Abstract
Digital nucleic acid amplification and detection methods provide excellent sensitivity and specificity and allow absolute quantification of target nucleic acids. Isothermal methods such as digital loop-mediated isothermal amplification (digital LAMP) have potential for use in rapid disease diagnosis in low-resource settings due to their speed and lack of thermal cycling. We previously developed a self-digitization (SD) chip, a simple microfluidics device that automatically digitizes a sample into an array of nanoliter wells, for use in digital LAMP. In this work, we improve the SD chip design to increase sample loading efficiency, speed, and completeness, and test a range of well volumes and numbers. We demonstrate the diagnostic capability of this platform by applying it to quantifying human papillomavirus 18 gene.
Introduction
Digital nucleic acid quantitation methods provide excellent sensitivity and specificity for pathogen detection and have potential for use in point-of-care (POC) clinical diagnostic devices.1 Polymerase chain reaction (PCR) is often used for DNA amplification in these methods,2,3 but is not ideal for POC applications due to its long reaction time and requirement for fine temperature control. Loop-mediated isothermal amplification (LAMP)4–7 provides a fast, low-cost alternative. LAMP is highly specific for target DNA sequences and can be fast under optimal conditions.8,9
Conventional quantitative real-time LAMP, which is dependent on the TTP (the time for fluorescence intensity to pass a certain threshold), is susceptible to variability in amplification efficiency, especially at low DNA target concentration, limiting its sensitivity and accuracy.10 Quantitative LAMP also requires an independent measurement of a standard (e.g. using UV-Vis absorbance). Digital LAMP (dLAMP) is an absolute quantitation method which provides greater sensitivity and accuracy. In dLAMP, a sample containing target DNA is diluted into many discrete volumes which initially contain either zero, or one or more, DNA molecules, and the DNA is amplified. Volumes that initially contain one or more molecules can result in DNA amplification and produce a fluorescent signal. The concentration of target DNA in the original sample is quantified based on the number of positive wells using Poisson statistics.11
Methods to digitize samples include using droplets within emulsions,12,13 the SlipChip platform,14–17 and microvalve-based arrays of reaction chambers.18–21 Though these platforms offer favorable performance, a simple and cost-effective method is still highly desired, especially in POC settings. To digitize samples in POC settings, we developed a self-digitization (SD) chip – a microfluidics device that spontaneously partitions samples into a large array of small chambers based on viscoelastic fluid phenomena driven by the geometric properties of the channels and chambers in the chip.22–26 Compared to other methods, SD technology results in digitization without the need for precise and complex external equipment and instrumentation.
In this study we modify our previous SD chip design,23 which resulted in significant improvements in digitization speeds, larger sampling volumes, and completeness of filling. We demonstrate that the modified SD chip design is compatible with a wide range of well volumes and numbers. We then apply the new SD chip design to the quantitative detection of human papillomavirus 18 (HPV-18) using dLAMP.
Experimental Section
Chemicals and reagents:
Bst 2.0 WarmStart® DNA Polymerase and Isothermal Amplification Buffer were purchased from New England BioLabs (Ipswich, MA). Calcein (high purity) and dNTPs were purchased from Thermo Fisher (Waltham, MA). Light mineral oil, betaine, bovine serum albumin (BSA), and MnCl2 were purchased from Sigma-Aldrich (St. Louis, MO). HPV-18 plasmid (full-genome length, 45152D) was purchased from American Type Culture Collection (Manassas, VA). HPV-16, 31 and 45 plasmids were from Seattle Children’s Research Institute. Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Abil WE 09 and Tegosoft DEC were from UPI Chem (Somerset NJ). Ssofast Evagreen Supermix and BioRad QX100 ddPCR oil were purchased from Bio-Rad Laboratories, Inc. (Hercules, CA).
