Abstract
Purpose
Alternations to the paternal epigenome, specifically the components of sperm chromatin, can lead to infertility in humans and potentially transmit aberrant information to the embryo. One key component of sperm chromatin is the post-translational modification of histones (PTMs). We previously identified a comprehensive profile of histone PTMs in normozoospermic sperm; however, only specific histone PTMs have been identified in abnormal sperm by antibody-based approaches and comprehensive changes to histone PTM profiles remain unknown. Here, we investigate if sperm with abnormalities of total motility, progressive motility, and morphology have altered histone PTM profiles compared to normozoospermic sperm samples.
Methods
Discarded semen samples from 31 men with normal or abnormal semen parameters were analyzed for relative abundance of PTMs on histone H3 and H4 by “bottom-up” nano-liquid chromatography-tandem mass spectrometry.
Results
Asthenoteratozoospermic samples (abnormal motility, forward progression, and morphology, n = 6) displayed overall decreased H4 acetylation (p = 0.001) as well as alterations in H4K20 (p = 0.003) and H3K9 methylation (p < 0.04) when compared to normozoospermic samples (n = 8). Asthenozoospermic samples (abnormal motility and progression, n = 5) also demonstrated decreased H4 acetylation (p = 0.04) and altered H4K20 (p = 0.005) and H3K9 methylation (p < 0.04). Samples with isolated abnormal progression (n = 6) primarily demonstrated decreased acetylation on H4 (p < 0.02), and teratozoospermic samples (n = 6) appeared similar to normozoospermic samples (n = 8).
Conclusion
Sperm samples with combined and isolated abnormalities of total motility, progressive motility, and morphology display distinct and altered histone PTM signatures compared to normozoospermic sperm. This provides evidence that alterations in histone PTMs may be important for normal sperm function and fertility.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1354-7) contains supplementary material, which is available to authorized users.
Keywords: Histones, Post-translational modifications, Paternal epigenetics, Sperm, Male infertility
Introduction
Infertility is a common problem in the USA, affecting up to 15% of couples attempting conception [1]. While the etiology of infertility is complex and often attributable to multiple causes, a male factor may contribute to 50% of cases [2]. A male factor is diagnosed when any semen parameter falls outside of the reference ranges of the semen analysis [3]. Indeed, in addition to obvious causes of infertility such as oligozoospermia, abnormalities in overall sperm motility, progressive motility, and morphology have been associated with poor clinical outcomes [4–8]. Current understanding of the molecular mechanisms leading to these defects and their reproductive implications is limited. Recently, however, the role of paternal epigenetics is emerging as a crucial factor in the etiology and reproductive consequences of male infertility.
One key epigenetic mechanism regulating male gamete formation and function involves the post-translational modification and retention of histones, which are critical protein components that provide the structural scaffold of chromatin. Chromosomal DNA is packaged into nucleosomes, each containing approximately 147 bp of DNA wrapped around a histone octamer containing two copies each of core histone H2A, H2B, H3, and H4 [9]. The covalent addition of post-translational modifications (PTMs), such as methylation (me), acetylation (ac), phosphorylation (ph), and crotonylation (cr), to predominantly the N-terminal tails of the core histones allows for activation or repression of underlying genes [10]. Histone PTMs are crucial to the regulation of cellular processes essential to spermatogenesis and sperm function, including transcription, DNA repair, DNA replication, and chromosome condensation [11]. Interestingly, the majority of histones (85–95%) are evicted during sperm maturation, which facilitates DNA compaction within the sperm nucleus. This unique process, termed spermiogenesis, begins in post-meiotic haploid male germ cells with the eviction of hyperacetylated histones and replacement with, first, slightly smaller, and positively charged transition proteins, and then ultimately by very small and highly arginine-rich protamines [12–14]. In human sperm, approximately 5–15% of histones are retained and proper histone-to-protamine exchange is critical for normal spermiogenesis, as aberrations in this replacement process are associated with male infertility and poor outcomes following in vitro fertilization [12, 15–22]. Initial genome-wide studies in human sperm suggested that the retention of histones occurs in a non-random manner, being enriched at genes encoding master regulators of early embryonic development [23–26]. Other reports, however, subsequently showed that histones are predominantly retained within distal intergenic regions and introns, and are associated with centromeric repeats and retrotransposons [27, 28]. This current controversy about the role of retained nucleosomes in sperm illustrates the importance of further understanding the histone signature in human sperm.
