Abstract
Objective:
Pyruvate kinases M (PKM), including the PKM1 and PKM2 isoforms, are critical factors in glucose metabolism. PKM2 promotes aerobic glycolysis, a phenomenon known as " the Warburg effect”. The purpose of this study was to identify the roles of PKM2 in regulating cellular metabolism.
Methods:
The CRISPR/Cas9 system was used to generate the PKM-knockout cell model to evaluate the role of PKM in cellular metabolism. Lactate levels were measured by the Vitros LAC slide method on an autoanalyzer and glucose levels were measured by the autoanalyzer AU5800. The metabolism of 13C6-glucose or 13C5-glutamine was evaluated by liquid chromatography/mass spectrometry analyses. The effects of PKM on tumor growth were detected in vivo in a tumor-bearing mouse model.
Results:
We found that both PKM1 and PKM2 enabled aerobic glycolysis, but PKM2 converted glucose to lactate much more efficiently than PKM1. As a result, PKM2 reduced glucose levels reserved for intracellular utilization, particularly for the production of citrate, and thus increased the α-ketoglutarate/citrate ratio to promote the generation of glutamine-derived acetyl-coenzyme A through the reductive pathway. Furthermore, reductive glutamine metabolism facilitated cell proliferation under hypoxia conditions, which supports in vivo tumor growth. In addition, PKM-deletion induced a reverse Warburg effect in tumor-associated stromal cells.
Conclusions:
PKM2 plays a critical role in promoting reductive glutamine metabolism and maintaining proton homeostasis. This study is helpful to increase the understanding of the physiological role of PKM2 in cancer cells.
Keywords: The Warburg effect, the reverse Warburg effect, PKM2, glucose metabolism, proton homeostasis
Introduction
Unlike non-proliferating cells, which predominantly use mitochondrial oxidative phosphorylation to generate ATP, proliferating cells including cancer cells tend to rely on aerobic glycolysis, a phenomenon known as “the Warburg Effect”1. Aerobic glycolysis is an inefficient method of generating energy, but may play a critical role in cell survival and proliferation in addition to ATP generation.In both non-proliferating and proliferating cells, glycolysis generates glycolytic intermediates including the end-product pyruvic acid, concomitantly with the production of NADH + H+ and ATP. In non-proliferating cells, pyruvic acid and NADH + H+ enter the mitochondria to generate ATP and are metabolized to water and carbon dioxide. In contrast, in proliferating cells, pyruvic acid is reduced to lactic acid; this process consumes all NADH + H+ produced in the glycolytic pathway2,3. In principle, the differential biochemical transformation of pyruvic acid, rather than common glycolytic intermediates, likely accounts for the beneficial effects of aerobic glycolysis in proliferating cells. In addition to ATP, the involved redox couples of NADH/NAD+ may be important. A recent study showed that the most essential function of mitochondrial respiration is to function as an electron acceptor to enable aspartate synthesis in proliferating cells4-6. When mitochondrial respiration is inhibited, pyruvic acid can be utilized as an alternative electron acceptor in a process that pyruvic acid is reduced to lactic acid. Thus, the Warburg effect may facilitate aspartate synthesis to support cell proliferation upon electron acceptor insufficiency. However, in most proliferating cells, including proliferating cancer cells, mitochondria are actively functioning3. Therefore, the Warburg effect may have other functions that favor cell proliferation. Our previous study showed that glucose catabolism can maintain intracellular proton homeostasis to support cell survival7. This indicates that the Warburg effect maintains extracellular proton homeostasis to support cell proliferation.
Pyruvate kinase catalyzes the last step of glycolysis, which is the conversion of phosphoenolpyruvate to pyruvate with ATP generation. Four pyruvate kinase isoforms exist. PKL and PKR are encoded by PKLR and expressed primarily in the liver and exclusively in erythrocytes, respectively8. The other two isoforms, PKM1 and PKM2, are encoded by alternatively spliced mRNAs of PKM and only differ by 22 amino acids9. PKM1 is hyper-expressed in normal tissues, whereas PKM2 expression is also found in some adult tissues, including the lung, liver, and spleen, as well as in all cancers examined to date10-12. PKM1 and PKM2 appear to exert differential effects on the fate of pyruvate. PKM1 is thought to direct pyruvate for mitochondrial oxidation, while PKM2 diverts pyruvate to lactate to support aerobic glycolysis13. Replacement of PKM2 with PKM1 has been shown to suppress aerobic glycolysis and tumor growth13. Therefore, PKM1 and PKM2 may differentially determine the metabolic fate of glucose. However, whether and how PKM affects glutamine metabolism remains unclear.
In this study, we used a PKM-knockout cancer cell model to evaluate the differential functions of PKM1 and PKM2 in maintaining metabolic homeostasis and regulating the metabolism of glucose and glutamine.
Materials and methods
Cell culture
HeLa and 4T1 cells were obtained from ATCC. Stable cell lines were generated by lentivirus infection. HeLa cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS; BioInd, Beit Haemek, Israel) and 50 IU penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere with 5% CO2 at 37°C. Hypoxia conditions were induced by exposure to 1.5% O2.
For cells cultured in the CO2-buffed incubator, the pH of the medium was adjusted using 2–120 mM sodium bicarbonate7.
Cell survival and proliferation assay
Cells were plated in triplicate in 12-well plates at 5 × 104 cells per well in 1 mL medium. After days as indicated in the figures, the wells were washed twice with phosphate-buffered saline (PBS) to remove dead cells, and then the entire contents of the well were trypsinized. The cell number was determined using a hemocytometer. For each well, the fold-change in cell number relative to Day0 was determined directly or on a log2 scale.
