Abstract
Despite advances in gene editing technologies, generation of tissue-specific knockout mice is time consuming. We used CRISPR/Cas9-mediated genome editing to disrupt genes in livers of adult mice in just a few months, which we refer to as somatic liver knockouts. In this system, Fah−/− mice are given hydrodynamic tail vein injections of plasmids carrying CRISPR/Cas9 designed to excise exons in Hpd; the Hpd-edited hepatocytes have a survival advantage in these mice. Plasmids that target Hpd and a separate gene of interest can therefore be used to rapidly generate mice with liver-specific deletion of nearly any gene product. We used this system to create mice with liver-specific knockout of argininosuccinate lyase, which develop hyperammonemia—observed in humans with mutations in this gene. We also created mice with liver-specific knockout of ATP binding cassette subfamily B member 11, which encodes the bile salt export pump. We found that these mice have a biochemical phenotype similar to that of Abcb11−/− mice. We then used this system to knock out expression of 5 different enzymes involved in drug metabolism within the same mouse. This approach might be used to develop new models of liver diseases and study liver functions of genes that are required during development.
While advances in gene editing technologies, particularly CRISPR/Cas9, allow for the generation of knockout mice faster than ever, this process still involves months of crossings and genotyping with no guarantee that the resulting mouse will be viable. Conditional knockouts using the Cre/loxP system can alleviate certain pitfalls, but this process is even more time-consuming, necessitating a more extensive breeding strategy and crossing of multiple mouse lines1, 2. To circumvent these issues, we have developed a method to rapidly knockout any gene from the livers of adult mice, utilizing selection pressure in the murine liver.
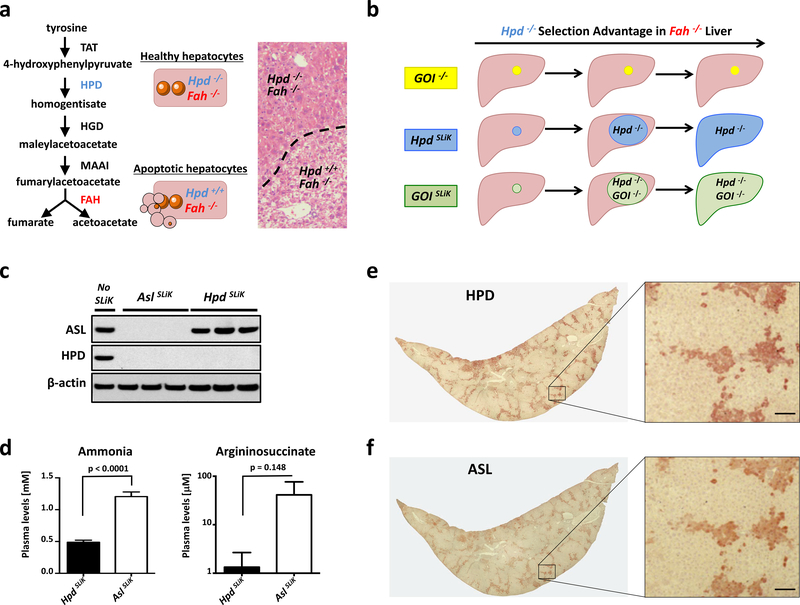
Hepatocytes in mice lacking the fumarylacetoacetate hydrolase (Fah) gene undergo apoptosis due to the accumulation of toxic tyrosine catabolites3–5. Fah−/− mice die as neonates due to hepatotoxicity, but can be rescued using the small molecule drug nitisinone, which inhibits the protein hydroxyphenylpyruvate dioxygenase (HPD) in order to prevent toxicity 6, 7. We previously demonstrated that using CRISPR-Cas9 to excise critical exons of the Hpd gene confers resistance to hepatotoxicity in Fah−/− mice, resulting in a selection advantage for Hpd-edited cells8 (Fig. 1a). We found the edited hepatocytes almost completely replaced the murine liver in as little as two months, and were maintained twelve months after genome editing with no obvious signs of liver injury (Supplementary Fig. 1-3). We reasoned that by simultaneously targeting Hpd and a separate gene of interest (GOI) in Fah−/− mice, we could use the selection advantage conferred by Hpd editing to rapidly generate a liver-specific knockout for nearly any desired GOI, a technique we refer to as Somatic Liver Knockout (SLiK) (Fig. 1b).
Figure 1.
Somatic Liver Knockout (SLiK) by multiplex CRISPR-Cas9 editing. (a) The toxic effect of Fah deficiency can be counteracted by deleting Hpd. (b) Schematic for SLiK showing the expansion of GOI- edited cells using the growth advantage of Hpd−/− hepatocytes in Fah−/− mice. (c) Western blot for HPD and ASL ten weeks after targeted editing in HpdSLiK (Hpd-deleted) or AslSLiK (Hpd- and Asl-deleted) mice. (d) Serum analysis of ammonia and argininosuccinate for HpdSLiK and AslSLiK mice (n=5). (e,f) Serial sections of AslSLiK liver lobes stained for either HPD (e) or ASL (f) in brown. Inset boxes are shown in higher magnification. Error bars indicate SEM. Scale bars are 50 μm.
To introduce CRISPR-Cas9 plasmids into the liver, SLiK uses hydrodynamic tail vein injection, a technique capable of transfecting up to 30% of hepatocytes without the need for viral or other complex delivery mechanisms9, 10. A growth advantage is conferred to hepatocytes in Fah−/− mice using two gRNAs targeted to introns flanking critical exons in the Hpd gene (Supplementary Fig. 4). Simultaneously, two gRNAs targeting early exons of the desired GOI are used to introduce exonic mutations. It is likely that every cell that gains a growth advantage from Hpd excision is also deficient in the GOI, since each GOI gRNA can cause a frameshift mutation, whereas both Hpd gRNAs must cut simultaneously and excise the intervening exons in order to gain the selective advantage of Hpd deletion8, 11. We performed proof-of-concept studies for SLiK using two different genes.
Argininosuccinate Lyase (ASL) is a critical protein in the urea cycle, and is produced mainly in the liver, and to a lesser amount in the kidneys. ASL deficiency causes increased ammonia concentration in the blood, leading to lethargy, seizures, and eventually death12. We generated AslSLiK mice by hydrodynamic injection of the CRISPR-Cas9 plasmids targeting both Hpd and Asl. In accord with the severity of the human disease, AslSLiK mice survived ~10 weeks after application of selection pressure, at which point they exhibited hyperammonemia and had to be euthanized with clinical symptoms (somnolence and abnormal behavior). This phenotype parallels that of the Asl knockout mouse (Asl−/−), which is neonatally lethal13. Western blot analysis confirmed liver protein depletion for both HPD and ASL in these mice (Fig. 1c), and biochemical analysis of plasma showed higher levels of ammonia and argininosuccinate, the substrate of ASL, in AslSLiK mice compared to HpdSLiK control mice (Fig. 1d). Argininosuccinate was also increased in the urine of AslSLiK mice (Supplementary Fig. 5). In contrast to the Asl−/− mouse, SLiK relies on the clonal expansion of edited cells, which have a growth advantage and replace non-edited apoptotic cells. We therefore provided residual ASL function by rescuing non-edited hepatocytes with nitisinone, thus generating mice with a nearly complete knockout for Asl while mitigating lethality (Fig. 1e,f). However, all HPD-negative cells were also negative for ASL, which suggests that, given sufficient time and a non-lethal phenotype, a complete liver knockout might be achievable.
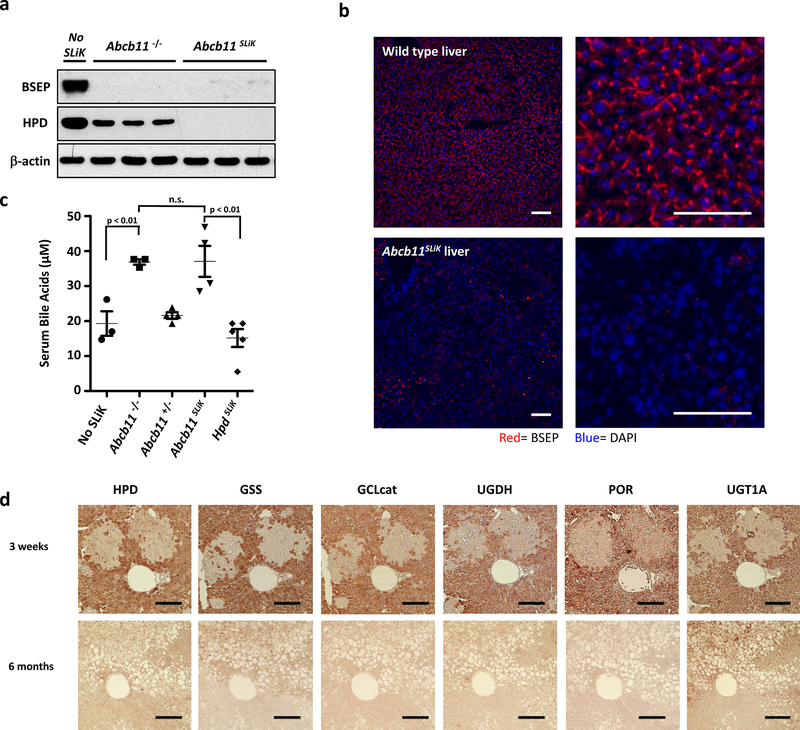
Progressive familial intrahepatic cholestasis-2 (PFIC2) is caused by deficiency of the Bile Salt Export Pump (BSEP), encoded by the Abcb11 gene 14. Abcb11−/− mice have been generated by traditional means, however, offspring of these mice often die due to maternal cholestatis15. As such, generating sufficient numbers of mice to study BSEP knockout is difficult. To bypass this hurdle, we used SLiK to eliminate BSEP from the livers of adult mice. Abcb11SLiK mice appear healthy, and both western blot and immunohistochemistry confirmed depletion of BSEP from the liver 8 weeks after injection with CRISPR-Cas9 constructs (Fig. 2a-b). Biochemically, serum bile acid concentrations in Abcb11SLiK mice achieved levels similar to those in conventionally generated Abcb11−/− mice (Fig. 2c). From these data, we can conclude that a full liver knockout is possible using SLiK, and the biochemical phenotype is comparable to the traditional knockout mouse model.
Figure 2.
SLiK of Abcb11 and multiplexed SLiK (a) Western blot of BSEP in Abcb11−/− knockout mice and Abcb11SLiK mice. (b) Immunofluorescent staining for BSEP (red) in wild-type (upper panels) and Abcb11SLiK (lower panels), counterstaining DAPI (blue). (c) Total serum bile acids in Abcb11−/− and Abcb11SLiK mice. (d) Multiplex SLiK of glutathione synthetase (GSS), glutamate cysteine ligase catalytic subunit (GCLcat), UDP-glucose-6’-dehydrogenase (UGDH), cytochrome P450 oxidoreductase (POR), and UDP glucuronosyltransferase family 1 member A (UGT1A), shown by immunostaining serial liver sections (brown) at 3 weeks and 6 months after nitisinone withdrawal (n=6 for each time point). Error bars represent SEM. Scale bars are 50 μm.
Traditional knockout models are impractical for multiple genes, however many hepatocellular functions depend on protein networks and redundant pathways. To demonstrate how SLiK can also be used to study such complex systems, we were able to simultaneously target five different enzymes implicated in drug metabolism within the same mouse (Fig. 2d). Immunohistochemistry confirmed that areas of expanding Hpd−/− hepatocytes were also negative for all five targeted proteins, and 6 month analysis showed not only extensive deletion of all genes, but also replicated the heterogeneous phenotype previously observed with hepatic Por deletion16.
In summary, SLiK is a novel technique to rapidly generate liver-specific knockout models without the need for embryonic gene targeting or prolonged breeding schemes. Furthermore, this technique does not require the expertise and laborious production of viral vectors. The degree of liver knockout can be controlled with a small molecule drug, and the method is amenable to multiplexing knockout of several genes simultaneously. It is important to note, however, that poor gRNA binding can influence GOI knockout efficiency, and that knockouts of genes required for cell survival or proliferation cannot be generated using SLiK. Despite these caveats, SLiK is a highly versatile method and should be valuable to biomedical research.
Supplementary Material
ACKNOWELDGMENTS
We thank C. Gillespie for critical comments on the manuscript. K.D.B. is supported by the National Heart Lung and Blood Institute (NHLBI) grant R01HL134510 and National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) grant R56DK115461. K.D.B. and D.D.M are supported by the Texas Hepatocellular Carcinoma Consortium (THCCC) (CPRIT #RP150587) and the Diana Helis Henry and Adrienne Helis Malvin Medical Research Foundations. W.R.L. and K.D.B are supported by the National Heart Lung and Blood Institute (NHLBI) grant HL132840. F.P.P. and B.B.C. were supported by T32HL092332, The DLDCC is supported by P30CA125123. CMM core facility of Texas Medical Center Digestive Disease Center (P30-DK56338).
Footnotes
COFLICT OF INTERESTS
The authors declare no financial, professional or personal conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A 1992;89:6861–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakso M, Sauer B, Mosinger B, Jr., et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci U S A 1992;89:6232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruppert S, Kelsey G, Schedl A, et al. Deficiency of an enzyme of tyrosine metabolism underlies altered gene expression in newborn liver of lethal albino mice. Genes Dev 1992;6:1430–43. [DOI] [PubMed] [Google Scholar]
- 4.Grompe M, al-Dhalimy M, Finegold M, et al. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev 1993;7:2298–307. [DOI] [PubMed] [Google Scholar]
- 5.Klebig ML, Russell LB, Rinchik EM. Murine fumarylacetoacetate hydrolase (Fah) gene is disrupted by a neonatally lethal albino deletion that defines the hepatocyte-specific developmental regulation 1 (hsdr-1) locus. Proc Natl Acad Sci U S A 1992;89:1363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grompe M, Lindstedt S, al-Dhalimy M, et al. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet 1995;10:453–60. [DOI] [PubMed] [Google Scholar]
- 7.Lindstedt S, Holme E, Lock EA, et al. Treatment of hereditary tyrosinaemia type I by inhibition of 4-hydroxyphenylpyruvate dioxygenase. Lancet 1992;340:813–7. [DOI] [PubMed] [Google Scholar]
- 8.Pankowicz FP, Barzi M, Legras X, et al. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun 2016;7:12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 1999;10:1735–7. [DOI] [PubMed] [Google Scholar]
- 10.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther 1999;6:1258–66. [DOI] [PubMed] [Google Scholar]
- 11.Canver MC, Bauer DE, Dass A, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem 2014;289:21312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagamani SC, Erez A, Lee B. Argininosuccinate lyase deficiency. Genet Med 2012;14:501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid Sutton V, Pan Y, Davis EC, et al. A mouse model of argininosuccinic aciduria: biochemical characterization. Mol Genet Metab 2003;78:11–6. [DOI] [PubMed] [Google Scholar]
- 14.Strautnieks SS, Bull LN, Knisely AS, et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet 1998;20:233–8. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Li F, Wang Y, et al. Maternal bile acid transporter deficiency promotes neonatal demise. Nat Commun 2015;6:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barzi M, Pankowicz FP, Zorman B, et al. A novel humanized mouse lacking murine P450 oxidoreductase for studying human drug metabolism. Nat Commun 2017;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.