While most previous studies of sperm histone PTMs utilized antibody-based approaches to evaluate a handful of specific PTMs, high-resolution “bottom-up” nano-liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) allows for a comprehensive assessment of histone PTM signatures over short peptides. Mass spectrometry has emerged as a high-throughput technology to analyze histones and PTMs without prior knowledge of specific modifications [29]. Given its unbiased and quantitative nature, this technology is valuable for comparison of specific histone PTMs between study groups. We previously utilized this approach to identify dynamic changes in histone PTMs during multiple stages of mouse spermatogenesis and also found conservation of histone H3 and H4 PTMs between mouse and human sperm [30]. Remarkably, we observed a striking consistency in the abundance of histone PTMs between normozoospermic sperm samples from different individuals [30].
Given increasing evidence of the importance of the paternal epigenome, the objective of this study was to determine whether semen pathologies correlate with altered histone PTM signatures. We first examined sperm samples displaying the most severe phenotype of abnormalities in total motility, progressive motility, and morphology (asthenoteratozoospermia, AT, mot/prog/morph). We subsequently investigated samples with less severe and isolated abnormalities including total motility and progressive motility (asthenozoospermia, AS, mot/prog), isolated abnormal progressive motility (PR, prog), and isolated abnormal morphology (teratozoospermia, TS, morph), for comparison to normozoospermic (NS) samples.
Materials and methods
Ethical approval
Approval for this study was obtained from the University of Pennsylvania Institutional Review Board (Protocol 815929).
Collection and processing of semen samples
De-identified semen samples were obtained from the University of Pennsylvania Fertility Center (Penn Fertility Care) from men presenting for routine semen analysis. Following 2–5 days of abstinence, semen samples were collected via masturbation into a sterile container. Samples were allowed to liquefy at room temperature for 30 min before analysis by trained andrology staff. Semen parameters were assessed according to the WHO reference values, 5th Edition [3] which uses the strict Kruger criteria for morphology assessment [31]. Semen samples grouped as normal were reported in our initial characterization of sperm histone PTMs [30] and met all of the following criteria: semen volume ≥ 1.5 mL, sperm concentration ≥ 15 million/mL, total motility ≥ 40%, progressive motility ≥ 32%, and morphology ≥ 4% [3]. Abnormal samples with a value below any of the above thresholds were grouped according to the abnormality (asthenoteratozoospermia, asthenozoospermia, abnormal progressive motility, and teratozoospermia, Table 1). As approximately 25–30 million total sperm are required for sufficient histone extraction for nanoLC-MS/MS analysis, samples with oligozoospermia were excluded from evaluation. Following semen analysis, de-identified semen samples were processed immediately. Briefly, total semen samples were first washed three times with PBS followed by centrifugation at 200 rcf to separate seminal fluid. The somatic cells in the resulting pellet were lysed (0.1% SDS, 0.5% Triton-X-100) for 30 min on ice. Samples were again centrifuged at 200 rcf for 10 min and the resulting sperm pellet resuspended in PBS. Microscopic inspection of the sperm was then performed to validate complete somatic cell lysis and total sperm count performed. In our previous study, we utilized pyrosequencing analysis of DNA methylation of maternal and paternal imprinted genes in mouse sperm to validate somatic cell lysis and purity of sperm chromatin samples [30].
Table 1.
Semen parameters of grouped samples including concentration, total motility, progressive motility (PR), and morphology. Values expressed as mean ± standard deviation
| Semen parameter | Normozoospermic (n = 8) | Asthenoteratozoospermic (n = 6) | Asthenozoospermic (n = 5) | Abnormal PR (n = 6) | Teratozoospermic (n = 6) |
|---|---|---|---|---|---|
| Concentration (× 106/mL) | 56.25 ± 16.11 | 34.33 ± 13.72 | 32.00 ± 22.2 | 21.33 ± 4.72 | 30.17 ± 6.37 |
| Total motility (%) | 53.63 ± 8.26 | 27.00 ± 7.40 | 28.80 ± 11.14 | 49.67 ± 9.16 | 51.67 ± 5.01 |
| Progressive motility (%) | 40.25 ± 5.78 | 13.17 ± 4.26 | 20.4 ± 21.67 | 21.67 ± 5.24 | 35.17 ± 1.82 |