Generation of PKM knockout 4T1 cell lines
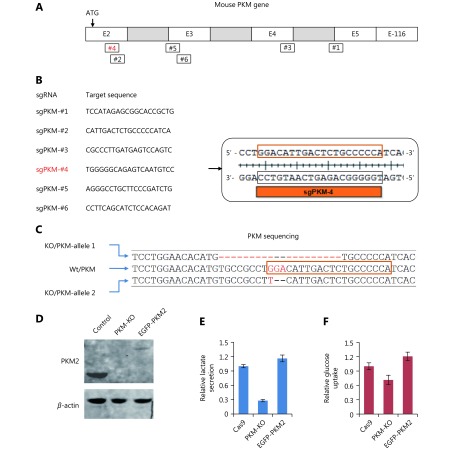
pCDH-Cas9-2A-GFP-BSD was used to express Cas9. Single-guide RNAs (sgRNAs) were cloned into the pLentiGuide-puro-Vector14 which had been linearized with BsmBI. Six target sequences were used for each gene based on the GeCKOv2 Library14. 5′-TCCATAGAGCGGCACCGCTG-3′, 5′-CATTGACTCTGCCCCCATCA-3′, 5′-CGCCCTTGATGAGTCCAGTC-3′, 5′-TGGGGGCAGAGTCAATGTCC-3′, 5′-AGGGCCTGCTTCCCGATCTG-3′ and 5′-CCTTCAGCATCTCCACAGAT-3′ were used for mouse PKM; 5′-ATCACTGCCTTCAGCCCGAG-3′, 5′-CAGCCACGTACCAACATTCA-3′, 5′-GACGAGCTGTCTGGGGATTC-3′, 5′-GGCTGTGCGCATGCAGCACC-3′, 5′-CATCAGGTTTGATGAAATCC-3′ and 5′-CCTGGAGCACATGTGCCGCC-3′ were used for human PKM. For the PKM knockout, the pCDH-Cas9-2A-GFP-BSD and single pLentiGuide-puro-sgPKM plasmids were co-transfected into HeLa and 4T1 cells in 6-well plates using Lipofectamine-3000. Cells were single-cell sorted with a flow cytometer based on green fluorescence into the wells of a 96-well plate containing 200 μL of RPMI supplemented with 10% FBS. Two plates of single cells were collected for each transfection. Cells were grown for three weeks, and the resultant colonies were trypsinized and expanded. Clones were validated for knockout of PKM by Western blot and sequencing.
Gene construction and lentivirus production
The human PKM1 and PKM2 cDNAs were cloned into lentiviral expression vector pCDH-Neo-CMV. Viral packaging was conducted as previously described15. Briefly, the expression plasmids pCDH-CMV-cDNA, pCMV-dR8.91, and pCMV-VSV-G were co-transfected into 293T cells by calcium phosphate coprecipitation at 20:10:10 μg (in a 10-cm dish). The transfection medium containing calcium phosphate and plasmid mixture was replaced with fresh complete medium after incubation for 5 h. Media containing the virus was collected at 48 h after transfection. Cancer cells were infected with medium containing viruses in the presence of polybrene (10 μg/mL) for 48 h, and then the cells were selected with neomycin.
Animal experiments
For animal experiments, 4–5-week-old, 19–20 g, female BALB/c mice were purchased from the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). All animal studies were conducted following an protocol approved by Tianjin Medical University Cancer Institute and Hospital and in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Laboratory Animals. Mice were administered with a standard diet and housed and maintained in pathogen-free cages with a 12-h:12-h light-dark cycle. Temperature and humidity were maintained at 24 ± 2°C and (50 ± 5)%, respectively.
BALB/c mice were subcutaneously injected with 2 × 106 4T1/Cas9 cells, PKM-KO cells, or PKM-KO cells stably expressing hPKM1 and hPKM2 into the left and right groins, respectively. All mice were sacrificed after 26 days and tumors were harvested, weighed, and fixed for further histological and molecular analyses. Statistical analyses were conducted by comparison to the control group with a paired Student’s t test.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections were stained with the following primary antibodies following antigen retrieval: PKM1 (Cell Signaling Technology, Danvers, MA, USA, #7067 at 1:300 dilution), PKM2 (Cell Signaling Technology, #4053 at 1:300 dilution) overnight at 4°C, then incubated with PV6001 (Zhongshan Goldbridge Biotechnology, Beijing, China) for 30 min at 37°C. Staining was performed using a DAB peroxidase substrate kit according to the manufacturer’s protocol (Zhongshan Goldbridge Biotechnology, Beijing, China). Stained sections were counterstained with hematoxylin. Images were acquired under an Olympus BX61 microscope (Tokyo, Japan).