Chip fabrication:
The microfluidic device flow layer was modified from a previous design using AutoCAD (Autodesk, San Rafael, CA), and was printed as three separate layers onto a Mylar photomask (Fine Line Imaging, Colorado Springs, CO). The fabrication process has been described in detail previously.22
Digitization experiments:
For the experiments shown in Figure 2 (A–C), the oil mixture used was 68.3% mineral oil, 31.3% hexadecane, 0.4% Tegosoft DEC, and 0.018% Abil WE 09. The aqueous mixture was 1x Ssofast Evagreen Supermix with 2 mg/mL BSA, 500 nM generic primer, and fluorescein in 0.3× PBS. In Figure 2D the oil mixture was a 2:1 ratio of BioRad QX100 ddPCR oil and FC-40, and the aqueous mixture was the same as in Figure 2 (A–C). The device was primed with oil by placing oil in the inlet and outlet reservoirs, placing the entire device under vacuum to pull air out of the array, then removing the device from vacuum to allow oil to flow into the array. To digitize a sample, aqueous sample (~125% of the total array volume) was loaded into the inlet reservoir. Vacuum pressure (−0.3 bar) was applied until all of the aqueous sample had been drawn through the device, followed by oil which displaced aqueous sample in the channels (2–10 minutes). Videos and bright field and fluorescence photographs were obtained on an Olympus MVX10 stereoscope with an Olympus MV PLAPO 0.63x objective.
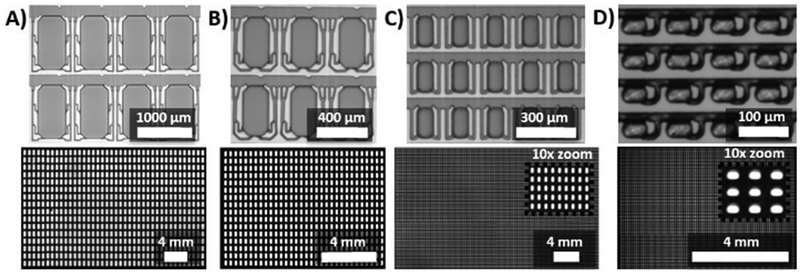
Figure 2.
The SD chip design digitizes samples over a range of well volumes and numbers. Top row: photographs of device masters; bottom row: fluorescence images of digitized samples containing fluorescein (A shows full array, B-D are partial arrays to facilitate visualization). Four SD chips were designed with different well numbers and dimensions: (A) 640 wells (1000 μm long × 520 μm wide × 200 μm tall per well) with individual well volumes of ~100 nL and a total volume of >60 μL; (B) 1024 wells (400 μm (l) × 200 μm (w) × 100 μm (h) per well) with individual well volumes of ~7.5 nL and a total volume of ~8 μL; (C) 25,600 wells (160 μm (l) × 80 μm (w) × 80 μm (h) per well) with a well volume of ~1 nL and a total volume of >25 μL; D) 10,240 wells (30 μm (l) × 50 μm (w) × 35 μm (h) per well) with a well volume of ~50 pL and a total volume of ~0.5 μL..
Reproducibility testing:
In Figure 3, the oil mixture used was a 6:3:1 ratio of BioRad QX100 ddPCR oil: FC-40: 1H,1H,2H,2H-perfluoro-1-octanol (PFO), and the aqueous solution was the same as in Figure 2. Images were acquired as in Figure 2; replicate images were combined into one image file for analysis.
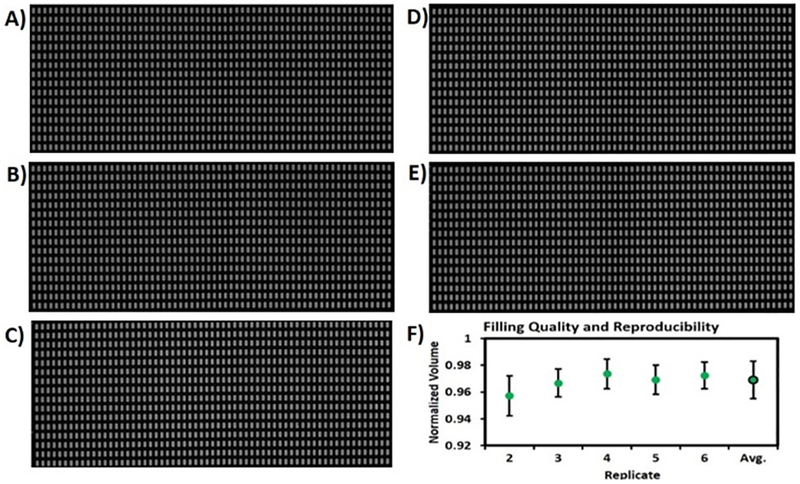
Figure 3.