| Morphology (% normal forms) | 5.00 ± 0.93 | 2.33 ± 0.82 | 5.00 ± 0.71 | 5.67 ± 1.63 | 1.67 ± 0.52 |
Acid extraction, propionic anhydride derivatization, and trypsin digestion of histones
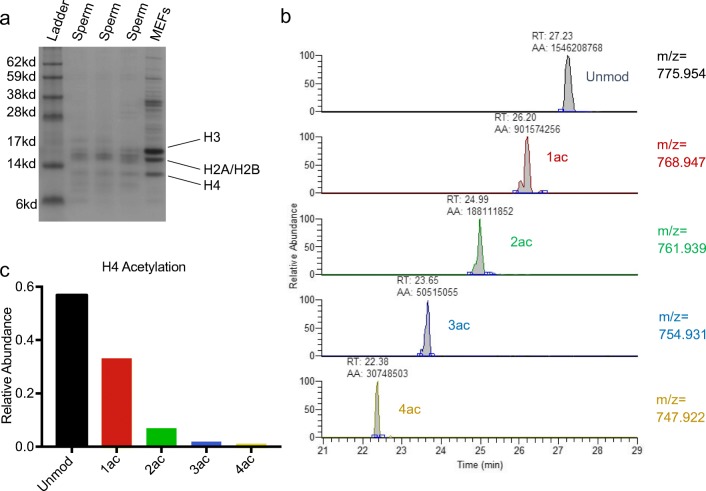
Acid extraction of histones for nanoLC-MS/MS analysis was conducted as previously described with minor modifications [30, 32, 33]. Briefly, following somatic cell lysis, sperm samples were treated with DTT (50 mM) for 30 min to aid in nuclear decondensation. Following centrifugation at 800 rcf for 5 min at 4 °C, the supernatant was removed and sperm cells were rotated at 4 °C for 30 min in hypotonic lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT with protease inhibitors). Pelleted nuclei (10,000 rcf, 10 min, 4 °C) were subsequently resuspended in 0.4 M sulfuric acid and rotated overnight at 4 °C. Following centrifugation (16,000 rcf, 10 min, 4 °C), the resulting supernatant was precipitated with 33% trichloroacetic acid (TCA) and incubated on ice for 4–6 h. Histones were pelleted by centrifuging at 16,000 rcf for 10 min at 4 °C and subsequently washed twice with cold acetone prior to resuspension in DNase/RNase free water and storage at − 80. To confirm adequate extraction, resuspended histones were quantified using the Pierce BCA Protein Assay Kit (ThermoScientific) and 1–2 μg of protein run on a 12% Bis-Tris protein gel, comparing to control histones extracted and purified from mouse embryonic fibroblasts (Fig. 1a).
Fig. 1.
Example of nanoLC-MS/MS Chromatogram of Histone H4 peptide aa 4–17 from human sperm and calculation of relative abundance. a Coomassie stained protein gel of acid extracted proteins from sperm samples and mouse embryonic fibroblasts (MEFs) demonstrating extraction of histones H3, H2A/H2B, and H4. b Chromatogram of histone H4 peptide aa 4–17 and its modified forms for 2+ charge. Peaks representing each modified form are identified via mass charge ratio (m/z) and retention time (RT). Automated area (AA) represents area under the curve calculated for each peak. c Bar graph demonstrating calculated relative abundance. The sum of the AA for all modified forms of the peptide in all three charge states is calculated and the relative abundance of each unmodified and modified form expressed as a percentage of the total
To prepare extracted histones for nanoLC-MS/MS, 20 μg of acid-extracted protein was used for propionylation and trypsin digestion. Each sample was treated twice with propionylation reagent (1-part propionic anhydride (Sigma), 3-parts 2-proponal) at pH 8 to block the ɛ-amino groups of unmodified and monomethyl lysine residues, which then restricts trypsin proteolysis to only the C-terminal side of arginine residues. Peptides were digested with 1 μg trypsin (1:20 trypsin/protein ratio, Invitrogen) for 6 h at 37 °C and subsequently treated with two additional rounds of propionylation. The resulting propionylated and trypsin digested peptides were purified via stage-tip desalting with C18 mini discs and stored at − 80 °C until nanoLC-MS/MS analysis.
Tandem mass spectrometry
Desalted histone peptides were loaded onto and separated by reversed-phase HPLC on a Thermo Scientific™ EASY-nLC 1000 system with a 75 μm i.d. × 15 cm Reprosil-Pur C18-AQ 3 μm nanocolumn run at 300 nL/min. Peptides were eluted with a gradient from 2 to 30% ACN (35 min) and to 98% ACN over 20 min in 0.1% formic acid. HPLC was coupled to a Thermo Scientific™ Q Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer. In each cycle, one full MS Orbitrap detection was performed with the scan range of 290 to 1600 m/z, a resolution of 70 K and AGC of 1 × e6. Then, data-dependent acquisition mode was applied with a dynamic exclusion of 30 s. MS2 scans were followed on parent ions from the most intense ones. Ions with a charge state of one were excluded from MS/MS. An isolation window of 3 m/z was used. Ions were fragmented using higher-energy collisional dissociation (HCD) with a collision energy of 24. The resolution was set to be 17.5 K with AGC of 1 × e5. Targeted scans were performed on a number of peptides to increase the identification of low-abundance modifications. Histone PTM quantification was performed by manual quantification utilizing retention time, MS1 and MS2 of ion peaks. The following peptides were analyzed for each sample: H3 aa 3–8 (TKQTAR), aa 9–17 (KSTGGKAPR), aa 18–26 (KQLATKAAR), aa 27–40 (KSAPATGGVKKPHR), aa 73–83 (EIAQDFKTDLR), and H4 aa 4–17 (GKGGKGLGKGGAKR), aa 20–23 (KVLR), and the miscleaved aa 18–23 (HRKVLR).
