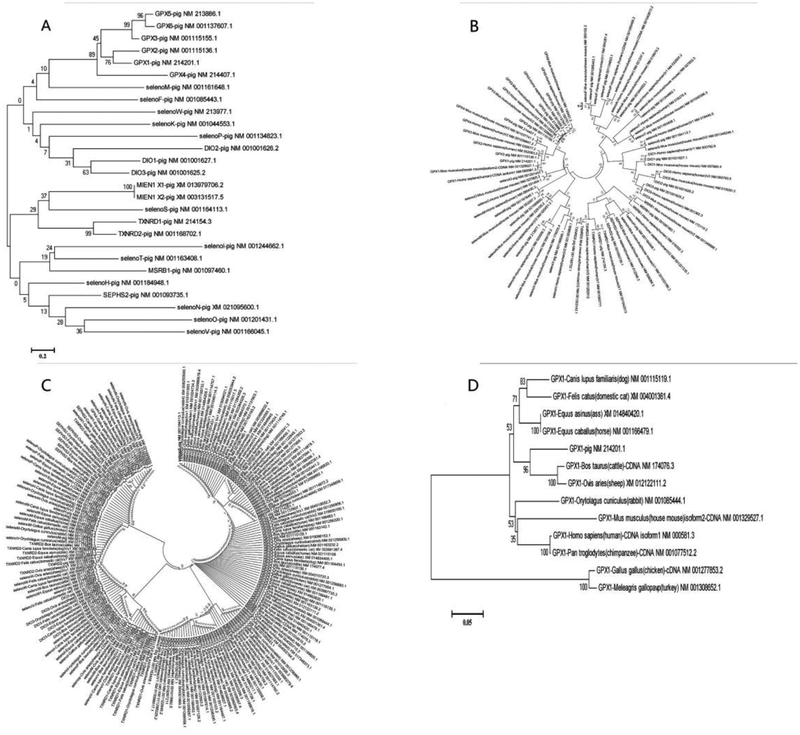
To assess the evolution of porcine selenogenome, we have performed searches of selenoprotein genes through SelenoDB 2.0 (selenoproteins database) and GenBank, and found out 25 such genes, which is consistent with the published reports [7,27]. Further we analyzed the selenogenome sequences by MEGA5 software as described previously [7, 28]. We could only analyze porcine selenoproteins at the cDNA level because no protein information can be acquired from the protein data bank, and Sec that is not included in the 20 amino acids cannot be recognized by MEGA5. We have searched all the sequences of pig selenoprotein genes, including DIO1, DIO2, DIO3, GPX1, GPX2, GPX3, GPX4, GPX5, GPX6, MSRB1 (SEPX1), SELENOF (SEP15), SELENOH (SELH), SELENOI (SELI), SELENOK (SELK), SELENOM (SELM), SELENON (SEPN1), SELENOO (SELO), SELENOP(SEPP1), SELENOS (SELS), SELENOT (SELT), SELENOV (SELV), SELENOW (SEPW), SEPHS2 (SPS2), TXNRD1, and TXNRD2, and analyzed them by maximum likelihood bootstrap 1000. The sequence homologous analysis allotted the porcine selenoproteins into two groups: DIOs, GPXs, and SELENOF, K, M, P and W belonged to one branch, and the remaining members belonged to the other branch (Fig. 1A). In addition, migration and invasion enhancer 1 (MIEN1), SELENOS, TXNRDs were diverted from the second branch. Thus, the porcine selenoproteins were actually evolved into three parallel branches. Furthermore, GPX4 was apart from the other GPX family members and became the most ancient GPX, whereas GPX5 and GPX6 were the most recently evolved GPX forms. Similarly, DIO2 was away from DIO1 and 3, and was the most ancient DIO family member.
Figure 1. Intraspecies and interspecies analyses of porcine selenogenome evolution.
A: Intraspecies comparison of porcine selenoproteins; B: Comparisons of porcine selenogenome with that of humans and rodents; C: Comparison of porcine selenogenome with that of domestic rabbit, sheep, chicken, cat, horse, ass, dog and cattle; D: Comparison of GPX1 among different species. The distance scale in substitutions per position is indicated at the bottom left in A, D.
d). Interspecies comparisons of porcine selenoprotein genes
We have also assessed the evolution relationship of 185 selenoprotein genes among more than 10 domestic species. Limited by the size of the phylogenic tree, we can illustrate herein the evolution relationships only between pigs and humans and rodents (Fig. 1B) as well as between pigs and 8 other domestic species including ass, cattle, chicken, domestic cat, dog, horse, rabbit and sheep (Fig. 1C).
All the 25 porcine selenoproteins also exist in humans, 24 of them exist in rodents except that rodent GPX6 is a Cys-homolog. The common selenoproteins in the three species share a very close evolutionary relationship and are highly conserved (Fig. 1B). Among all the selenoprotein genes, SELENOT displayed the highest similarity among the three species and was the most conserved selenoprotein gene. However, the similarity was very low between different selenoprotein families, suggesting that these families were independent in the evolution.