Metabolic profiling and tracing assay
Liquid chromatography (LC)/mass spectrometry (MS) analyses were conducted on a TSQ Quantiva triple quadrupole mass spectrometer networked to a Dionex UltiMate 3,000 UPLC system (Thermo Fisher Scientific, Waltham, MA, USA) at the Metabolomics Facility at Tsinghua University Branch of China National Center for Protein Sciences (Beijing, China). Multiple reaction monitoring mode was conducted using chemical standards. Experiments were performed in medium containing 10% dialyzed FBS (Gibco, Grand Island, NY, USA). Medium contained 100% of either the 13C6-glucoseor 13C5-glutamine pool or unlabeled other pool. DMEM lacking glucose, glutamine, and pyruvate was prepared from powder (Sigma, St. Louis, MO, USA), and then supplemented with labeled-glucose or labeled-glutamine. All reconstituted experimental media contained 10 mM glucose and 1 mM glutamine. Cells were grown in 60-mm dishes until 80% confluence, and then rinsed with PBS and cultured with 2 mL isotope-containing medium for 8 h under the different experimental conditions as the figure showing. Cells were extracted by freeze-thawing three times in 0.5 mL 80% methanol (pre-chilled to –80°C). Macromolecules and debris were removed by centrifugation at 14,000 × g for 20 min at 4–8°C, and the metabolite-containing supernatants were dried under nitrogen gas. Dried samples were stored at –80°C and then resuspended in 50 μL 80% methanol and prepared for LC/MS analyses. Next, 1 μL of each sample was injected onto a Synergi Hydro-RP 100A 2.1 × 100 mm column (Phenomenex, Torrance, CA, USA) for metabolite separation at a column temperature of 35°C. Mobiles phases A and B were 10 mM thiobarbituric acid in aqueous buffer at pH 5 and 100% methanol respectively. The chromatographic gradient using mobile phase B was as follows: 0–3.5 min: 1% B; 3.5–22 min: from 1% to 70% B; 22–23 min: from 70% to 90% B; 23–25 min: 90% B; 25–30 min: 1% B. Data were acquired using a positive/negative switching method. Spray voltages of 3.5 and 2.5 kV were applied for positive and negative mode, respectively. Q1 and Q3 resolution was set to 0.7, and a 1-s cycle time was used. Multiple reaction monitoring data were analyzed using Tracefinder (Thermo Fisher Scientific) to quantify the metabolites for flux analysis. Retention times and mass fragmentation signatures of all metabolites were validated using pure standards. Ion pairs with various isotope labels were derived based on the chemical structures of the precursors and fragments. The abundance of each mass isotopomer was then mathematically corrected to eliminate naturally abundant isotopes and finally converted into a percentage of the total pool.
For metabolite profiling experiments, cells were grown in 60-mm dishes until 80% confluence, and then rinsed with PBS and cultured in 2 mL DMEM containing 10% dialyzed FBS for 8 h. To determine the relative abundance of intracellular metabolites across samples, the cells was extracted with 0.5 mL 80% pre-chilled methanol. Samples were prepared and analyzed as described above. The areas of the ion peaks of interest were corrected by the cell number. Finally, the relative abundance of metabolites was compared between samples.
Glucose uptake and lactate excretion assay
Cells (2 × 105) were plated in 12-well plates and maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (BioInd) and 100 IU penicillin/streptomycin (Invitrogen) in a humidified atmosphere with 5% CO2 at 37°C. After 24 h, the medium was exchanged with 0.5 mL defined DMEM (10 mM glucose, 1 mM glutamine, without pyruvate) containing 10% dialyzed fetal bovine serum and 100 IU/mL penicillin/streptomycin. Cells were incubated for 8 h, after which the culture medium was collected to measure the glucose and lactate concentrations. Lactate levels were measured by the Vitros LAC Slide method on an autoanalyzer (VITROS 5600 Integrated System, Ortho-Clinical Diagnostics, Raritan, NJ, USA). Glucose levels were measured by the autoanalyzer AU5800 (Beckman Coulter, Brea, CA, USA). The increased levels of lactate and reduced levels of glucose in the medium, normalized to the cell number, were determined to reflect lactate production and glucose uptake by the cells.
The conversion efficiency of glucose to lactate (λ) was calculated as follows:
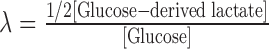 |
[Glucose] is glucose uptake by cells. [Glucose-derived lactate] is the excretory lactate produced from glucose, and was calculated using the following equation:
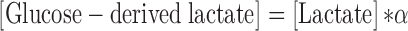 |
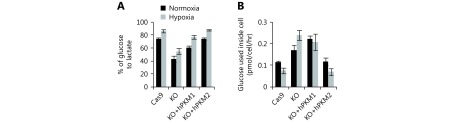
[Lactate] is lactate production by cells, and α is the fraction of glucose-labeled lactate/total lactate and be determined by measuring the enrichment of 13C6-glucose in lactate. As shown in Figure 1F, lactate was predominantly generated from glucose, and the α values for 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells were (95.53 ± 0.40)%, (90.93 ± 3.24)%, (92.50 ± 1.10)%, and (93.50 ± 1.07)%, respectively.
1.
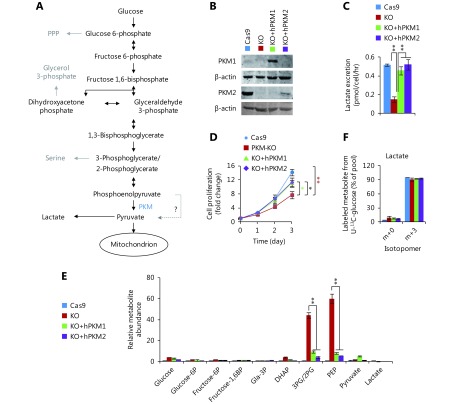
Generation of PKM-knockout cell model. (A) Glycolytic pathway. (B) Western blot of PKM in 4T1 cell lines. 4T1/Cas9 is the control cell line; 4T1/PKM-KO is the PKM-knockout cell line; 4T1/KO+hPKM1 and 4T1/KO+hPKM2 are PKM-knockout cells re-expressing human PKM1 and PKM2. (C) Lactate excretion of 4T1 cell lines. (D) Proliferation rate of 4T1 cell lines. (E) Relative metabolite abundance of 4T1 cell lines. (F) Mass isotopomer analysis of lactate in 4T1 cell lines cultured with 13C6-glucose for 8 h. In all panels, error bars indicate SD (n = 3). *P < 0.05, ** P < 0.01.