Statistical analysis
To quantify the relative abundance of histone PTMs, the area under each peak in the MS chromatogram was measured for the [M + H]+, [M + 2H]2+, and [M + 3H]3+ ions. The sum of all modified forms was designated as 100% and the relative quantity of each PTM calculated by dividing the area of each modified form by the total area. Given the small sample size in each group, the non-parametric Mann-Whitney test was used to assess for differences in the relative abundance of normozoospermic vs. abnormal groups. A p value < 0.05 was considered statistically significant. Data analysis was performed using STATA version 13.1 (StataCorp, College Station, TX, USA).
Results
Semen samples
A total of 31 semen samples were included for analysis. As described in the “Materials and methods,” a formal semen analysis was performed by the Penn Andrology Laboratory on each sample according to the WHO 5th edition guidelines prior to processing for histone analysis. Baseline semen characteristics for each group are summarized in Table 1 and semen parameters for each individual are presented in Supplemental Table 1. Individual samples were assigned to one of the following groups: normozoospermic (n = 8), asthenoteratozoospermic (AT, n = 6), asthenozoospermic (AS, n = 5), isolated abnormal progressive motility (PR, n = 6), or teratozoospermic (TS, n = 6).
Quantitative analysis of histone PTMs utilizing mass spectrometry
We utilized nanoLC-MS/MS to perform an unbiased and quantitative comparison of particular histone PTMs between normozoospermic samples and sperm samples with various clinical abnormalities. Figure 1 demonstrates an example of this methodology for the calculation of relative abundance of specific PTMs. Following gel-electrophoretic confirmation of proper acid extraction of histones (Fig. 1a), samples were prepared for nanoLC-MS/MS. The resulting chromatogram was inspected and the correct peak identified via mass charge ratio (m/z), retention time (RT), MS1 and MS2, as indicated for an example of acetylated forms of the histone H4 peptide 4–13 (Fig. 1b). The area under the curve (AA) was calculated for each modified and unmodified form of the peptide and summed to obtain total peptide abundance. The relative abundance of each form was expressed as a percentage of the total (Fig. 1c).
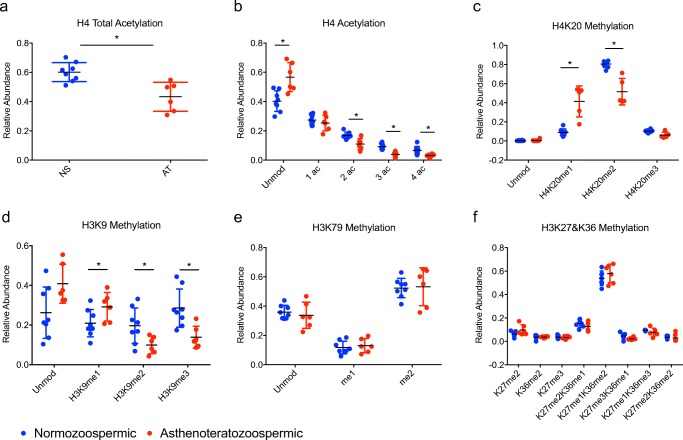
Histone PTM profile in normozoospermic vs. asthenoteratozoospermic samples
We first compared the most severe phenotype of asthenoteratozoospermia (AT, mot/prog/morph) with normozoospermic samples (NS) (Fig. 2). Interestingly, we found a significant decrease in overall acetylation on histone H4 aa 4–17 (p = 0.001) (Fig. 2a). Of note, when examining specifically modified amino acids, a significant decrease in di-, tri-, and quadruple acetylation was noted on this peptide (p = 0.01, 0.002, 0.007, respectively) as well as a corresponding increase in the unmodified form (p = 0.007) (Fig. 2b). In addition, a significant increase in the monomethylated form of H4K20 (H4K20me1) and a decrease in the dimethylated form H4K20me2 (p = 0.003, 0.003) was detected (Fig. 2c). On histone H3, there was a significant increase in monomethylated K9 (H3K9me1) (p = 0.04) and a decrease in di- and tri-methylated K9 (H3K9me2, H3K9me3) (p = 0.01, 0.005, respectively) (Fig. 2d). Although there was a trend towards an increase in the unmodified form of the H3K9 peptide in the AT (mot/prog/morph) group, this did not reach statistical significance (p = 0.07). There was also no difference in the acetylated form of H3K14 between AT (mot/prog/morph) and NS (data not shown). The relative abundance of the remainder of the modifications examined [including H3K79, H3K27, H3K36, and H3K79 (Fig. 2e, f) and on H3K4, H3K18, H3K23 (data not shown)] was not different between AT and normal sperm, showing clear specificity for the changes noted above.