The completeness and uniformity of well filling were assessed using an SD chip with 1024 wells of 7.5 nL each. A-E) Fluorescence stereoscope images of five replicates of arrays loaded with fluorescein solution. F) The average normalized well volume for each replicate was calculated based on normalized integrated pixel intensities. The five replicates showed near complete fill fractions (95–97%) and high monodispersity (CV <1.4%).
Well volume calculations:
Well heights were measured using a custom-built, white light interferometer.27 Based on the regular spacing of the wells in the arrays, images were calibrated to a specific μm/pixel scale. In the images shown in Figure 3, the scale was 7.4 μm/pixel and there were 1369.6 expected pixels/well. Since the depth of field of the stereoscope was greater than the height of the wells, the images capture all of the fluorescence within a vertical column, represented by each pixel. For a uniformly fluorescent solution, the relative pixel intensity should correlate with the relative height of the fluorescent solution within the well. A significant portion of the wells showed the maximized pixel intensity due to aqueous sample completely filling the well vertically, observed as a plateau in a line scan. For a given image, each well was analyzed at two different threshold intensities, a low threshold intended to collect virtually all of the fluorescent signal, and a high threshold that collects only signal from pixels in the plateaus. A line scan of a section of these wells (Figure S1) shows this plateau. The average value of the pixels above the high threshold represents the expected intensity (at the plateau). This value was multiplied by the expected pixel area to obtain the expected integrated intensity per well. Each well was treated individually to correct for illumination nonuniformity. The actual integrated intensity, measured at the low threshold for each well, was divided by the expected integrated well intensity to obtain a normalized actual filled volume for each well (Figure 3F).
LAMP assay (bulk):
The LAMP assay was performed using the following conditions per 25 μL reaction: 8 units Bst 2.0 WarmStart® DNA Polymerase, 1 × Isothermal Amplification Buffer, 4 mM MgSO4, 50 μM calcein, 1.4 mM of each dNTP, 0.8 M betaine, 1.2 mg/mL BSA, 1 mM MnCl2, varying concentrations of HPV-18 plasmid DNA, and HPV-18 LAMP primers (0.2 μM F3, 0.2 μM B3, 1.6 μM FIP, 1.6 μM BIP, 0.8 μM LF, 0.8 μM LB). The primer sequences were F3: 5’ – CGC GTC CTT TAT CAC AGG – 3’; B3: 5’ – TGG AAT CCC CAT AAG GAT – 3’; FIP: 5’ – GGC ACC ATA TCC AGT ATC TAC CAT AAT TGC CCC CCT TTA GAA CT – 3’; BIP: 5’ – TGC AAG ATA CTA AAT GTG AGG TAC CGC AGA CAT TTG TAA ATA ATC A – 3’; LF: 5’ – TCA CCA TCT TCC AAA ACT G – 3’; LB: 5’ – ATT GGA TAT TTG TCA GTC T – 3’. Calcein was mixed with manganese chloride (MnCl2) at a molar ratio of 1:20 before addition to the reaction mix. The reaction mix was incubated at 63°C for 90 min in a Bio-Rad CFX96 Real-Time PCR instrument and fluorescent signal was recorded at 60 sec intervals at a wavelength of 560–580 nm in real-time.
Digital LAMP:
The oil composition was 0.04% by mass Abil WE 09, 3% by mass light mineral oil in Tegosoft DEC. The device was primed with oil mixture as described above, and double-sided Kapton tape was applied to cover the outlet reservoir. A custom PDMS adaptor connected to vacuum was aligned to the outlet reservoir and secured onto the double-sided tape. 50 minutes after oil loading, a LAMP sample was pipetted into each inlet reservoir, and vacuum was turned on. Aluminum foil was used to cover the device to prevent photobleaching. After aqueous sample was no longer visible in the inlet reservoir (about 1–3 mins), oil was allowed to flow through the device for several minutes to remove excess aqueous solution from the main channels. The chip was put on the thermal control instrument with an in situ adapter, and heated to and maintained at 63°C for 90 min.