Glucose retained for intracellular utilization for functions other than lactate production was calculated as follows:
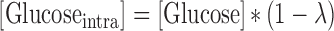 |
Western blot
After different treatments, the cells were washed twice with PBS and lysed in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium vanadate, 1 mg/mL leupeptin, 1 mM phenylmethyl-sulfonylfluoride). Protein concentration was measured by the Bradford method. Equal amounts of protein (50 μg) were loaded onto 10% SDS-PAGE gels and blotted onto a polyvinylidene fluoride membrane. To block the membrane, 5% fat-free milk was added, followed by incubation with primary antibody for 1 h and secondary antibody for 1 h at room temperature. Western blot detection was carried out using a Li-Cor Odyssey image reader (Li-Cor, Lincoln, NE, USA). Goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG secondary antibodies were obtained from Li-Cor. The following antibodies were used in this study: PKM1 (Cell Signaling Technology, #7067at 1:1000 dilution), PKM2 (Cell Signaling Technology, #4053at 1:1000 dilution), and β-actin (Santa Cruz, Cat #sc-1616at 1:5000 dilution) as an internal control.
Statistical analysis
Data are presented as the means ± SD. Statistical analyses were performed using unpaired, two-tailed Student’s t test to compare two groups, and analysis of variance was used for experiments involving more than two groups. Asterisks in the figure indicate statistical significance levels (*, P < 0.05; **, P < 0.01).
Results
Lactate excretion is mediated by both PKM1 and PKM2
To further examine the role of PKM in cancer cells (Figure 1A), we generated a cell line with a PKM deletion. We used the CRISPR/Cas9 system14 to knockout PKM which encodes both pyruvate kinases, PKM1 and PKM2, via differentially spliced mRNA isoforms9. Six sgRNA target sequences were used in human HeLa and mouse 4T1 cells (Supplementary Figure S1A, B). In 96 colonies selected for each sgRNA, PKM expression and lactate production were measured. However, we only obtained one 4T1 colony with no detectable PKM1 and PKM2 proteins and significantly reduced lactate excretion (Figure 1B, C). Sequencing analysis revealed that the reading frame of PKM was shifted (Supplementary Figure S1C). 4T1/PKM-KO cells proliferated much slower than control 4T1/Cas9 cells (Figure 1D). Therefore, PKM knockout appears to be disadvantageous for cell survival. This may explain why it was difficult to obtain PKM-knockout cells. 4T1/PKM-KO cells exhibited dramatic accumulation of phosphoenolpyruvate and 3/2-phosphoglycerate, the precursors of pyruvate (Figure 1E). Re-expression of human PKM1 or PKM2 in 4T1/PKM-KO cells remarkably eliminated the accumulated precursors of pyruvate (Figure 1E) and restored lactate production (Figure 1C) and cell proliferation (Figure 1D). Thus, PKM deletion led to biological changes in 4T1/PKM-KO cells. Unexpectedly, only a very small quantity of PKM2 protein, compared to that in control 4T1/Cas9 cells, was sufficient to maintain lactate production and glucose uptake (Supplementary Figure S1D–F).
S1.
PKM-knockout in 4T1 cells. (A) Schematic for PKM gene. Exon 1 is not shown here. # indicates sgRNA target position in PKM gene. (B) sgRNA target sequences for mouse PKM gene. sgPKM-#4 successfully knockout PKM gene. (C) Sequencing of PKM gene in 4T1/PKM-KO cells targeted by sgPKM-#4 as shown in (B). Two alleles are differentially damaged, one with 20bp deletion, the other containing 2bp loss. (D) Western blot of PKM2 in 4T1/Cas9, 4T1/PKM-KO and 4T1/KO+EGFP-PKM2 cells. 4T1/KO+EGFP-PKM2 cells are 4T1/PKM-KO cells expressing EGFP-PKM2. (E) (F) Relative lactate excretion and relative glucose uptake of 4T1/Cas9, 4T1/PKM-KO and 4T1/KO+EGFP-PKM2.
Interestingly, in addition to glycolytic intermediates, pyruvate and lactate in 4T1/PKM-KO cells were predominantly derived from glucose, as revealed by measuring the enrichment 13C from 13C6-glucose (Figure 1F and Supplementary Figure S2A). This suggests the presence of compensation pathways to overcome PKM deletion. Indeed, it was recently reported that the glycolytic enzyme phosphoglycerate mutase can act as a phosphate acceptor to enable the conversion of phosphoenolpyruvate to pyruvate, bypassing PKM16.
S2.
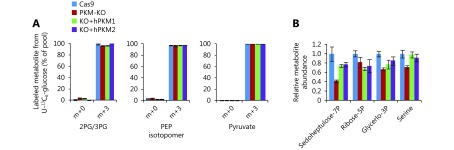
Effect of PKM on glucose-associated metabolites. (A) Mass isotopomer analysis of 3-phosphoglycerate/2-phosphoglycerate, phosphoenolpyruvate and pyruvate in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells cultured with 13C6-glucose for 8h. (B) Relative metabolite abundance in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells.
