Abstract
Purpose
The quantification of mtDNA in cumulus granulosa cells (CGCs) surrounding an oocyte has been positively linked with morphological embryonic quality. In the present study, we evaluated the link between the amount of mtDNA in CGCs surrounding an oocyte and the chances for the corresponding embryo of implanting and leading to an ongoing pregnancy.
Methods
This is an observational study, performed on 84 oocyte-cumulus-complexes (OCCs) having led to the replacement of an embryo in the maternal uterus, retrieved from 71 patients undergoing IVF with intracytoplasmic sperm. The OCCs were classified in two groups, one including 26 OCCs having led to an implanted embryo and the other including 58 OCCs having led to a non-implanted embryo. The average mtDNA content of CGCs was assessed by using a quantitative real-time PCR technique.
Results
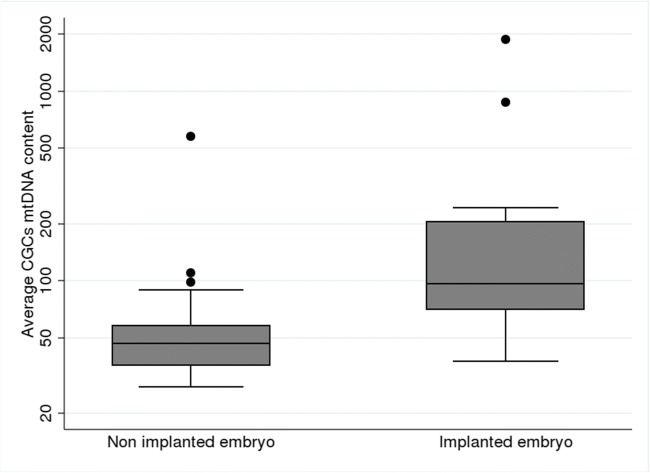
Significantly higher mtDNA copy numbers in CGCs were associated with implanted embryos than with non-implanted embryos (mean 215 [sd 375] and 59 [sd 72], respectively; p < 104). Multivariate analysis, taking into account the women’s age, the embryo quality, and the AMH level, suggests an independent relationship between the mtDNA content of CGCs and the potential of embryo implantation.
Conclusion
During in vitro fertilization (IVF) procedures, the probability of the implantation of the embryo appears to be closely correlated to the mtDNA copy numbers in the CGCs. Our results highlight the interest of mtDNA quantification in GCGs as a biomarker of the potential of embryo implantation.
Keywords: Cumulus cells, Granulosa cells, Embryo implantation, Mitochondria, Mitochondrial DNA
Introduction
Assisted reproductive technologies (ART) have made considerable progress over the years, but the overall success rates of in vitro fertilization (IVF) still remain relatively low. Although there are variations from country to country, the overall delivery rate with IVF is estimated at 20.3% per transfer of fresh embryo [1].
The possibility of choosing the best embryo for transfer may be expected to allow selection of the embryo with the highest developmental potential so as to increase the chances of pregnancy and limit the risk of multiple pregnancies. Currently, this choice is based on the morphology of the embryo and on the kinetics of development that reflect embryo viability [2]. However, these criteria remain rather limited and are not completely informative [3]. Today, the gold standard of ART practitioners would be to find reliable, non-invasive biomarkers of the potential of embryo implantation [4].
Embryogenesis stems from oogenesis, and the scalability of the embryo largely depends on oocyte competence. The assessment of oocyte competence may therefore be a tool for improving embryo selection. But, apart from the poorly informative morphological analysis, it is difficult to study the oocyte without altering its integrity. It would thus appear reasonable to focus on indirect markers of oocyte quality. The study of the cumulus granulosa cells (CGCs), surrounding the oocyte, is considered to be one of the best non-invasive approaches for evaluating oocyte quality and the developmental potential of the embryo [5].
Oocyte-cumulus-complex interactions orchestrate carbohydrate, lipid, and protein metabolisms to ensure the appropriate energy balance required for the meiosis and fertilization of the oocyte, and for support during early embryogenesis. Thus, CGC mitochondria, which are central agents of the energetic metabolic pathways, are directly involved in the establishment of oocyte competence during oogenesis [6]. Consequently, the study of mitochondria in CGCs would offer a promising means of assessing oocyte quality.