Chip imaging:
Devices were imaged using a Typhoon FLA9000 imaging system (GE Healthcare, Pittsburgh, PA, USA). Calcein solutions were excited at 473 nm and images were obtained using a long pass filter (510LP) with a pixel resolution of 10 μm. Images were analyzed using ImageJ (http://rsbweb.nih.-gov).
Data analysis:
Well volumes were determined using images taken before amplification. Absolute well volumes were determined using a well height of 100 μm, and 628.3 pixels per well based on a measured 10.17 μm/pixel and expected well dimensions of 350×200 μm with a 50 μm bevel on each corner. Normalized well volumes were calculated as described in the “Well volume calculations” section. Wells that shrunk during amplification (mainly those near the outer edge of the array) were removed from analysis. The entire outer ring of wells was always eliminated, and wells showing <50% of the expected well area or that showed a decrease in area during amplification of >20% were excluded. The shrinkage of the digitized volume was caused by evaporation through the porous PDMS during heating. However, this moderate reduction in the total number of wells analyzed did not decrease appreciably the quantification ability of the device. Each well was assigned an index value to enable comparison of images before and after amplification. On average, 77% of wells were used in analysis. To calculate the absolute concentration of a specific target sequence within a DNA sample, positive wells must be distinguished from negative wells after amplification. For experiments using calcein, the baseline signal was not stable from before to after amplification, and the signal before amplification could not be used to establish a baseline or threshold. Thus, the average well intensity was used to produce a bimodal histogram of well intensity, and the minimum value between the two modes was used to identify an initial threshold value. This value was used to find the average intensity of positive wells and negative wells. The midpoint of these two averages was used to set a new threshold and this process was repeated until the rate of positives and negatives converged (1 or 2 iterations).
Measured sample concentrations were calculated blind (without knowing the expected concentrations) by one author (Jason Kreutz). A different author (Allison Sheen) performed the dLAMP experiment.
Results and Discussion
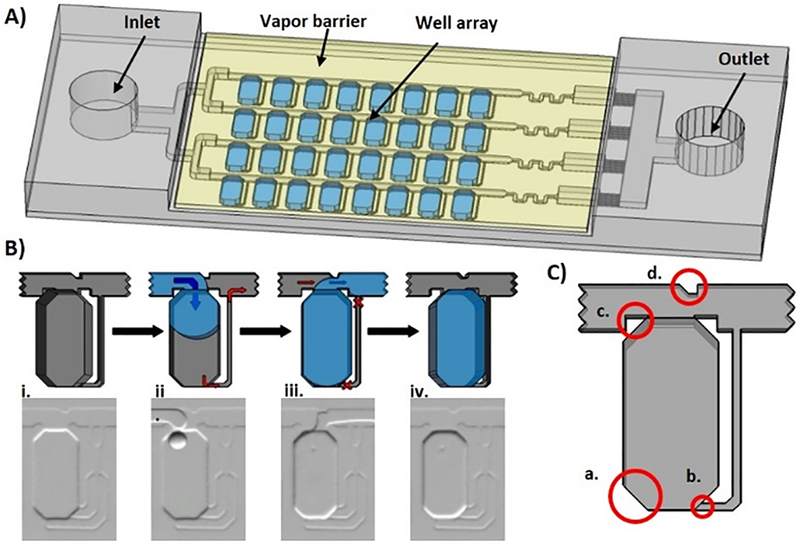
The SD chip, which is compatible with dLAMP and other digital nucleic acid quantification methods, is shown schematically in Figure 1A. The sample loading process is shown in Figure 1B. One refinement in the SD chip design was to optimize the height difference between the channels and wells to drive fluid into the wells with a “Greek key” drainage channel to more efficiently displace oil from the wells during sample filling. Additional refinements, shown in Figure 1C, include (a) using beveled well corners to reduce high energy distortions of aqueous droplets in wells, (b) using a height difference between wells and drainage channels to drain oil more efficiently, and (c, d) introducing notches in the channels to improve oil breakoff during digitization and sample filling. These parameters were optimized for each SD chip design shown in Figure 2; for example, larger digitized volumes (e.g. Figure 2A) benefited from the use of larger and more drainage channels than smaller digitized volumes (e.g. Figure 2D). The full digitization process can be seen in Supporting Video 1.