PKM2 suppresses the entry of glucose into acetyl-coenzyme A (Ac-CoA)
Re-expression of PKM1 and PKM2 in 4T1/PKM-KO cells had similar effects on lactate production and growth in vitro (Figure 1C, D). We further investigated the metabolism of glucose in these cells.
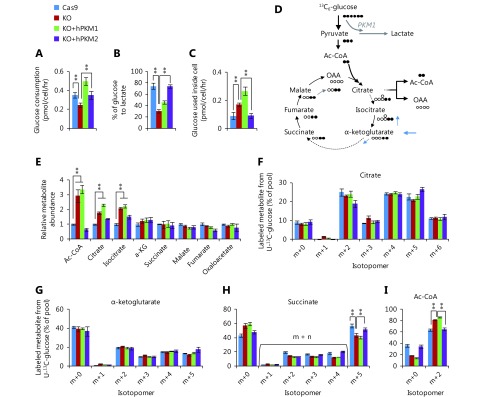
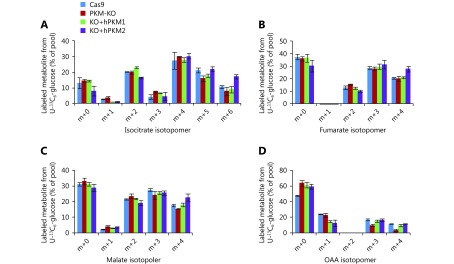
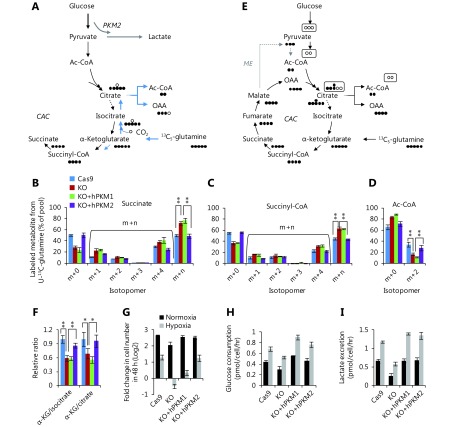
Glucose uptake was reduced in PKM-knockout 4T1 cells, but was reversed in PKM2-re-expressing cells and even enhanced in PKM1-re-expressing cells (Figure 2A). Furthermore, PKM-knockout led to a low conversion efficiency of glucose to lactate, which was completely restored by PKM2-re-expression but only slightly by PKM1-re-expression (Figure 2B). More glucose was retained for cellular processes other than lactate production in PKM2-null cells, including 4T1/KO and 4T1/KO+hPKM1 cells, compared to that in PKM2-expressing cells, such as 4T1/Cas9 and 4T1/KO+hPKM2 cells (Figure 2C). Glucose typically supports the biosynthesis of ribose 5-phosphate through the pentose phosphate pathway, glycerol 3-phosphate, and serine from glycolytic intermediates (Figure 1A). However, the abundance of these metabolites was not increased in PKM2-null cells (Supplementary Figure S2B). Therefore, we measured the cellular abundance of downstream metabolites of pyruvate, including intermediates in the citric acid cycle (CAC) and Ac-CoA (Figure 2D). Consistent with the glucose utilization inside cells (Figure 2C), cellular Ac-CoA, citrate, and isocitrate were found to be significantly increased in PKM2-null cells (Figure 2E). Interestingly, the 13C6-glucose-labeled fraction of metabolites, except for succinate and Ac-CoA, showed no corresponding changes (Figure 2F, G and Supplementary Figure S3A–D). The 13C6-glucose-labeled succinate was significantly decreased, while labeled Ac-CoA was enhanced in PKM2-null cells (Figure 2H, I). These results demonstrate that the metabolic pathway of glucose is diverted to Ac-CoA rather than the CAC in PKM2-null cells, including PKM1-expressing cells, suggesting suppression of PKM2 by glucose-derived Ac-CoA.
2.
PKM2-null promotes metabolic flux of glucose to Ac-CoA. (A) Glucose consumption of 4T1 cell lines, 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells. (B) Conversion efficiency of glucose to lactate in 4T1 cell lines. (C) Glucose kept for intracellular utilization other than lactate production in 4T1 cell lines. (D) Schematic for metabolic flux of glucose to the citric acid cycle (CAC) and Ac-CoA. (E) Relative metabolite abundance of 4T1 cell lines. (F–I) Mass isotopomer analysis of citrate, α-ketoglutarate, succinate, and Ac-CoA in 4T1 cell lines cultured with 13C6-glucose for 8 h. In all panels, error bars indicate SD (n = 3). **P < 0.01.
S3.
Labeled glucose-derived metabolites. (A-D) Mass isotopomer analysis of isocitrate, fumarate, malate and oxaloacetate in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells cultured with 13C6-glucose for 8h.
