Abstract
Vitamin D has been connected with increased intramyocellular lipid (IMCL) and has also been shown to increase mitochondrial function and insulin sensitivity. Evidence suggests that perilipin 2 (PLIN2), a perilipin protein upregulated with calcitriol treatment, may be integral to managing increased IMCL capacity and lipid oxidation in skeletal muscle. Therefore, we hypothesized that PLIN2 is required for vitamin D induced IMCL accumulation and increased mitochondrial oxidative function. To address this hypothesis, we treated C2C12 myotubes with 100 nM calcitriol (the active form of vitamin D) and/or PLIN2 siRNA in a four group design and analyzed markers of IMCL accumulation and metabolism using qRT-PCR, cytochemistry, and oxygen consumption assay. Expression of PLIN2, but not PLIN3 or PLIN5 mRNA was increased with calcitriol, and PLIN2 induction was prevented with siRNA knockdown without compensation by other perilipins. PLIN2 knockdown did not appear to prevent lipid accumulation. Calcitriol treatment increased mRNA expression of triglyceride synthesizing genes DGAT1 and DGAT2 and also lipolytic genes ATGL and CGI-58. PLIN2 knockdown decreased the expression of CGI-58 and CPT1, and was required for calcitriol-induced upregulation of DGAT2. Calcitriol increased oxygen consumption rate while PLIN2 knockdown decreased oxygen consumption rate. PLIN2 was required for a calcitriol-induced increase in oxygen consumption driven by mitochondrial complex II. We conclude that calcitriol increases mitochondrial function in myotubes and that this increase is at least in part mediated by PLIN2.
Keywords: vitamin D, PLIN2, skeletal muscle, mitochondria, metabolism, lipid droplet, IMCL, C2C12
1. Introduction
The ability to store lipid as potential energy is one of the oldest and most highly conserved adaptations of life on Earth [1–3]. In mammals, adipocytes are evolved to store large quantities of lipid, but appreciable lipid stores are also found in the skeletal muscle where they are made available for β-oxidation in mitochondria. Intramyocellular lipid (IMCL) is stored in lipid droplets (LD), highly specialized and tightly regulated organelles that play important roles in lipid accumulation, storage, and lipolysis. Lipid droplets are comprised of a phospholipid monolayer studded with several dozen different proteins, including perilipins (PLINs), that surrounds a neutral lipid core of triacylglycerides (TAG), diacylglycerides (DAG), and cholesteryl esters (CE) [4–6]. PLINs are essential for LD formation and function [7]. Of the 5 members of the perilipin family, PLIN2, PLIN3, and PLIN5 (also referred to as ADRP, TIP47, and OXPAT, respectively) are found in skeletal muscle. Although some have reported high levels of PLIN4 expression in skeletal muscle [8], its expression is not widely recognized [9]. While each PLIN seems to play an important and independent role in IMCL regulation, PLIN2 is the most highly expressed PLIN in skeletal muscle and is thought to serve primarily as a scaffolding protein that modulates access of adipose triglyceride lipase (ATGL) to its enzymatic substrate, TAG [10, 11]. While some have reported that PLIN2 knockout is protective against pathological lipid accumulation in some tissues [12, 13], others have suggested that overexpression of PLIN2 increases oxidative capacity and improves metabolic function in skeletal muscle [14, 15]. Recent work from our group indicates that treatment with calcitriol (1,25-dihydroxyvitamin D3, the active form of vitamin D) increases PLIN2 expression in muscle cells in vitro [16].
Dietary vitamin D supplementation and calcitriol treatment have both been shown to support healthy skeletal muscle function [17–19]. Vitamin D is associated with increased IMCL content in both clinical and basic research [16, 20]. Increased IMCL, especially in association with increased body fat mass, is often associated with increased inflammation, metabolic dysfunction, and insulin resistance. Pathological lipid processing is seen in many tissues and is broadly referred to as lipotoxicity [21–24]. Although vitamin D increases IMCL, it has also been associated with decreased inflammation and improved insulin sensitivity, mitochondrial activity, and functional capacity in skeletal muscle [18, 19, 25–27].
This paradoxical increase in IMCL and mitochondrial function following vitamin D supplementation may be a product of an increase in the efficiency with which muscle accumulates, stores, and oxidizes lipid, a process often referred to as “lipid flux”. Increased rates of lipid flux and well-regulated storage of lipids as TAG in lipid droplets may help to prevent lipotoxicity [28–30] as modeled in the athlete’s paradox, a condition observed in endurance athletes characterized by both increased IMCL, high mitochondrial efficiency, and insulin sensitivity [31–33]. Increased expression of PLIN2 has been connected to increased TAG storage and oxidation [14, 15], and may be a mechanism through which vitamin D increases lipid flux in skeletal muscle. This study sought to determine the role of PLIN2 in vitamin D mediated increases in IMCL accumulation and mitochondrial function. We hypothesized that PLIN2 is required for vitamin D-induced IMCL accumulation and increased mitochondrial oxidative function. To investigate this, we used a C2C12 murine muscle cell line treated with calcitriol. The role of PLIN2 in changes in lipid content and metabolism was determined by knocking down PLIN2 expression using siRNA.
2. Methods
2.1. Cell culture
C2C12 myoblasts were obtained from American Type Culture Consortium (ATCC; Manassas, Virginia, USA) and grown to a maximum of 60% confluence. At appropriate confluence, cells were trypsinized and seeded overnight in growth medium (GM) consisting of DMEM containing 1000 mg/L glucose with L-glutamine and sodium bicarbonate (MilliporeSigma, Burlington, MA, USA; #D6046) supplemented with 10% FBS (Gemini Bio-Products, West Sacramento, CA, USA; #100–106) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, USA;Scientific, Waltham, MA, USA; #15140–122) in a humidified incubator kept at 37°C and 10% CO2 (Day 0). Following overnight seeding, GM was replaced with differentiation medium (DM) consisting of DMEM (same as above) supplemented with 2% horse serum (Day 1). DM was changed every other day.
2.2. Treatment with PLIN2 siRNA and Calcitriol
On Day 5 in DM, differentiated myotubes were treated with 10 nM Thermo Fisher Stealth siRNA against PLIN2 (Thermo Fisher Scientific Waltham, MA, USA; #132001) or medium GC content scramble Stealth siRNA (Thermo Fisher Scientific Waltham, MA, USA; #12935300) as previously published [14]. All siRNA was prepared in DM with 0.2% Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific Waltham, MA, USA; #13778) and 20% Opti-MEM (Thermo Fisher Scientific Waltham, MA, USA; #31985). Cells were treated with siRNA for a total of 48 hours. On Day 7, cells were treated with vehicle control (0.1% ethanol) (CTL) or 100 nM calcitriol (MilliporeSigma, Burlington, MA, USA; #D1530) (VitD) for 24 hours. Cells with PLIN2 knockdown are represented in text as siCTL and siVitD. Cell growth and treatment is summarized in Fig. A.1.
2.3. Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Cells were seeded 50,000/well in a 24 well culture plate and treated as described above. After treatment, media was removed and cells were washed in phosphate buffered saline (PBS) then scraped off the plate in 150 μL QIAzol Reagent (Qiagen, Hilden, Germany; #79306). Three wells receiving the same treatment were combined and lysed in a bead homogenizer. RNA was isolated using an ethanol precipitation on a RNA elution column (Enzymax, Lexington, KY, USA; #EZCR101). RNA was then reverse transcribed with a qScript cDNA synthesis kit (Quanta Biosciences, Beverly, MA; 101414–106) according to the manufacturer’s recommendations. Relative gene expression was measured using PowerUp SYBR (Thermo Fisher Scientific, Waltham, MA; #A25778) in a QuantStudio 3 real time PCR machine (Thermo Fisher Scientific, Waltham, MA). The geometric mean of three housekeeping genes (RER1, VCP, and EMC7) was used as an endogenous control. Expression was quantified using the 2ΔΔ-Ct method. Values were normalized to the CTL for each respective treatment and reported as fold change. Primers were purchased through Integrated DNA Technologies and primer sequences used in this study are listed in Table. A.1.
2.4. Oil Red O Staining
To assess neutral lipid accumulation, myotubes were fixed in 4% paraformaldehyde (PFA) and washed with PBS, then stained with oil red O (ORO) (MilliporeSigma, Burlington, MA USA; #O-0625) prepared in triethylphosphate according to the manufacturer’s specifications for 30 minutes at 37°C with occasional rocking. Cells were then washed and imaged. PLIN2 knockdown myotubes were treated with 100 μM palmitate for 3 hours prior to ORO staining. For all cytochemistry experiments, cells were imaged using a Zeiss AxioObserver D1 inverted fluorescent microscope (Jena, Germany). Micrographs were taken using a Zeiss AxioCam MR camera and analyzed and annotated using AxioVision SE64 Rel. 4.9.1 software (Zeiss, Jena, Germany).
2.5. Succinate Dehydrogenase Activity Staining
Myotubes were washed with PBS, fixed with 4% PFA and then incubated in 1.2 mM nitro blue tetrazolium chloride (NBT) (MilliporeSigma, Burlington, MA, USA; #N6876) with 275 mM succinic acid (MilliporeSigma, Burlington, MA, USA; #224731) in PBS at 37°C for 120 minutes. Cells were then washed 3 times with PBS and imaged as described above.
2.6. Seahorse Oxygen Consumption Rate (OCR) Assay
Myoblasts were plated 10,000 per well in a Seahorse XFe24 assay plate (Agilent, Santa Clara, CA, USA) overnight in GM, then differentiated, and treated as described above. On the day of the assay, fresh Seahorse XF Assay Medium (Agilent, Santa Clara, CA, USA; #102365) was supplemented with 5 mM glucose and 1 mM pyruvate and pH adjusted to 7.4. Myotubes were washed twice with XF Assay Medium then incubated for one hour in 100 μL of XF Assay Medium in a humidified chamber at 37°C with atmospheric CO2. Vehicle and calcitriol treatments were maintained throughout incubation and the assay. Following incubation, XF Assay Medium was added to a final volume of 525 μL. OCR and Extracellular Acidification Rate (ECAR) were measured at 37°C using Seahorse XFe24 Analyzer (Agilent, Santa Clara, CA, USA). During the assay, each treatment was injected sequentially to achieve the following final concentrations: 2.5 μM oligomycin (Biomol, Hamburg, Germany; #CM-111), 4 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (Biomol, Hamburg, Germany; #CM120), 10 mM succinate (MilliporeSigma, Burlington, MA, USA; #S-7501) with 0.8 μM rotenone (Biomol, Hamburg, Germany; #CM-117), and 1 μM antimycin A (MilliporeSigma, Burlington, MA, USA; #A8674). OCR was normalized to the average basal rate among all treatments for each given experiment. Specific rates based on equations suggested by Agilent Biosciences (listed in Table A.2) were used to calculate the following OCRs: basal, oligomycin, FCCP, rotenone/succinate, antimycin A, ATP-linked, maximum, reserve, complex I, complex II, leak, non-mitochondrial, and mitochondrial area under the curve (AUC).
2.7. Statistical Analysis
The Student’s t-test was used to determine the effects of calcitriol alone. A 2 × 2 factorial ANOVA was used to determine the effects of calcitriol (VitD effect), PLIN2 knockdown (siPLIN2 effect), and the interaction between treatments. When there was a significant interaction between VitD and siPLIN2, the Fisher’s LSD post-hoc test was applied. All tests were two-tailed, with statistical significance defined as p < 0.05. All data were normally distributed. All quantitative results are shown as mean ± SEM of no fewer than 3 independent experiments. Statistical calculations were performed using JMP 12 (SAS Institute, Cary, NC, USA).
3. Results
3.1. Vitamin D drives changes in PLIN2 expression and mitochondrial activity in C2C12 myotubes consistent with increased lipid flux
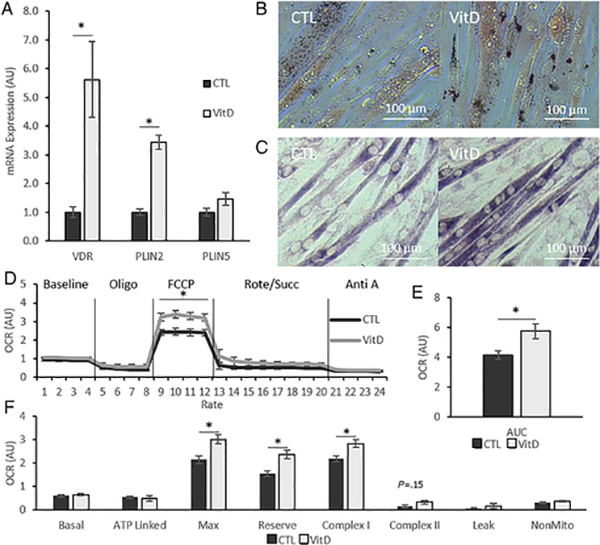
We first examined the effect of VitD alone on the expression of genes associated with IMCL accumulation and mitochondrial metabolism. Using qRT-PCR, we measured gene expression of vitamin D receptor (VDR) and PLIN2 in C2C12 myotubes treated with VitD or CTL for 24 hours and found that the expression of both genes was increased after VitD treatment (p < 0.05) (Fig 1A). Because PLIN5 is commonly associated with increased lipid storage and increased mitochondrial function [34, 35], we also measured its expression with VitD treatment. RT-PCR analysis showed that PLIN5 did not change with VitD treatment (Fig. 1A). To connect increased PLIN2 expression with increased IMCL accumulation, we labeled neutral lipid content in myotubes using ORO staining. Consistent with other publications [16, 20], we found that IMCL content in C2C12 myotubes appeared to be increased following VitD treatment (Fig. 1B).
Fig. 1. Calcitriol increases lipid storage and mitochondrial activity in C2C12 myotubes.
(A) Gene expression of vitamin D receptor (VDR), perilipin 2 (PLIN2) and perilipin 5 (PLIN5) normalized to CTL in differentiated C2C12 myotubes. (B) Representative brightfield micrographs depicting oil red O staining. (C) Representative brightfield micrographs depicting succinate dehydrogenase activity staining. (D) Oxygen consumption rate (OCR) of myotubes throughout experiment with addition of oligomycin, FCCP, rotenone/succinate, and antimycin A. (E) Total mitochondrial OCR. (F) OCR calculated using equations based on those provided by Agilent Biosciences. All data are represented as mean ± SEM, n = 5. All micrographs obtained at 32x magnification. Scale bars = 100 μm. CTL = 0.1% ethanol, 24 h ours; VitD = 100 nM calcitriol, 24 hours. n = 5; bars represent mean ± SEM; * p < 0.05, independent t-test. Oligo, Oligomycin; FCCP, Carbonyl cyanide-4-trifluoromethoxy)phenylhydrazone; Rote/Succ, Rotenenone & Succinate; Anti A, Antimycin A.
Having confirmed increased PLIN2 expression and IMCL accumulation, we next moved to measure changes in mitochondrial metabolism. We completed an initial assessment of mitochondrial function by SDH activity staining and found that VitD treatment appeared to increase the intensity of SDH activity staining in myotubes (Fig. 1C). For a detailed, quantitative analysis of the VitD’s impact on muscle cell mitochondrial metabolism, we analyzed OCR by Seahorse extracellular flux analyzer. Results show that VitD increased OCR throughout the experiment with statistically significant increases in OCR after the administration of FCCP (Fig. 1D & E). To further characterize changes in OCR, OCR specific to ATP-production and mitochondrial complexes were measured. We found that increases were attributed to OCR driven by complex I and maximal OCR. These changes were accompanied by a trend to increase complex II OCR, however, this failed to reach statistical significance (p = 0.15) (Fig. 1F).
3.2. PLIN2 knockdown does not impact mRNA expression of other perilipin genes
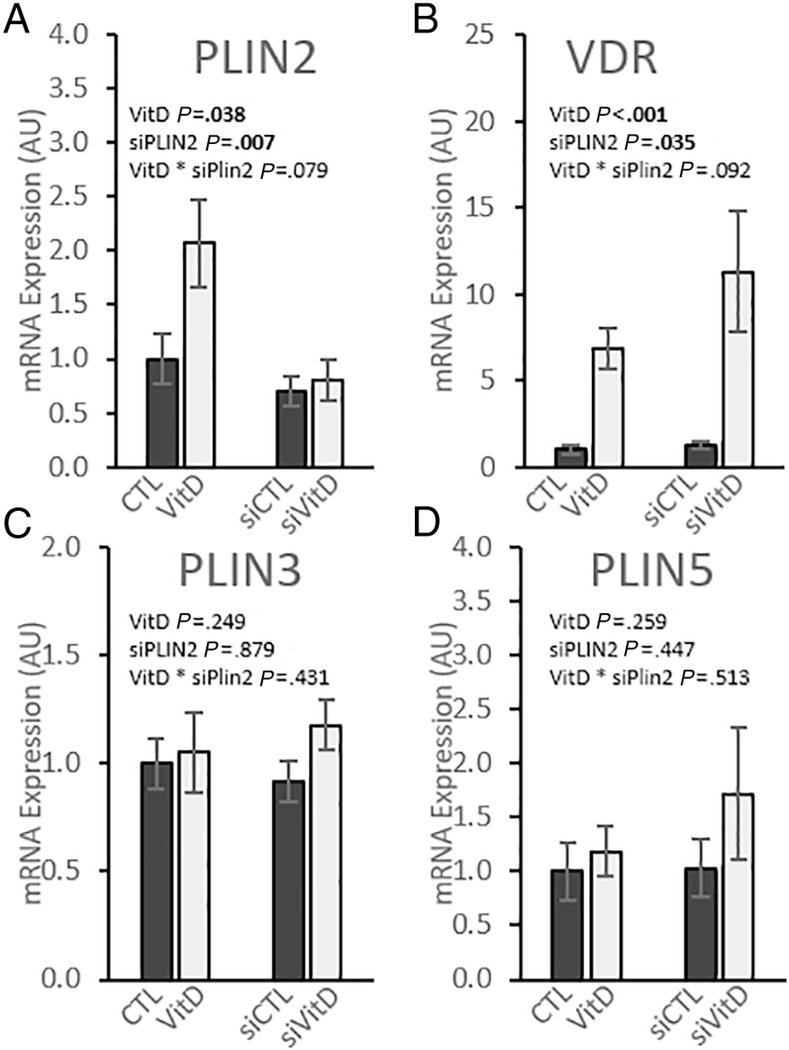
siRNA knockdown was used to investigate the role that PLIN2 plays in regulating VitD-induced changes in IMCL and mitochondrial function. RT-PCR quantification of mRNA expression verified that siRNA successfully decreased PLIN2 mRNA expression (p < 0.01); cells treated with siVitD decreased by 69% and siCTL decreased by 29% compared to their respective scramble controls (Fig. 2A). While VitD treatment caused increased VDR expression (p < 0.001), there was no effect of siRNA and no interaction between the treatments (Fig 2B). Perilipins other than PLIN2, namely PLIN3 and PLIN5, have been connected with increased mitochondrial activity in skeletal muscle [35–37]. We measured expression of these genes to ensure that these perilipins were not upregulated to compensate for decreased PLIN2 expression and found that neither was significantly increased under any treatment condition (Fig. 2C-D).
Fig. 2. PLIN2 knockdown decreases PLIN2 expression without compensation by other perilipin genes.
Gene expression in differentiated C2C12 myotubes relative to vehicle control with scramble siRNA. Cells were treated with either scramble siRNA and vehicle control (CTL), scramble siRNA and 100 nM calcitriol (VitD), PLIN2 siRNA and vehicle control (siCTL), or PLIN2 siRNA and 100 nM calcitriol (siVitD). All data are represented as mean ± SEM, n = 6. Values not sharing letters are significantly different (Fisher’s LSD). p values represent 2-way ANOVA.
3.3. PLIN2 knockdown does not prevent lipid accumulation
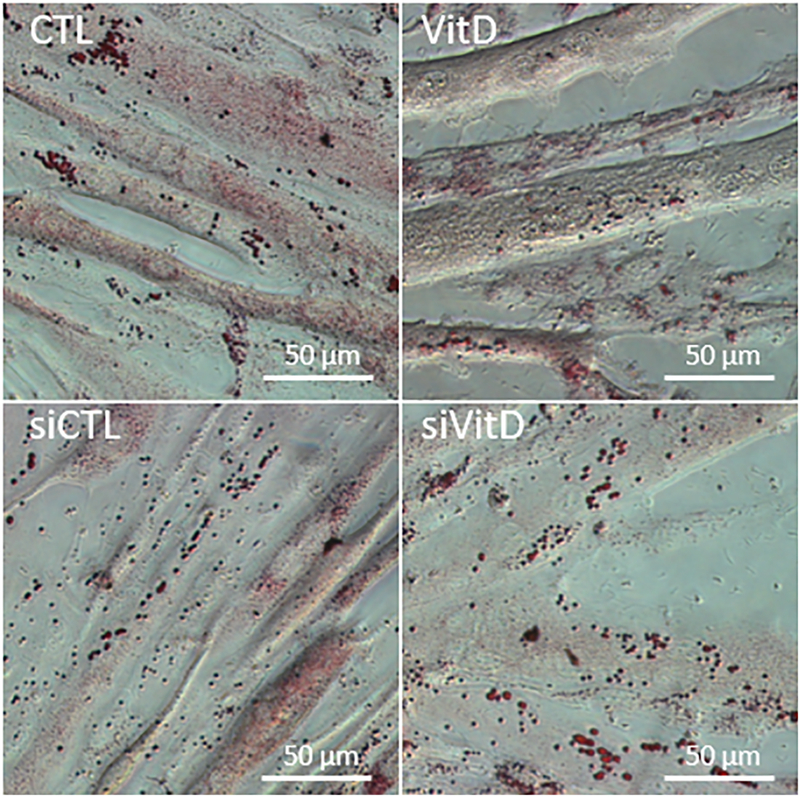
Overexpressing PLIN2 has been shown to increase lipid accumulation in skeletal muscle [14, 38], while knockdown prevents lipid accumulation in multiple tissues [12, 39, 40]. To see how PLIN2 knockdown impacts IMCL accumulation in our model, we treated differentiated myotubes with palmitate and assessed neutral lipid accumulation with ORO staining. Micrographs showed that PLIN2 knockdown did not appear to prevent lipid accumulation in C2C12 myotubes (Fig. 3).
Fig. 3. PLIN2 knockdown does not appear to prevent neutral lipid accumulation in C2C12 myotubes.
Oil Red O micrographs show ample lipid accumulation in C2C12 myotubes after 3h of treatment with 100 μM palmitate despite PLIN2 knockdown. Images acquired at 40x magnification, scale bar = 50 μm.
3.4. PLIN2 knockdown and VitD exert opposing effects on genes regulating lipid flux
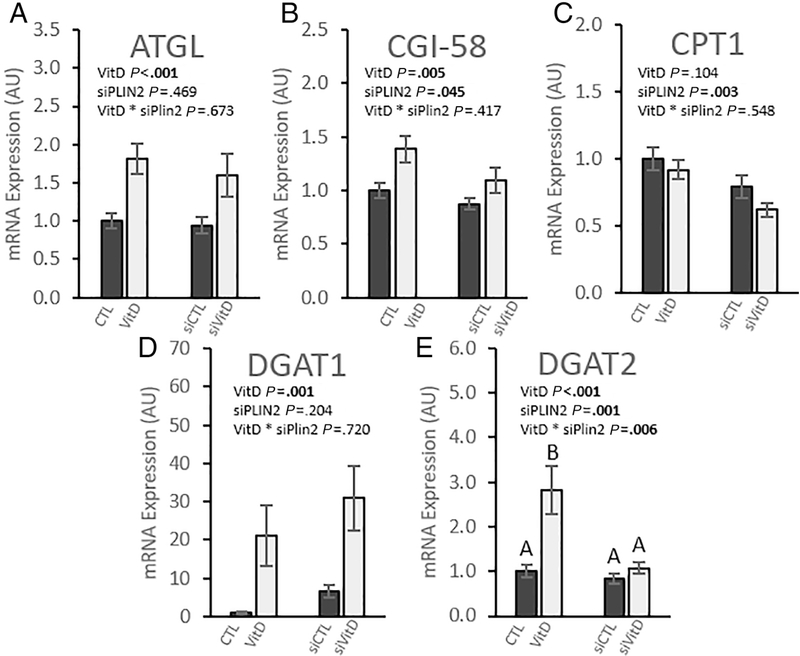
IMCL storage and lipolysis are tightly regulated at the gene level and are associated with both the expression of PLIN2 [9, 41] and VDR [16]. We therefore measured the expression of key regulators of triglyceride storage and lipolysis to determine how they are impacted by simultaneous PLIN2 knockdown and VitD treatment. We found that VitD increased the expression of ATGL (p < 0.001) with no effect of siPLIN2 (Fig. 4A). Comparative gene identification-58 (CGI-58) mRNA expression was upregulated by VitD but decreased after PLIN2 knockdown (p = 0.045) (Fig. 4B). PLIN2 knockdown downregulated carnitine palmityltransferase 1 (CPT1) gene expression (p = 0.003) with VitD associated with a trend to slightly decrease expression (p = 0.104) (Fig. 4C). These results show a pattern of VitD increasing the expression and PLIN2 knockdown decreasing the expression of key genes that regulate lipolysis and β-oxidation.
Fig. 4. PLIN2 knockdown and vitamin D exert opposing effects on genes regulating lipolysis and lipid storage.
Gene expression in differentiated C2C12 myotubes relative to vehicle control with scramble siRNA. Cells were treated with either scramble siRNA and vehicle control (CTL), scramble siRNA and 100 nM calcitriol (VitD), PLIN2 siRNA and vehicle control (siCTL), or PLIN2 siRNA and 100 nM calcitriol (siVitD). All data are represented as mean ± SEM, n = 6. Values not sharing letters are significantly different (Fisher’s LSD). p values represent 2-way ANOVA.
We next examined the expression of genes integral to triglyceride storage in lipid droplets. We found that VitD had a very strong positive effect on diglyceride O-acyltranferase 1 (DGAT1) expression (p < 0.001). While comparisons did not reach statistical significance, PLIN2 knockdown was associated with a mean increase in DGAT1 expression by approximately 500% compared to CTL and 50% compared to VitD (Fig. 4D). Diglyceride O-acyltranferase 2 (DGAT2) was also strongly upregulated with VitD treatment (p < 0.001). We observed an interaction effect between VitD and siPLIN2 (p = 0.006) (Fig. 4E). Post-hoc analysis revealed that DGAT2 expression in VitD treated myotubes was decreased to CTL levels after PLIN2 knockdown.
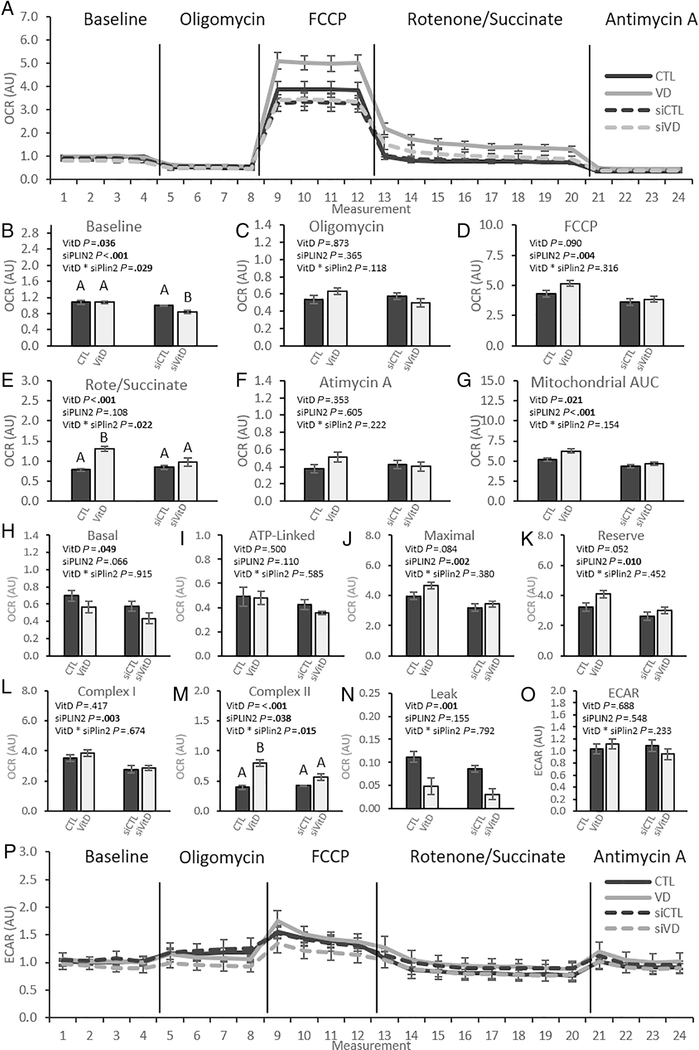
3.5. PLIN2 knockdown prevents VitD induced increases in OCR
To obtain detailed, quantitative analysis of changes in mitochondrial function after treatment with VitD and PLIN2 knockdown, we measured OCR using a Seahorse XFe24 extracellular flux analyzer (Fig. 5A). Simplified values obtained from the full OCR trace (Fig 5A) are shown as bar graphs for ease of interpretation (Figs 5B-F). At baseline, both VitD and siPLIN2 had significant effects on OCR. There was a significant interaction effect (p = 0.029) (Fig 5B). Post-hoc analysis indicated that OCR was decreased by siVitD, with no other differences between groups. Neither VitD nor siPLIN2 produced a significant effect in OCR following the administration of oligomycin (Fig. 5C). VitD did not affect OCR following FCCP injection, but siPLIN2 caused a significant decrease in OCR (Fig. 5D). VitD, but not PLIN2, produced a significant effect following the administration of rotenone and succinate with a significant interaction effect (p = 0.022) (Fig 5E). Post-hoc analysis indicated that VitD increased OCR in comparison to CTL, but the addition of siPLIN2 decreased OCR in the siVitD cells to levels equivalent to siCTL and CTL cells. There were no significant changes in response to either treatment after administration of Antimycin A (Fig 5F).
Fig. 5. Vitamin D increases mitochondrial function and efficiency dependent on PLIN2 upregulation.
C2C12 myotubes were treated with 100 nM calcitriol and PLIN2 siRNA or their respective controls in a 2 × 2 design and OCR was measured by Seahorse XFe24 flux analyzer. (A) OCR was increased throughout measurements after treatment with calcitriol (grey lines) while siPLIN2 blunts OCR (hashed lines). Differences in OCR after each injection were quantified and compared for raw OCR (B-F), OCR specific to mitochondrial activity (H-N), and ECAR (O-P). All data are represented as mean ± SEM, n = 3–4. p values calculated by 2-way ANOVA. Bars not sharing letters are significantly different (Fisher’s LSD). OCR, Oxygen Consumption Rate; ECAR, Extracellular Acidification Rate; FCCP, Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone.
More specific analyses focusing on mitochondrial specific OCR were performed (Figs 5G-N). Following quantification of the area under the curve (AUC) specific to mitochondrial respiration, we found that VitD increased mitochondrial OCR while siPLIN2 decreased OCR (Fig. 5G). VitD decreased basal mitochondrial respiration (p = 0.049), and siPLIN2 produced a trend towards decrease (p = 0.066) (Fig 5H). No significant responses to treatment were observed in ATP-Linked OCR, although siPLIN2 produced a trend towards decreased OCR (p = 0.110) (Fig 5I). VitD produced a trend towards increased Maximal OCR that failed to reach significance (p = 0.084) (Fig 5J). siPLIN2 significantly decreased Maximal OCR. Similarly, VitD yielded a trend towards increased Reserve OCR (p = 0.052), while siPLIN2 caused a significant decrease (Fig 5K). Complex I was unaffected by VitD but was decreased by siPLIN2 (Fig 5L). Both VitD and siPLIN2 had a significant effect on Complex II OCR. Co-treatment of VitD and siPLIN2 produced an interaction effect at complex II wherein VitD treatment alone increases OCR with either siCTL or siRNA, but siPLIN2 treatment decreases the magnitude of the VitD effect (Fig. 5M). VitD decreased mitochondrial leak, an indicator of mitochondrial uncoupling, while siPLIN2 caused a trend towards decreased mitochondrial leak (p = 0.115) (Fig 5N). There were no changes in extracellular acidification rate (ECAR), a measurement of glycolysis (Figs. 5O & 5P). In summary, VitD produced trends towards increased maximal and reserve mitochondrial OCR, a significant increase of OCR driven by complex II, and decreased basal OCR and mitochondrial leak. Conversely, siPLIN2 decreased maximal and reserve OCR, OCR driven by both complex I and complex II, and a trend towards decreased basal OCR. siPLIN2 reversed the VitD-mediated increase of OCR driven by complex II.
4. Discussion
This study was the first to examine the impact of calcitriol or PLIN2 knockout on mitochondrial function in skeletal muscle myotubes and advances our understanding of how these two factors modulate muscle bioenergetics. Data supported the hypothesis that calcitriol improves oxidative metabolism in differentiated skeletal muscle myotubes and that these benefits were partially mediated by the lipid packaging protein PLIN2. We showed that PLIN2 knockdown in myotubes decreased mitochondrial function, but did not appear to prevent IMCL accumulation. Our working hypothesis is illustrated in Fig. 6. This study validates PLIN2 as an important component of mitochondrial metabolism in skeletal muscle, and bioenergic findings reported here should be considered in future studies investigating PLIN2 knockdown or knockout. We speculate that changes in mitochondrial metabolism were driven by increased fatty acid oxidation. Future investigations should examine the relationship between lipolysis, acyl chain import into mitochondria, and β-oxidation in reference to PLIN2 in skeletal muscle.
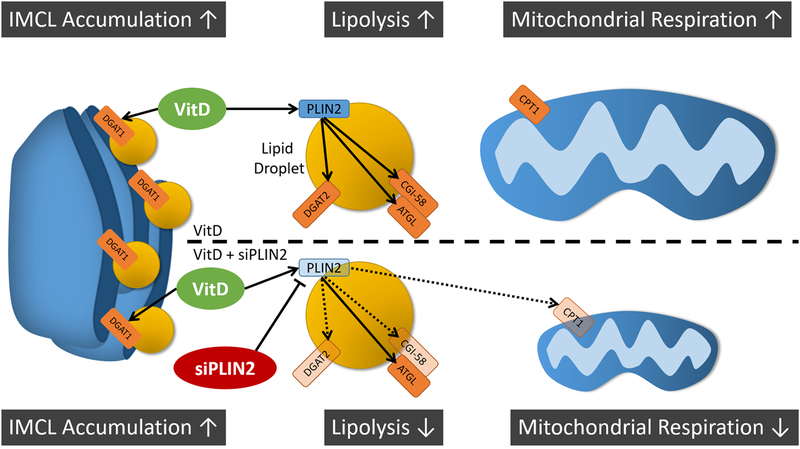
Fig. 6. Graphical abstract and working hypothesis.
VitD treatment increases IMCL accumulation, mRNA associated with both lipid storage and lipolysis, and mitochondrial respiration in skeletal muscle, but increases in lipolytic gene expression and mitochondrial respiration are dependent on PLIN2 expression. Calcitriol treatment (VitD) increases gene expression of lipid droplet proteins PLIN2, ATGL, and CGI-58 (top). VitD also increases mRNA expression for DGAT proteins that localize to both the endoplasmic reticulum for new lipid droplet synthesis (DGAT1) and to mature lipid droplets (DGAT2). These genes together are associated with increased capacity for IMCL accumulation and lipolysis and correspond with increased mitochondrial respiration. PLIN2 knockdown with siRNA before VitD treatment (bottom) decreases the expression of not only PLIN2 but also lipolytic cofactor CGI-58 and lipid droplet refilling gene DGAT2 (indicated by faded shapes and hashed lines). This is associated with decreases in CPT1 mRNA expression and mitochondrial respiration. VitD = Calcitriol; PLIN2 = Perilipin 2; DGAT2 = diglyceride O-acyltransferase 2; DGAT1 = diglyceride O-acyltransferase 1; ATGL = adipose triglyceride lipase; CGI-58 = comparative gene identifier 58; CPT-1 = carnitine palmitoyltransferase 1.
We first showed that calcitriol increases PLIN2 mRNA expression and neutral lipid accumulation in accord with previously published clinical and in vitro studies from our group [16, 20]. Curiously, this appears to be in opposition to a recent publication by Li et al. [42], who showed that a high-vitamin D dietary intervention in mice reduced PLIN2 expression and prevented IMCL accumulation. However, the observed difference reported by Li et al. may be a product of pathological lipid accumulation incited by vitamin D deficiency instead of decreased IMCL with vitamin D supplementation. While vitamin D deficiency and supplementation may have similar effects on PLIN2 expression, evidence suggests the physiological conditions that underlie these scenarios are substantially different. Without markers of mitochondrial activity or β-oxidation, it is difficult to gauge the impact of IMCL on mitochondrial health.
Data obtained in this study through Seahorse oxygen consumption and SDH activity staining support the hypothesis that calcitriol treatment drives improved mitochondrial bioenergetics and that increased PLIN2 expression and IMCL accumulation after calcitriol treatment is not detrimental to mitochondrial function. Others have shown that vitamin D treatments improve mitochondrial function in both clinical and in vitro models. Sinha et al. [25] showed that calcitriol decreases the half time of creatine phosphorylation, a marker of mitochondrial function, in a clinical model. Ryan et al. have shown that calcitriol treatment in human primary myoblasts increases OCR in both healthy and cancerous models [26, 43]. However, differentiation of skeletal muscle triggers substantial changes in mitochondrial substrate management and bioenergetic remodeling [44], and mechanisms in myoblasts cannot be assumed to drive metabolic changes in myotubes after the same treatment.
Consistent with previous work form our group, we showed that calcitriol treatment increased the gene expression of ATLG, CGI-58, DGAT1, and DGAT2 [16]. These genes represent the rate limiting steps of both lipolysis and TAG acylation, and their upregulation suggests that calcitriol increases lipid flux [45]. This potential increase in lipid flux may have beneficial implications for muscle lipid storage that decrease the risk of lipotoxicity [28]. While vitamin D is generally accepted to contribute to the health and function of a variety of tissues, the role of PLIN2 in lipid homeostasis is hotly debated. Many studies suggest that PLIN2 enables steatosis and lipotoxicity [12, 39, 46], but others show that it may have a more beneficial effect, especially in skeletal muscle [14]. The divergence in the impact of PLIN2 may be associated with the role of lipids in the target tissues, specifically, how efficiently lipid is stored and used. We show that PLIN2 knockdown had no impact on the expression of several key lipid management genes when comparing CTL to siCTL myotubes, however, PLIN2 knockdown prevented calcitriol induced expression of CGI-58, CPT1, and DGAT2. CGI-58 is a potent regulator of lipolysis that acts both in conjunction with and independent from ATGL [47–49], suggesting that PLIN2 knockdown impairs lipolytic capacity. On the other hand, others have reported an increase in lipid oxidation in response to PLIN2 knockdown in skeletal muscle [14], which implies an increase in lipolysis. It is of note that PLIN2 also binds to ATGL and prevents the association of CGI-58, thereby decreasing ATGL activity [41]. However, excess ATGL activity is known to increase DAG and ceramide abundance and incite metabolic dysfunction [29]. Therefore, it may be important to increase the expression of DGAT in harmony with ATGL to prevent the accumulation of bioactive signaling lipids.
Our results revealed that both DGAT1 and DGAT2 were upregulated with calcitriol treatment; however, PLIN2 knockdown prevented calcitriol-induced upregulation of DGAT2. Although both DGAT enzymes are important regulators of lipid metabolism, DGAT2 plays a larger role in TAG homeostasis [50] and likely contributes more directly to mitochondrial metabolism. DGAT1 is thought to mediate TAG acylation in the ER and nascent lipid droplets, whereas DGAT2 is found in the ER, cytosolic lipid droplets, and associated with mitochondria [51–53]. This, combined with decreased expression of CPT1, suggests that PLIN2 knockdown may make acyl chains less available for oxidation in mitochondria and exacerbate lipotoxic effects of aberrant ATGL activity.
Changes in the expression of lipolytic and lipid storage genes in response to PLIN2 knockdown and calcitriol imply changes in IMCL accumulation in myotubes. However, while not quantitative, ORO staining presented in this study did not support our hypothesis that PLIN2 is required for IMCL accumulation. Although previous studies have shown that PLIN2 knockout prevents lipid accumulation in multiple tissues [12, 39, 40], we note that many of studies are in vivo studies using complete knockout models. Also, mRNA expression of siCTL vs CTL is only reduced by approximately 30% in this study, a much less substantial decrease in expression than the approximately 75% knockdown observed in previous PLIN2 knockout in C2C12 cells [14]. This level of knockdown may not be sufficient to prevent new LD formation or dramatically reduce IMCL accumulation; however, it does allow us to interpret changes as those produced by the increase in PLIN2 expression opposed to the simple presence of PLIN2.
Our investigation into oxygen consumption uncovered dramatic effects of calcitriol supplementation that were highly dependent on PLIN2 upregulation. This is most easily observed in the increase on OCR measured by AUC after calcitriol treatment that returned to baseline levels with PLIN2 knockdown. Strong trends towards increase at maximal (p = 0.084) and reserve OCR (p = 0.052) provide evidence that calcitriol increases the mitochondrial oxidative capacity of C2C12 myotubes. These increases were both reduced to magnitudes below CTL after PLIN2 knockdown. Altogether, we observed siPLIN2 effects on basal mitochondrial OCR, maximal OCR, reserve OCR, and complexes I and II. Furthermore, all changes in mitochondrial respiration that failed to reach significance after PLIN2 knockdown trended to decease OCR. This provides evidence that increases in OCR observed after treatment with calcitriol are dependent on PLIN2 upregulation. Building on this, because vitamin D drives increases in ATGL and CGI-58 mRNA, and no change in ECAR in response to treatments, we hypothesize that increases in OCR are driven by increased rates of fatty acid oxidation.
The dramatic decrease in mitochondrial leak as a result of calcitriol treatment was combined with no change in the ATP-linked OCR, suggesting that the VitD effect observed at basal mitochondrial OCR may have been driven by decreases in proton leak. As a result, VitD increased the efficiency with which mitochondria use oxygen to produce ATP. Decreased rates of electron leak are also associated with decreased rates of oxidative stress in mitochondria [54], and calcitriol has been shown to decrease oxidative stress and damage in skeletal muscle [55, 56]. Future research should investigate molecular connections between calcitriol and mitochondrial oxidative stress.
We acknowledge the limitation that it is difficult to make strong claims regarding the direct impact of lipid flux in the observed changes in mitochondrial metabolism without direct measurement of fatty acid oxidation. There was also no notable increase in IMCL with ORO staining after VitD treatment as is claimed by previous research [16]. This could be a product of minor differences in lipid availability or more efficient lipid clearance with calcitriol treatment, resulting in less IMCL accumulation with a similar level of palmitate import. There is growing evidence that lipid species are perhaps more important to maintaining cellular function than total lipid abundance, and vitamin D has been shown to change the lipid profile in muscle [16, 42]. Finally, this study did not assess any markers of cellular stress, limiting our ability to make claims regarding the impact of shifts in lipid management after calcitriol treatment.
We conclude that calcitriol treatment in myotubes increases both IMCL storage and mitochondrial function, and that the upregulation of PLIN2 is required to realize metabolic improvements, but not IMCL accumulation. Although PLIN2 is not required for increased IMCL accumulation, mitochondrial function is markedly impaired after PLIN2 knockdown. Our data suggest that PLIN2 knockdown is detrimental to metabolic function in skeletal muscle. These findings contribute to the understanding of how vitamin D regulates mitochondrial function and the roles of PLIN2 in skeletal muscle mitochondrial metabolism.
Supplementary Material
C2C12 cells are seeded overnight then differentiated for a total of 6 days in DMEM with 2% horse serum before treatment with calcitriol for 24 hours. All cells are treated with either 10 nM PLIN2 siRNA or scrambled control siRNA with 1:500 Lipofectamine RNAiMAX starting day 5 for a total of 48 hours.
RER1 = Retention in Endoplasmic Reticulum Sorting Receptor 1; VCP = Valosin Containing Protein; EMC7 = Endoplasmic Reticulum Membrane Protein Complex Subunit 7; VDR = Vitamin D Receptor; ATGL = Adipose Triglyceride Lipase; DGAT1 = Diacylglyceride O-Acyltransferase 1; CGI-58 = Comparative Gene Identification-58; CPT-1 = Carnitine Palmitoyltransferase 1; PLIN2 = Perilipin 2; PLIN3 = Perilipin 3; PLIN5 = Perilipin 5.
Calculations used to derive OCR rates from Seahorse XFe24.
Highlights.
Vitamin D increases PLIN2 gene expression in skeletal muscle
PLIN2 knockdown blunts upregulation of lipid storage and lipolysis genes
Vitamin D increases mitochondrial function and oxygen consumption in skeletal muscle
Augmentation of mitochondrial function by vitamin D is partially dependent on PLIN2
Acknowledgments
Grants, Sponsors, and Funding Sources:
This work was supported by the National Institutes of Health National Institute on Aging [grant number 1R21AG046762-01A1] and Institute of General Medical Sciences [grant number 1P20GM121327-01], and received pilot project funding from the University of Kentucky Center for Muscle Biology. The authors would also like to thank Kate Kosmac, PhD, from the Center for Muscle Biology for assistance with cytochemistry and cell imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy DJ, The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma, 2012. 249(3): p. 541–85. [DOI] [PubMed] [Google Scholar]
- 2.Waltermann M and Steinbuchel A, Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J Bacteriol, 2005. 187(11): p. 3607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Na H, et al. , Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim Biophys Acta, 2015. 1853(10 Pt A): p. 2481–91. [DOI] [PubMed] [Google Scholar]
- 4.Billecke N, et al. , Perilipin 5 mediated lipid droplet remodelling revealed by coherent Raman imaging. Integr Biol (Camb), 2015. 7(4): p. 467–76. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh K, et al. , Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci, 2012. 125(Pt 17): p. 4067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuerschner L, Moessinger C, and Thiele C, Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic, 2008. 9(3): p. 338–52. [DOI] [PubMed] [Google Scholar]
- 7.Sztalryd C and Kimmel AR, Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie, 2014. 96: p. 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pourteymour S, et al. , Perilipin 4 in human skeletal muscle: localization and effect of physical activity. Physiological Reports, 2015. 3(8): p. e12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacPherson REK and Peters SJ, Piecing together the puzzle of perilipin proteins and skeletal muscle lipolysis. Applied Physiology, Nutrition & Metabolism, 2015. 40(7): p. 641–651. [DOI] [PubMed] [Google Scholar]
- 10.Bell M, et al. , Consequences of Lipid Droplet Coat Protein Downregulation in Liver Cells. Diabetes, 2008. 57(8): p. 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Listenberger LL, et al. , Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res, 2007. 48(12): p. 2751–61. [DOI] [PubMed] [Google Scholar]
- 12.McManaman JL, et al. , Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res, 2013. 54(5): p. 1346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte M, et al. , Perilipin 2 and Age-Related Metabolic Diseases: A New Perspective. Trends Endocrinol Metab, 2016. [DOI] [PubMed] [Google Scholar]
- 14.Bosma M, et al. , Perilipin 2 Improves Insulin Sensitivity in Skeletal Muscle Despite Elevated Intramuscular Lipid Levels. Diabetes, 2012. 61(11): p. 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd SO, et al. , Preferential utilization of perilipin 2-associated intramuscular triglycerides during 1 h of moderate-intensity endurance-type exercise. Exp Physiol, 2012. 97(8): p. 970–80. [DOI] [PubMed] [Google Scholar]
- 16.Jefferson GE, et al. , Calcitriol concomitantly enhances insulin sensitivity and alters myocellular lipid partitioning in high fat-treated skeletal muscle cells. J Physiol Biochem, 2017. 73(4): p. 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visser M, Deeg DJ, and Lips P, Low vitamin D and high parathyroid hormone levels as determinants of loss of muscle strength and muscle mass (sarcopenia): the Longitudinal Aging Study Amsterdam. J Clin Endocrinol Metab, 2003. 88(12): p. 5766–72. [DOI] [PubMed] [Google Scholar]
- 18.Bischoff-Ferrari HA, et al. , Higher 25-hydroxyvitamin D concentrations are associated with better lower-extremity function in both active and inactive persons aged > or =60 y. Am J Clin Nutr, 2004. 80(3): p. 752–8. [DOI] [PubMed] [Google Scholar]
- 19.Pojednic RM and Ceglia L, The emerging biomolecular role of vitamin D in skeletal muscle. Exerc Sport Sci Rev, 2014. 42(2): p. 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redzic M, Powell DK, and Thomas DT, Vitamin D status is related to intramyocellular lipid in older adults. Endocrine, 2014. 47(3): p. 854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slawik M and Vidal-Puig AJ, Lipotoxicity, overnutrition and energy metabolism in aging. Ageing Res Rev, 2006. 5(2): p. 144–64. [DOI] [PubMed] [Google Scholar]
- 22.Han J and Kaufman RJ, The role of ER stress in lipid metabolism and lipotoxicity. Journal of Lipid Research, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symons JD and Abel ED, Lipotoxicity contributes to endothelial dysfunction: a focus on the contribution from ceramide. Rev Endocr Metab Disord, 2013. 14(1): p. 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garbarino J and Sturley SL, Saturated with fat: new perspectives on lipotoxicity. Curr Opin Clin Nutr Metab Care, 2009. 12(2): p. 110–6. [DOI] [PubMed] [Google Scholar]
- 25.Sinha A, et al. , Improving the vitamin D status of vitamin D deficient adults is associated with improved mitochondrial oxidative function in skeletal muscle. J Clin Endocrinol Metab, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Ryan ZC, et al. , 1alpha,25-Dihydroxyvitamin D3 Regulates Mitochondrial Oxygen Consumption and Dynamics in Human Skeletal Muscle Cells. J Biol Chem, 2016. 291(3): p. 1514–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta D, et al. , Serum vitamin-D predicts insulin resistance in individuals with prediabetes. Indian J Med Res, 2013. 138(6): p. 853–60. [PMC free article] [PubMed] [Google Scholar]
- 28.Funai K and Semenkovich CF, Skeletal muscle lipid flux: running water carries no poison. American Journal of Physiology - Endocrinology And Metabolism, 2011. 301(2): p. E245–E251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt MJ, Storing up trouble: does accumulation of intramyocellular triglyceride protect skeletal muscle from insulin resistance? Clin Exp Pharmacol Physiol, 2009. 36(1): p. 5–11. [DOI] [PubMed] [Google Scholar]
- 30.Listenberger LL, et al. , Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA, 2003. 100(6): p. 3077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dube JJ, et al. , Exercise-induced alterations in intramyocellular lipids and insulin resistance: the athlete’s paradox revisited. Am J Physiol Endocrinol Metab, 2008. 294(5): p. E882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodpaster BH, et al. , Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J Clin Endocrinol Metab, 2001. 86(12): p. 5755–61. [DOI] [PubMed] [Google Scholar]
- 33.Pruchnic R, et al. , Exercise training increases intramyocellular lipid and oxidative capacity in older adults. Am J Physiol Endocrinol Metab, 2004. 287(5): p. E857–62. [DOI] [PubMed] [Google Scholar]
- 34.Bosma M, et al. , The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem Cell Biol, 2012. 137(2): p. 205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosma M, et al. , Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. Biochim Biophys Acta, 2013. 1831(4): p. 844–52. [DOI] [PubMed] [Google Scholar]
- 36.Covington JD, et al. , Skeletal Muscle Perilipin 3 and Coatomer Proteins Are Increased following Exercise and Are Associated with Fat Oxidation. PLoS ONE, 2014. 9(3): p. e91675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gemmink A, et al. , Dissociation of intramyocellular lipid storage and insulin resistance in trained athletes and type 2 diabetes patients; involvement of Perilipin 5? J Physiol, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conte M, et al. , Increased Plin2 expression in human skeletal muscle is associated with sarcopenia and muscle weakness. PLoS One, 2013. 8(8): p. e73709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carr RM, et al. , Absence of perilipin 2 prevents hepatic steatosis, glucose intolerance and ceramide accumulation in alcohol-fed mice. PLoS One, 2014. 9(5): p. e97118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libby AE, et al. , Perilipin-2 Deletion Impairs Hepatic Lipid Accumulation by Interfering with Sterol Regulatory Element-binding Protein (SREBP) Activation and Altering the Hepatic Lipidome. J Biol Chem, 2016. 291(46): p. 24231–24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacPherson REK, et al. , Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology, 2013. 304(8): p. R644–R650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, et al. , Vitamin D prevents lipid accumulation in murine muscle through regulation of PPARgamma and perilipin-2 expression. J Steroid Biochem Mol Biol, 2017. [DOI] [PubMed] [Google Scholar]
- 43.Ryan ZC, et al. , 1alpha,25-dihydroxyvitamin D3 mitigates cancer cell mediated mitochondrial dysfunction in human skeletal muscle cells. Biochem Biophys Res Commun, 2018. 496(2): p. 746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malinska D, et al. , Changes in mitochondrial reactive oxygen species synthesis during differentiation of skeletal muscle cells. Mitochondrion, 2012. 12(1): p. 144–8. [DOI] [PubMed] [Google Scholar]
- 45.Marcotorchino J, et al. , Vitamin D protects against diet-induced obesity by enhancing fatty acid oxidation. J Nutr Biochem, 2014. 25(10): p. 1077–83. [DOI] [PubMed] [Google Scholar]
- 46.Chen E, et al. , PLIN2 is a Key Regulator of the Unfolded Protein Response and Endoplasmic Reticulum Stress Resolution in Pancreatic beta Cells. Sci Rep, 2017. 7: p. 40855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweiger M, et al. , Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem, 2006. 281(52): p. 40236–41. [DOI] [PubMed] [Google Scholar]
- 48.Lass A, et al. , Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab, 2006. 3(5): p. 309–19. [DOI] [PubMed] [Google Scholar]
- 49.Lord CC, et al. , Regulation of Hepatic Triacylglycerol Metabolism by CGI-58 Does Not Require ATGL Co-activation. Cell Rep, 2016. 16(4): p. 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yen CL, et al. , Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res, 2008. 49(11): p. 2283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu N, et al. , The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol, 2012. 198(5): p. 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McFie PJ, et al. , Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J Biol Chem, 2011. 286(32): p. 28235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stone SJ, et al. , The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem, 2009. 284(8): p. 5352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cadenas S, Mitochondrial uncoupling, ROS generation and cardioprotection. Biochim Biophys Acta, 2018. [DOI] [PubMed] [Google Scholar]
- 55.Ke CY, et al. , Vitamin D3 Reduces Tissue Damage and Oxidative Stress Caused by Exhaustive Exercise. Int J Med Sci, 2016. 13(2): p. 147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dzik K, et al. , Vitamin D supplementation attenuates oxidative stress in paraspinal skeletal muscles in patients with low back pain. Eur J Appl Physiol, 2018. 118(1): p. 143–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C2C12 cells are seeded overnight then differentiated for a total of 6 days in DMEM with 2% horse serum before treatment with calcitriol for 24 hours. All cells are treated with either 10 nM PLIN2 siRNA or scrambled control siRNA with 1:500 Lipofectamine RNAiMAX starting day 5 for a total of 48 hours.
RER1 = Retention in Endoplasmic Reticulum Sorting Receptor 1; VCP = Valosin Containing Protein; EMC7 = Endoplasmic Reticulum Membrane Protein Complex Subunit 7; VDR = Vitamin D Receptor; ATGL = Adipose Triglyceride Lipase; DGAT1 = Diacylglyceride O-Acyltransferase 1; CGI-58 = Comparative Gene Identification-58; CPT-1 = Carnitine Palmitoyltransferase 1; PLIN2 = Perilipin 2; PLIN3 = Perilipin 3; PLIN5 = Perilipin 5.
Calculations used to derive OCR rates from Seahorse XFe24.