Figure 1. The self-digitization (SD) chip.
(A) The SD chip consists of an array of channels and wells that spontaneously compartmentalizes aqueous samples into defined volumes for digital nucleic acid quantification assays. (B) The sample loading process is illustrated in single well schematics (top) and photographs (bottom). (i) The device is primed with an oil mixture to eliminate air. (ii) Aqueous sample is loaded, travels through channels, and expands into wells to lower the surface energy. Hydrophobic drainage channels assist sample filling by allowing oil to drain from the wells but are too small to allow aqueous sample to transfer. (iii) Additional oil is loaded and displaces aqueous solution from the channels but not from the wells, isolating the aqueous solution in compartments (iv). (C) Enhancements of the SD chip design include (a) using beveled well edges to reduce high-energy distortions of aqueous droplets; (b) using “Greek key” drainage channels with a height difference between the wells and drainage channels to improve oil drainage; (c) notches in the channels on both sides of the wells to facilitate oil breakoff during digitization; and (d) a notch in the channel opposite the well to direct aqueous solution into the well during filling.
One major challenge in digital nucleic acid quantification is being able to process the thousands of individual small volumes (e.g. pL-nL) while also analyzing large enough total volumes to enable relevant assays. One advantage of the SD chip platform is its functionality across a range of well volumes and numbers, to fit any assay requirement. We designed SD chips spanning a range of well volumes (~ 100, 7.5, 1, and 0.05 nL), well numbers (640, 1024, 25,600, and 10,240 wells, respectively), covering total volumes from 0.5->60 μL (Figure 2). All four SD chips showed excellent sample digitization (>99.9% digitized in 2A, B and D and 98.7% digitized in 2C), as observed in fluorescence images (Figure 2, bottom), indicating that the SD chip design was scalable over a wide range of well volumes and numbers. We have also demonstrated that the digitization works across a range of aqueous and oil samples, as we used both mineral oil based (Figure 2 A–C and Figure 4), and fluorinated oil (Figure 2 D and Figure 3) based oil systems, and using both PCR mix (Figure 2 and Figure 3), and LAMP mix (Figure 4) based aqueous solutions. The motivation for this series of experiments was to demonstrate the broad compatibility of the SD process and chip with different oil and aqueous compositions.
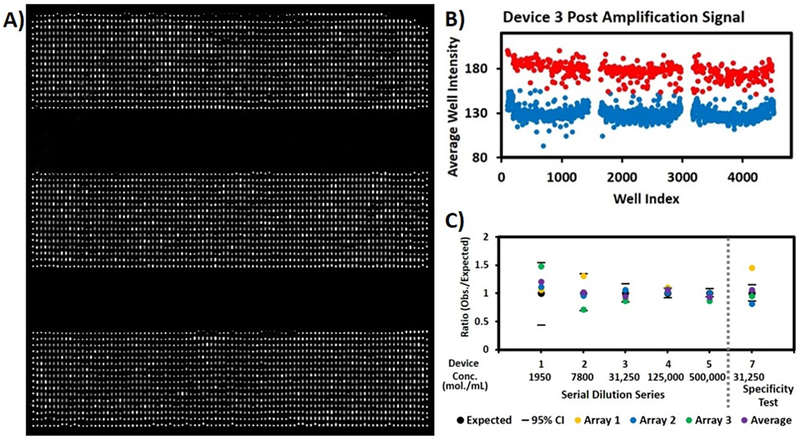
Figure 4.