PKM2 promotes the entry of glutamine into Ac-CoA
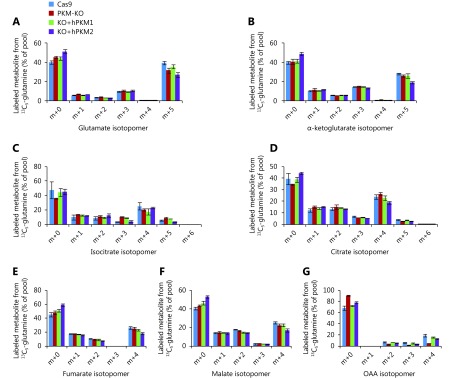
Citrate and α-ketoglutarate are the direct precursors of Ac-CoA and succinate, but showed different 13C6-glucose-labeled patterns, suggesting that other metabolites can be converted to these compounds competitively with glucose. Glutamine is used as the second-line carbon source to glucose in cells and can replenish the CAC (Figure 3A); thus, we measured the enrichment of 13C in metabolites from 13C5-glutamine. We observed that 13C5-glutamine-labeled succinate/succinyl-CoA (m + n) was increased, while labeled Ac-CoA was decreased in PKM2-null (PKM-KO and KO+hPKM1) cells (Figure 3B–D). In contrast, the other labeled metabolites did not show a pattern associated with the status of PKM2 (Supplementary Figure S4A–G). 13C5-Glutamine can label Ac-CoA through the forward CAC and malic enzymes that convert 13C5-glutamine-derived malate to labeled pyruvate, the precursor of Ac-CoA (Figure 3E). If this occurs, 13C5-glutamine-derived citrate m+6 and isocitrate m+6 should be detected; however, we did not detect these compounds (Supplementary Figure S4C, D). Alternatively, 13C5-glutamine can be reductively metabolized to produce labeled Ac-CoA m+217,18 (Figure 3A), concomitantly with the production of isocitrate m+5 and citrate m+5 which were detected (Supplementary Figure S4C, D). These results indicate that glutamine-derived Ac-CoA is repressed in PKM2-null cells and support that PKM2 promoted glutamine-derived Ac-CoA via the reductive pathway.
3.
PKM2-null reduces metabolic flux of glutamine to Ac-CoA and sensitizes cells to hypoxia conditions. (A) Schematic for metabolic flux of glutamine to Ac-CoA through the reductive pathway via the backward CAC. (B–D) Mass isotopomer analysis of succinate, succinyl-CoA, and Ac-CoA in 4T1 cell lines, 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells cultured with 13C5-glutamine for 8 h. (E) Schematic for metabolic flux of glutamine to Ac-CoA through the forward CAC and malic enzyme (ME). (F) Relative ratio of α-ketoglutarate/isocitrate and α-ketoglutarate/citrate in 4T1 cell lines. (G) Proliferation rate of 4T1 cell lines, 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells under normoxia and hypoxia conditions. (H) Glucose uptake of 4T1 cell lines, 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells under normoxia and hypoxia conditions. (I) Lactate production of 4T1 cell lines, 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells under normoxia and hypoxia conditions. In all panels, error bars indicate SD (n = 3). *P < 0.05, ** P < 0.01.
S4.
Labeled glutamine-derived metabolites. (A-G) Mass isotopomer analysis of glutamate, α-ketoglutarate, isocitrate, citrate, fumarate, malate and oxaloacetate in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells cultured with 13C5-glutamine for 8 h.
It was previously reported that reductive glutamine metabolism was determined from the α-ketoglutarate/citrate ratio19. Therefore, we measured the α-ketoglutarate/citrate and α-ketoglutarate/isocitrate ratios in these cells. Our results showed that compared to PKM2-null (PKM-KO and KO+hPKM1) cells, PKM2-expressing (4T1/Cas9 and 4T1/KO+hPKM2) cells had higher ratios (Figure 3F), which may have driven the reductive glutamine utilization.
Glutamine-derived Ac-CoA for fatty acid synthesis through reductive carboxylation supports cell proliferation in hypoxia. Here, we measured the proliferation rate of these cell lines during hypoxia. PKM2-null cells were much more sensitive to hypoxia than PKM2-expressing cells (Figure 3G). Additionally, hypoxia increased glucose consumption, lactate production, and the conversion efficiency of glucose to lactate in all cell lines (Figure 3H, I and Supplementary Figure S5A). However, hypoxia conditions only reduced intracellular glucose utilization in PKM2-expressing cells but not in PKM2-loss cells (Supplementary Figure S5B).
S5.
Effect of hypoxia on glucose metabolism. (A) Conversion efficiency of glucose to lactate in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells under normoxia and hypoxia. (B) Glucose kept for intracellular utilization other than lactate production in 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1 and 4T1/KO+hPKM2 cells under normoxia and hypoxia.