Fig. 2.
Relative abundance of histone PTMs on H4 and H3 in normozoospermic vs. asthenoteratozoospermic sperm samples. Dot plot comparing the relative abundance of PTMs on histones H4 and H3 between normozoospermic (NS) vs. asthenoteratozoospermic (AT) sperm samples: a) Total acetylation on H4 b) Number of acetyl groups on H4 c) H4K20 methylation d) H3K9 methylation e) H3K79 methylation f) H3K27 & K36 methylation. *p < 0.05
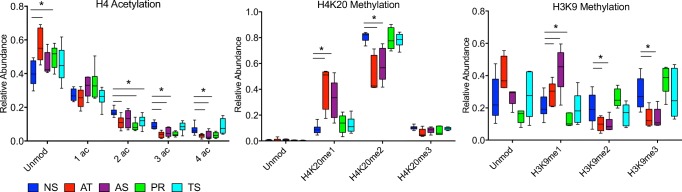
Histone PTM profile in asthenozoospermic, teratozoospermic, and samples with isolated abnormal progressive motility
Given the striking changes in relative abundance observed in H4 acetylation, H4K20 methylation, and H3K9 methylation between asthenoteratozoospermic (AT, mot/prog/morph) and normozoospermic (NS) samples, we next investigated whether changes in these modifications were associated with other isolated and less severe combinations of semen abnormalities. We performed a similar analysis on samples with asthenozoospermia (AS, mot/prog), isolated abnormal progressive motility (PR, prog), and teratozoospermia (TS, morph) and compared the PTM profile in each group to the normozoospermic (NS) samples (Fig. 3). Similar to the AT (mot/prog/morph) group, both the AS (mot/prog) and PR (prog) groups showed a significant decrease in tri- and quadruple acetylation on H4 (AS, p = 0.03, 0.04; PR, p = 0.002, 0.02 respectively) (Fig. 3a). The PR (prog) group also demonstrated a significant increase in the unmodified form (p = 0.02) and a significant decrease in the di-acetylated form of H4 (p = 0.003). Sperm with abnormal morphology (TS) exhibited histone PTM profiles similar to the NS group with the exception of a significant decrease in the di-acetylated form of the H4 peptide (p = 0.01). When examining H4K20 methylation, the AS (mot/prog) group was strikingly similar to the AT (mot/prog/morph) group, exhibiting a significant increase in H4K20me1 and a decrease in H4K20me2 (p = 0.005, 0.005) (Fig. 3b). There was no difference in the relative abundance of histone PTMs from the PR (prog) and TS (morph) groups for the H4K20 peptide.
Fig. 3.
Relative abundance of histone PTMs on H4 and H3 in normozoospermic vs. samples with multiple and isolated abnormalities of motility and progression. Box and whisker plot comparing relative abundance of histone PTMs on H4 aa 4–17, H4 aa 20–23, and H3 aa 9–17 in normozoospermic vs. asthenoteratozoospermic (AT), asthenozoospermic (AS), isolated abnormal progressive motility (PR), and teratozoospermic (TS) sperm samples. *p < 0.05
Analysis of the H3K9 peptide revealed the most distinct changes occurring in the AS (mot/prog) group (Fig. 3c). H3K9me1 increased in the AS (mot/prog) group (p = 0.01), and H3K9me2 and me3 were decreased compared to the NS group (p = 0.04, 0.01, respectively). While an increase in H3K9me1 was observed in the PR (prog) group (p = 0.02), there were no other differences in the abundance of the modified or unmodified forms of the H3K9 peptide. Furthermore, no differences were observed between the TS (morph) and NS group on this peptide. Hence, specific classes of semen abnormalities appear to have distinctly altered histone PTM signatures.