A) Three replicates of dLAMP HPV-18 assay. The HPV-18 concentration of the original sample was about 3.1×104 molecules/mL. The arrays consisted of 1,536 wells with a well volume of 6.5 nL and a total volume of 10 μL. B) A plot of the average well intensities from images in (A). Red dots represent wells defined as positive (HPV-18 detected); blue dots represent wells defined as negative (no HPV-18). C) Absolute quantification results: the ratio of measured and expected HPV-18 concentrations for six devices loaded with different HPV concentrations. Dashes in the figure indicate 95% confidence intervals based on expected concentrations. For Devices 1–4, the ratio of measured to expected concentration fell within the 95% confidence interval. For Device 5, two of the three ratios fell within the 95% confidence interval. A negative control containing no template was also performed, which had only two false-positive wells over three replicates. Devices 7 tested the specificity of the assay. Control experiment for specificity was also performed containing HPV 16, 31, and 45 plasmids at 3.1×104 molecules/mL each, and the expected concentration for the HPV-18 target is zero so the ratio is undefined. The three control replicates had zero, one, and three false-positive wells, an overall false-positive rate of ~0.1% (data not shown). Device 7 contained HPV 16, 18, 31, and 45 plasmids at 3.1×104 molecules/mL each. One of three Device 7 arrays showed a ratio within the 95% confidence interval, but the average of the three also fell within the interval.
We examined the completeness and uniformity of well filling using the 1024-well SD chip shown in Figure 2B. Fluorescein spiked PCR solution was loaded into five different devices, and fluorescence images were obtained using a stereoscope (Figure 3A–E). To analyse whether well filling was complete both vertically and laterally within the wells, we measured the volume in each well based on normalized integrated pixel intensities, as detailed in the Experimental Section. Because the depth of field of the stereoscope was greater than the well height, the images capture all of the fluorescence within a vertical column, represented by the corresponding pixel. Therefore, the relative intensity of each pixel should correlate with the relative height of the fluorescent solution in the well (Detail in “data analysis” in Experimental Section). Figure S1 shows a line scan of well fluorescence. Plateaus indicate that the well is fully filled vertically. Analysis of the normalized volume in each well showed that the completeness of well filling was 95–97%, with a coefficient of variation (CV) of <1.4% (Figure 3F), indicating high monodispersity. These results indicate that the SD chip can achieve precise, complete, and reproducible well filling.
We applied the SD chip design to quantifying HPV-18 using dLAMP. HPV infection is the principal cause of cervical carcinoma,28 and high-risk HPV subtypes, including HPV-18, have been identified as etiologic agents responsible for most cervical cancers worldwide. HPV quantitation and genotyping help in evaluating the risk of carcinogenesis and the effectiveness of treatment.29,30 Previous studies have used LAMP for HPV detection,31–35 but none has used digital LAMP. Here we quantify HPV-18 using digital LAMP with an SD chip with 1536 wells of 6.5 nL each. The concentration of a plasmid containing the HPV-18 gene was determined using UV-Vis absorbance, and samples with a range of concentrations from 1,950 to 500,000 copies/mL were prepared by serial dilution. Figure 4A shows combined fluorescence images of three SD chip replicates that had been loaded with a sample containing 31,250 HPV18 plasmid copies/mL and subjected to LAMP. Positive wells, in which amplification has occurred, appear brighter than negative wells. A plot of the average well fluorescence intensity is shown in Figure 4B. Details of the data analysis are shown in the Experimental Section. The number of positive wells was counted and the sample concentration was calculated based on Poisson statistics. As not all wells may be suitable for analysis; for example, the well might have shrunk during amplification or there might be optical defects or dust that interfered with imaging. Thus, the analysis utilized the actual number and volume of wells suitable for analysis as per the protocol described under Detail in Data analysis. The measured concentration showed excellent agreement with the expected concentration at all five concentrations (Figure 4C). We also performed the negative control containing no template, which had only two false-positive wells over three replicates, giving an overall false-positive rate of <0.1%, corresponding to a concentration much lower than the lowest tested concentration. These results indicate that dLAMP using the new SD chip design allowed accurate HPV-18 quantitation with a wide dynamic range.