CGC mtDNA copy numbers were shown to be good predictors of embryo quality in IVF procedures, with positive and negative predictive values of 84.4% and 82.1%, respectively [7]. More recently, we confirmed this finding with a study on 202 embryos showing a significant link between the quantity of mtDNA in CGCs surrounding an oocyte and embryo quality [8]. However, embryo quality is a subjective notion and merely a surrogate endpoint. The main objective in IVF is the search for markers that would indicate embryos with the best chances of implantation leading to pregnancy.
In the present study, we focused on 84 embryos transferred in 71 women to evaluate the link between the amount of mtDNA in CGCs surrounding an oocyte and the chances for the corresponding embryo of implanting and leading to an ongoing pregnancy.
Materials and methods
Sample size calculation
Since there is no data available in the literature regarding the relationship between the implantation potential of an embryo and the mtDNA content of CGCs, we based our sample size calculation on the expected effect size.
To highlight a large sample size (corresponding to a sample size of 0.8 based on the Cohen classification), considering a type I error and a power set, respectively at 0.05 and 0.8, a 2:1 ratio between non-implanted and implanted embryos and a two-sided test, we had to recruit at least 60 patients (20 in the implanted group and 40 in the non-implanted group) [9].
Patient characteristics
CGCs were obtained from 71 patients undergoing IVF with intracytoplasmic sperm injection (ICSI) at the ART Center of the University Hospital of Angers, France, from April 2017 to April 2018. Patients were supported by ICSI for male infertility indications, which represent about 50% of the IVF procedures in our facility.
Our goal being to study the embryonic implantation, we were interested in women for whom a single embryo transfer (SET) was expected “a priori” (with some “a posteriori” exceptions). According to the procedure of our center, they were young women (under 35) coming for a first attempt, or women coming for a second request (with a previous history of pregnancy), or finally, couples refusing the transfer of two embryos for personal reasons.
The characteristics of the patients as well as the products used for the suppression of pituitary gonadotrophin release (Cétrorelix [Cetrotide®, Merck-Serono, Geneva, Switzerland]; Ganirelix [Orgalutran®, Organon, Oss, Netherlands]; Triptoréline [Decapeptyl®, Ipsen Pharma, Paris, France]) and follicular growth stimulation (uFSH + uLH: Ménotropine [Menopur®, Ferring Pharmaceuticals, Copenhagen, Denmark]; rFSH: Follitropine alpha [Gonal-f®, Merck-Serono, Geneva, Switzerland] or Follitropine beta [Puregon®, Organon, Oss, Netherlands]) are summarized in Table 1.
Table 1.
Characteristics of the patients according to embryo implantation
| Overall embryos n = 84 (%) |
Implanted embryos n = 26 (%) |
Non-implanted embryos n = 58 (%) |
Univariate analysis p value |
Multivariate analysis p value | |
|---|---|---|---|---|---|
| Age (years) | 31.3 ± 4.3 | 30 ± 4.3 | 32 ± 4.2 | 0.12 | 0.23 |
| BMI | 23.8 ± 4.6 | 22.8 ± 3.3 | 24.3 ± 5.0 | 0.41 | |
| Tobacco | 0.07 | ||||
| Non smokers | 61 | 15 | 46 | ||
| Smokers | 19 | 10 | 9 | ||
| Information missing | 4 | 1 | 3 | ||
| FSH (UI/l) | 7.8 ± 2.5 | 7.2 ± 1.9 | 8.1 ± 2.70 | 0.17 | |
| E2 (pg/ml) | 40.4 ± 22.5 | 37.0 ± 17.2 | 42.0 ± 24.7 | 0.3 | |
| AMH (ng/ml) | 3.4 ± 3.4 | 4.1 ± 3.4 | 3.1 ± 2.4 | 0.0236 | 0.29 |
| AFC | 18.2 ± 10.1 | 20.7 ± 12.8 | 17.1 ± 8.5 | 0.39 | |
| Total dose of FSH administered | 2130 ± 828 | 2090 ± 692 | 2148 ± 887 | 0.69 | |
| Number of OCCs retrieved | 9.2 ± 5.3 | 9.4 ± 4.7 | 9.0 ± 4.7 | 0.41 | |
| Number of cleaved embryos | 5.1 ± 3.5 | 5.46 ± 3.0 | 4.9 ± 3.7 | 0.18 | |
| LH Suppression | 1 | ||||
| Agonist | 74 | 23 (88.5) | 51 (87.9) | ||
| Antagonist | 10 | 3 (11.5) | 7 (1.1) | ||
| Stimulation | 0.74 | ||||
| FSH | 77 | 25 | 52 | ||
| FSH + LH | 7 | 1 | 6 | ||
| Embryo quality | 0.1 | 0.16 | |||
| Good | 32 (38.1) | 13 (50.0) | 19 (32.8) | ||
| Fair | 45 (53.6) | 13 (50) | 32 (55.2) | ||
| Poor | 7 (8.3) | 0 (0) | 7 (12.0) | ||
| mtDNA copy number per CGC | 108 ± 226 | 215 ± 375 | 59 ± 72 | < 10−4 | 0.02 |
The ovulation was induced with Ovitrelle® (Choriogonadotropine alfa, Merck-Serono, Geneva, Switzerland), and the oocytes were retrieved transvaginally under ultrasound guidance.