Discussion
Here, we employ highly sensitive nanoLC-MS/MS and reveal a novel and irregular histone PTM signature associated with clinically defined abnormal semen samples. We compared the histone PTM profile of normozoospermic sperm samples with abnormal samples displaying combined or isolated abnormalities of total motility, progressive motility, and morphology. We identified striking differences in the relative abundance of H4 acetylation as well as H4K20 and H3K9 methylation in asthenoteratozoospermic (mot/prog/morph) samples exhibiting all three abnormalities. Further investigation of sperm samples with less severe combined and isolated abnormalities demonstrated that decreased histone acetylation on H4 was associated with asthenozoospermia (mot/prog) and isolated abnormal progressive motility. Alterations of H3K9 and H4K20 methylation were also found in asthenozoospermic (mot/prog) samples. In contrast, teratozoospermic sperm samples were found to have similar PTM profiles to normozoospermic samples.
Decreases in histone H4 acetylation were noted with almost every semen abnormality evaluated in this study. Histone acetylation is traditionally considered a PTM associated with transcriptional activation; however, in sperm, hyperacetylation of histones is also critical to the histone-to-protamine exchange process during spermiogenesis [34–40]. Furthermore, decreased acetylation of H4 has been described in men with impaired spermatogenesis including those with both qualitatively normal appearing sperm on testicular biopsy as well as those with more severe impairments of spermiogenesis [40]. This is in agreement with our findings of decreased H4 acetylation in clinically abnormal appearing sperm and underscores the importance of proper histone acetylation for normal spermiogenesis and fertility. Of note, due to our experimental and analytical design that relies upon relative histone abundance, we are unable to determine if this decrease in histone acetylation is due to an overall increase in unmodified histone retention or a decrease in histone acetylation. Future experiments that utilize alternative means to assess histone retention/protamine incorporation in sperm followed by nanoLC-MS/MS are needed to definitively answer this question.
We further identified significant changes in the abundance of H4K20 and H3K9 methylation in both asthenoteratozoospermic (mot/prog/morph) and asthenozoospermic (mot/prog) sperm samples. H4K20 methylation is implicated in a diverse number of biological processes, including DNA replication and the DNA damage response. Furthermore, alterations with this PTM are associated with a variety of disease states including cancer and developmental disorders [41, 42]. H4K20 and H3K9 methylation are also considered hallmarks of heterochromatin with the combination of H4K20 and H3K9 methylation leading to a highly condensed and transcriptionally repressed state [11, 43]. Proper methylation and de-methylation of H3K9 are also suggested to be essential for the normal progression of spermatogenesis and for the incorporation of both transition proteins and protamines [35]. This has been demonstrated through mouse models with either loss of function of specific demethylases or mutations in methyltransferases resulting in severe spermatogenic defects or complete meiotic arrest [44, 45]. Importantly, recent evidence suggests that H3K9me3 in association with H4K20me3 from paternal heterochromatin is transmitted from the sperm to the embryo in the human [46]. Our identification of altered H3K9me and H4K20me in asthenoteratozoospermic (mot/prog/morph) and asthenozoospermic (mot/prog) sperm samples further highlights the potential importance of these specific histone PTMs as well as the paternal epigenome on fertility and embryogenesis in both normal and pathologic clinical conditions.
The majority of research on histones in the setting of fertility has focused on proper histone-to-protamine exchange and the establishment of protamine 1:2 ratios [16–19]. There is, however, minimal data specifically investigating the role of histone PTMs in the setting of human infertility, and the available studies are generally limited to a handful of specific PTMs. Shifts in the distribution of H3K9 acetylation have been described at selected genes between the sperm of fertile and infertile men [47]. In addition, H4K12 acetylation has also been examined in the setting of male infertility, with a loss of binding sites demonstrated in select developmentally important promoters in sperm from subfertile men [48]. Interestingly, in a group of seven patients with either poor embryogenesis following IVF or altered protamination, a more dispersed pattern of histones was found when compared with control sperm from fertile men. Furthermore, while the localization of modified histones (H3K4me and H3K27me) was overall similar to controls, subtle differences between the two groups included either loss or significant reduction of developmental promoters with H3K4me and H3K27me, as well as the loss of H3K4me3 from three paternally imprinted loci in infertile men [49]. While we were unable to determine the clinical outcomes of included subjects due to the de-identified nature of our study, our analysis similarly did not reveal a difference in the overall abundance of H3K27 methylation in normozoospermic samples when compared to samples with abnormalities.
