Abstract
Purpose
The purpose of the study was to compare the morphokinetic parameters of embryos carrying balanced chromosomal translocations with those carrying unbalanced chromosomal translocations using time-lapse microscopy.
Methods
The study group included 270 embryos that underwent biopsies on day 3 for preimplantation genetic diagnosis (PGD) for chromosomal translocations in our unit between 2013 and 2015. All embryos were incubated under time-lapse microscopy and evaluated for timing of developmental events up to day 5. The timing of these events was compared between balanced and unbalanced embryos, potentially viable and nonviable variants, and maternal versus paternal inheritance of the translocation.
Results
The PGD analysis found that 209 (77%) of the 270 biopsied embryos carried an unbalanced translocation. Embryos carrying unbalanced translocations, which are expected to lead to implantation failure or miscarriage, cleaved less synchronously and were delayed in time of cleavage to the 4-cell stage (t4) and in time of start of blastulation (tSB) compared with balanced embryos (P < 0.05). Furthermore, embryos carrying nonviable translocations demonstrated a significant delay at the time of pronuclei fading (tPNf) compared with those carrying potentially viable translocations (P < 0.05). Embryos whose unbalanced translocations were of maternal origin were significantly delayed in most of the morphokinetic parameters (including tPNf, t2, t3, t4, t6, t7, t8, cc2, s2, and tSB) compared with embryos carrying balanced translocations (P < 0.05).
Conclusions
Embryos carrying unbalanced chromosomal translocations mainly of maternal origin undergo delayed development and asynchronous cleavage that may lead to implantation failure or miscarriage.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1361-8) contains supplementary material, which is available to authorized users.
Keywords: Time-lapse imaging, Morphokinetic parameters, Chromosomal translocations, Preimplantation development
Introduction
Chromosomal translocations are abnormalities caused by the rearrangement of large segments of DNA between non-homologous chromosomes. A chromosomal translocation is classified as being “unbalanced” when the exchange of chromosomal material results in either additional or missing genetic material (i.e., aneuploidy) or as being “apparently balanced” when there is no loss of genetic materials. Both male and female carriers of balanced translocations typically do not exhibit any adverse phenotypic effect themselves, but more than 60% of their gametes are predicted to be unbalanced, leading to abnormal embryos. These individuals are therefore at risk for infertility, repeated implantation failures, pregnancy loss, and birth defects [1–4]. Indeed, while balanced translocations are found in only 0.2% of the neonatal population, the incidence is higher among infertile couples (0.6%) and in women with recurrent miscarriages (9.2%). Moreover, 3.2% of couples who failed more than 10 in vitro fertilization (IVF) cycles are carriers of balanced translocations [5]. Risks associated with chromosomal translocations are also dependent upon the gender of the translocation carrier, the chromosomes involved and the position of the chromosomal breakpoints [4, 6, 7]. Preimplantation genetic diagnosis (PGD) is used as an alternative for carriers of balanced translocation in order to select euploid embryos for transfer and maximize the chance for a healthy offspring. PGD for chromosomal translocations is performed on either single blastomeres biopsied from cleavage-stage embryos combined with fluorescence in situ hybridization (FISH) [8], or on trophectoderm cells biopsied from day 5 blastocysts combined with chromosomal microarray analysis (CMA) [9].
The traditional embryo assessment based on morphology has been shown to have very low correlation to its ploidy [10]. Time-lapse imaging provides the embryologists with more information on embryo dynamics over multiple embryo developmental stages [11, 12]. The continuous monitoring of morphokinetic parameters, such as pronuclear fading (tPNf), timing of cleavage events, synchronicity of cell divisions, cell cycle intervals, and initiation of blastulation (tSB), provides valuable data that can be considered as markers for embryo quality and implantation potential [13–18]. Numerical chromosomal abnormalities (aneuploidy) have already been shown to play a major role in implantation failure and early miscarriage. Although regular morphological parameters do not differ between euploid and aneuploid embryos, recent studies have reported that aneuploid embryos display delayed initiation of blastocyst formation compared to euploid embryos due to numerical chromosomal abnormality [19–21]. Time-lapse analysis of embryos carrying structural chromosomal abnormalities (chromosomal translocations), however, has not yet been examined in this setting. The aim of the present study was to use time-lapse microscopy to evaluate whether unbalanced embryos show different morphokinetics compared to balanced/normal embryos.