Isolation of oocytes and cumulus cells
OCCs were retrieved 36 h after treatment with human chorionic gonadotropin and washed in multiple dishes with Flushing medium® (Origio-France, Limonest, France) to eliminate the remaining mural granulosa cells, blood cells, and cellular debris. The OCCs were subsequently incubated for 1 h in Ferticult® culture media (Fertipro, Beernem, Belgium). For each patient, the oocytes were individually stripped using hyaluronidase (80 IU, Fertipro, Beernem, Belgium) and gently pipetted with a 125-μm diameter stripper pipette (Origio-France, Limonest, France). The CGCs associated with mature oocytes used for ICSI were recovered and isolated. CGCs were recovered in 500 μl of physiological serum, and centrifuged at 10,000g for 5 min. The supernatant was removed, and the CGC pellets immediately frozen at − 80 °C until nucleic acid extraction.
DNA extraction
Total DNA extraction, from isolated CGCs, was carried out using the Nucleospin Tissue Kit® (Macherey-Nagel, Düren, Germany) according to the manufacturer’s recommendations, as previously described [8].
Quantification of mtDNA
We used the technique already published in 2017 [8]. Briefly, the mean mtDNA copy number in CGCs was determined by real-time quantitative PCR (Q-PCR) using SYBR green DNA intercalator on the CFX connect System® (Biorad, Hercules, CA, USA). The pairs of primers selected were respectively MT-CO1 (mtDNA nucleotide positions 7017–7036 and 7205–7224) to quantitate mtDNA, and B2M (beta 2 microglobulin, exon 3 – 112 to − 93 and exon 3 + 84 to + 113) to quantitate nuclear DNA in follicular cells.
The average CGC mtDNA was determined by calculating the ratio between the mtDNA copy number and half the nuclear DNA copy number (reflecting the copy number per diploid genome).
Embryo culture and transfer
After sperm injection, embryos were cultivated in Global medium® (Life Global, Guilford, USA) under an atmosphere of CO2. They were observed 18 h post-injection to objectify the presence of the two pronuclei and 48 h post-injection to objectify embryo cleavage. The embryos were scored at day 2 (48 h post-injection) and day 3 (72 h post-injection if not transferred at day 2) according to the ESHRE consensus [10] defining three groups of embryos, i.e., “good quality,” “fair quality,” or “poor quality” embryos.
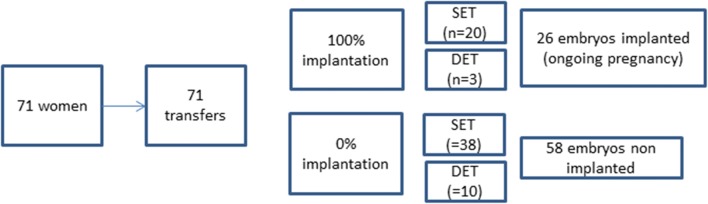
Only the CGCs of the oocytes that led to an embryo replaced in the maternal uterus were preserved. Fifteen days after embryo transfer, an assay of BHCG in the maternal blood was performed, and in case of positivity, an ultrasound examination at 12 weeks allowed objectification of the cardiac activity signaling ongoing pregnancy. In the majority of cases, as expected, a SET had been performed. But in some cases, the poor embryo quality or the patient’s wish led us to transfer two embryos (DET = double embryo transfer). In these cases, only the transfers that led to the simultaneous implantation or non-implantation of the two embryos (dizygotic twin pregnancy or absence of pregnancy) were retained for the study. Biochemical pregnancies and early miscarriages were discarded from the study.
The flow chart (Fig. 1) summarizes the women and the embryos retained for the study.
Fig. 1.
Embryo flow chart
Statistical analysis
All continuous variables (age, BMI, FSH basal, AMH, AFC, total dose of FSH administered, and mtDNA) were described with mean values and standard deviations and were compared using the Mann-Whitney test. All binary and qualitative variables (smoking status, type of LH suppression, type of stimulation, embryo quality, and the two outcomes: implantation or implantation failure) were summarized with their counts or percentages and were compared using Fisher’s exact test. For the purposes of statistical analysis, the association between outcomes and the factors potentially linked with implantation (age and embryo quality, notably) and the mtDNA content of CGCs were investigated using generalized estimating equations models to take into account the cluster of oocytes belonging to individual mothers, and the p values corresponding to the factors studied were recorded. All the statistical tests were bilateral, and the differences were considered significant at p < 0.05. All analyses were computed using Stata 13.1.
Ethical approval
All participants gave their written informed consent, and the study was approved by the Ethical Committee of the University Hospital of Angers, France (Numbers DC-2014-2224 and AC-2016-2799).
Results
Patients
As shown in Table 1, there was no difference between the “implanted” and “non-implanted” groups with respect to the patients’ characteristics (age, BMI, and smoking status), their treatment (type of stimulation, LH suppression, and dose of FSH administered), or the progress of their attempt (number of oocytes retrieved, number of embryos obtained, embryonic quality at day two).
Only the evaluation of the ovarian reserve before treatment, although showing equivalent FSH levels and AFC values, showed a significantly higher AMH level in the “implanted” group (p = 0.024).
CGCs mtDNA
The average number of mtDNA copies in the CGCs was significantly higher in the “implanted” group in comparison with the “non-implanted” group (p < 10−4) (Table 1 and Fig. 2).
Fig. 2.
Average mtDNA per CGC for implanted and non-implanted embryos
Moreover, the multiparametric study taking into account the factors identified in the study as having a link with the implantation (AMH and mtDNA of the CGCs), as well as the factors classically recognized as being related to oocyte quality and the chances of implantation (age and embryo quality), showed no significant difference between implanted and non-implanted embryos except for the mtDNA content in CGCs (Table 1).
Discussion
Although endometrial receptivity may be involved in embryo implantation failures, the vast majority of these are related to embryonic factors. Embryo selection is still largely dependent on a morpho-kinetic evaluation based on a static observation of the embryos. The recent development of techniques such as time lapse microscopy (TLM) or preimplantation genetic testing for aneuploidies (PGT-A) promise improved results through the dynamic evaluation of embryo development and the elimination of aneuploid embryos [4, 11]. Nevertheless, these techniques are not always feasible for reasons of cost or ethical regulations nor do they fully reflect the physiological status of the embryo. The evolution of biochemistry and molecular biology techniques has also been used to determine the mechanisms governing embryonic viability and to define biomarkers of implantation, either in culture media or by trophoectoderm biopsy of the embryo, or as mentioned above, in CGCs or follicular fluid [4]. But the results of these approaches have not yet proved clinically useful.
It is now recognized that the mitochondrial mass of the oocyte is of fundamental importance for early embryonic development, especially since the mtDNA does not gainfully replicate during early embryogenesis, implying that the mitochondrial mass of the fertilizable oocyte must be sufficient for its distribution among the several embryonic blastomeres to ensure the optimal functioning of each cell until the resumption of mtDNA replication, which occurs at the blastocyst stage in humans [12]. Moreover, we have reported a link between the oocyte mitochondrial mass and the mean mtDNA content of CGCs [13]. Further, we showed a positive relationship between the mtDNA content of CGCs and the embryonic quality defined by morpho-kinetic criteria [8]. In this study, we extend the results obtained on embryo quality to implantation potential, showing a significantly higher mean mtDNA content in the CGCs surrounding oocytes that led to successfully implanted embryos. The absence of a significant difference between the implanted group and the non-implanted group in terms of embryonic quality testifies to the value added by the mtDNA quantification of CGCs in the selection of embryos. Moreover, the multiparametric study taking into account the factors identified in the study as being linked with the implantation (AMH and mtDNA of the CGCs), as well as the factors classically recognized as being related to oocyte competence, argues in favor of a close independent relationship between the mtDNA content of CGCs and embryo implantation potential. We previously showed that CGCs mtDNA is positively related to embryonic quality [8]; here, we show that for embryos of equivalent quality, this amount of mtDNA is related to embryonic implantation potential. This suggests that the quantitation of mtDNA of CGCs may be an additional biomarker of oocyte competence and thus a guide to select, among a cohort, the best embryo to transfer during IVF procedures.
In view of the importance of mitochondria in supplying embryonic energy, several authors have already considered the quantitation of mtDNA as a biomarker of embryo viability. Thus, a study focusing on the embryo secretome showed that embryos that successfully developed into blastocysts showed a significantly higher mtDNA/gDNA ratio in the culture medium compared with those that stopped developing or evolved more slowly [14]. Unfortunately, the possibility of the culture media being contaminated by exogenous DNA or mtDNA from CGCs present around the embryo makes the use of this biomarker rather delicate [15].
Another topic of debate is the predictive value of the mtDNA level in embryonic trophoectoderm cells. Recent reports of retrospective and prospective studies suggest that the larger the amount of mtDNA, the less likely was the blastocyst to implant successfully [16, 17]. These results implying “the less the mtDNA the better” would seem to disagree with our own findings. But it should be mentioned that the results published by Fragouli et al. 2017 have been debated by other teams [18], and furthermore, our findings are not necessarily in total contradiction. Indeed, our work bears on CGCs, i.e., in cells reflecting the process of oogenesis that is upstream of embryogenesis. A higher level of oocyte mtDNA could testify to oocyte competence and independently, later at the embryo stage, a high level of mtDNA could testify to abnormal mitochondrial activation due to an energetic need potentially linked with abnormalities of embryonic developmental or an insufficient mitochondrial pool in the early embryo. This is all the more plausible, as an earlier study [19] showed that the amount of mtDNA in the early embryo (unlike the blastocyst) is higher in young women than in older women in whom the embryo quality is worse. Moreover, the mitochondrial mass is related to the metabolic status of a cell. CGCs have a rather glycolytic metabolism, whereas trophoectoderm cells are the first cells in the embryo exhibiting mitochondrial differentiation and increased activity of oxidative phosphorylation [20]. An adaptive process of these cells in response to impaired oocyte or embryo competence would probably have a different translation on the mitochondrial mass.
At least, other authors have demonstrated a positive relationship between the mtDNA content of CGCs and that of peripheral blood cells [7] itself related to the fertility of a woman [21, 22]. Thus, the impaired fertility for women with low levels of blood mtDNA [22] may be related to low CGC mtDNA levels associated with decreased chances of embryo implantation.
Despite the small size of our population, the significance of our results highlights the interest of mtDNA quantification in GCGs as a biomarker of the potential of embryo implantation. The simplicity and rapidity of the technique make possible to use this biomarker in routine clinical practice, without having to freeze the embryos and postpone the transfer. Indeed, the mtDNA content may be easily quantified in CGCs during the in vitro embryo development in order to select and transfer the embryos with the highest mtDNA content in their corresponding follicular cells.
Larger and prospective studies will be required to confirm these preliminary results and before considering any use in clinical practice.
Acknowledgments
We are grateful to Kanaya Malkani for his critical reading and comments on the manuscript.
Funding
This study was supported by a grant from the “Agence de la Biomédecine” – Appel d’Offre Recherche “AMP, diagnostic prenatal et diagnostic génétique” 2017.
Compliance with ethical standards
All participants gave their written informed consent, and the study was approved by the Ethical Committee of the University Hospital of Angers, France (Numbers DC-2014-2224 and AC-2016-2799).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Ishihara O, Adamson GD, Dyer S, de Mouzon J, Nygren KG, Sullivan EA, Zegers-Hochschild F, Mansour R. International committee for monitoring assisted reproductive technologies: world report on assisted reproductive technologies, 2007. Fertil Steril. 2015;103:402–413.e11. doi: 10.1016/j.fertnstert.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril. 2012;98:1481–1489.e10. doi: 10.1016/j.fertnstert.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol. 2008;20:234–241. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 4.Gardner DK, Meseguer M, Rubio C, Treff NR. Diagnosis of human preimplantation embryo viability. Hum Reprod Update. 2015;21:727–747. doi: 10.1093/humupd/dmu064. [DOI] [PubMed] [Google Scholar]
- 5.Feuerstein P, Puard V, Chevalier C, Teusan R, Cadoret V, Guerif F, Houlgatte R, Royere D. Genomic assessment of human cumulus cell marker genes as predictors of oocyte developmental competence: impact of various experimental factors. PLoS One. 2012;7:e40449. doi: 10.1371/journal.pone.0040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumesic DA, Guedikian AA, Madrigal VK, Phan JD, Hill DL, Alvarez JP, Chazenbalk GD. Cumulus cell mitochondrial resistance to stress in vitro predicts oocyte development during assisted reproduction. J Clin Endocrinol Metab. 2016;101:2235–2245. doi: 10.1210/jc.2016-1464. [DOI] [PubMed] [Google Scholar]
- 7.Ogino M, Tsubamoto H, Sakata K, Oohama N, Hayakawa H, Kojima T, Shigeta M, Shibahara H. Mitochondrial DNA copy number in cumulus cells is a strong predictor of obtaining good-quality embryos after IVF. J Assist Reprod Genet. 2016;33:367–371. doi: 10.1007/s10815-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desquiret-Dumas V, Clément A, Seegers V, Boucret L, Ferré-L’Hotellier V, Bouet PE, Descamps P, Procaccio V, Reynier P, May-Panloup P. The mitochondrial DNA content of cumulus granulosa cells is linked to embryo quality. Hum Reprod Oxf Engl. 2017;32:607–614. doi: 10.1093/humrep/dew341. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J. Power analysis for the behavioral sciences. 2. Mahwah: Lawrence Erlbaum Associates Editors; 1988. [Google Scholar]
- 10.ASRM and ESHRE Special Interest Group of Embryology. Balaban B. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 11.Rosenwaks Z. Biomarkers of embryo viability: the search for the “holy grail” of embryo selection. Fertil Steril. 2017;108(5):719–721. doi: 10.1016/j.fertnstert.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 12.St John J. The control of mtDNA replication during differentiation and development. Biochim Biophys Acta. 2014;1840:1345–1354. doi: 10.1016/j.bbagen.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 13.Boucret L, de la Barca JM C, Morinière C, Desquiret V, Ferré-L’Hôtellier V, Descamps P, Marcaillou C, Reynier P, Procaccio V, May-Panloup P. Relationship between diminished ovarian reserve and mitochondrial biogenesis in cumulus cells. Hum Reprod Oxf Engl. 2015;30:1653–1664. doi: 10.1093/humrep/dev114. [DOI] [PubMed] [Google Scholar]
- 14.Stigliani S, Persico L, Lagazio C, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA in day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Mol Hum Reprod. 2014;20:1238–1246. doi: 10.1093/molehr/gau086. [DOI] [PubMed] [Google Scholar]
- 15.Hammond ER, McGillivray BC, Wicker SM, Peek JC, Shelling AN, Stone P, Chamley LW, Cree LM. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertil Steril. 2017;107:220–228.e5. doi: 10.1016/j.fertnstert.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Ravichandran K, McCaffrey C, Grifo J, Morales A, Perloe M, Munne S, Wells D, Fragouli E. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod Oxf Engl. 2017;32:1282–1292. doi: 10.1093/humrep/dex070. [DOI] [PubMed] [Google Scholar]
- 17.Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo JA, Munné S, Wells D. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum Reprod Oxf Engl. 2017;32:2340–2347. doi: 10.1093/humrep/dex292. [DOI] [PubMed] [Google Scholar]
- 18.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, Viotti M. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril. 2017;107:34–42.e3. doi: 10.1016/j.fertnstert.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel C-E, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11:e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. Int J Dev Biol. 2012;56(10–12):799–808. doi: 10.1387/ijdb.120140ec. [DOI] [PubMed] [Google Scholar]
- 21.Bonomi M, Somigliana E, Cacciatore C, Busnelli M, Rossetti R, Bonetti S, Paffoni A, Mari D, Ragni G, Persani L. Italian Network for the study of Ovarian Dysfunctions. Blood cell mitochondrial DNA content and premature ovarian aging. PLoS One. 2012;7(8):e42423. doi: 10.1371/journal.pone.0042423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busnelli A, Lattuada D, Rossetti R, Paffoni A, Persani L, Fedele L, et al. Mitochondrial DNA copy number in peripheral blood: a potential non-invasive biomarker for female subfertility. J Assist Reprod Genet. 2018. [DOI] [PMC free article] [PubMed]