Reverse Warburg effect in stromal cells is induced by PKM loss
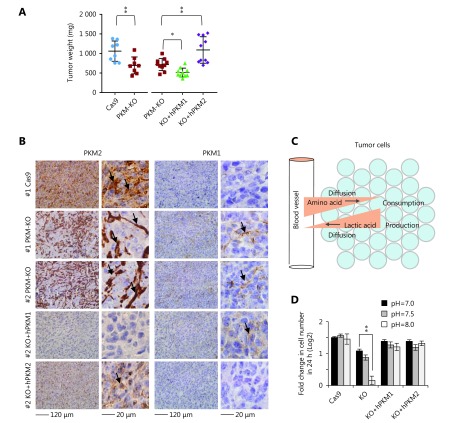
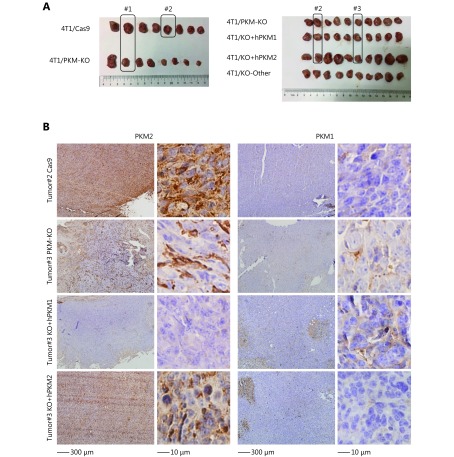
We next inoculated these cells in BALB/c mice to test the effects of PKM on in vivo tumor growth. 4T1/PKM-KO tumors grew significantly slower than control 4T1/Cas9 tumors (Figure 4A and Supplementary Figure S6A, B). Compared to 4T1/PKM-KO tumors, 4T1/KO+hPKM2 tumors expanded faster, while 4T1/KO+hPKM1 tumors grew more slowly (Figure 4A). This suggests that PKM2, rather than PKM1, supports tumor growth in vivo, in agreement with previous observations13. We next detected PKM expression in fixed tumor sections by immunohistochemistry. Consistent with previous reports13,20, most cells in 4T1/Cas9 tumors were positively stained for PKM2 but not for PKM1 (Figure 4B and Supplementary Figure S6B), which supported the Warburg effect. Similarly, 4T1/KO+hPKM2 tumors showed PKM2 staining in tumor cells (Figure 4B and Supplementary Figure S6B). In contrast, 4T1/KO+hPKM1 tumors displayed PKM1 staining in tumor cells, with no obvious PKM2 staining in tumorous or stromal tissues (Figure 4B and Supplementary Figure S6B), suggesting that PKM1 in tumor cells also maintains the Warburg effect in vivo. Interestingly, 4T1/PKM-KO tumors showed strong PKM2 staining in tumor-associated stromal cells but not in tumor cells (Figure 4B and Supplementary Figure S6B), a phenomenon known as the reverse Warburg effect21,22. Notably, weak PKM1 staining was observed in stromal cells of 4T1/PKM-KO tumors (Figure 4B and Supplementary Figure S6B), suggesting that PKM1 also enables the reverse Warburg effect23.
4.
PKM maintains proton homeostasis and is differentially expressed in tumors. (A) Tumor weight of 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells inoculated in BALB/c mice for 26 days.*P < 0.05, ** P < 0.01. (B) Representative IHC staining images of 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 tumors for PKM2 or PKM1. # indicates tumor number, see Supplementary Figure S6A for more details. Micrographs of the same tumors are shown on the left (200 ×) and right (400 ×) Arrows point to positive staining. (C) A model showing proton balancing between amino acid consumption and lactic acid production in tumor tissues. (D) Proliferation rate of 4T1/Cas9, 4T1/PKM-KO, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells cultured with medium with different pH, as indicated, for 24 h. Error bars indicate SD (n = 3). **P < 0.01.
S6.
PKM is differentially expressed in tumors.
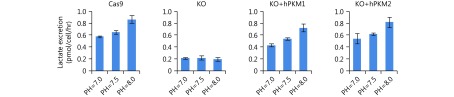
The reverse Warburg effect may provide nutrients for tumor cells21,22 and have an additional role in maintaining the micro-environment. Proliferating cells must absorb large quantities of amino acids for their active anabolism, particularly for the synthesis of proteins, which account for approximately 60% of the cell mass24. Amino acid-associated protons are finally converted into water molecules during protein synthesis, which requires proton-balancing compensation. This is particularly important in vivo because of the limited extracellular space. Such compensation can be optimally provided by lactic acid from aerobic glycolysis through the Warburg effect (Figure 4C). To examine whether lactate production maintains proton homeostasis, alkaline medium was used to deprive the cells of protons. We observed that alkaline medium triggered lactate production in PKM-expressing cells, which is consistent with our previous results7, but not in PKM-KO cells (Supplementary Figure S7). 4T1 cells normally generating lactate, including 4T1/Cas9, 4T1/KO+hPKM1, and 4T1/KO+hPKM2 cells, grew well in alkaline medium, whereas 4T1/PMK-KO cells had nearly lost the ability to proliferate (Figure 4D). These results indicate that robust lactate production protects cells from proton deprivation. When PKM in tumor cells is inactivated, the Warburg effect may be conferred by tumor-associated stromal cells. The “so-called” reverse Warburg effect was recently reported in cancer-associated fibroblasts21,22.
S7.
Effect of PKM on lactate excretion in the alkaline medium.
Discussion
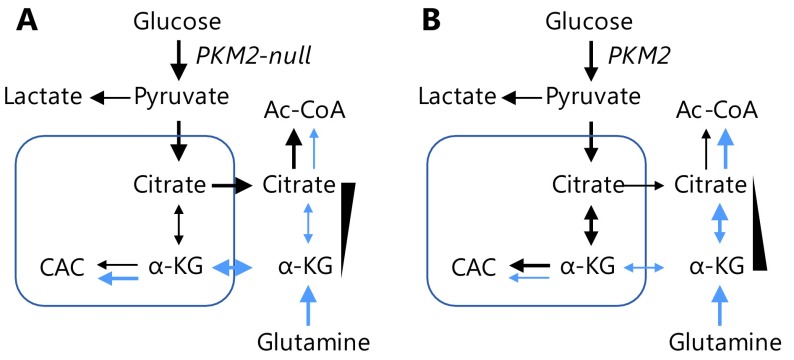
In the current study, we use a PKM-knockout cell model to clearly demonstrate that PKM2 catabolizes glucose to secretory lactate much more efficiently than PKM1. This high efficiency endows PKM2-expressing cells with the ability to reduce intracellular utilization of glucose and production of citrate. This results in an increased α-ketoglutarate/citrate ratio that promotes reductive glutamine metabolism to generate glutamine-derived Ac-CoA, the metabolic foundation for fatty acid synthesis (Figure 5). The reductive catabolism of glutamine to provide a carbon source for lipogensis to support cell proliferation under hypoxia conditions17, commonly exhibited by in vivo tumor cells, has been well-characterized. Therefore, our results provide a mechanistic explanation for the important physiological role of PKM2 in cancer cells, which may underlie the in vivo advantage of PKM2 in tumor growth.