Several selenoprotein genes have an in frame TGA encoding Sec in pig, but not in other species (Fig.2C). Examples include no SELENOS in ass, no SELENOT in sheep, and no MIEN1 in chicken. Pig MIEN1 consisted of two splicing variants: MIEN1 X1 and MIEN1 X2 that did not contain Sec, but were classified as conditional selenoproteins [29]. The mammalian selenoproteins can be broadly classified into two classes: housekeeping selenoproteins and stress-related selenoproteins [30]. Housekeeping selenoproteins are less affected by dietary Se status and often serve functions critical to cell survival, whereas stress-related selenoproteins are not essential for survival and often show decreased expression in Se-deficient conditions [7]. Additionally, some conditional selenoproteins may not contain Sec but belong to selenoproteins in some condition. For example, in bacteria and some mammals including pigs, SELENOT, SELENOW and SELENOH are selenoproteins, which were found containing CXXU domain model [15]. However, in other species, SELENOT, SELENOW and SELENOH do not contain Sec because Sec in CXXU domain model is replaced by Cys, and becomes CXXC motif. The selenoproteins with CXXU domain also contain an insert of about 70 amino acids, which is thought as a signal changing the second Cys in CXXC motif into Sec in CXXU domain. Therefore, these examples suggest that either Sec or Cys for conditional selenoproteins might depend on the role of natural selection or the variability of Se abundance in the environment, which are important driving forces in the evolution of Se utilization traits as described above.
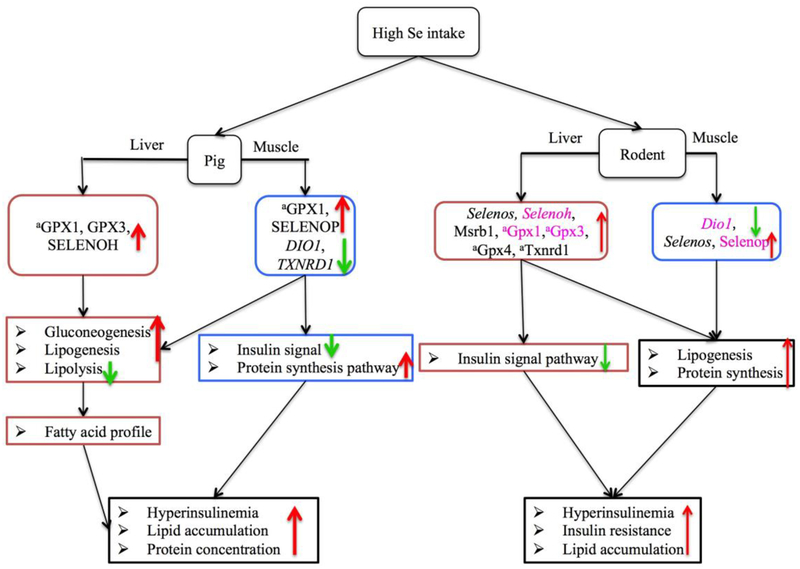
Figure 2. Scheme of regulatory pathways and mechanisms for the diabetogenic potential of high Se intakes in comparison with adequate or deficient Se intakes in pigs and rodents.
↑ Activation or increase; ↓ inhibition or decrease; Dio1, iodothyronine deiodinase 1; GPX1, glutathione peroxidase 1; GPX3, glutathione peroxidase 3; GPX4, glutathione peroxidase 4; Msrb1, selenoprotein X; SELENOH, selenoprotein H; SELENOP, selenoprotein P; Selenos, selenoprotein S; TXNRD1, thioredoxin reductase 1.
After comparing the porcine selenogenome with that of eight other domestic species, we have found an obvious convergent evolution. Specifically, pig SEPHS2 had a high similarity to chicken SELENOW1, TXNRD2, and GPX4. This type of convergence also existed between pig SEPHS2 and house cat SELENOH and horse SELENOW1 or between pig SELENOS and rabbit TXNRD2. During evolution, selenoprotein Pb (SELENOPb) and selenophosphate synthetase 2a (SEPHS2a) have had Sec lost or replaced by Cys. Overall, pigs exhibited a distant relationship with domestic fowl, but were closely related to livestock, especially sheep and cattle (Fig. 1C).
e). Comparison of GPX1 among different species
Figure 2A indicates a relatively low or distant similarity among different porcine selenoprotein genes by intraspecies comparisons, although a few family selenoprotein genes exhibited a close relationship. Nevertheless, we have found relatively higher similarities for the same selenoprotein genes between pigs and other species. Due to the information volume and the space limitation, we have presented only the evolutionary relationship of GPX1, as a representative, between pigs and 12 other domestic species (Fig. 1D). Likewise, porcine GPX1 had a higher similarity to that of cattle and sheep than the other domestic species. This once again suggests a closer relationship between pig and cattle and sheep, but the farthest relationship with domestic fowls.
3. Regulations of porcine selenogenome by dietary Se and fat
a). Dietary Se requirements
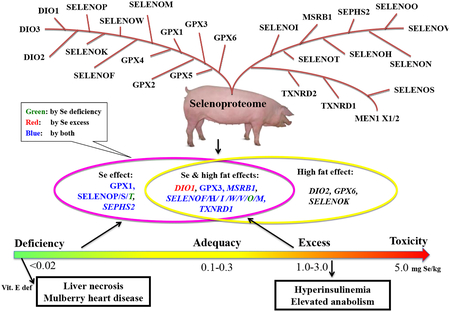
Three levels of dietary Se concentrations (mg Se/kg diet): deficiency (< 0.02), adequacy (0.1 – 0.3), and excess (0.4 – 5), have been often used to determine regulations of selenogenome and selenproteome expression by Se. Dietary Se requirements are 0.15–0.4 mg/kg for rodents, pigs [31], fish [32], beef cattle [33], and dairy cattle [34]. These requirements were established based on saturation of glutathione peroxidase activity in blood and tissues, health, and physiological states. Dietary concentrations > 0.4 mg Se/kg is often considered to exceed nutrient requirements by many species, to be wasteful, or even to act as a potential hazard to animals and environment.