In conclusion, we utilized nanoLC-MS/MS to identify distinct differences in the abundance of histone PTMs in sperm with both combined and isolated abnormalities of total motility, progressive motility, and morphology when compared to normozoospermic sperm samples. The identification of a unique PTM signature associated with particular clinical abnormalities is of significant translational importance because sperm with phenotypic abnormalities may carry improperly marked chromatin into the oocyte. Additional studies utilizing next-generation sequence techniques such as MNase or ChIP-seq are necessary to determine the specific alterations in genomic locations of the altered histone PTMs identified in this study. Localization of these modifications to genomic features, i.e., at promoter or enhancers, will shed further light onto the role of these modifications in embryogenesis. The increased use of assisted reproductive technologies such as IVF and ICSI to circumvent male infertility highlights the importance of improving our understanding of the paternal epigenome in sperm function and pathology. While further studies are needed to investigate the functional significance of our findings, the definition of histone PTM profiles is critical at this early stage of understanding of abnormalities and provide an avenue for additional research. Indeed, future research will elucidate the dynamic mechanisms involved in the establishment of these PTMs (i.e., enzyme HATs and HDACs) and how these mechanisms are altered in spermatogenesis and infertility.
Electronic supplementary material
(DOCX 19 kb)
Acknowledgements
We would like to thank all of the staff at the Penn Fertility Care Andrology laboratory for their assistance, time, and effort in the de-identification of samples used in this study. In addition, we would like to thank The Penn Center for the Study of Epigenetics in Reproduction.
Funding
This work was funded by the National Institutes of Health Grants P50HD06817 (S.L.B./M.S.B./C.C/L.J.L), T32HD040135 (S.B.S/C.C), 5K12HD065257-07 (S.B.S), F32HD086939 (L.J.L.), and GM110174 (B.A.G).
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Approval for this study was obtained from the University of Pennsylvania Institutional Review Board (Protocol 815929).
Informed consent
As all samples were discarded and de-identified, the University of Pennsylvania Institutional Review Board determined that this study was exempt from requiring informed consent.
Contributor Information
Samantha B. Schon, Email: sschon@umich.edu
Shelley L. Berger, Phone: 215-746-3106, Email: bergers@upenn.edu
References
- 1.Stephen EH, Chandra A. Declining estimates of infertility in the United States: 1982–2002. Fertil Steril. 2006;86:516–523. doi: 10.1016/j.fertnstert.2006.02.129. [DOI] [PubMed] [Google Scholar]
- 2.Thonneau P, Marchand S, Tallec A, Ferial ML, Ducot B, Lansac J, Lopes P, Tabaste JM, Spira A. Incidence and main causes of infertility in a resident population (1,850,000) of three French regions (1988-1989) Hum Reprod. 1991;6:811–816. doi: 10.1093/oxfordjournals.humrep.a137433. [DOI] [PubMed] [Google Scholar]
- 3.Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HWG, Behre HM, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 4.Larsen L. Computer-assisted semen analysis parameters as predictors for fertility of men from the general population. Hum Reprod. 2000;15:1562–1567. doi: 10.1093/humrep/15.7.1562. [DOI] [PubMed] [Google Scholar]
- 5.Nallella KP, Sharma RK, Aziz N, Agarwal A. Significance of sperm characteristics in the evaluation of male infertility. Fertil Steril. 2006;85:629–634. doi: 10.1016/j.fertnstert.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Gunalp S, Onculoglu C, Gurgan T, Kruger TF, Lombard CJ. A study of semen parameters with emphasis on sperm morphology in a fertile population: an attempt to develop clinical thresholds. Hum Reprod. 2001;16:110–114. doi: 10.1093/humrep/16.1.110. [DOI] [PubMed] [Google Scholar]
- 7.Vawda AI, Gunby J, Younglai EV. Andrology: semen parameters as predictors of in-vitro fertilization: the importance of strict criteria sperm morphology. Hum Reprod. 1996;11:1445–1450. doi: 10.1093/oxfordjournals.humrep.a019417. [DOI] [PubMed] [Google Scholar]
- 8.Verheyen G, Tournaye H, Staessen C, De Vos A, Vandervorst M, Van Steirteghem A. Controlled comparison of conventional in-vitro fertilization and intracytoplasmic sperm injection in patients with asthenozoospermia. Hum Reprod. 1999;14:2313–2319. doi: 10.1093/humrep/14.9.2313. [DOI] [PubMed] [Google Scholar]
- 9.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7:a025064–a025022. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res. 1978;117:347–356. doi: 10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- 13.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 14.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 15.Gatewood J, Cook G, Balhorn R, Bradbury E, Schmid C. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X. Sperm nuclear histone to protamine ratio in fertile and infertile men: evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl. 2006;27:414–420. doi: 10.2164/jandrol.05171. [DOI] [PubMed] [Google Scholar]