5.
Model showing the effect of PKM2 on metabolic flux of glucose and glutamine to Ac-CoA. (A) In PKM2-null cells, glucose-derived pyruvate is inefficiently catabolized to lactate and thus diverted to citrate biosynthesis, which reduces the α-ketoglutarate/citrate ratio to suppress the metabolic flux of glutamine to Ac-CoA. (B) In the presence of PKM2, cells convert glucose efficiently to lactate and inefficiently to citrate, thus increasing the α-ketoglutarate/citrate ratio to promote the metabolic flux of glutamine to Ac-CoA.
As revealed in the current study, a very low level of PKM2 maintains full cellular functions (Supplementary Figure S1D). This suggests that inhibition of PKM2 is not a preferential strategy for treating cancers25, as it is difficult to completely block PKM2 activity using inhibitors in vivo. Therefore, activating PKM2 to gain PKM1-like activity may be a suitable method for treating cancer26.
Our previous study demonstrated that glucose catabolism can maintain intracellular proton homeostasis to support cell survival7. Proliferating cancer cells exhibit active anabolism, particularly protein synthesis, which consumes large numbers of protons near cells. Through aerobic glycolysis, glucose can be metabolized to lactic acid for excretion from the cell to maintain cellular proton homeostasis. Therefore, it is vital for proliferating cancer cells to efficiently produce lactic acid. Unlike PKM2, PKM1 does not promote in vivo tumor growth but can maintain proton homeostasis. Therefore, when tumoral PKM2 is inactivated, tumor cells may use PKM1 or the reverse Warburg effect provided by tumor-associated stromal cells to maintain the microenvironment to survive.
However, tumor cells often suffer from glucose deficiency and thus lack a sufficient number of protons. To survive under these hash conditions, cancer cells may absorb proteins rather than amino acids. During the synthesis of proteins from amino acids, amino acid-associated protons are converted to water, which requires balancing of proton homeostasis. In contrast, conversion of proteins involves the re-arrangement of amino acids and does not involve proton consumption. One example demonstrating the advantage of proteins as amino acid sources are fertilized bird eggs. The fertilized eggs are closed systems and predominantly consist of proteins rather than amino acids. Upon rapid proliferation, cells in these systems transform nutritional proteins around them to intracellular proteins to support embryonic development. From a nutritional perspective, tumors are also relatively closed systems because of their poor vasculature to support rapid tridimensional cell proliferation. Therefore, extracellular proteins may be an important amino acid source for cancer cell proliferation in vivo, as observed recently27-29.
Collectively, our results demonstrate that PKM2 can efficiently maintain cellular proton homeostasis and promote reductive glutamine metabolism.
Acknowledgements
This work is supported by the funds from National Natural Science Foundation of China (Grant No. 81672762, 81622037 and 81602446).
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Warburg O On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Dayton TL, Jacks T, Vander Heiden MG PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–30. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liberti MV, Locasale JW The warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–8. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 2015;162:552–63. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162:540–51. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio . Science. 2016;352:231–5. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui YF, Wang YY, Liu M, Qiu L, Xing P, Wang X, et al Determination of glucose deficiency-induced cell death by mitochondrial ATP generation-driven proton homeostasis. J Mol Cell Biol. 2017;9:395–408. doi: 10.1093/jmcb/mjx011. [DOI] [PubMed] [Google Scholar]
- 8.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262:14366–71. [PubMed] [Google Scholar]
- 9.Noguchi T, Inoue H, Tanaka T The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing . J Biol Chem. 1986;261:13807–12. [PubMed] [Google Scholar]
- 10.Chaneton B, Gottlieb E Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37:309–16. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Wong N, Ojo D, Yan J, Tang D PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–91. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 12.Mazurek S Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–80. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 14.Sanjana NE, Shalem O, Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li BH, Gordon GM, Du CH, Xu JH, Du W Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell. 2010;17:469–80. doi: 10.1016/j.ccr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang JJ, Hiller K, et al Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, et al Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nature Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 22.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe R, Witkiewicz AK, et al Mitochondrial metabolism in cancer metastasis: visualizing tumor cell mitochondria and the "reverse Warburg effect" in positive lymph node tissue. Cell Cycle. 2012;11:1445–54. doi: 10.4161/cc.19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiavarina B, Whitaker-Menezes D, Martinez-Outschoorn UE, Witkiewicz AK, Birbe R, Howell A, et al Pyruvate kinase expression (PKM1 and PKM2) in cancer-associated fibroblasts drives stromal nutrient production and tumor growth. Cancer Biol Ther. 2011;12:1101–13. doi: 10.4161/cbt.12.12.18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, et al Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. 2016;36:540–9. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortés-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, et al M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci USA. 2013;110:489–94. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anastasiou D, Yu YM, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, et al Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23:235–41. doi: 10.1038/nm.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu WY, Grabocka E, et al Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]