b). Regulation of porcine selenogenome by dietary Se only
Overall, 16 porcine selenoproteins were affected by dietary Se, and five of which responded to only dietary Se, and 11 responded to both dietary Se and fat (Table 1) [35–38]. Among the five selenoproteins responding to only dietary Se, GPX1, SELENOP, and SELENOS had their expressions decreased in multiple tissues (blood, heart, kidney, liver, muscle, pituitary, spleen, testes, or thyroid), and SELENOT had its expression increased in heart, liver, and muscle in dietary Se deficiency compared with the Se adequacy. However, dietary Se deficiency caused diverse responses of SEPHS2 in various tissues. Dietary Se excess caused diverse responses of GPX1, SELENOP, SELENOS, and SEPHS2, but had no effect on SELENOT in heart, liver, and muscle [35–37]. Compared with the Se-adequate (0.3 mg/kg) diet, the Se-excess (3 mg/kg) diet did not alter the mRNA or protein abundances of GPX1 in heart, kidney, liver, muscle, and thyroid [35–37], but actually elevated GPX activities in the liver, muscle, and thyroid [35,37]. In comparison with 0.17 mg Se/kg, 0.5 mg Se/kg enhanced GPX1 activity in muscle [39]. The Se excess diet did not alter protein levels of SELENOP or SELENOS in the heart, kidney, and liver, but elevated SELENOP in the muscle [36], testes [40], and thyroid [35], and SELENOS in the thyroid [35]. Thus, GPX1 and SELENOP, the two abundantly expressed selenoproteins, were readily affected by dietary Se in pigs. In addition to the five selenoproteins listed in Table 1, the mRNA abundance of liver SELENON in Se deficiency or excess was not affected when compared with Se adequacy, but it was higher in Se deficiency than in Se excess [37].
Table 1.
Responses of selenogenome expression to dietary Se and fat in pigs1
| Name | Location | < 0.02 mg Se/kg2 | 0.4 – 3 mg Se/kg2 | High Fat Diet |
|---|---|---|---|---|
| Responding to Only Se | ||||
| GPX13 | Cyto/Mito | H/K/aL/M/Pi/S/aTh↓ | H/K/Th↔; aL/aM/aTh↑; Pi/S↓ | |
| SELENOP3 | EC | K/L/M/Te/Th↓ | H/K/L↔; M/Te/Th↑ | |
| SELENOS3 | ER/Memb | B/K/L/Pi/S/Th↓ | B/H/K/L/M↔; Th↑ | |
| SELENOT | ER | H/L/M↑ | H/L/M↔ | |
| SEPHS2 | Cyto | L/Th↔; H↑; HT/Pi↓ | H/L↔; HT/Pi/Th↓ | |
| Responding to Only Fat | ||||
| DIO2 | Memb4 | Th↑ | ||
| GPX6 | EC4 | M↑ | ||
| SELENOK | ER/Memb4 | Pa↓ | ||
| Responding to Both Se and Fat | ||||
| DIO1 | Memb | L/Pi/Te/Th↔ | M↓ | Pa↓ |
| GPX3 | EC | aPl/Pi/Te↓ | aPl↔; L/M↑ | HT↑ |
| MSRB1 | Cyto/Nu | S↓ | S↓ | A↓ |
| SELENOF3 | ER | K/Pi/S/Th↓ | Pi/S/Te/Th↓ | L↑ |
| SELENOH3 | Nu | K/L/Pi/S/Th↓ | K/Th↔; L↑; S↓ | Pa↓ |
| SELENOI3 | Memb4 | Pi/S/Th↓ | Pi/S/Th↓ | A/HT/Pa/Pi↓ |
| SELENOM | ER/Gol4 | S/Th↓ | S↓ | A/K↓; Pi↑ |
| SELENOO | Mito4 | H/HT↑ | H/HT↔ | K↑ |
| SELENOV | ? | HT/K/M↑; Th↓ | HT/K/M↔; Th↓ | A/L↓; M↑ |
| SELENOW | Cyto | L↓ | L↓ | Pa↓ |
| TXNRD1 | Cyto/Nu | L↓; Te↔ | M/Te↓; L↔ | Th↑; HT/Pi↓ |
Abbreviations and arrows: superscript a (a), enzyme activity of the selenoprotein; Cyto, cytosol; EC, extracellular; ER, endoplasmic reticulum; A, adipose tissue; B, blood; Gol, Golgi apparatus; H, heart; HT, hypothalamus; K, kidney; L, liver; M, muscle; Memb, membrane; Mito, mitochondria; Nu, nucleus; Pa, pancreas; Pl, Plasma; Pi, pituitary; S, spleen; Se, selenium; Te, testes; Th, thyroid;; ↑, increase; ↓, decrease; ↔, no differences.
Compared with 0.1 – 0.3 mg Se/kg diet.
Changed in many tissues
Supported by the database of uniprot (http://www.uniprot.org/uniprot/).
Comparatively, eight of the assayed selenoprotein genes in rodents were affected by only dietary Se in the adipose tissue and liver (Table 2). Specifically, Gpx1, and Selenot were overlapped with those in pigs [35–37; 41]. Among those eight rodent selenoproteins, Gpx1, Gpx3, Selenot, Selenow, Txnrd1, Txnrd2, and Txnrd3 were all down-regulated in the liver by Se deficiency, but most of them remained unchanged by excessive dietary Se [40,42–45].
Table 2.
Responses of selenogenome expression to dietary Se and fat in rodents1
| Name | Location | Se Deficiency2 | 1 mg Se/kg2 | 3 – 5 mg Se/kg2 | High Fat Diet |
|---|---|---|---|---|---|
| Responding to Only Se | |||||
| Dio1 | Memb | A↔ | A↓ | A↔ | |
| aGpx1 | Cyto/Mito | L↓ | L↓ | L↑ | |
| Selenow | Cyto | L↓ | L↓ | L↔ | |
| Txnrd1 | Cyto/Nu | L↓ | L↔ | L↔ | |
| Txnrd2 | Cyto | L↓ | L↔ | L↔ | |
| Txnrd3 | Mito | L↓ | L↔ | L↔ | |
| Gpx3 | EC | aPl/L↓ | aPl/L↑ | aPl/L↔ | |
| Selenot | ER | L↓ | |||
| Responding to Both Se and Fat | |||||
| Dio2 | ER | A/L↓ | L↑ | ||
| Selenof | ER | A↓; L↑ | A/L↑ | ||
| Selenok | ER | A↓ | A↓ | ||
| Selenom | ER | A/L↓ | A↓; L↑ | ||
| Selenos | ER | M↓ | L↑ | A↓; L↑ | |
| Gpx4 | Cyto/Nu/Mito | L↓ | A↓ | A↓ | |
| Msrb1 | Cyto/Nu | aL↓ | A↓ | L↔ | L↑ |
| Selenoh | Nu | L↓ | A↓ | L↑ | A↓; L↑ |
| Dio3 | Memb | A↓ | A/L↑ | ||
| Selenoi | Memb3 | A↓ | A↓; L↑ | ||
| Gpx6 | EC3 | A↓ | A↓; L↑ | ||
| Selenop | EC | A↑ | |||
Abbreviations and arrows: superscript a (a), enzyme activity of the selenoprotein; Cyto, cytosol; EC, extracellular, ER, endoplasmic reticulum; A, adipose tissue; Memb, membrane; Mito, mitochondria; Nu, nucleus; Pl, Plasma; Se, selenium; ↑, increase; ↓, decrease; ↔, no differences.
Compared with 0.1 – 0.3 mg Se/kg diet.
Supported by the database of uniprot (http://www.uniprot.org/uniprot/).
c). Regulation of porcine selenogenome by both dietary Se and fat
Among the 11 porcine selenoproteins responding to both dietary Se and fat (Table 1), the mRNA abundances of most of the genes (except DIO1, SELENOO, SELENOV, and TXNRD1) in various tissues were decreased by dietary Se deficiency (< 0.02 mg/kg), compared with the Se adequacy (0.3 mg/kg). DIO1 transcripts in the liver, pituitary, testis, and thyroid were unchanged in dietary Se deficiency [35], but in the muscle and pancreas was decreased by high dietary Se [36] and fat [38], respectively (Table 1). In multiple porcine tissues including pituitary, spleen, and thyroid, both dietary Se deficiency and excess decreased the expression of SELENOF and SELENOI [35]. In the Se deficient pigs, the nucleus-localized SELENOH also had its gene expression decreased in the tissues like those for SELENOF [35]. Dietary fat not only elevated the expressions of GPX3 in the hypothalamus, SELENOF in the liver, SELENOM in the pituitary, SELENOO in the kidney, SELENOV in the muscle, and TXNRD1 in the thyroid, but also decreased the expressions of some selenoprotein genes in multiple endocrine organs, like adipocyte tissue, hypothalamus, pancreas, and pituitary [38]. Compared with the Se adequacy, the Se deficiency decreased the GPX3 expression in the pituitary and testes [35], and Se excess up-regulated its expression in the liver and muscle [35]. Plasma GPX3 activity of pigs was significantly decreased by Se deficiency, but remained unchanged at week 8 [35, 37] or was elevated at week 16 after the treatment of the high Se diet [35]. However, GPX3 expression in hypothalamus was increased by high fat diet [38].
Overall, 12 rodent selenoprotein genes were affected by both dietary Se and fat (Table 2). Among them, five members were endoplasmic reticulum (ER)-resident proteins, i.e. Dio2, Selenof, Selenok, Selenom, and Selenos. In the liver and adipose tissues, their expressions were saturated at 0.3 mg Se/kg, but were differentially affected by the high fat diet [41]. Compared with 0.1 mg Se/kg diet, the Se-deficient diet down-regulated enzyme activity and protein abundance of Msrb1 in mouse liver, but supplementing Se at 0.4 to 2.25 mg/kg failed to enhance the protein abundance [42]. It is interesting to note that the high fat diet up- and down-regulated the expression of most of the 12 selenoprotein genes in the liver and adipose tissue, respectively [41].
d). Regulation of porcine selenogenome by dietary fat only
Fewer selenoprotein genes were affected by dietary fat only than dietary Se only in pigs (Table 1). Only DIO2, GPX6 and, SELENOK expression responded in that way. The high diet up-regulated the gene expressions of DIO2 in the thyroid and GPX6 in the muscle, but down-regulated SELENOK expression in the pancreas [38].
e). Regulation of porcine selenogenome by neither dietary Se nor fat
Changes in dietary Se concentrations ranged from deficiency to 3 mg/kg [35–37] or in dietary fat ranged from < 0.82 – 7% [38] did not affect the expressions of DIO3, GPX2, GPX4, TXNRD2, and TXNRD3 in various pig tissues. Likewise, Gpx2, Selenon, Selenoo, Selenov, and Sephs2 had no responses to dietary Se or fat changes in the rodent tissues, while the other selenoproteins had varied responses to either dietary Se or fat [40, 43–45].
f). Tissue- or subcellular-specific responses of selenogenome to dietary Se & fat
Expressions of several porcine selenoprotein genes in the pituitary, testes, and thyroid were changed in a similar manner: down-regulated by both dietary Se deficiency and excess. Hypothalamus had three selenoprotein genes affected by dietary fat (Table 1). Expression of selenoproteins mainly residing in cytosol in both pigs and rodents was readily affected by dietary Se and fat, and the same seemed to be true for those ER-resident selenoproteins in rodents. Because of the role of ER in lipid metabolism [47], altering the ER-resident selenoproteins may impair protein folding [48, 49] that partially explains the effects of dietary Se or fat on body lipid metabolism [50]. In addition, the affected selenoproteins, except DIO1, were largely down-regulated by Se deficiency in various tissues. Some of them, such as GPX1, GPX3, MSRB1, SELENOH, SELENOP, SELENOS, SELENOT, and TXNRDs, were saturated in selected tissues (e.g. kidney and liver) in Se adequacy when comparing dietary Se excess and adequacy.
4. Regulation of porcine glucose, lipid, protein metabolism by Se and selenoproteins
a). Hyperinsulinemia and insulin resistance induced by excess Se
Supranutritional Se intakes exhibited a pro-diabetic potential in pigs, as growing pigs fed 3.0 mg Se/kg diet (as Se-yeast) for 11 weeks developed hyperinsulinemia and insulin resistant [36] (Fig. 2). Consistently, feeding pigs with 1.0 mg Se/kg diet (Se-yeast) for 2 weeks also induced mild hyperinsulinemia [51]. Similar effects of the excess Se intakes were also shown in rodents [44, 52].
b). Alterations of lipid, fatty acid, and amino acid profiles by excess Se
Pigs fed 3.0 mg Se/kg diet had higher concentrations of total cholesterol (TC), triglyceride (TG), and (or) non-esterified fatty acid (NEFA) in both liver and adipose tissue compared with the controls [36]. Similarly, higher hepatic TG concentration was also reported in rats receiving 75 and 150 μg/kg Se than those fed the Se deficient diet [53]. Mice with Gpx1 overexpression developed obesity [52]. Moreover, feeding pigs 3.0 mg Se/kg diet led to higher concentrations of hepatic saturated fatty acids (myristic, palmitic, stearic, and behenic acids) and lower concentrations of polyunsaturated fatty acids (dihomo-γ-linolenic, oleic, α-linolenic, and eicosatrienoic acids) [36] in the liver and(or) adipose tissue (Fig. 2). In addition, the excess Se diet produced moderate increases in the protein concentrations of the liver and muscle in pigs, along with increases in aspartic acid, glutamine, and alanine [36].
c). Selenoproteins on insulin signaling, lipogenesis, and protein synthesis
Up-regulated GPX1, GPX3, SEENOLH, SELENOP and SELENOV and down-regulated DIO1 and TXNRD1 in pig tissues, similar to those in rodents, were associated with decreased gene expression related to insulin signaling (INSR and AKT), glycolysis (glucokinase), gluconeogenesis (PEPCK), lipogenesis (FOXO1, SREBP1, ACC and FAS), and protein synthesis pathway (mTOR and RPS6/S6) [35–39, 44, 54]. Pigs fed the high Se diet had elevated hepatic GPX1 and GPX3 as well as increased SELENOP and decreased DIO1 and TXNRD1 mRNA abundance in liver or muscle, which was associated with insulin resistance and enhanced lipogenesis and protein synthesis [36] (Fig. 2). Meanwhile, an increased Selenos was also correlated with impaired insulin signaling, lipogenesis, and protein synthesis in rats but not in pigs [44]. Pancreatic SELENOV might be correlated with insulin secretion and hepatic cholesterol synthesis [44, 55, 56]. Other selenoproteins including GPX4, MSRB1 and TXNRD1 were also found to affect glucose metabolism and diabetes risk [53, 57] (Fig. 2).
d). Genetically-altering Gpx1 and Selenop on insulin signaling and lipogenesis
Overexpression of Gpx1 in mice enhanced glycolysis and lipogenesis, suppressed insulin signaling [52], and aggravated glucose-stimulated insulin secretion (GSIS) [58, 59], whereas knockout of Gpx1 in mice led to lower fasting plasma insulin concentration, attenuated GSIS, and altered expression of pertaining genes [60]. Knockout of Selenop in mice improved insulin signaling [61]. In addition, other selenoprotein genes including Dio1, Gpx3, Gpx4, Selenoh, Msrb1, Selenos, Selenov, Txnrd1 have also been shown to be involved in Se-mediated regulation of energy metabolism in rodents [44, 53, 57]. We are developing similar porcine selenoprotein gene knockout models to explore their role in body glucose, lipid, and protein metabolism of pigs.
5. Concluding remarks
Our phylogenetic analyses place the 25 porcine selenoproteins into two primitive groups and three parallel branches. The porcine selenogenome shared a high similarity to that of humans and rodents, and exhibited a close evolutionary relationship with that of sheep and cattle. While SELENOT was the most conserved selenooprotein among all of the identified members, different selenoprotein families shared low similarities and thus were rather independent in the evolution. There were convergent or divergent evolutions among selenoproteins between the pigs and eight other domestic species.
Expression of 12 out of the 25 porcine selenoprotein genes in a number of tissues was affected by dietary Se, and only half of these genes responded to dietary fat as well. Seemingly, this affected gene number was fewer than that in the rodents, and the affected genes were most abundantly expressed and(or) the pertaining proteins were mainly cytosolic. Although the expression constancy or fluctuation of selected selenoproteins in tissues at different dietary Se concentrations may reflect their biological importance or negligence, underlying mechanisms for the species-, tissue-, and subcellular location-specific regulation of their expression remain elusive.
Linking the regulation of selenoprotein expression by dietary fat to their impacts on glucose, fat, and protein metabolism in the pigs mayl help unveil a bilateral mechanism between the macro- and micro-nutrients interactions in the body. This link will offer us new concepts and approaches in understanding the homeostasis of nutrient metabolism and pathogenesis of metabolic diseases. High dietary Se intakes induced hyperinsulinemia, insulin resistance, lipogenesis, lipolysis, gluconeogenesis, and tissue accumulations of lipid and protein in pigs. These responses were consistent with the observed pro-diabetogenic potential of supranutrition of Se in humans [62–65]. The pig research has revealed specific effects of dietary excess Se on tissue distributions of several fatty acids and amino acids. Because pigs are an excellent model of human nutrition and medicine, the pig data will help guide us in accurately assessing body Se status and appropriate supplementation of Se in foods for humans. As Se is a required feed additive to swine ration, dietary Se deficiency or excess can be a practical concern in swine production [66, 67]. Meanwhile, pork is consumed as one of the major animal proteins. Appropriate Se concentrations in pork are important for the public health [68]. Altogether, further elucidating the evolution, regulation, and function of the porcine selenogenome and selenoproteome will have a broad significance.
Highlights:
The 25 porcine selenoproteins fall into two groups and three branches.
Porcine selenogenome is phylogenetically closest to that of sheep and cattle.
Expression of partial porcine selenogenome is regulated by dietary Se & fat.
Dietary Se intake affects glucose, lipid, and protein metabolism of pigs.
High Se intake induces hyperinsulinemia and alters tissue lipid & protein in pigs.
Acknowledgements
The research in the authors’ laboratory was supported in part by a grant from the Major International (Regional) Joint Research Program of the Natural Science Foundation of China (No. 31320103920), the 111 Project from the Education Ministry of China (No. B18053), a grant of NIH DK 53018 (to XL), Natural Science Foundation of China (31572382), and National Key Research and Development Program of China (Project No. 2017YFA010320).
Abbreviations
- ACC
acetyl CoA carboxylase
- AKT
protein kinase B
- Cys
cysteine
- DIO1
type 1 iodothyronine deiodinase
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- FOXO1
forkhead box protein O1
- GPX
glutathione peroxidase
- INSR
insulin receptor
- MIEN
migration and invasion enhancer
- MSRB
methionine-R-sulfoxide reductase B
- mTOR
mammalian target of rapamycin
- NEFA
non-esterified fatty acid
- PEPCK
phosphoenolpyruvate carboxykinase
- RPS6/S6
ribosomal protein S6
- Se
selenium
- Sec
selenocysteine
- SELENO
selenoprotein
- SEPHS2a
selenophosphate synthetase 2a
- SREBP1
sterol regulatory element-binding protein 1
- TC
total cholesterol
- TG
triglyceride
- TXNRD
thioredoxin reductase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Whanger P;Vendeland S;Park YC;Xia Y Metabolismofsubtoxiclevels of selenium in animals and humans. Ann. Clin.Lab.Sci 26:99–113;1996. [PubMed] [Google Scholar]
- [2].Allaway WH An overview of distribution patterns of trace elements in soils and plants. Ann. N. Y. Acad. Sci 199:17–25; 1972. [DOI] [PubMed] [Google Scholar]
- [3].Franke KW A new toxicant occurring naturally in certain samples of plant foodstuffs. I. Results obtained in preliminary feeding trials. J. Nutr 8:597; 1934. [Google Scholar]
- [4].Casteel SW; Osweiler GD; Cook WO; Daniels G; Kadlec R Selenium toxicity in swine. Am. Vet. Med. J 186:1084–1085; 1985. [PubMed] [Google Scholar]
- [5].Yang GQ; Wang SZ; Zhou RH et al. Endemic selenium intoxication of humans in China. Am J Clin Nutr 37:872–881; 1983. [DOI] [PubMed] [Google Scholar]
- [6].Rederstorff M; Krol A & Lescure A Understanding the importance of selenium and selenoproteins in muscle function. Cell Mol Life Sci 63:52–59; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mariotti M; Ridge PG; Zhang Y; Lobanov AV; Pringle TH; Guigo R; Hatfield DL; Gladyshev VN Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS One 7:e33066; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lobanov AV; Hatfield DL; Gladyshev VN Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol 9: R62; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kryukov GV; Castellano S; Novoselov SV; Lobanov AV; Zehtab O; Guigó R; Gladyshev VN Characterization of Mammalian Selenoproteomes. Science 300:1439–1443; 2003. [DOI] [PubMed] [Google Scholar]
- [10].Ashton K; Hooper L; Harvey LJ; Hurst R; Casgrain A; Fairweather-Tait SJ Methods of assessment of selenium status in humans: a systematic review. Am J Clin Nutr 89:2025S–2039S; 2009. [DOI] [PubMed] [Google Scholar]
- [11].Méplan C Selenium and chronic diseases: a nutritional genomics perspective. Nutrients 7:3621–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].“Sources of Meat”. Food and Agriculture Organization (FAO) 25 November 2014. Retrieved 19 November 2016. [Google Scholar]
- [13].Patterson JK;Lei XG;Miller DD The pig as an experimental model for elucidating the mechanisms governing dietary influence on mineral absorption. Exp. Biol.Med.(Maywood) 233:651–664; 2008. [DOI] [PubMed] [Google Scholar]
- [14].Spurlock ME and Gabler NK. The development of porcine models of obesity and the metabolic syndrome. J Nutr 138:397–402; 2008. [DOI] [PubMed] [Google Scholar]
- [15].Groenen MA, Archibald AL, Uenishi H, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lobanov AV; Hatfield DL; and Gladyshev VN Eukaryotic Selenoproteins and Selenoproteomes. Biochim Biophys Acta 1790: 1424–1428; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lobanov AV; Fomenko DE; Zhang Y; Sengupta A; Hatfield DL, et al. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol 8: R198; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mangiapane E; Pessione A; Pessione E Selenium and selenoproteins: an overview on different biological systems. Curr Protein Pept Sci 15:598–607; 2014. [DOI] [PubMed] [Google Scholar]
- [19].Castellano S On the unique function of selenocysteine — Insights from the evolution of selenoproteins. BBA - General Subjects 1790: 1463–1470; 2009. [DOI] [PubMed] [Google Scholar]
- [20].Kimura M Evolutionary rate at the molecular level. Nature 217: 624–626; 1968. [DOI] [PubMed] [Google Scholar]
- [21].Romero H; Zhang Y; Gladyshev VN and Salinas G Evolution of selenium utilization traits. Genome Biol 6: R66; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peng T; Lin J; Xu YZ; and Zhang Y Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 10: 2048–2059; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rother M; Mathes I; Lottspeich F; Bock A Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J Bacteriol 185: 107–114; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fordyce FM Selenium deficiency and toxicity in the environment. Essentials of Medical Geology pp375–416; 2013. [Google Scholar]
- [25].Voight BF;Kudaravalli S; Wen X; Pritchard JK A map of recent positive selection in the human genome. PLoS Biol 4: e72; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hancock AM; Rienzo AD Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci USA 107: 8924–8930; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sun LH; Pi DA; Zhao L; Wang XY; Zhu LY; Qi DS; Liu YL Response of Selenium and Selenogenome in Immune Tissues to LPS-Induced Inflammatory Reactions in Pigs. Biol Trace Elem Res 177:90–96; 2017. [DOI] [PubMed] [Google Scholar]
- [28].Kumar S; Tamura K; Jakobsen IB; Nei M MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245; 2001. [DOI] [PubMed] [Google Scholar]
- [29].Birney E; Hudson TJ; Green ED Prepublication data sharing. Nature 461: 168–170; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carlson BA; Xu XM; Gladyshev VN; Hatfield DL Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem 280: 5542–5548; 2005. [DOI] [PubMed] [Google Scholar]
- [31].National Research Council (US) Subcommittee on Laboratory Animal Nutrition. Nutrient Requirements of Laboratory Animals: Fourth Revised Edition, 1995[J]. [PubMed] [Google Scholar]
- [32].National Research Council (US) Subcommittee on Laboratory Animal Nutrition. Nutrient Requirements of Laboratory Animals, 1993[J].
- [33].National Research Council (US) Subcommittee on Laboratory Animal Nutrition. Nutrient requirements of beef cattle: Seventh Revised Edition, 2000[J]. [Google Scholar]
- [34].National Research Council (US) Subcommittee on Laboratory Animal Nutrition. Nutrient requirements of dairy cattle: Seventh Revised Edition, 2001[J]. [Google Scholar]
- [35].Liu Y; Zhao H; Zhang QS; Tang JY; Li K; Xia XJ; Wang KN; Li K; Lei XG Prolonged dietary selenium deficiency or excess does not globally affect selenoprotein gene expression and/or protein production in various tissues of pigs. J Nutr 142:1410–1416; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhao Z; Barcus M; Kim J; Lum KL; Mills C; Lei XG High dietary selenium intake alters lipid metabolism and protein synthesis in liver and muscle of pigs. J Nutr 146: 1625–1633; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou JC; Zhao H; Li JG; Xia XJ; Wang KN; Zhang YJ; Liu Y; Zhao Y; Lei XG Selenoprotein gene expression in thyroid and pituitary of young pigs is not affected by dietary selenium deficiency or excess. J Nutr 139: 1061–1066; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao H; Li K; Tang JY; Zhou JC; Wang KN; Xia XJ; Lei XG Expression of selenoprotein genes is affected by obesity of pigs fed a high-fat diet. J Nutr 145: 1394–1401; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pinto A; Juniper DT; Sanil M; Morgan L; Clark L; Sies H; Rayman MP; Steinbrenner H Supranutritional selenium induces alterations in molecular targets related to energy metabolism in skeletal muscle and visceral adipose tissue of pigs. J Inorg Biochem 114: 47–54; 2012. [DOI] [PubMed] [Google Scholar]
- [40].Zhou JC; Zheng S; Mo J; Liang X; Xu Y; Zhang H; Gong C; Liu XL; Lei XG Dietary selenium deficiency or excess reduces sperm quality and testicular mrna abundance of nuclear glutathione peroxidase 4 in rats. J Nutr 147: 1947–1953; 2017. [DOI] [PubMed] [Google Scholar]
- [41].Novoselov SV; Kim HY; Hua D et al. Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid Redox Signal 12: 829–38; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Raines AM; and Sunde RA Selenium toxicity but not deficient or super-nutritional selenium status vastly alters the transcriptome in rodents. BMC Genomics 12: 26; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sunde RA, and Raines AM. Selenium regulation of the selenoprotein and nonselenoprotein transcriptomes in rodents. Adv Nutr 2: 138–50; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zeng MS; Li X; Liu Y; Zhao H; Zhou JC; Li K; Huang JQ; Sun LH; Tang JY; Xia XJ; Wang KN; Lei XG A high-selenium diet induces insulin resistance in gestating rats and their offspring. Free Radic Biol Med 52: 1335–1342; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Han SJ, et al. Characterization of mammalian selenoprotein o: a redox-active mitochondrial protein. PLoS One 9: e95518; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhao ZP; Zhou JC; Lei XG Differential regulation of transcriptional factors of selenoprotein genes by moderately high dietary concentrations of Se and fat in liver and adipose tissue of wildtype and gpx1−/− mice. FASEB J Published online 1 Apr; 2017 (Abstract). [Google Scholar]
- [47].Han J; Kaufman RJ The role of ER stress in lipid metabolism and lipotoxicity. J Lipid Res 57: 1329–38; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shchedrina VA; Zhang Y; Labunskyy VM; Hatfield DL; Gladyshev VN Structure-function relations, physiological roles, and evolution of mammalian ER-residentselenoproteins. Antioxid Redox Signal 12: 839–49; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Labunskyy VM; Hatfield DL; Gladyshev VN The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life 59: 1–5; 2007. [DOI] [PubMed] [Google Scholar]
- [50].Wang S; Kaufman RJ How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr Opin Lipidol 25: 125–132; 2014. [DOI] [PubMed] [Google Scholar]
- [51].Liu F; Celi P; Cottrell JJ; Chauhan SS; Leury BJ; Dunshea FR Effects of a short-term supranutritional selenium supplementation on redox balance, physiology and insulin-related metabolism in heat-stressed pigs. J Anim Physiol Anim Nutr (Berl) 102: 276–285; 2018. [DOI] [PubMed] [Google Scholar]
- [52].McClung JP; Roneker CA; Mu WP; Lisk DJ; Langlais P; Liu F; Lei XG Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. P Natl Acad Sci USA 101: 8852–8857; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mueller AS; Klomann SD; Wolf NM; Schneider S; Schmidt R; Spielmann J; Stangl G; Eder K; Pallauf J Redox regulation of protein tyrosine phosphatase 1b by manipulation of dietary selenium affects the triglyceride concentration in rat liver. J Nutr 138: 2328–2336; 2008. [DOI] [PubMed] [Google Scholar]
- [54].Wang XD; Vatamaniuk MZ; Wang SK; Roneker CA; Simmons RA; Lei XG Molecular mechanisms for hyperinsulinaemia induced by overproduction of selenium-dependent glutathione peroxidase-1 in mice. Diabetologia 51: 1515–1524; 2008. [DOI] [PubMed] [Google Scholar]
- [55].Nassir F; Moundras C; Bayle D; Serougne C; Gueux E; Rock E; Rayssiguier Y; Mazur A Effect of selenium deficiency on hepatic lipid and lipoprotein metabolism in the rat. Brit J Nutr 78:493–500; 1997. [DOI] [PubMed] [Google Scholar]
- [56].Mazur A; Nassir F; Gueux E; Moundras C; Bellanger J; Grolier P; Rock E; Rayssiguier Y Diets deficient in selenium and vitamin E affect plasma lipoprotein and apolipoprotein concentrations in the rat. Brit J Nutr 76: 899–907; 1996. [DOI] [PubMed] [Google Scholar]
- [57].Labunskyy VM; Lee BC; Handy DE; Loscalzo J; Hatfield DL; Gladyshev VN Both maximal expression of selenoproteins and selenoprotein deficiency can promote development of type 2 diabetes-like phenotype in mice. Antioxid Redox Signal 14: 2327–2336; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yan X; Pepper MP; Vatamaniuk MZ; Roneker CA; Li L; Lei XG Dietary selenium deficiency partially rescues type 2 diabetes-like phenotypes of glutathione peroxidase-1-overexpressing male mice. J Nutr 142: 1975–1982; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pepper MP; Vatamaniuk MZ; Yan X; Roneker CA; Lei XG impacts of dietary selenium deficiency on metabolic phenotypes of diet-restricted gpx1-overexpressing mice. Antioxid Redox Signal 14: 383–390; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang X; Vatamaniuk MZ; Roneker CA; Pepper MP; Hu LG; Simmons RA; and Lei XG Knockouts of SOD1 and GPX1 exert different impacts on murine islet function and pancreatic integrity. Antioxid Redox Signal 14: 391–401; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mita Y; Nakayama K; Inari S; Nishito Y; Yoshioka Y; Sakai N; Sotani K; Nagamura T; Kuzuhara Y; Inagaki K; Iwasaki M; Misu H; Ikegawa M; Takamura T; Noguchi N; Saito Y Selenoprotein P-neutralizing antibodies improve insulin secretion and glucose sensitivity in type 2 diabetes mouse models. Nat Commun 8: 1658; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bleys J; Navas-Acien A; Guallar E Serum selenium and diabetes in U.S. adults. Diabetes Care 30: 829–834; 2007. [DOI] [PubMed] [Google Scholar]
- [63].Gao S; Jin Y; Hall KS; Liang C; Unverzagt FW; Ji R; Murrell JR; Cao J; Shen J; Ma F; et al. Selenium level and cognitive function in rural elderly Chinese. Am J Epidemiol 165: 955–965; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Laclaustra M; Navas-Acien A; Stranges S; Ordovas JM; Guallar E Serum selenium concentrations and diabetes in U.S. adults: National Health and Nutrition Examination Survey (NHANES) 2003–2004. Environ Health Perspect 117: 1409–1413; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Faghihi T; Radfar M; Barmal M; Amini P; Qorbani M; Abdollahi M; Larijani B A randomized, placebo-controlled trial of selenium supplementation in patients with type 2 diabetes: effects on glucose homeostasis, oxidative stress, and lipid profile. Am J Ther 21: 491–495; 2014. [DOI] [PubMed] [Google Scholar]
- [66].Surai PF; Fisinin VI Selenium in pig nutrition and reproduction: boars and semen quality-a review. Asian-Australas J Anim Sci. 28: 730–46; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhan X; Qie Y; Wang M; Li X; Zhao R Selenomethionine: an effective selenium source for sow to improve Se distribution, antioxidant status, and growth performance of pig offspring. Biol Trace Elem Res 142: 481–91;2011. [DOI] [PubMed] [Google Scholar]
- [68].Fisinin VI; Papazyan TT; Surai PF Producing selenium-enriched eggs and meat to improve the selenium status of the general population. Crit Rev Biotechnol 29: 18–28; 2009. [DOI] [PubMed] [Google Scholar]