Abstract
Background:
Autism spectrum disorders (ASD) involve neurodevelopmental dysregulations that lead to visible symptoms at early stages of life. Many ASD-related mechanisms suggested by animal studies are supported by demonstrated improvement in autistic-like phenotypes in adult animals following experimental reversal of dysregulated mechanisms. However, whether such mechanisms also act at earlier stages to cause autistic-like phenotypes is unclear.
Methods:
We used Shank2−/− mice carrying a mutation identified in human ASD (exons 6 and 7 deletion) and combined electrophysiological and behavioral analyses to see if early pathophysiology at pup stages is different from late pathophysiology at juvenile and adult stages, and correcting early pathophysiology can normalize late pathophysiology and abnormal behaviors in juvenile and adult mice.
Results:
Early correction of a dysregulated mechanism in young mice prevents manifestation of autistic-like social behaviors in adult mice. Shank2−/− mice, known to display N-methyl-D-aspartate receptor (NMDAR) hypofunction and autistic-like behaviors at post-weaning stages after postnatal day 21 (P21), show the opposite synaptic phenotype—NMDAR hyperfunction—at an earlier pre-weaning stage (~P14). Moreover, this NMDAR hyperfunction at P14 rapidly shifts to NMDAR hypofunction after weaning (~P24). Chronic suppression of the early NMDAR hyperfunction by the NMDAR antagonist memantine (P7-21) prevents NMDAR hypofunction and autistic-like social behaviors from manifesting at later stages (~P28 and P56).
Conclusions:
Early NMDAR hyperfunction leads to late NMDAR hypofunction and autistic-like social behaviors in Shank2−/− mice, and early correction of NMDAR dysfunction has the long-lasting effect of preventing autistic-like social behaviors from developing at later stages.
Keywords: autism, treatment, synapse, Shank2, NMDA receptor, memantine
Introduction
Autism spectrum disorders (ASD), characterized by social deficits and repetitive behaviors, exhibit a strong genetic influence. While the number of candidate ASD-risk genes is rapidly increasing (1), our understanding of the underlying mechanisms has lagged, partly because of the intrinsic heterogeneity and complexity of causal mechanisms in different brain regions at different developmental times.
Many ASD-related mechanisms suggested by animal studies are supported by demonstrated improvement in autistic-like phenotypes in adult animals following experimental reversal of dysregulated mechanisms (2-11). However, whether such mechanisms also act at earlier stages to cause autistic-like phenotypes is unclear (12,13).
Shank is a family of postsynaptic scaffolding proteins known to regulate excitatory synapse development and function (14, 15) and involved in various psychiatric disorders, including ASD and Phelan McDermid Syndrome (4, 7, 16, 17). Shank2, the second member of the family also known as ProSAP1, has also been implicated in ASD and other disorders, including intellectual disability, developmental delay, and schizophrenia (18-31).
More recently, studies on mice lacking Shank2 in the whole brain or specific cell types have further enhanced our understanding of the normal functions of Shank2 as well as ASD-related pathophysiology (32-45). For example, mice lacking exons 6-7 and exon 7of Shank2 display autistic-like behavioral abnormalities that are associated with altered N-methyl-D-aspartate receptor (NMDAR) function (37, 38).
More specifically, Shank2−/− mice with exons 6-7 deletion show decreased NMDAR-mediated synaptic currents and NMDAR-dependent synaptic plasticity (38). Importantly, the treatment of adult Shank2−/− mice with D-cycloserine (NMDAR agonist) rapidly normalizes social interaction, suggesting that suppressed NMDAR function may underlie the social deficits. However, it remains unclear whether the NMDAR hypofunction observed in adult Shank2−/− mice (exons 6-7) also acts at early stages to play important roles in the development of social deficits.
In the present study, we found an increase in NMDAR function at an earlier stage (~ postnatal day or P14), which sharply contrasts with the decreased NMDAR function at ~P21. Importantly, suppression of the early NMDAR hyperfunction by the treatment of Shank2−/− mice with memantine (NMDAR antagonist) during P7-21 normalized NMDAR function and social interaction at juvenile (~P28) as well as adult (~P56) stages. These results suggest that early NMDAR hyperfunction induces NMDAR hypofunction and social deficits at later stages, and that early correction of NMDAR function has long- lasting effects.
Methods and materials
Animals
Shank2−/− mice lacking the exons 6-7 of the Shank2 gene have been previously reported (38). Shank2−/− mice lacking exon 7 (37) and exon 24 (33) have also been previously reported. All mutant and their WT littermates of Shank2−/− mice lacking exons 6-7 were generated on and backcrossed to a C57BL/6N background for more than 20 generations. Shank2−/− mice lacking exons 6-7 under a different background (C57BL/6J) were generated by backcrossing the original mice in the genetic background of C57BL/6N with C57BL/6J for more than six generations. Shank2−/− mice lacking exon 7 have been previously published (37), and backcrossed to and maintained in a C57BL/6J background for more than 5 generations.
Results
Distinct transcriptomic profiles in Shank2−/− brains at P14 and P25
Mice lacking Shank2/ProSAP1 (Shank2−/− mice; exons 6-7; global deletion) display autistic-like social deficits that are rapidly improved by acute D-cycloserine (NMDAR agonist) treatment at juvenile and adult stages (38). However, excitatory synaptogenesis in mice is most active during the first three postnatal weeks (46), and expression levels of Shank2 protein in the whole brain peak at ~P14 (Figure 1A), although Shank2 mRNA levels did not show a similar pattern (Figure 1B).
Figure 1. Distinct transcriptomic profiles in Shank2−/− brains at P14 and P25.
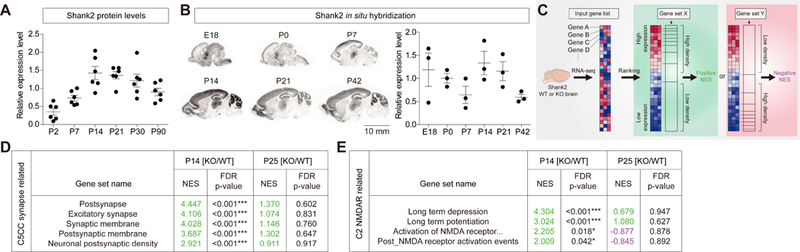
(A) Levels of Shank2 proteins in whole mouse brains increase during postnatal (P) days, reaching a plateau at ~P14. For quantification, Shank2 signals at each developmental stages were normalized to those of α-tubulin and also to the average values of each age groups. (n = 6 male mice for each stage). (B) Levels of Shank2 mRNAs in the mouse brain during development, revealed by in situ hybridization of sagittal sections. The limited correlation between Shank2 protein and mRNA levels might be attributable to that the transcripts are differentially translated or that the in situ images do not cover the whole brain. For quantification, Shank2 signals normalized to the brain area at each developmental stages were normalized to the average values of each age groups. (n = 3 male mice for each stage). Scale bar, 10 mm. (C) A schematic of gene set enrichment analysis (GSEA; software.broadinstitute.org/gsea/). The direction (positive/negative) and amplitude of enrichments reflect the extent to which the genes in a target gene set falls at the top (positive) or bottom (negative) of the ranked input gene list. (D) GSEA using KO/WT-P14 and KO/WT-P25 gene lists from Shank2−/− mice and target gene sets in the C5-CC category (gene ontology_cellular component). NES, normalized enrichment score accounting for the degree of overrepresentation at the top or bottom of a ranked list; FDR, false discovery rate. Positive and negative NES values are indicated in green and violet, respectively. (n = 6 mice for WT and KO at P14 and P25, ***FDR < 0.001; see Table S4 for details). (E) GSEA using KO/WT-P14 and KO/WT-P25 gene lists from Shank2−/− mice and NMDAR-related gene sets in the C2-all category. ‘Activation of NMDA receptor...’ indicates ‘Activation of NMDA receptor upon glutamate binding and postsynaptic event’. (n = 6 mice for WT and KO at P14 and P25, *FDR < 0.05, ***FDR < 0.001; see Table S3 for details).
We thus sought to identify phenotypes that are most prominent at P14 through comparative RNA-Seq transcriptomic analysis of wild-type (WT) and Shank2−/− brains at P21, a stage around weaning where active synaptogenesis is largely completed (Table S1). We found 16 differentially expressed genes (DEGs; FDR < 0.05, fold change > 2.0) in naïve six pairs of WT versus Shank2−/− mice at P14, and 35 DEGs at P25 (Table S2). However, because the small number of the identified DEGs did not yield significant gene ontology terms or pathways, we next performed gene set enrichment analysis (GSEA; software.broadinstitute.org/gsea/) (Figure 1C), which aims to achieve unbiased results by testing the enrichment of a list of ranked, whole genes, rather than a subset above an arbitrary cutoff (fold-change or significance), in pre-curated gene sets (47, 48).
The two lists of genes from Shank2−/− versus WT (KO/WT) mice at P14 and P25, ranked by differential expression, were distinctly enriched for a large number of target gene sets in the C2 category (curated gene sets) (Table S3). Importantly, the top 10 enriched target gene sets for P14 and P25 were largely distinct; and in the case of the four overlapping gene sets associated with ribosome/translational terms, the directions of enrichments were opposite (Figure S1).
Additional GSEA using target gene sets in the C5 category (gene ontology) revealed that the KO/WT-P14, but not KO/WT-P25, gene list was enriched for gene sets associated with excitatory postsynaptic compartments, including excitatory synapse, postsynaptic membrane, and postsynaptic density (Figure 1D; Figure S2; Table S4).
Because Shank2−/− mice show autistic-like behaviors accompanied by reduced NMDAR function (38), we examined NMDAR-related enrichments. The KO/WT-P14 but not KO/WT-P25 gene list was enriched for gene sets associated with NMDAR activation and NMDAR-dependent synaptic plasticity (long-term potentiation and long-term depression) (Figure 1E; Figure S2). Collectively, these results suggest that Shank2 deletion in mice leads to distinct transcriptomic changes at P14 and P25.
Reversal of NMDAR function in Shank2−/− mice during P14 and P25
Based on these results, we next measured excitatory synaptic transmission mediated by NMDARs and AMPARs (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors) in the hippocampus, a brain region implicated in ASD (49-51), in Shank2−/− mice at P14 and P25. We found an increase in the ratio of NMDAR/AMPAR-mediated synaptic transmission at Schaffer collateral-CA1 pyramidal (SC-CA1) synapses at ~P14, as compared with that at WT synapses (Figure 2A; all electrophysiology data summarized in Table S5).
Figure 2. Reversal of NMDAR function in the Shank2−/− hippocampus during P14 and P25.
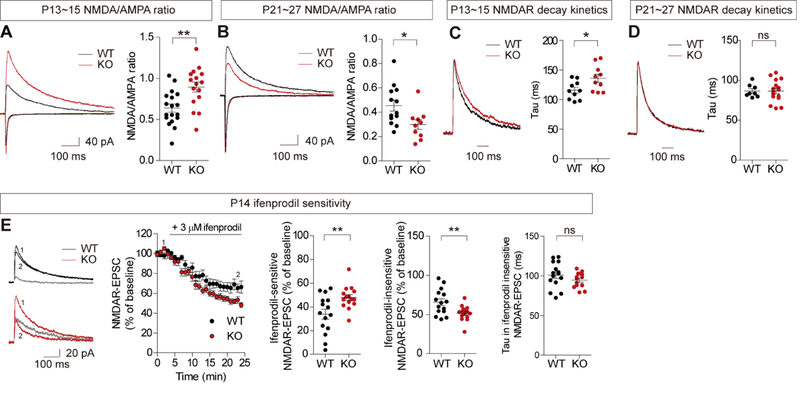
(A and B) NMDAR function is enhanced at Shank2−/− SC-CA1 synapses at ~P13-15, but reduced at ~P21-27, as shown by the NMDA/AMPA ratio. (n = 18 neurons from 11 mice for WT and 16 (11) for KO (Shank2−/− at P13-15, and 13 (8) for WT and 10 (6) for KO at P21-27, *P < 0.05, **P < 0.01, Student t-test; see Table S5 for the summary of electrophysiology results, and Table S6 for statistical details). (C and D) Reduced decay of NMDAR currents at Shank2−/− SC-CA1 synapses at ~P13-15, but not at ~P21-27. (n = 10 (4) for WT and 10 (4) for KO at P13-15, and 8 (6) for WT and 16 (9) for KO at P21-27, *P < 0.05, ns, not significant, Student t-test). (E) NMDAR currents at P14 Shank2−/− SC-CA1 synapses are more sensitive to ifenprodil (GluN2B antagonist) relative to those at WT synapses, which, in combination with the data in (A), predicts a ~2.07-fold and ~1.06-fold increases in ifenprodil-sensitive GluN2B and ifenprodil-insensitive GluN2A components, respectively. Numbers 1 and 2 in WT and KO traces indicate NMDAR currents before and after ifenprodil treatment, respectively, and grey lines in WT and KO traces indicate ifenprodil-sensitive (or GluN2B-dependent) component. Note that the decay constant of the residual NMDAR- EPSCs lacking the GluN2B component is comparable between genotypes. (n = 15 (9) for WT and 15 (7) for KO, **P < 0.01, ns, not significant, Student t-test).
In contrast, basal synaptic transmission, mainly mediated by AMPARs, and paired-pulse ratio (a measure of presynaptic release probability) were normal at Shank2−/− SC-CA1 synapses (Figure S3). In addition, there were no changes in miniature excitatory postsynaptic currents (mEPSCs), mediated by AMPARs, and miniature inhibitory postsynaptic currents (mIPSCs) in Shank2−/− CA1 pyramidal neurons (Figure S4). These results suggest that NMDAR-, but not AMPAR-, mediated synaptic transmission is increased at P14 Shank2−/− SC-CA1 synapses.
At ~P24 (P21-27), however, the NMDA/AMPA ratio at Shank2−/− SC-CA1 synapses was decreased, as compared with that at WT synapses (Figure 2B), consistent with the results from the previous study (38). This study also demonstrated normal basal transmission at Shank2−/− SC-CA1 synapses and normal mEPSCs in Shank2−/− CA1 pyramidal neurons. These results suggest that NMDAR-, but not AMPAR-, mediated synaptic transmission is reduced at P25 Shank2−/− SC-CA1 synapses.
This age-dependent shift in NMDAR function was similar in males and females (Figure S5). In addition, the increased NMDAR function at P14 Shank2−/− synapses involved mainly the GluN2B subunit of NMDARs, as supported by the slower decay kinetics of the Shank2−/− NMDAR currents at P14, but not at ~P24 (Figure 2C-D), and the higher sensitivity of the currents to the GluN2B-specific inhibitor ifenprodil, as compared with those in WT mice (Figure 2E).
In addition to the hippocampus, layer II/III pyramidal neurons in the medial prefrontal cortex (mPFC), another ASD-related brain region (52), showed a similar switch of NMDAR function: an increased NMDA/AMPA ratio at Shank2−/− synapses at P14, but a decreased ratio at ~P24 (Figure S6).
When mEPSCs were measured in these mPFC neurons, the amplitude of mEPSCs in P14 Shank2−/− neurons was normal (Figure S7A), suggesting, together with the increased NMDA/AMPA ratio, that NMDAR-mediated transmission is selectively increased. The mEPSC frequency, however, was decreased in these Shank2−/− mPFC neurons, which, together with the normal paired pulse ratio (Figure S7B), suggests that excitatory synapse number is decreased in P14 Shank2−/− mPFC neurons.
These results collectively suggest that Shank2 deletion induces a rapid temporal switch of NMDAR function from hyper- to hypofunction across the period of P14 to P24 in the hippocampus and mPFC.
To explore the mechanisms underlying the distinct NMDAR function at P14 and P25 Shank2−/− synapses, we examined total and phosphorylation levels of NMDAR subunits in Shank2−/− brains. Phosphorylation, but not total, levels of NMDAR subunits (GluN2B-Ser1303 and GluN1-Ser896), which have been shown to regulate surface and synaptic trafficking of GluN2B (53), were increased at P14 but decreased at P21 (Figure S8A-B).
Other glutamate receptor (GluA and mGluR) subunits were largely normal in total levels in both P14 and P21 Shank2−/− brains. In addition, signaling molecules were also largely normal, with moderate changes in the phosphorylation of ERK1, CREB, and p38 (Figure S8C-D).
Early memantine normalizes NMDAR function in juvenile and adult Shank2−/− mice
We next tested whether the early NMDAR hyperfunction at P14 is causally associated with NMDAR hypofunction at later stages by chronically suppressing NMDAR hyperfunction at ~P14 (P7-21) and examining NMDAR phenotypes in juvenile (~P28) and adult (~P56) Shank2−/− mice.
We first tested whether early memantine treatment rescues NMDAR function in adult Shank2−/− mice. Young Shank2−/− mice orally administered the NMDAR antagonist memantine (20 mg/kg, twice daily) for 2 weeks before weaning (P7-21) showed normalized NMDA/AMPA ratio at Shank2−/− hippocampal SC-CA1 synapses at P25-31 (Figure 3A), without affecting mEPSCs (Figure S9).
Figure 3. Early memantine treatment normalizes NMDAR function in the juvenile and adult Shank2−/− hippocampus.
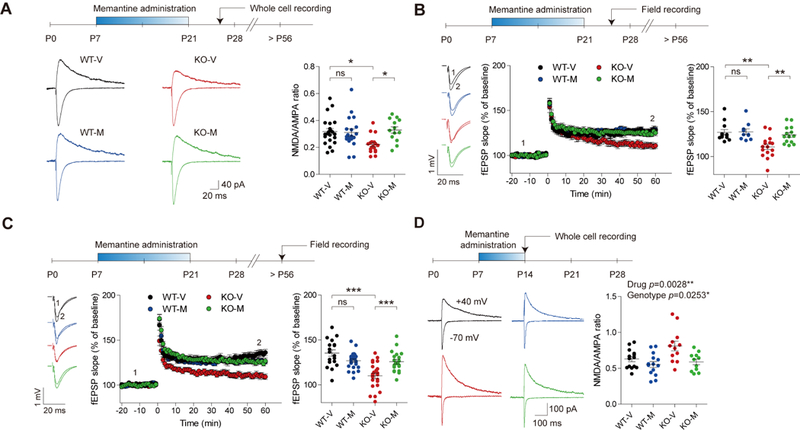
(A) Early memantine treatment (P7-21) normalizes the NMDA/AMPA ratio at juvenile (~P25-31) Shank2−/− SC-CA1 synapses. Young Shank2−/− mice were orally administered memantine (20 mg/kg) twice daily for 2 weeks during P7-21 followed by measurements of NMDA/AMPA ratio at ~P25-31. (n = 22 neurons from 9 mice for WT- V, 19 (6) for WT-M, 18 (9) for KO-V, and 13 (3) for KO-M, *P < 0.05, ns, not significant, two-way ANOVA with Holm-Sidak test). (B and C) Early memantine treatment (P7-21) normalizes LTP induced by high- frequency stimulation at juvenile (~P25-31; B) and adult (> P56; C) Shank2−/− SC-CA1 synapses. Numbers 1 and 2 indicate fEPSP slopes during the baseline and last 5-min recordings, respectively. (Juvenile, n = 12 slices from 6 mice for WT-V, 10 (5) for WT-M, 16 (8) for KO-V, and 14 (8) for KO-M; adult, n = 18 (9) for WT-V, 25 (12) for WT-M, 23 (9) for KO-V, and 20 (7) for KO-M, **P < 0.01, ***P < 0.001, ns, not significant, two-way ANOVA with Holm-Sidak test). (D) Early memantine treatment for a shorter period (P7-14; 20 mg/kg oral/day) normalizes the NMDA/AMPA ratio at ~P15 at Shank2−/− SC-CA1 synapses. (n = 14 neurons from 8 mice for WT-V, 14 (4) for WT-M, 12 (7) for KO-V, and 11 (6) for KO-M, *P < 0.05, **P < 0.05, two-way ANOVA).
In addition, early memantine rescued NMDAR-dependent long-term potentiation (LTP) induced by high frequency stimulation at Shank2−/− SC-CA1 synapses, both immediately after memantine treatment (P25-31) and at an adult stage (>P56) (Figure 3B-C), with no difference between males and females (Figure S10). In contrast, early memantine treatment had no effect on LTP at WT SC-CA1 synapses at either stage.
Early memantine treatment for a shorter period (P7-14) could rescue the NMDA/AMPA ratio even at P14, without affecting mEPSCs (Figure 3D; Figure S11), indicative of a strong and immediate effect of early memantine treatment on the normalization of NMDAR function. These results collectively suggest that early memantine treatment induces immediate and long-lasting normalization of NMDAR function.
Early memantine improves social interaction in juvenile and adult Shank2−/−mice
We next tested whether early memantine treatment can rescue abnormal behaviors in Shank2−/− mice at later (juvenile and adult) stages. Early memantine treatment (20 mg/kg, twice daily; P7-21) significantly improved social interaction of juvenile (P28-35) Shank2−/− mice in the direct interaction test (Figure 4A-B). Social isolation or weaning of mice before direct social-interaction tests had no effect on the results (Figure S12). In contrast, early memantine treatment did not affect social interaction in WT mice (Figure 4B). Memantine also improved social interaction of adult (~P56) Shank2−/− mice in the three-chamber test (54) (Figure 4C-E; Figure S13), indicative of a long-lasting effect.
Figure 4. Early memantine treatment improves social interaction in juvenile and adult Shank2−/− mice.
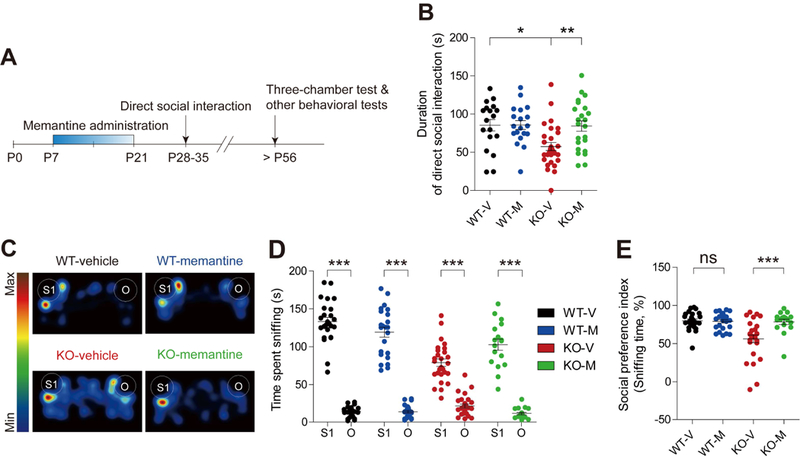
(A) Schematic depiction of early memantine treatment. Young Shank2−/− mice were orally administered memantine (20 mg/kg) twice daily for 2 weeks from P7 to P21, followed by measurements of social interactions at juvenile (P28-35) and adult (P56) stages. (B) Early memantine treatment improves social interaction of juvenile Shank2−/− mice (P28-35), as indicated by time spent in social interaction in the direct interaction test. (n = 18 mice (WT-V/vehicle), 19 (WT-M/memantine), 29 (KO-V), and 23 (KO-M), *P < 0.05, **P < 0.01, two-way ANOVA with Bonferroni test). (C-E) Early memantine treatment improves social interaction of adult Shank2−/− mice (> P56), as indicated by time spent sniffing S1/O (stranger mouse/object; D) and the social preference index based on sniffing time (the numerical difference between the time spent sniffing S1 and O divided by their sum x 100) (E) in the three-chamber test. (n =25 mice (WT-V), 24 (WT-M), 27 (KO-V), and 17 (KO-M), ***P < 0.001, Student’s t-test (D-WT/V and D-WT/M), Wilcoxon matched-pairs signed rank test (D-KO/V and D-KO/M), and Mann Whitney U test (E; two-way ANOVA was not performed because the social preference index is a normalized value)).
When we tested olfactory function and social hierarchy, known to significantly affect social interaction in mice (55-60), olfactory function was similar between genotypes, as reported previously (38), whereas Shank2−/− mice were submissive to WT mice in both tube and urine tests (Figure S14A-C), suggesting that Shank2 deletion affects social hierarchy in mice. This submissiveness, however, did not correlate with social interaction at the individual mouse level (Figure S14D-G). In addition, this submissiveness was not improved by early memantine treatment (Figure S14B-C).
Early memantine treatment modestly improved hyperactivity in adult Shank2−/− mice in the open-field test (Figure S15A-C). However, increasing habituation to a new environment eliminated the drug effect (Figure S15 D-G). In addition, the hyperactivity of Shank2−/− mice in a familiar environment was not normalized by early memantine treatment (Figure S16). Early memantine treatment did not rescue other behavioral abnormalities of Shank2−/− mice, including reduced ultrasonic vocalizations, enhanced repetitive behaviors, and anxiolytic-like behavior (Figure S17).
Early D-cycloserine or late memantine fails to normalize social interaction in Shank2−/− mice
Importantly, treating Shank2−/− mice with D-cycloserine instead of memantine from P7 to P21 did not improve social interaction or hyperactivity, similar to the results from WT mice (Figure 5A-C; Figure S18). Intriguingly, early memantine-treated WT mice showed decreased NMDAR function at ~P21-25, as shown by the decreased NMDA/AMPA ratio and normal mEPSC amplitude (Figure S19); the mEPSC frequency was also reduced, likely through compensatory mechanisms. These results suggest that artificially elevated NMDAR function in WT mice during an early stage (P7-21) is sufficient to induce NMDAR hypofunction in juvenile mice (~P21-25) but not abnormal behaviors in adult mice.
Figure 5. Early D-cycloserine or late memantine treatment does not improve social interaction in adult Shank2−/− mice.
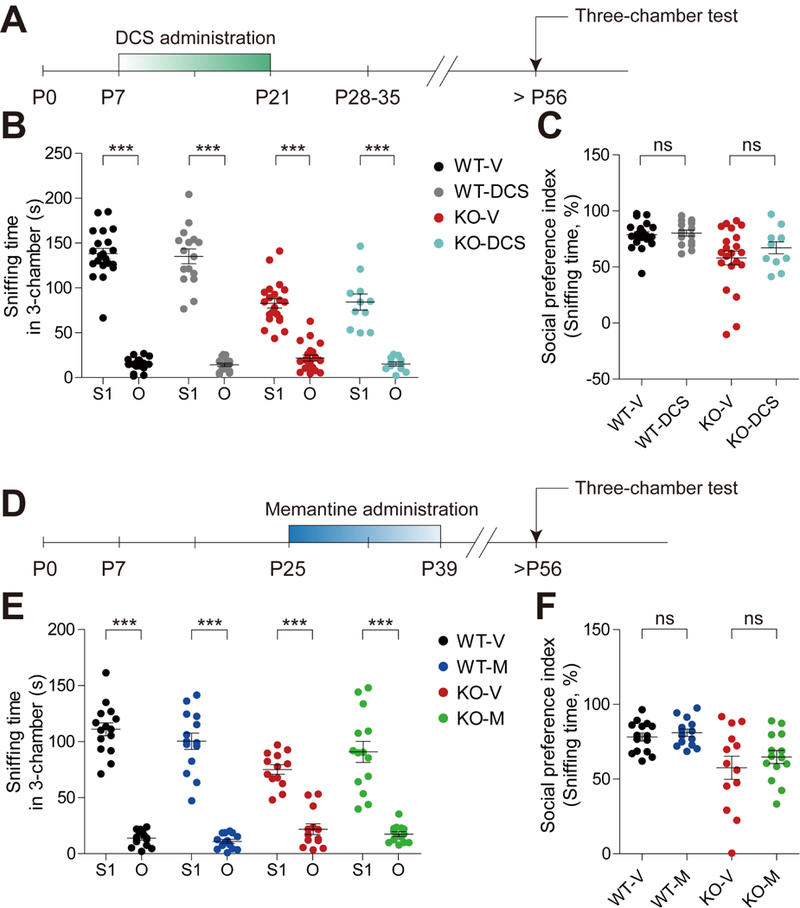
(A) Schematic depiction of early D-cycloserine (DCS) treatment and behavioral tests. Young Shank2−/− mice were orally administered D-cycloserine (40 mg/kg) twice daily for two weeks during P7-21, followed by consecutive measurements of open-field hyperactivity and three-chamber social interaction during P56-70. (B and C) Early D-cycloserine treatment fails to improve social interaction of adult Shank2−/− mice, as indicated by time sniffing S1/O and the social preference index based on sniffing time in the three-chamber test. (n = 21 mice for WT-V, 16 (WT-M), 21 (KO-V), and 11 (KO-M), ns, not significant, ***P < 0.001, ns, not significant, paired Student’s t-test or Wilcoxon matched pairs signed rank test (B), Mann Whitney U test (C). (D) Schematic depiction of late chronic memantine treatment and behavioral tests. Shank2−/− mice were orally administered memantine (20 mg/kg) twice daily for 2 weeks during P25-39, followed by consecutive measurements of three-chamber social interaction and open-field hyperactivity during P56-90. (E and F) Late chronic memantine treatment fails to improve social interaction in Shank2−/− mice, as indicated by time sniffing S1/O and the social preference index based on sniffing time in the three-chamber test. (n = 15 (WT-V), 14 (WT-M), 13 (KO-V), and 14 (KO-M), ns, not significant, ***P < 0.001, ns, not significant, paired Student’s t- test or Wilcoxon matched pairs signed rank test (E), unpaired Student’s t-test (F).
Treating Shank2−/− mice with memantine at a late stage (P25-39) failed to improve social interaction or hyperactivity (Figure 5D-F; Figure S20). Acute memantine treatment at P56 also failed to correct social interaction or hyperactivity (Figure S21). These results collectively suggest that early NMDAR hyperfunction in Shank2−/− mice should be corrected early and in the right direction.
Developmental switch of NMDAR function is not observed in other Shank2−/− mutant mice
We next tested if the temporal shift in NMDAR function in our Shank2−/− mice also occur in other Shank2-mutant mouse lines (33, 37). Shank2−/− mice lacking exon 24 (Shank2- dEx24) showed reduced NMDAR function at P21, similar to the reported NMDAR hypofunction at ~2-4 months (33), but normal NMDAR function at P14 (Figure S22). In addition, Shank2−/− mice lacking exon 7 (Shank2-dEx7) showed enhanced NMDAR function at P21, similar to the reported NMDAR hyperfunction at ~P24 (37), but normal NMDAR function at P14 (Figure S23).
Moreover, a switch in the background of our Shank2−/− mice lacking exons 6-7 from C57BL/6N to C57BL/6J had no effect on the reduced NMDAR function at ~P24 (Figure S24). Therefore, temporal patterns of NMDAR functions in different Shank2−/− mouse lines seem to be distinct.
Lastly, we sought to identify additional molecular differences between Shank2−/− mice lacking exons 6-7 and those lacking exon 7, which show opposite NMDAR functions at ~P21 (37, 38) and minimally overlapping transcriptomic profile (34). Intriguingly, Shank2−/− mice lacking exons 6-7 had undetectable levels of the shorter Shank2a splice variant, whereas Shank2−/− mice lacking exon 7 had markedly increased (~6-fold) levels of the Shank2a variant, while both mouse had similarly decreased levels of the longer Shank2b splice variant (Figures S25 and S26). Whether this differential splice variant expression leads to differences in NMDAR function remains to be determined. Our molecular modeling, however, predicted that the short peptide (57 aa- long) produced from the Shank2b splice variant in Shank2-dEx7 mice by protein truncation may form a structure that partly mimics the N-terminal region of the Shank2 PDZ domain and compete with the normal Shank2 PDZ domain for binding to the C- terminal PDZ-binding domains of GKAPs/SAPAPs in a dominant-negative manner (Figure S27).
Discussion
Our study provides early NMDAR hyperfunction at ~P14 as a causal mechanism for the NMDAR hypofunction and social deficits in Shank2−/− mice at juvenile (~P28) and adult (~P56) stages. In support of this, Shank2−/− mice display a rapid reversal of NMDAR function during P14 and P21 in the hippocampus (Figure 2; Figures S3-5) as well as in the mPFC (Figures S6 and S7). Importantly, early memantine treatment (P7-21) normalizes NMDAR function and social interaction without affecting other behaviors at juvenile and adult stages (Figures 3 and 4; Figures S9-17). In addition, pharmacological correction of NMDAR function in a wrong direction (early D- cycloserine), or during a wrong time window (late memantine), does not normalize social interaction in Shank2−/− mice (Figure 5; Figures S18 and S20). These results strongly suggest that early NMDAR hyperfunction induces late NMDAR hypofunction and social deficits in Shank2−/− mice, and early correction of NMDAR function has long- lasting effects on NMDAR function and social interaction.
Regarding how early memantine treatment induces long-lasting influences on electrophysiology and behavior in Shank2−/− mice, it is possible that once the NMDAR function is normalized by early chronic memantine treatment at the juvenile stage, the continuing lack of Shank2 expression after the end of early memantine treatment period (P7-21) may not induce a further change in NMDAR function. This hypothesis could be tested by inducing a Shank2 KO by a genetic method that starts from ~P21 after the completion of normal brain development during embryonic and early postnatal stages. Our results are reminiscent of a recent report that showed early correction of serotonin levels in infants through mother milk in a mouse model of autism carrying the human 15q11-13 duplication has long-lasting effects, rescuing serotonin levels as well as social behaviors in adults (61), which, together with our results, strongly suggest that early corrections are important.
Early memantine treatment of WT mice does not affect NMDAR function (Figures 3 and 4). This might be attributable to that the daily and total doses of memantine are in the range that can normalize NMDAR function in Shank2−/− mice but that do not suppress the normal NMDAR function in WT mice. Intriguingly, early D- cycloserine treatment of WT mice induces a decrease in NMDAR function at the juvenile stage (Figures S19), although it does not affect behaviors at the adult stage (Figure 5A-C). This could be attributable to that the doses of D-cycloserine may be just enough to induce a decrease in NMDAR activity but not to impair behaviors in WT mice, which may require a continuing deficit such as Shank2 KO. Alternatively, D-cycloserine may affect all NMDAR-expressing neurons, whereas Shank2 KO may mainly affect Shank2-expressing neurons.
Our data also provide evidence suggesting that the GluN2B component contributes to the distinct NMDAR functions at P14 and P21 Shank2−/− synapses (Figure 2). Specifically, P14 Shank2−/− synapses show markedly increased GluN2B component, whereas P21 Shank2−/− synapses show normal levels of GluN2B component. Therefore, the increased NMDAR-mediated currents at P14 may be mainly mediated by GluN2B-containing NMDARs with slower decay kinetics, whereas the decreased NMDAR-mediated currents at P21, where both GluN2A and GluN2B are reduced, may involve both GluN2B-dependent and -independent mechanisms.
Regarding further details, our immunoblot analyses suggest candidate mechanisms involving GluN1/GluN2B phosphorylation (Figure S8). It is known that GluN1-Ser896 phosphorylation promotes the ER-to-plasma membrane trafficking of GluN1 (62). In addition, CaMKII/PKC-dependent GluN2B-Ser1303 phosphorylation promotes synaptic retention of GluN2B through mechanisms involving the binding of GluN2B to MAGUKs (i.e. PSD-95) and AP2-dependent endocytosis of GluN2B (63-66). Therefore, the increased phosphorylation of GluN1-Ser896 and GluN2B-Ser1303 at P14 Shank2−/− synapses may promote surface and synaptic localization of NMDARs, whereas the decreased phosphorylation of GluN1-Ser896 and GluN2B-Ser1303 at P21 Shank2−/− synapses may suppress synaptic retention of NMDARs. These mechanisms might contribute to the differential contributions of the GluN2B component to P14 and P21 Shank2−/− NMDAR currents. However, it should be pointed out that these biochemical changes could reflect those from many different types of cells with some limitations in the interpretation.
Developmental switches of NMDAR function were not observed in other Shank2−/− mouse mutant lines. Shank2-dEx7 mice show normal NMDAR function at ~P14 but show increased NMDAR function at ~P21, as reported previously (67). In addition, Shank2-dEx24 mice showed reduced NMDAR function at ~P21, as reported previously (33), but normal NMDAR function at ~P14, partly similar to our mouse line. This discrepancy might be attributable to the differential expression of Shank2 splice variants in these mouse lines (Figures S25-26). For instance, we found a marked difference in the levels of the Shank2a transcript in Shank2-dEx6-7 and Shank2-dEx7 lines and, by molecular modeling (Figure S27), the possibility that a short truncated peptide derived from the Shank2a transcript in Shank2-dEx7 mice inhibits the normal interaction between Shank2 and SAPAPs, which may affect NMDAR activity or signaling, in a dominant-negative manner, although the presence of such short peptide could not be directly determined by immunoblot analyses. In addition, the exon 24 deleted in Shank2-dEx24 mice encodes the proline-rich region containing the Homer- and cortactin-binding sites (33), known to mediate the formation of a polymeric network structure and regulate the actin cytoskeleton in the postsynaptic density (68, 69). Although further details remain to be studied, these results are at the very least in line with that, in humans, different SHANK2 mutations, i.e. exons 6-7 and exon 7 deletions, are associated with distinct ASD symptoms (37). Similarly, two different mutations of SHANK3 (a SHANK2 relative) from ASD and schizophrenia patients, respectively, cause ASD- and schizophrenia-like phenotypes in mice (70).
NMDAR dysfunction, which would suppress normal brain development and function, has been suggested as an important candidate mechanism that may underlie ASD (71). Disturbances in NMDAR function, as a subset of excitatory synaptic function, would also induce the imbalance between synaptic and neuronal excitation and inhibition, another key mechanism implicated in ASD (72, 73). Given that NMDAR function is an important target for dynamic regulation and fine-tuning for normal brain development and function (53, 74), our results suggest that NMDAR dysfunctions observed in many other animal models of ASD (71, 75-90) may need to be further examined for their temporal changes. This might eventually help us develop more cautious treatment strategies for ASD patients with distinct temporal patterns of NMDAR dysfunctions.
In conclusion, our results suggest that Shank2 deletion in mice leads to NMDAR hyperfunction at an early stage that is causally associated with NMDAR hypofunction and social deficits at later stages. In addition, our study suggests that early correction of NMDAR hyperfunction in Shank2−/− mice has long-lasting effects, preventing synaptic and social abnormalities from manifesting at later stages.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation (NRF) grant (2013M3C7A1056732 to H.K.), NRF-2013-Fostering Core Leaders of the Future Basic Science Program (to C.C.), Global PhD Fellowship Program (NRF-2013H1A2A1032785 to S.H. and NRF-2015H1A2A1033937 to R.K.), the National Honor Scientist Program (NRF-2012R1A3A1050385 to B.K.K.), the National Institute of Health grants (MH098114, MH104316, and HD087795 to Y.H.J.), Korea Institute of Science and Technology Information (K-17-L03-C02-S01 to H.K.), and the Institute for Basic Science (IBS-R002-D1 to E.K.).
Footnotes
Further information on methods is available in Supplementary Information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. (2013): SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Molecular autism. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudhof TC (2008): Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoghbi HY, Bear MF (2012): Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harbor perspectives in biology. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang YH, Ehlers MD (2013): Modeling autism by SHANK gene mutations in mice. Neuron. 78:8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeron T (2015): From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature reviews Neuroscience. 16:551–563. [DOI] [PubMed] [Google Scholar]
- 6.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH (2016): Advancing the understanding of autism disease mechanisms through genetics. Nature medicine. 22:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sala C, Vicidomini C, Bigi I, Mossa A, Verpelli C (2015): Shank synaptic scaffold proteins: keys to understanding the pathogenesis of autism and other synaptic disorders. Journal of neurochemistry. 135:849–858. [DOI] [PubMed] [Google Scholar]
- 8.Volk L, Chiu SL, Sharma K, Huganir RL (2015): Glutamate synapses in human cognitive disorders. Annual review of neuroscience. 38:127–149. [DOI] [PubMed] [Google Scholar]
- 9.Ehninger D, Silva AJ (2011): Rapamycin for treating Tuberous sclerosis and Autism spectrum disorders. Trends in molecular medicine. 17:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser T, Feng G (2015): Modeling psychiatric disorders for developing effective treatments. Nature medicine. 21:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleijer KT, Schmeisser MJ, Krueger DD, Boeckers TM, Scheiffele P, Bourgeron T, et al. (2014): Neurobiology of autism gene products: towards pathogenesis and drug targets. Psychopharmacology. 231:1037–1062. [DOI] [PubMed] [Google Scholar]
- 12.Aceti M, Creson TK, Vaissiere T, Rojas C, Huang WC, Wang YX, et al. (2015): Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biological psychiatry. 77:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, et al. (2012): Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 151:709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng M, Hoogenraad CC (2007): The Postsynaptic Architecture of Excitatory Synapses: A More Quantitative View. Annual review of biochemistry. 76:823–847. [DOI] [PubMed] [Google Scholar]
- 15.Sheng M, Kim E (2011): The postsynaptic organization of synapses. Cold Spring Harbor perspectives in biology. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossa A, Giona F, Pagano J, Sala C, Verpelli C (2017): SHANK genes in autism: Defining therapeutic targets. Progress in neuro-psychopharmacology & biological psychiatry [DOI] [PubMed] [Google Scholar]
- 17.Monteiro P, Feng G (2017): SHANK proteins: roles at the synapse and in autism spectrum disorder. Nature reviews Neuroscience. 18:147–157. [DOI] [PubMed] [Google Scholar]
- 18.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. (2012) :Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS genetics. 8:e1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, et al. (2011): Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Human molecular genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, et al. (2013) :Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. Journal of medical genetics. 50:144–150. [DOI] [PubMed] [Google Scholar]
- 21.Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, et al. (2012): A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda). 2:1665–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. (2012): De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 485:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. (2010): Functional impact of global rare copy number variation in autism spectrum disorders. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chilian B, Abdollahpour H, Bierhals T, Haltrich I, Fekete G, Nagel I, et al. (2013): Dysfunction of SHANK2 and CHRNA7 in a patient with intellectual disability and language impairment supports genetic epistasis of the two loci. Clinical genetics. 84:560–565. [DOI] [PubMed] [Google Scholar]
- 25.Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. (2014): Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS genetics. 10:e1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. (2012): Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 380:1674–1682. [DOI] [PubMed] [Google Scholar]
- 27.Wischmeijer A, Magini P, Giorda R, Gnoli M, Ciccone R, Cecconi L, et al. (2011): Olfactory Receptor-Related Duplicons Mediate a Microdeletion at 11q13.2q13.4 Associated with a Syndromic Phenotype. Molecular syndromology. 1:176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peykov S, Berkel S, Degenhardt F, Rietschel M, Nothen MM, Rappold GA (2015): Rare SHANK2 variants in schizophrenia. Molecular psychiatry. 20:1487–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peykov S, Berkel S, Schoen M, Weiss K, Degenhardt F, Strohmaier J, et al. (2015): Identification and functional characterization of rare SHANK2 variants in schizophrenia. Molecular psychiatry. 20:1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Homann OR, Misura K, Lamas E, Sandrock RW, Nelson P, McDonough SI, et al. (2016) : Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Molecular psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costas J (2015): The role of SHANK2 rare variants in schizophrenia susceptibility. Molecular psychiatry. 20:1486. [DOI] [PubMed] [Google Scholar]
- 32.Kim R, Kim J, Chung C, Ha S, Lee S, Lee E, et al. (2018): Cell-Type-Specific Shank2 Deletion in Mice Leads to Differential Synaptic and Behavioral Phenotypes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 38:4076–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pappas AL, Bey AL, Wang X, Rossi M, Kim YH, Yan H, et al. (2017): Deficiency of Shank2 causes mania-like behavior that responds to mood stabilizers. JCI Insight. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim CS, Kim H, Yu NK, Kang SJ, Kim T, Ko HG, et al. (2017): Enhancing inhibitory synaptic function reverses spatial memory deficits in Shank2 mutant mice. Neuropharmacology. 112:104–112. [DOI] [PubMed] [Google Scholar]
- 35.Ha S, Lee D, Cho YS, Chung C, Yoo YE, Kim J, et al. (2016): Cerebellar Shank2 Regulates Excitatory Synapse Density, Motor Coordination, and Specific Repetitive and Anxiety-Like Behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 36:12129–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peter S, Ten Brinke MM, Stedehouder J, Reinelt CM, Wu B, Zhou H, et al. (2016): Dysfunctional cerebellar Purkinje cells contribute to autism-like behaviour in Shank2- deficient mice. Nature communications. 7:12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel V, Kuebler A, et al. (2012) : Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. [DOI] [PubMed] [Google Scholar]
- 38.Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. (2012): Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 486:261–265. [DOI] [PubMed] [Google Scholar]
- 39.Heise C, Preuss JM, Schroeder JC, Battaglia CR, Kolibius J, Schmid R, et al. (2018): Heterogeneity of Cell Surface Glutamate and GABA Receptor Expression in Shank and CNTN4 Autism Mouse Models. Frontiers in molecular neuroscience. 11:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Lee E, Kim R, Kim J, Lee S, Park H, et al. (2018): Shank2 Deletion in Parvalbumin Neurons Leads to Moderate Hyperactivity, Enhanced Self-Grooming and Suppressed Seizure Susceptibility in Mice. Frontiers in molecular neuroscience. 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ey E, Torquet N, Le Sourd AM, Leblond CS, Boeckers TM, Faure P, et al. (2013): The Autism ProSAP1/Shank2 mouse model displays quantitative and structural abnormalities in ultrasonic vocalisations. Behavioural brain research. 256:677–689. [DOI] [PubMed] [Google Scholar]
- 42.Sato M, Kawano M, Mizuta K, Islam T, Lee MG, Hayashi Y (2017): Hippocampus-Dependent Goal Localization by Head-Fixed Mice in Virtual Reality. eNeuro. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon SY, Kwon SG, Kim YH, Yeo JH, Ko HG, Roh DH, et al. (2017): A critical role of spinal Shank2 proteins in NMDA-induced pain hypersensitivity. Mol Pain. 13:1744806916688902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferhat AT, Torquet N, Le Sourd AM, de Chaumont F, Olivo-Marin JC, Faure P, et al. (2016): Recording Mouse Ultrasonic Vocalizations to Evaluate Social Communication. Journal of visualized experiments: JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko HG, Oh SB, Zhuo M, Kaang BK (2016): Reduced acute nociception and chronic pain in Shank2−/− mice. Mol Pain. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo Y, Lin A, Chang P, Gan WB (2005): Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 46:181–189. [DOI] [PubMed] [Google Scholar]
- 47.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. (2005): Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP (2007): GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 23:3251–3253. [DOI] [PubMed] [Google Scholar]
- 49.Schumann CM, Hamstra J, Goodlin-Jones BL, Lotspeich LJ, Kwon H, Buonocore MH, et al. (2004): The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. The Journal of neuroscience : the official journal of the Society for Neuroscience. 24:6392–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endo T, Shioiri T, Kitamura H, Kimura T, Endo S, Masuzawa N, et al. (2007): Altered chemical metabolites in the amygdala-hippocampus region contribute to autistic symptoms of autism spectrum disorders. Biological psychiatry. 62:1030–1037. [DOI] [PubMed] [Google Scholar]
- 51.Felix-Ortiz AC, Tye KM (2014): Amygdala inputs to the ventral hippocampus bidirectionally modulate social behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 34:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, et al. (2011): Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 477:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lussier MP, Sanz-Clemente A, Roche KW (2015): Dynamic Regulation of N- Methyl-d-aspartate (NMDA) and alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors by Posttranslational Modifications. The Journal of biological chemistry. 290:28596–28603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang M, Silverman JL, Crawley JN (2011): Automated three-chambered social approach task for mice Current protocols in neuroscience /editorial board, Crawley Jacqueline N[et al. ]. Chapter 8:Unit 8 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverman JL, Yang M, Lord C, Crawley JN (2010): Behavioural phenotyping assays for mouse models of autism. Nature reviews Neuroscience. 11:490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferguson JN, Young LJ, Hearn EF, Matzuk MM, Insel TR, Winslow JT (2000): Social amnesia in mice lacking the oxytocin gene. Nature genetics. 25:284–288. [DOI] [PubMed] [Google Scholar]
- 57.Wang F, Zhu J, Zhu H, Zhang Q, Lin Z, Hu H (2011): Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science. 334:693–697. [DOI] [PubMed] [Google Scholar]
- 58.Yang M, Lewis F, Foley G, Crawley JN (2015): In tribute to Bob Blanchard: Divergent behavioral phenotypes of 16p11.2 deletion mice reared in same-genotype versus mixed-genotype cages. Physiology & behavior. 146:16–27. [DOI] [PubMed] [Google Scholar]
- 59.Spencer CM, Alekseyenko O, Serysheva E, Yuva-Paylor LA, Paylor R (2005): Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes, brain, and behavior. 4:420–430. [DOI] [PubMed] [Google Scholar]
- 60.Kalbassi S, Bachmann SO, Cross E, Roberton VH, Baudouin SJ (2017): Male and Female Mice Lacking Neuroligin-3 Modify the Behavior of Their Wild-Type Littermates. eNeuro 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakai N, Nagano M, Saitow F, Watanabe Y, Kawamura Y, Kawamoto A, et al. (2017) : Serotonin rebalances cortical tuning and behavior linked to autism symptoms in 15q11–13 CNV mice. Sci Adv. 3:e1603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD (2001): An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. The Journal of neuroscience : the official journal of the Society for Neuroscience. 21:3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanz-Clemente A, Gray JA, Ogilvie KA, Nicoll RA, Roche KW (2013): Activated CaMKII couples GluN2B and casein kinase 2 to control synaptic NMDA receptors. Cell reports. 3:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lavezzari G, McCallum J, Lee R, Roche KW (2003): Differential binding of the AP-2 adaptor complex and PSD-95 to the C-terminus of the NMDA receptor subunit NR2B regulates surface expression. Neuropharmacology. 45:729–737. [DOI] [PubMed] [Google Scholar]
- 65.Prybylowski K, Chang K, Sans N, Kan L, Vicini S, Wenthold RJ (2005): The synaptic localization of NR2B-containing NMDA receptors is controlled by interactions with PDZ proteins and AP-2. Neuron. 47:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanz-Clemente A, Matta JA, Isaac JT, Roche KW (2010): Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron. 67:984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmeisser MJ (2015): Translational neurobiology in Shank mutant mice--model systems for neuropsychiatric disorders. Annals of anatomy = Anatomischer Anzeiger: official organ of the Anatomische Gesellschaft. 200:115–117. [DOI] [PubMed] [Google Scholar]
- 68.Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, et al. (2009): The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 137:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. (1999): Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD- 95/GKAP complex and cortactin. Neuron. 23:569–582. [DOI] [PubMed] [Google Scholar]
- 70.Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, et al. (2016): Mice with Shank3 Mutations Associated with ASD and Schizophrenia Display Both Shared and Distinct Defects. Neuron. 89:147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee EJ, Choi SY, Kim E (2015): NMDA receptor dysfunction in autism spectrum disorders. Current opinion in pharmacology. 20C:8–13. [DOI] [PubMed] [Google Scholar]
- 72.Lee E, Lee J, Kim E (2017): Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biological psychiatry. 81:838–847. [DOI] [PubMed] [Google Scholar]
- 73.Nelson SB, Valakh V (2015): Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 87:684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paoletti P, Bellone C, Zhou Q (2013): NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature reviews Neuroscience. 14:383–400. [DOI] [PubMed] [Google Scholar]
- 75.Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, et al. (2010): Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 30:2115–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung W, Choi SY, Lee E, Park H, Kang J, Park H, et al. (2015): Social deficits in IRSp53 mutant mice improved by NMDAR and mGluR5 suppression. Nature neuroscience. [DOI] [PubMed] [Google Scholar]
- 77.Huang TN, Chuang HC, Chou WH, Chen CY, Wang HF, Chou SJ, et al. (2014): Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nature neuroscience. 17:240–247. [DOI] [PubMed] [Google Scholar]
- 78.Lee EJ, Lee H, Huang TN, Chung C, Shin W, Kim K, et al. (2015): Trans-synaptic zinc mobilization improves social interaction in two mouse models of autism through NMDAR activation. Nature communications. 6:7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duffney LJ, Wei J, Cheng J, Liu W, Smith KR, Kittler JT, et al. (2013): Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. The Journal of neuroscience : the official journal of the Society for Neuroscience. 33:15767–15778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. (2015): Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators. Cell reports. 11:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaramillo TC, Liu S, Pettersen A, Birnbaum SG, Powell CM (2014): Autism- related neuroligin-3 mutation alters social behavior and spatial learning. Autism research : official journal of the International Society for Autism Research. 7:264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, et al. (2012): Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. The Journal of neuroscience : the official journal of the Society for Neuroscience. 33:18448–18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rinaldi T, Kulangara K, Antoniello K, Markram H (2007): Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proceedings of the National Academy of Sciences of the United States of America. 104:13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang J, Kim E (2015): Suppression of NMDA receptor function in mice prenatally exposed to valproic acid improves social deficits and repetitive behaviors. Frontiers in molecular neuroscience. 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim KC, Lee DK, Go HS, Kim P, Choi CS, Kim JW, et al. (2014): Pax6- dependent cortical glutamatergic neuronal differentiation regulates autism-like behavior in prenatally valproic acid-exposed rat offspring. Molecular neurobiology. 49:512–528. [DOI] [PubMed] [Google Scholar]
- 86.Takeuchi K, Gertner MJ, Zhou J, Parada LF, Bennett MV, Zukin RS (2013): Dysregulation of synaptic plasticity precedes appearance of morphological defects in a Pten conditional knockout mouse model of autism. Proceedings of the National Academy of Sciences of the United States of America. 110:4738–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM (2016): Altered Striatal Synaptic Function and Abnormal Behaviour in Shank3 Exon4–9 Deletion Mouse Model of Autism. Autism research : official journal of the International Society for Autism Research. 9:350–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, Gupta N, et al. (2015): Autism-Associated Insertion Mutation (InsG) of Shank3 Exon 21 Causes Impaired Synaptic Transmission and Behavioral Deficits. The Journal of neuroscience : the official journal of the Society for Neuroscience. 35:9648–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Espinosa F, Xuan Z, Liu S, Powell CM (2015): Neuroligin 1 modulates striatal glutamatergic neurotransmission in a pathway and NMDAR subunit-specific manner. Frontiers in synaptic neuroscience. 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. (2010): Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Molecular autism. 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