- 17.Chevaillier P, Mauro N, Feneux D, Jouannet P, David G. Anomalous protein complement of sperm nuclei in some infertile men. Lancet. 1987;2:806–807. doi: 10.1016/S0140-6736(87)92547-5. [DOI] [PubMed] [Google Scholar]
- 18.de Mateo S, Ramos L, van der Vlag J, de Boer P, Oliva R. Improvement in chromatin maturity of human spermatozoa selected through density gradient centrifugation. Int J Androl. 2010;34:256–267. doi: 10.1111/j.1365-2605.2010.01080.x. [DOI] [PubMed] [Google Scholar]
- 19.Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl. 2001;22:604–610. [PubMed] [Google Scholar]
- 20.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update. 2007;13:313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 21.Aoki VW, Emery BR, Liu L, Carrell DT. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl. 2006;27:890–898. doi: 10.2164/jandrol.106.000703. [DOI] [PubMed] [Google Scholar]
- 22.Aoki V, Liu L, Jones K, Hatasaka H, Gibson M, Peterson C, et al. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil Steril. 2006;86:1408–1415. doi: 10.1016/j.fertnstert.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AHFM. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 26.Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausió J, Taylor J, et al. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 2017;18:1366–1382. doi: 10.1016/j.celrep.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev Cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Carone BR, Hung J-H, Hainer SJ, Chou M-T, Carone DM, Weng Z, et al. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 2014;1–20. Elsevier Inc. [DOI] [PMC free article] [PubMed]
- 29.Yuan ZF, Arnaudo AM, Garcia BA. Mass spectrometric analysis of histone proteoforms. Annu Rev Anal Chem (Palo Alto Calif) 2014;7:113–128. doi: 10.1146/annurev-anchem-071213-015959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luense LJ, Wang X, Schon SB, Weller AH, Shiao EL, Bryant JM, et al. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin. 2016;9:1–15. doi: 10.1186/s13072-016-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruger TF, Acosta AA, Simmons KF, Swanson RJ, Matta JF, Oehninger S. Predictive value of abnormal sperm morphology in in vitro fertilization. Fertil Steril. 1988;49:112–117. doi: 10.1016/S0015-0282(16)59660-5. [DOI] [PubMed] [Google Scholar]
- 32.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 33.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. [DOI] [PMC free article] [PubMed]
- 34.Govin J, Lestrat C, Caron C, Pivot-Pajot C, Rousseaux S, Khochbin S. Histone acetylation-mediated chromatin compaction during mouse spermatogenesis. Ernst Schering Res Found Workshop. 2006;57:155–72. [DOI] [PubMed]
- 35.Boissonnas CC, Jouannet P, Jammes H. Epigenetic disorders and male subfertility. Fertil Steril. 2013;99:624–631. doi: 10.1016/j.fertnstert.2013.01.124. [DOI] [PubMed] [Google Scholar]
- 36.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 1839;2014:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Goudarzi A, Shiota H, Rousseaux S, Khochbin S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol. 2014;426:3342–3349. doi: 10.1016/j.jmb.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 38.Dada R, Kumar M, Jesudasan R, Fernández JL, Gosálvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012;29:213–223. doi: 10.1007/s10815-012-9715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faure AK. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod. 2003;9:757–763. doi: 10.1093/molehr/gag101. [DOI] [PubMed] [Google Scholar]
- 40.Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34:384–390. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- 41.van Nuland R, Gozani O. Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol Cell Proteomics. 2016;15:755–764. doi: 10.1074/mcp.R115.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 44.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 45.Peters A, O'Carroll D, Scherthan H, Mechtler K. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 46.van de Werken C, van der Heijden GW, Eleveld C, Teeuwssen M, Albert M, Baarends WM, Laven JSE, Peters AHFM, Baart EB. Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nat Commun. 2014;5:5868. doi: 10.1038/ncomms6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steilmann C, Paradowska A, Bartkuhn M, Vieweg M, Schuppe HC, Bergmann M, et al. Presence of histone H3 acetylated at lysine 9 in male germ cells and its distribution pattern in the genome of human spermatozoa. Reprod Fertil Dev. 2011;23:997–915. doi: 10.1071/RD10197. [DOI] [PubMed] [Google Scholar]
- 48.Vieweg M. Methylation analysis of histone H4K12ac-associated promoters in sperm of healthy donors and subfertile patients. Clin Epigenetics. 2015;7:1–17. doi: 10.1186/s13148-015-0058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb)