ABSTRACT
Salmonellosis outbreaks associated with sprouted legumes have been a food safety concern for over two decades. Despite evidence that Salmonella enterica triggers biotic plant defense pathways, it has remained unclear how plant defenses impact Salmonella growth on sprouted legumes. We used Medicago truncatula mutants in which the gene for the flagellin receptor FLS2 was disrupted to demonstrate that plant defenses triggered by FLS2 elicitation do not impact the growth of Salmonella enterica serovar Typhimurium ATCC 14028S. As a control, we tested the growth of Salmonella enterica serovar Typhimurium LT2, which has a defect in rpoS that increases its sensitivity to reactive oxygen species. LT2 displayed enhanced growth on M. truncatula FLS2 mutants in comparison to wild-type M. truncatula. We hypothesize that these growth differences are primarily due to differences in 14028S and LT2 reactive oxygen species sensitivity. Results from this study show that FLS2-mediated plant defenses are ineffective in inhibiting growth of Salmonella entrica 14028S.
Keywords: Salmonella, flagellin, FLS2, plant–microbe interactions
This study demonstrates that FLS2-mediated plant defenses are ineffective in preventing growth of Salmonella enterica strain 14028S.
INTRODUCTION
Salmonella enterica contamination of sprouted legume seeds, such as alfalfa, has been a food safety concern for decades (Mahon et al.1997; Dechet et al.2014). Seed stocks are often implicated as the source of contamination (Mahon et al.1997) and the favorable growth conditions provided by the sprout production environment lead to a rapid increase in Salmonella numbers (Howard and Hutcheson 2003). Salmonella serovars display multiple logs of growth on sprouting legume seeds and in the surrounding sprouted seed exudate, which contains a rich mixture of nutrients, including sugars and amino acids (Howard and Hutcheson 2003; Jayaraman et al.2014; Kwan et al.2015). Inhibition of attachment factor production, such as O-antigens, aggregative fimbria or cellulose, reduces Salmonella attachment and subsequent growth on sprouting legumes indicating that Salmonella closely associates with sprouting legumes (Barak et al.2005, 2007). Furthermore, Salmonella can establish persistent endophytic populations in sprouted legume seedlings with inocula containing as few as two colony forming units (Jayaraman et al.2014).
The presence of Salmonella triggers transcriptional changes in the plant, including the activation of plant defense genes. In Arabidopsis thaliana, inoculation with Salmonella activates defense pathways (Schikora et al.2008). Salmonella inoculation of the model legume Medicago truncatula also induces significant transcriptional changes, including the up-regulation of multiple defense genes (Jayaraman et al.2014). The interaction between Salmonella and plants has been further characterized on a mechanistic level through investigation of the role that Salmonella flagellin plays in triggering plant responses to Salmonella. An epitope of bacterial flagellin (referred to as flg22) is a Microbe Associated Molecular Pattern (MAMP) that is recognized by a variety of plant species through the LRR receptor-like kinase FLS2. (Felix et al.1999; Gómez-Gómez and Boller 2000). Recognition of this eliciting epitope stimulates the Pathogen Triggered Immunity response in plants (Jones and Dangl 2006). Stimulation of Pathogen Triggered Immunity through the FLS2 receptor in Arabidopsis triggers many plant defense responses, including the production of reactive oxygen species (ROS) (Apel and Hirt 2004), deposition of callose (Luna et al.2011) and closure of stomata (Zeng and He 2010). Work by Meng and colleagues demonstrated that synthetic flg22 peptide representing the FLS2 eliciting portion of Salmonella flagellin elicits an immune response, including the production of ROS, in tomato and Nicotiana benthamiana (Meng, Altier and Martin 2013). Immune response stimulation by Salmonella flg22 has also been demonstrated in Arabidopsis (Garcia et al.2014); however, it has not been shown if recognition of flagellin through the FLS2 receptor impacts Salmonella growth in sprouted legume seeds.
While multiple studies have demonstrated that Salmonella activates plant defense pathways, the high levels of Salmonella growth in the sprouted legume seed environment suggest that this immune response may not significantly impact Salmonella proliferation. Here, we used M. truncatula mutants, in which the gene for the FLS2 receptor has been disrupted by transposon insertion, to elucidate the impact that the exposure of M. truncatula to Salmonella flagellin has on Salmonella growth on sprouted legume seedlings. By manipulating the presence of the plant FLS2 receptor, we were able to investigate this interaction without adding the confounding variable of bacterial motility. Defects in bacterial motility reduce plant colonization by growth-promoting bacteria (de Weger et al.1987; Broek, Lambrecht and Vanderleyden 1998; Gao et al.2016; Rossi et al.2016). Motility enhances the invasiveness of the plant pathogen Pseudomonas syringae (Haefele and Lindow 1987). Conflicting results of the role of flagellar motility in Salmonella plant colonization have been reported. In A. thaliana, Medicago sativa and lettuce, Salmonella colonization or internalization is reduced in motility mutants (Cooley, Miller and Mandrell 2003; Kroupitski et al.2009; Cowles et al.2016); however, a study done in M. truncatula found enhanced colonization by motility mutants (Iniguez et al.2005). Due to the demonstrated importance of bacterial motility in plant colonization and the conflicting findings regarding the role of motility in Salmonella plant colonization, the role of FLS2 triggered plant defenses using M. truncatula fls2 mutants instead of Salmonella flagella mutants were investigated.
MATERIALS AND METHODS
Bacterial serovars and inoculum preparation
Strains used in this work were Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028S and Salmonella enterica subsp. enterica serovar Typhimurium LT2. Strains were stored in nutrient broth (BD Biosciences, Sparks, MD) with 15% (vol/vol) glycerol at −70°C. Working stocks were maintained on Luria-Bertani (LB) agar (BD Biosciences) at 4°C.
Single colonies were stab inoculated into motility agar and cultured at 37°C for use in plant colonization assays. Plant inoculum was prepared by dissolving four 5 mm punches of agar from a motility agar plate into 50 mL sterile water.
Plant material
M. truncatula seeds were acid scarified using sulfuric acid, surface sterilized for 2 minutes using commercial bleach, and germinated on 1% agar plates supplemented with 1 μg/ml gibberellic acid. Seeds were vernalized at 4°C for 1–3 days before germination. Colonization assays and formazan precipitation assays were performed with 1-day-old seedlings. M. truncatula R108 seeds were used as wild type.
M. truncatula FLS2 was identified based on homology to A. thaliana FLS2. M. truncatula fls2 mutants were identified by PCR-based reverse genetics screening (Cheng et al.2014) a population of M. truncatula transposon (Tnt1) insertion lines (Tadege et al.2008). Mutants were self-pollinated, and the F2 generation was screened by PCR for homozygous mutants. Tnt1 insertion site was mapped by PCR and confirmed by Sanger sequencing using a primer embedded in the Tnt1 and a primer in the flanking chromosome region (Fig S1, Supporting Information).
Sprouted seedling Salmonella growth assay
One-day-old M. truncatula seedlings were placed in 1.5 ml of inoculum prepared from Salmonella grown in motility agar. The Salmonella suspension was used to inoculate four to six replicates containing four seedlings each of either R108, fls2–1 or fls2–2. The Salmonella inoculum was prepared fresh for each trial and contained an average of 106 colony forming units (CFU). Replicate colonization was normalized to the average colonization of R108 for each respective inoculum. Inoculated seedlings were kept at 25°C without light for 18 hours. These conditions were chosen to emulate the conditions in which commercial legume sprouts are produced. Following incubation, the seedlings were collected, weighed and homogenized in 1 × PBS, pH 7.2, with 20% glycerol. Serial dilution in 1 × PBS, pH 7.2 with 20% glycerol and enumeration of CFU were performed to determine colonization levels. Colonization levels were normalized to seedling weight. In colonization assays using flg22, the peptide was used at a concentration of 10 μM.
Hydrogen peroxide survival assay
14028S and LT2 were grown in sprouting seed exudates for 18 hours at 25°C. Cultures were diluted in sterile water to a concentration of approximately 107 CFU/ml and challenged with 30 mM hydrogen peroxide at 25°C without shaking. CFU enumeration was performed before the addition of hydrogen peroxide and at 30 and 60 minutes after the addition of hydrogen peroxide. Dilution and plating were conducted as described above.
Nitroblue tetrazolium (NBT) detection of reactive oxygen species
M. truncatula production of ROS was detected using a modified NBT staining protocol (Grellet Bournonville and Díaz-Ricci 2011). Seedlings were suspended in 50 mM potassium phosphate buffer pH 7.4 with 2 mM NBT +/− 10 μM flg22 peptide. Seedlings were incubated in the dark at 25°C for 18 hours. Following incubation, the supernatant containing precipitated formazan was collected, and absorbance was read at 600 nm.
Flagella stain
Flagella were visualized using Flagella stain (Hardy Diagnostics, Santa Maria, CA). 14028S and LT2 were grown overnight in motility agar. Plugs of agar were removed from these plates and suspended in LB for 15 minutes. Wet mounts were made by gently transferring cells to glass slides and applying coverslips. Flagella stain was applied to the cells through capillary action by placing the stain on the edge of the coverslip. Cells were visualized using an Olympus BHS microscope (Center Valley, PA) at 1000× magnification.
Statistical methods
Colonization numbers in each trial were normalized to the wild-type control and are reported as fold change relative to the wild-type control. Outliers were identified using Grubb's test. Student's T-test for independent groups was used to evaluate statistical significance (P < 0.05). The standard error of the mean is shown in all figures.
DISCUSSION AND RESULTS
Validation of M. truncatula fls2 lines
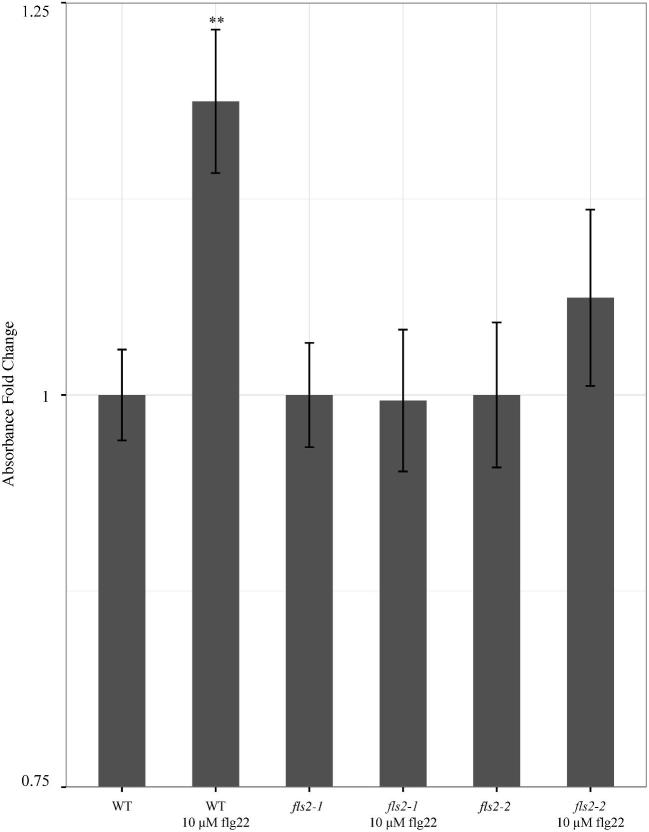
Stimulation of the FLS2 receptor with bacterial flagellin leads to production of ROS (Apel and Hirt 2004). Following standard genotyping (Fig S1, Supporting Information), we validated our M. truncatula fls2 lines using a NBT precipitation assay to assess ROS production (Grellet Bournonville and Díaz-Ricci 2011). Wild-type M. truncatula plants showed an increase in precipitated formazan when elicited with flg22, indicating an increase in the production of ROS. The two independent Tnt1 insertion lines, M. truncatula fls2–1 and fls2–2, did not exhibit an increase in precipitated formazan when exposed to flg22, indicating little to no ROS production (Fig 1). This confirms that FLS2-dependent ROS production is inactivated in both M. truncatula fls2 mutant lines.
Figure 1.
The absorbance of precipitated formazan following elicitation with 10 μM flg22 for 24 hours was used to determine production of ROS in wild-type M. truncatula R108, fls2–1 mutant and fls2–2 mutant. For each genotype, fold change in absorbance was calculated between plants in control solution (2 mM NBT) and plants in eliciting solution (2 mM NBT with 10 μM flg22). **Indicates P value < 0.01 between control and flg22 treatment. N ≥ 15 for all genotypes and treatments.
Salmonella triggers a FLS2 dependent immune response, but Salmonella 14028S is not susceptible to this response
Inoculation with Salmonella stimulates immune responses in both A. thaliana and M. truncatula (Schikora et al.2008; Jayaraman et al.2014), and treatment with Salmonella flg22 stimulates immune responses in tomato, N. benthamiana and A. thaliana (Meng, Altier and Martin 2013; Garcia et al.2014). It is unclear what impact this activation of plant defense pathways has on Salmonella on sprouted legumes. Our data suggest that growth of Salmonella 14028S on sprouted legumes is not influenced by defenses triggered by bacterial flagellin through the plant FLS2 receptor.
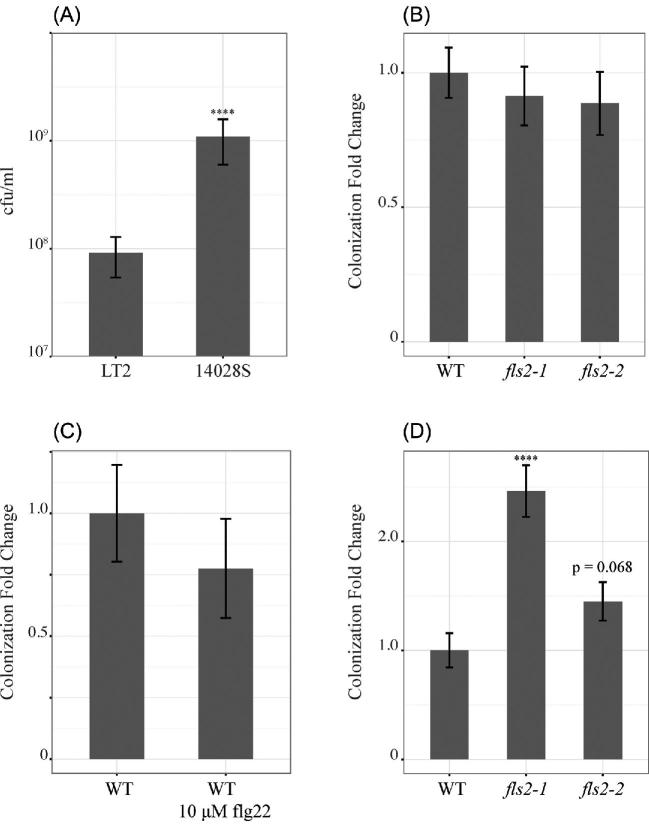
Salmonella 14028S numbers on both M. truncatula fls2 lines were equivalent to those on wild-type M. truncatula (Fig 2B). To ensure that this was not a result of 14028S failing to stimulate the FLS2 receptor, we treated wild-type M. truncatula with 10 μM flg22 peptide in conjunction with 14028S inoculation. This concomitant elicitation and inoculation did not result in a change in 14028S final numbers (Fig 2C). However, treatment of A. thaliana with flg22 results in significantly reduced growth of P.s syringae pv. tomato DC3000 (Zipfel et al.2004), demonstrating that exogenous elicitation of the FLS2 receptor can impact bacteria that are sensitive to FLS2 triggered defense reactions. In contrast to the sprouted legume seedlings used in this work, growth of syringe inoculated Salmonella was reduced on N. benthamiana leaves that were pretreated by syringe infiltration of flg22 (Meng, Altier and Martin 2013).
Figure 2.
Growth of Salmonella enterica 14028S or LT2 on M. truncatula R108, fls2–1 mutant and fls2–2 mutant was determined by colony enumeration. (A)Salmonella enterica 14028S growth compared to LT2 growth on wild-type M. truncatula R108. Growth shown in colony forming units per gram of fresh plant weight; N ≥ 10. (B)Salmonella enterica 14028S growth on M. truncatula fls2–1 or M. truncatula fls2–2 as compared to growth on wild-type M. truncatula; N = 15. (C) Growth of Salmonella enterica 14028S on M. truncatula R108 treated with 10 μM flg22 as compared to growth on untreated plants; N = 10. (D) Growth of Salmonella enterica LT2 on M. truncatula fls2–1 and fls2–2 as compared to growth on wild-type M. truncatula; N ≥ 15. ****Indicates P value < 0.0001.
As an additional control, both to validate our fls2 lines and to ensure that Salmonella is recognized by M. truncatula through the FLS2 receptor, we conducted in planta growth experiments with Salmonella enterica LT2. LT2 is more than 98% identical in sequence to 14028S, differing mainly in prophage regions, but is avirulent due to a disruption in rpoS, which encodes for the general stress response transcriptional regulator σs (Swords, Cannon and Benjamin 1997; Jarvik et al.2010). Since σs promotes transcription of oxidative stress response genes, in addition to other stress response genes, LT2 was used as a control strain for FLS2-triggered plant defenses. The LT2 strain had greater growth on M. truncatula fls2 lines (P < 0.0001 on fls2–1 and p = 0.068 on fls2–2) than on the wild-type plant, demonstrating that it is sensitive to FLS2-triggered defenses (Fig 2D). This result further validated disruption of FLS2 in the M. truncatula fls2 lines and shows that Salmonella triggers FLS2-dependent defense responses.
Flagella staining of both LT2 and 14028S cells confirmed the presence of flagellated cells (Fig S2, Supporting Information). However, the peptide sequence of certain bacterial flagellin, including the flagellin of the plant pathogen Agrobacterium tumefaciens and the plant symbiont Sinorhizobium meliloti, do not elicit a response from the FLS2 receptor (Felix et al.1999). Salmonella produces two distinct antigenic forms of flagella by switching between the expression of flagella genes fliC and fljB (Aldridge et al.2006). Analysis of both flagella variants (fliC and fljB) in 14028S and LT2 showed homology within the FLS2 eliciting domain (Fig S3, Supporting Information), confirming that 14028S is not avoiding elicitation of FLS2 through antigenic variation of flagella (McClelland et al.2001; Jarvik et al.2010).
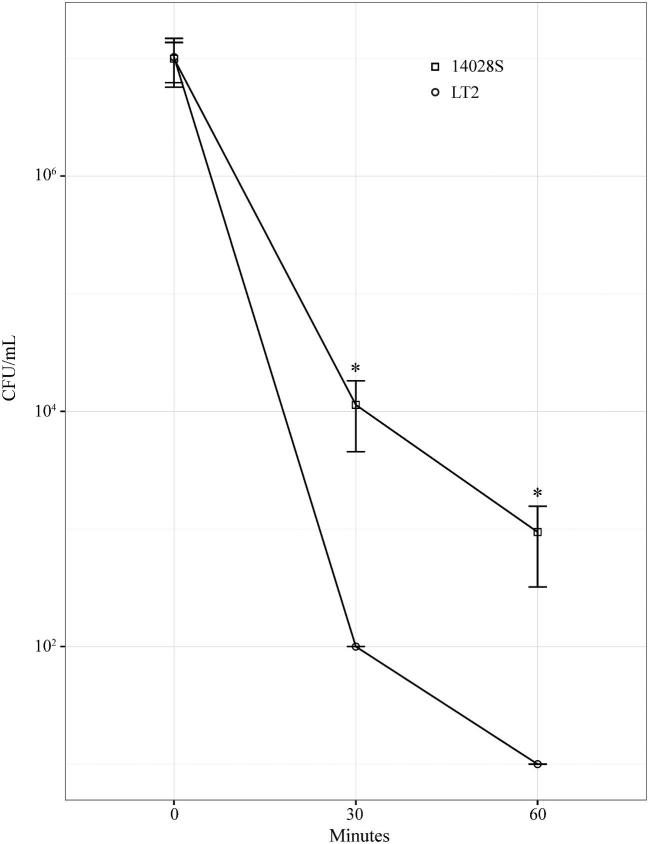
We hypothesize that the susceptibility to FLS2-triggered defenses displayed by LT2 is due to reduced tolerance for ROS (Fig 3). Furthermore, we suggest that the highly efficient and redundant mechanisms that Salmonella possesses for neutralizing ROS are one factor that allows it to proliferate to high levels in sprouts, despite triggering the plant immune response (Hébrard et al.2009). In addition to the production of ROS (Apel and Hirt 2004), FLS2 stimulation triggers the deposition of callose (Luna et al.2011) and stomata closure (Zeng and He 2010), although these defenses are unlikely to have a significant impact on Salmonella in sprouted seedlings. Callose deposition is a strategy for fortifying internal plant tissue through the production of high molecular weight polymers that can contain antimicrobial compounds (Luna et al.2011). While Salmonella does grow endophytically, it displays higher growth epiphytically and does not produce the enzymes required to degrade plant cell walls (Cooley, Miller and Mandrell 2003; Teplitski, Barak and Schneider 2009; Jayaraman et al.2014). Callose deposition would be largely ineffective against epiphytic Salmonella growth, and Salmonella does not display the degradative endophytic growth that callose deposition would protect against. The impact of stomata closure would also be minimal, as Salmonella displays preferential growth in the rhizosphere (Cooley, Miller and Mandrell 2003). For these reasons, it is probable that the production of ROS is the primary FLS2 triggered defense response that is relevant to Salmonella growth on plants.
Figure 3.
Salmonella enterica 14028S and LT2 grown in sprouted seedling exudates for 18 hours were challenged with 30 mM hydrogen peroxide. Survival was determined by culture sampling and colony enumeration. LT2 measured below the limit of detection at 30 and 60 minutes, and was scored at the limit of detection. *Indicates P value < 0.05. LT2 N = 3, 14 028 N = 6.
As an intracellular human pathogen capable of replicating within macrophage where it will encounter oxidative stress, Salmonella is well equipped to neutralize the ROS produced through elicitation of the FLS2 receptor. The Salmonella genome encodes for three catalases (KatE, KatG and KatN), two alkyl hydroperoxide reductases (AhpC and TsaA) and two superoxide dismutases (SodC1 and SodC2) which allow Salmonella to proliferate even when challenged with FLS2-triggered ROS (Ammendola et al.2005; Hébrard et al.2009). Furthermore, it is possible that the oxidative stress encountered on sprouted seedlings could impact virulence of Salmonella. While Salmonella 14028S grown on Arabidopsis did not display an increase in mammalian cell invasiveness as compared to Salmonella 14028S grown in rich media (Schikora et al.2011), FLS2-triggered ROS production may select for cells with greater ROS tolerance in heterogeneous Salmonella populations.
The human pathogen Salmonella enterica shows rapid growth in the sprouted legume environment and has been responsible for outbreaks of salmonellosis involving the consumption of contaminated sprouts. While many groups have demonstrated that plants perceive Salmonella and activate defense pathways in response to it, these defenses do not influence the in planta growth of Salmonella 14028S. Defenses such as callose deposition and stomata closure are probably ineffective because they target bacteria with growth strategies and lifestyles that differ from Salmonella, while the defense offered by the production of ROS is ineffective against Salmonella due to ROS-inactivating enzymes.
Supplementary Material
Acknowledgements
We would like to thank Dhileepkumar Jayaraman, Matthew Crook, Shane Bernard and Josh Lensmire for thoughtful discussion and advice, and Jared Godfrey for technical assistance.
FUNDING
This work was funded by USDA Hatch Grant WIS01694. Generation of M. truncatula Tnt1 mutants was funded from NSF Plant Genome Grants (DBI-0703285 and IOS-1127155).
Conflict of interest. None declared.
REFERENCES
- Aldridge PD, Wu C, Gnerer J et al.. Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica. Proc Natl Acad Sci 2006;103:11340–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammendola S, Ajello M, Pasquali P et al.. Differential contribution of sodC1 and sodC2 to intracellular survival and pathogenicity of Salmonella enterica serovar Choleraesuis. Microbes Infect 2005;7:698–707. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 2004;55:373–99. [DOI] [PubMed] [Google Scholar]
- Barak JD, Jahn CE, Gibson DL et al.. The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol Plant Microbe Interact 2007;20:1083–91. [DOI] [PubMed] [Google Scholar]
- Barak JD, Lisa G, Naraghi-Arani P et al.. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol 2005;71:5685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek A, Lambrecht M, Vanderleyden J. Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasilense. Microbiology 1998;144:2599–606. [DOI] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK et al.. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol 2014;201:1065–76. [DOI] [PubMed] [Google Scholar]
- Cooley MB, Miller WG, Mandrell RE. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl Environ Microbiol 2003;69:4915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles KN, Willis DK, Engel TN et al.. Diguanylate cyclases AdrA and STM1987 regulate Salmonella enterica exopolysaccharide production during plant colonization in an environment-dependent manner. Appl Environ Microbiol 2016;82:1237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weger LA, van Der Vlugt CIM, Wijfjes AHM et al.. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol 1987;169:2769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechet AM, Herman KM, Chen Parker C et al.. Outbreaks caused by sprouts, United States, 1998-2010: lessons learned and solutions needed. Foodborne Pathog Dis 2014;11:635–44. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S et al.. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 1999;18:265–76. [DOI] [PubMed] [Google Scholar]
- Gao S, Wu H, Yu X et al.. Swarming motility plays the major role in migration during tomato root colonization by Bacillus subtilis SWR01. Biol Control 2016;98:11–17. [Google Scholar]
- Garcia AV, Charrier A, Schikora A et al.. Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol Plant 2014;7:657–74. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2. Mol Cell 2000;5:1003–11. [DOI] [PubMed] [Google Scholar]
- Grellet Bournonville CF, Díaz-Ricci JC. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem Anal 2011;22:268–71. [DOI] [PubMed] [Google Scholar]
- Haefele DM, Lindow SE. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol 1987;53:2528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébrard M, Viala JPM, Méresse S et al.. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol 2009;191:4605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MB, Hutcheson SW. Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl Environ Microbiol 2003;69:548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez AL, Dong Y, Carter HD et al.. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact 2005;18:169–78. [DOI] [PubMed] [Google Scholar]
- Jarvik T, Smillie C, Groisman EA et al.. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 2010;192:560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Valdés-López O, Kaspar CW et al.. Response of Medicago truncatula seedlings to colonization by Salmonella enterica and Escherichia coli O157:H7. PLoS One 2014;9:e87970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature 2006;444:323–9. [DOI] [PubMed] [Google Scholar]
- Kroupitski Y, Golberg D, Belausov E et al.. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl Environ Microbiol 2009;75:6076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan G, Pisithkul T, Amador-Noguez D et al.. De novo amino acid biosynthesis contributes to Salmonella enterica growth in alfalfa seedling exudates. Appl Environ Microbiol 2015;81:861–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J et al.. Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 2011;24:183–93. [DOI] [PubMed] [Google Scholar]
- Mahon BE, Pönkä A, Hall WN et al.. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seeds. J Infect Dis 1997;175:876–82. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J et al.. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001;413:852–6. [DOI] [PubMed] [Google Scholar]
- Meng F, Altier C, Martin GB. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ Microbiol 2013;15:2418–30. [DOI] [PubMed] [Google Scholar]
- Rossi FA, Medeot DB, Liaudat JP et al.. In Azospirillum brasilense, mutations in flmA or flmB genes affect polar flagellum assembly, surface polysaccharides, and attachment to maize roots. Microbiol Res 2016;190:55–62. [DOI] [PubMed] [Google Scholar]
- Schikora A, Carreri A, Charpentier E et al.. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One 2008;3:e2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Virlogeux-Payant I, Bueso E et al.. Conservation of Salmonella infection mechanisms in plants and animals. PLoS One 2011;6:e24112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords WE, Cannon BM, Benjamin WH. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun 1997;65:2451–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J et al.. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 2008;54:335–47. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Barak JD, Schneider KR. Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr Opin Biotechnol 2009;20:166–71. [DOI] [PubMed] [Google Scholar]
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 2010;153:1188–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L et al.. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004;428:764–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.