The specificity of the dLAMP HPV-18 assay (using HPV-18 primers) was tested using plasmid containing HPV-16, 31, and 45 genes (Figure 4C, Device 7). We performed a control experiment, which contained HPV-16, 31, and 45 at ~3.1×104 molecules/mL each, three HPV types that are not targeted by the HPV-18 assay and thus should not be detectable. The three negative control replicates showed zero, one, and three false-positive wells, an overall false-positive rate of ~0.1% (data not shown). Device 7 contained HPV 16, 31, 18, and 45 at ~3.1×104 molecules/mL each. We also performed bulk real-time LAMP on the same set of HPV samples, using primers against HPV-18 (Figure S2). Samples with HPV-16, 31 and 45 showed no amplification. The sample containing all four HPV including HPV-18 at 3.1×104 molecules/mL showed an increase in fluorescence a little later than the sample containing only HPV-18 at 3.1×104 molecules/mL, indicating a difference in reaction kinetics when multiple HPV types are present. Agarose gel electrophoresis results further confirmed LAMP amplification in samples containing HPV-18 plasmid including in mixed samples but not in samples containing off-target plasmids only (Figure S3). These results highlight the specificity of dLAMP and its ability to provide accurate quantitation in a mixed sample.
Conclusions
In this study, we show an improved design of our SD chip with increased efficiency, completeness, and reproducibility of sample filling resulting in fast, accurate and specific digital nucleic acid quantitation. We demonstrate that the SD chip is compatible with a wide range of well volumes and numbers, and thus is compatible with a variety of applications. In a dLAMP assay designed to quantify HPV-18, the assay showed excellent sensitivity and specificity for HPV-18 versus other HPV genes. This dLAMP SD chip assay is simple to perform, with a fast reaction time, and requires minimal equipment. With these characteristics, this platform can be further developed for use in rapid disease diagnosis in low-resource settings. Specifically, we are currently developing a portable instrument with a simple thermal control and imaging system for use with the SD chip.
Supplementary Material
Acknowledgements
We gratefully acknowledge support from the National Institutes of Health (R01EB021150) and the Bill & Melinda Gates Foundation.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Dietel M, Jöhrens K, Laffert M, Hummel M, Bläker H, Müller BM, Lehmann A, Denkert C, Heppner FL, Koch A, Sers C and Anagnostopoulos I, Cancer Gene Ther, 2013, 20, 211. [DOI] [PubMed] [Google Scholar]
- 2.Petralia S and Conoci S, ACS Sensors, 2017, 2, 876–891. [DOI] [PubMed] [Google Scholar]
- 3.Nolan T, Hands RE and Bustin SA, Nat Protocols, 2006, 1, 1559. [DOI] [PubMed] [Google Scholar]
- 4.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N and Hase T, Nucleic Acids Res, 2000, 28, e63–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagamine K, Watanabe K, Ohtsuka K, Hase T and Notomi T, Clin Chem, 2001, 47, 1742–1743. [PubMed] [Google Scholar]
- 6.Nagamine K, Hase T and Notomi T, Mol Cell Probe, 2002, 16, 223–229. [DOI] [PubMed] [Google Scholar]
- 7.Tomita N, Mori Y, Kanda H and Notomi T, Nat. Protocols, 2008, 3, 877–882. [DOI] [PubMed] [Google Scholar]
- 8.Tanner NA and Evans TC, Curr Protoc Mol Biol, 2014, 15, 11–14. [DOI] [PubMed] [Google Scholar]
- 9.Schoepp NG, Schlappi TS, Curtis MS, Butkovich SS, Miller S, Humphries RM and Ismagilov RF, Sci Transl Med, 2017, 9, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nixon G, Garson JA, Grant P, Nastouli E, Foy CA and Huggett JF, Anal Chem, 2014, 86, 4387–4394. [DOI] [PubMed] [Google Scholar]
- 11.Dorazio RM and Hunter ME, Anal Chem, 2015, 87, 10886–10893. [DOI] [PubMed] [Google Scholar]
- 12.Gutiérrez-Aguirre I, Rački N, Dreo T and Ravnikar M, Methods Mol Biol, 2015, 1302, 331–347. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, Link DR and Larson JW, Lab Chip, 2011, 11, 2167–2174. [DOI] [PubMed] [Google Scholar]
- 14.Shen F, Du W, Davydova EK, Karymov MA, Pandey J and Ismagilov RF, Anal Chem, 2010, 82, 4606–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen F, Du W, Kreutz JE, Fok A and Ismagilov RF, Lab Chip, 2010, 10, 2666–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selck DA, Karymov MA, Sun B and Ismagilov RF, Anal Chem, 2013, 85, 11129–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun B, Shen F, McCalla SE, Kreutz JE, Karymov MA and Ismagilov RF, Anal Chem, 2013, 85, 1540–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottesen EA, Hong JW, Quake SR and Leadbetter JR, Science, 2006, 314, 1464–1467. [DOI] [PubMed] [Google Scholar]
- 19.Heyries KA, Tropini C, VanInsberghe M, Doolin C, Petriv OI, Singhal A, Leung K, Hughesman CB and Hansen CL, Nat Meth, 2011, 8, 649. [DOI] [PubMed] [Google Scholar]
- 20.Jensen EC, Bhat BP and Mathies RA, Lab on a Chip, 2010, 10, 685–691. [DOI] [PubMed] [Google Scholar]
- 21.White AK, Heyries KA, Doolin C, VanInsberghe M and Hansen CL, Anal Chem, 2013, 85, 7182–7190. [DOI] [PubMed] [Google Scholar]
- 22.Cohen DE, Schneider T, Wang M and Chiu DT, Anal Chem, 2010, 82, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gansen A, Herrick AM, Dimov IK, Lee LP and Chiu DT, Lab Chip, 2012, 12, 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider T, Yen GS, Thompson AM, Burnham DR and Chiu DT, Anal Chem, 2013, 85, 10417–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson AM, Gansen A, Paguirigan AL, Kreutz JE, Radich JP and Chiu DT, Anal Chem, 2014, 86, 12308–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Q, Gao Y, Yu B, Ren H, Qiu L, Han S, Jin W, Jin Q and Mu Y, Lab Chip, 2012, 12, 4755–4763. [DOI] [PubMed] [Google Scholar]
- 27.Yen GS, Fujimoto BS, Schneider T, Huynh DTK, Jeffries GDM and Chiu DT, Lab Chip, 2011, 11, 974–977. [DOI] [PubMed] [Google Scholar]
- 28.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC and Wacholder S, Lancet, 2007, 370, 890–907. [DOI] [PubMed] [Google Scholar]
- 29.Posner MR, Lorch JH, Goloubeva O, Tan M, Schumaker LM, Sarlis NJ, Haddad RI and Cullen KJ, Ann Oncol, 2011, 22, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP and Gillison ML, N Engl J Med, 2010, 363, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo L, Nie K, Yang MJ, Wang M, Li J, Zhang C, Liu HT and Ma XJ, J Clin Microbiol, 2011, 49, 3545–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saetiew C, Limpaiboon T, Jearanaikoon P, Daduang S, Pientong C, Kerdsin A and Daduang J, J Virol Methods, 2011, 178, 22–30. [DOI] [PubMed] [Google Scholar]
- 33.Satoh T, Matsumoto K, Fujii T, Sato O, Gemma N, Onuki M, Saito H, Aoki D, Hirai Y and Yoshikawa H, J Virol Methods, 2013, 188, 83–93. [DOI] [PubMed] [Google Scholar]
- 34.Livingstone DM, Rohatensky M, Mintchev P, Nakoneshny SC, Demetrick DJ, van Marle G and Dort JC, J Clin Virol, 2016, 75, 37–41. [DOI] [PubMed] [Google Scholar]
- 35.Rohatensky MG, Livingstone DM, Mintchev P, Barnes HK, Nakoneshny SC, Demetrick DJ, Dort JC and van Marle G, BMC Cancer, 2018, 18, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.