Materials and methods
Study population and design
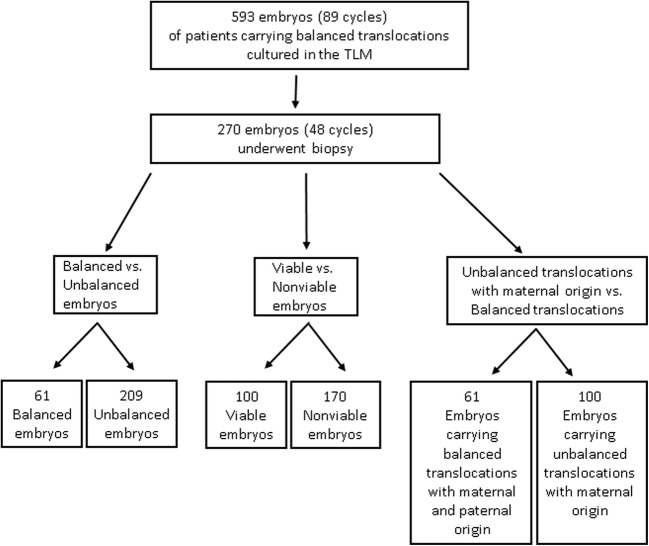
The study group consisted of all embryos from women who underwent PGD for chromosomal translocations at our unit between April, 2013 and December, 2015 (89 cycles, 593 embryos). All of the embryos in this study were fertilized by intracytoplasmic sperm injection (ICSI) and cultured in the EmbryoScope™ time-lapse microscope. Inclusion criteria were all embryos from those PGD cycles that underwent blastomere biopsy on day 3 (at 67–73 h following ICSI; mean = 70.4 h) and those that were at a ≥ 6-cell stage at the time of biopsy and had been cultured in the EmbryoScope™ until embryo transfer. Exclusion criteria were low-grade embryos that were not suitable for blastomere biopsy at day 3 (i.e., < 6 cells or with > 20% fragmentation). The final study group for all analyses included 270 biopsied embryos from 48 cycles (Fig. 1). The baseline demographic features and ovarian stimulation outcomes of the women whose embryos were studied are presented in Supplemental Table 1.
Fig. 1.
Flowchart showing the formation of the study group. The original group included 593 embryos from 89 cycles that underwent PGD-FISH for chromosomal translocations. The final study group for all analyses included 270 biopsied embryos from 48 cycles. Comparisons between balanced and unbalanced embryos, potentially viable and nonviable variants, and parental inheritance were performed by means of time-lapse technology
Ovarian stimulation, fertilization, and embryo culture
Controlled ovarian stimulation was carried out by the long gonadotropin releasing hormone (GnRH) agonist, the short GnRH agonist, or GnRH antagonist protocols. The long protocol began with the administration of subcutaneous injections of 0.1 mg/day of the GnRH-α triptorelin (Decapeptyl; Ferring, Kiel, Germany) for at least 14 days, followed by concomitant recombinant follicle-stimulating hormone [rFSH; Gonal F (Serono, Geneva, Switzerland) or Puregon (Organon, Oss, The Netherlands)], human menopausal gonadotrophin (hMG; Menogon, Ferring, Kiel, Germany), or highly purified human menopausal gonadotropin (Menopur, Ferring Pharmaceuticals, Geneva, Switzerland). The short protocol began with the administration of the GnRH-α from the first day of the cycle followed by concomitant daily r-FSH and GnRH-α from day 3 of the cycle. Stimulation in the antagonist protocol started with the administration of gonadotropins from days 2–3 of the cycle. GnRH antagonist (cetrorelix acetate 0.25 mg, Cetrotide®, Serono or Ganirelix, Orgalutran®, Merck, and Co., Inc.) was started when the leading follicle was ≥ 12 mm or the estradiol level was > 450 pg/ml and continued until the day of human chorionic gonadotropin (hCG) administration. Choriogonadotropin α 250 mcg (Ovitrelle; Serono, Geneva, Switzerland) was administered when at least three follicles achieved a diameter of 18 mm. Ovum pickup was performed 36 h later.
The cumulus-oocyte complexes were isolated into Multipurpose Handling Medium–Complete (MHM-C) (Irvine Scientific). Oocytes were denuded of cumulus cells by hyaluronidase and a fine pipette. Sperm samples were loaded on sperm separation medium (Isolate, Irvine Scientific, USA) and washed with MHM-C medium. ICSI was performed at 2–4 h following oocyte retrieval by means of a Nikon inverted microscope (Nikon) with Narishige and Eppendorf micromanipulators. Each embryo was incubated in a separate droplet of human embryo culture medium covered with paraffin oil in an EmbryoSlide® culture dish (Fertilitech) to allow individual assessment and documentation. Incubation in the EmbryoScope™ incubator lasted from day 0 following ICSI and continued up to day 5 of development.
Blastomere biopsy for PGD
Blastomere biopsy for PGD was performed 67–73 h after ICSI. Prior to biopsy, the embryos were incubated for 2–5 min in Ca2+/Mg2+-free bicarbonate-buffered medium [Quinn’s Advantage Medium with HEPES (Ca/Mg free), SAGE, USA)] in order to loosen cell-to-cell adhesions. A laser (Zilos™, Hamilton Thorne Research, wavelength 1480 nm) was used to create a ~ 30 μm opening in the zona pellucida, and two blastomeres were gently biopsied by an aspiration pipette through the hole. It is well accepted that the accuracy of FISH analysis on a single cell is approximately 85%. Therefore, biopsy and analysis of two cells would increase the accuracy of the analysis to reach ~ 98% (probability calculation 1-[0.15 × 0.15] = 0.9775). After biopsy, the embryos were immediately returned to the EmbryoScope™ (Vitrolife, Sweden) until transfer.
Fluorescence in situ hybridization
Single blastomeres were spread onto a Superfrost Plus glass slide (Kindler GmbH, Freiburg, Germany) using 0.01 M HCl/0.1% Tween 20 solution [22]. A set of specific fluorescence probes was designed for each translocation, and FISH analysis was performed according to the manufacturer’s recommendations. In brief, 10 μl of probe mixture (Vysis, Inc., Downer’s Grove, IL, USA) was added to the slides embedded with nuclei, denatured for 5 min at 75 °C, and hybridized overnight at 37 °C in a moist chamber, as previously described [23]. The nuclei were then examined using a fluorescence microscope with the appropriate filter sets (Olympus BX51). The FISH images were captured using a computerized system (FISHView; Applied Spectral Imaging, Migdal HaEmek, Israel). An embryo(s) found to have a normal/balanced translocation was transferred back to the uterus for implantation. It is important to note that FISH analysis cannot differ between embryos carrying a balanced translocation (similar to the parental carrier undergoing PGD) and a normal embryo with no translocation. However, we treat the embryos in both cases as healthy and eligible for transfer, since the phenotype of the balanced translocation is normal and due to the fact that all parts of the chromosomes are present in both and only their location is different (translocated to another chromosome). The embryos were excluded from the study in cases of an inconclusive result due to hybridization failure or to an indistinguishable fluorescent signal, as well as in cases of mosaicism (where each blastomere gives different results).
Time-lapse monitoring of embryo morphokinetics
All embryos were incubated in the integrated EmbryoScope™ time-lapse monitoring system (EmbryoScope™; Vitrolife, Sweden) from the time of ICSI until embryo transfer on days 4–5. During the study period (April 2013–December 2015), the embryos were cultured in two IVF media: CSC (Continuous Single Culture® Complete with HSA, Irvine Scientist) or GT (Global Total, Life Global). Since we were aware of the possible impact of the media on embryo development, and according to our policy that requires continuous quality control and analysis of the data, we had already compared the dynamics of the embryos cultures in these two media. This analysis included 513 treatment cycles in which the embryos were cultured in the CSC medium, and 668 treatment cycles in which the embryos were cultured in the GT medium. Clinical pregnancy rates of both groups were similar (22.4 in CSC vs. 21.7% in GT, P < 0.01; unpublished data). Therefore, embryonic development in these two culture media should also be similar.
Embryo scoring and selection with time-lapse monitoring were performed by analysis of time-lapse images of each embryo with software developed specifically for image analysis (EmbryoViewer workstation; UnisenseFertilitech A/S). Embryo morphology and developmental events were recorded to demonstrate the precise timing of the observed cell divisions in correlation to the timing of ICSI: specifically, time of pronuclei fading (tPNf), time of cleavage to a 2-blastomere (t2), a 3-blastomere (t3), a 4-blastomere (t4), and so forth until reaching a 9-blastomere (t9) embryo. The lengths of the second and the third cell cycles (cc2 and cc3, respectively), and the synchrony in the division from 3 to 4 cell (s2) and 5 to 8 cells (s3) were documented. The timing of first signs of compaction that indicated the first time point at which cell boundaries could no longer be clearly seen was also recorded (tStart CM). The first signs of the beginning of blastulation (tSB) were recorded as soon as a star-shaped space was demonstrated in the compacted embryo. All the assessments and annotations of the embryos were performed by senior embryologists, thereby ensuring a very low interobserver variation.
Statistical analysis
Normality of the morphokinetic embryo outcome measures was checked first by the Kolmogorov-Smirnov test. If the result of the test was significant, we used the following rule suggested by Kim HY [24]: Z = Skewvalue/SEskewness, Z = kurtosis/SE kurtosise, if absolute z-scores for either skewness or kurtosis are larger than 3.29, which corresponds with an alpha level 0.05, then reject the null hypothesis and conclude that the distribution of the sample is not normal. Accordingly, the following measures were found to be normally distributed: tPNf, t2, t3, t4, t5, t6, t9+, tStart CM, and tSB, and are therefore reported by mean and standard deviation. The distribution of the following morphokinetic outcome measures was found to be abnormal: t7, t8, cc2, cc3, s2, s3, and is therefore reported by median and interquartile range. For measures that did not follow the normal distribution, we decided to use the rank transformation in the regressions models.
In order to compare the outcome measures between balanced and unbalanced embryos or between potentially viable and nonviable embryos, we used the generalized linear model Glimmix, with Gaussian link function for the morphokinetic parameters and with the Logit link for comparing blastocyst morphology [high (yes vs. no), moderate (yes vs. no) and low (yes vs. no)]. All models included age as a covariate. The Glimmix procedure takes into account the dependency of the embryos upon their mothers, i.e., the two-level hierarchical structure of the data, embryos clustered within mothers. Comparisons of morphokinetics parameters were performed between the unbalanced embryos with maternal (Table 3) or paternal (Supplemental Table 2) origin and all balanced embryos (with maternal and paternal origin); therefore, these observations are considered independent. In order to compare the outcome measures between groups while adjusting to mother’s age, we used multivariate linear regression for the morphokinetic measures, and logistic multivariate regression for the morphology of blastocysts. A P value < 0.05 was considered significant. Statistical analysis was performed by SAS for Windows version 9.4.
Table 3.
Comparison between embryos with potentially viable and nonviable translocations
| Parameter | Potentially viable (n = 100) | Nonviable (n = 170) | P value | ||
|---|---|---|---|---|---|
| Mean | std | Mean | std | ||
| tPNf | 23.35 | (3.33) | 24.21 | (3.42) | 0.03 |
| t2 | 26.01 | (3.78) | 26.74 | (3.48) | 0.08 |
| t3 | 37.20 | (4.72) | 37.97 | (4.72) | 0.30 |
| t4 | 38.57 | (4.67) | 39.58 | (4.84) | 0.049 |
| t5 | 50.91 | (6.98) | 50.84 | (7.52) | 0.95 |
| t6 | 53.26 | (6.71) | 54.06 | (7.05) | 0.26 |
| t7, median (Q1, Q3) | 52.9 | (48.8, 56.6) | 53.2 | (49.8, 58.4) | 0.16 |
| t8, median (Q1, Q3) | 54.9 | (51.0, 59.9) | 55.7 | (52.0, 61.7) | 0.15 |
| t9+ | 71.97 | (10.05) | 74.16 | (11.94) | 0.08 |
| cc2, median (Q1, Q3) | 11.7 | (10.7, 12.3) | 11.7 | (10.7, 13.0) | 0.27 |
| cc3, median (Q1, Q3) | 13.6 | (11.9, 16.0) | 13.7 | (11.7, 15.8) | 0.99 |
| s2, median (Q1, Q3) | 0.7 | (0.0, 1.0) | 0.7 | (0.3, 1.3) | 0.06 |
| s3, median (Q1, Q3) | 6.3 | (3.0, 13.3) | 7.5 | (4.2, 16.1) | 0.09 |
| tStart CM | 85.89 | (11.90) | 86.16 | (11.30) | 0.87 |
| tSB | 100.76 | (8.53) | 104.25 | (9.07) | 0.01 |
Pronuclei fading (tPNf),t2, t3, t4, t5, t6, t7, t8, t9 = time (h) between ICSI and pronuclei fading, two-, three-, four-, five-, six-, seven-, eight-, and nine-cell stage, respectively; cc2, cc3 = length (h) of the second and third cell cycle, respectively; s2, s3 = synchrony (h) in the division from three to four and five to eight cells, respectively. Initiation of compaction (tStart CM) and initiation of blastulation (tSB) is time (h) between ICSI and initiation of compaction and initiation of blastulation, respectively. A p value < 0.05 was considered significant
Ethics
The study was approved by the ethics committee of the Tel Aviv Medical Center and institutional review board approval for retrieving IVF data was obtained (748/15).
Results
A total of 270 embryos from 48 cycles (mean female age 32 years, range 25–41) that were biopsied and analyzed for chromosomal translocations by FISH were included in this study. Two-hundred and nine of them (77%) were diagnosed with unbalanced translocations and 61 (23%) were chromosomally balanced.
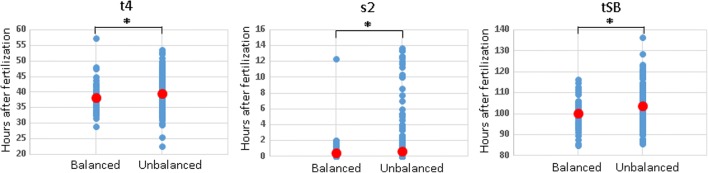
We first compared the morphokinetic parameters of embryos carrying unbalanced translocations to those of embryos carrying balanced translocations. The timing of cleavage to t4 and tSB initiation for the unbalanced embryos was significantly delayed compared with the balanced ones (38.22 h vs 39.49 h and 103.71 h vs. 100.14 h, respectively, P < 0.05). In addition, the unbalanced embryos were less synchronized (i.e., S2 = t4-t3 was longer) than the balanced embryos (0.7 h vs. 0.4 h, respectively, P < 0.05). The timing of all other tested morphokinetic parameters was not significantly different between the groups (Table 1, Fig. 2). Our results also showed that the number of high-quality blastocysts (blastocysts or hatched blastocysts with compact inner cell mass (ICM) and high trophectoderm morphology grade, i.e., AA/AB/BA) was significantly lower and that the number of low-quality blastocysts with dispersed ICM cells and loose trophectoderm (grade 7CA, 7CC, 4CC, 3CC), as well as those with degenerative cells/vacuoles was significantly higher in unbalanced embryos (P < 0.05; Table 2).
Table 1.
The morphokinetic parameters of embryos carrying balanced and unbalanced translocations
| Parameter | Balanced (n = 61) | Unbalanced (n = 209) | P value | ||
|---|---|---|---|---|---|
| Mean | std | Mean | std | ||
| tPNf, | 23.35 | (3.58) | 24.06 | (3.34) | 0.11 |
| t2 | 25.90 | (3.83) | 26.64 | (3.53) | 0.13 |
| t3 | 37.42 | (4.78) | 37.76 | (4.72) | 0.84 |
| t4 | 38.22 | (4.51) | 39.49 | (4.84) | 0.03 |
| t5 | 50.74 | (7.22) | 50.90 | (7.35) | 0.73 |
| t6 | 53.10 | (6.53) | 53.96 | (7.03) | 0.31 |
| t7, median (Q1, Q3) | 54.9 | (51.3, 58.9) | 55.6 | (51.4, 61.5) | 0.10 |
| t8, median (Q1, Q3) | 58.2 | (52.6, 65.5) | 60.2 | (54.3, 66.6) | 0.25 |
| t9+ | 71.97 | (9.85) | 73.77 | (11.71) | 0.20 |
| cc2, median (Q1, Q3) | 11.7 | (10.7, 12.0) | 11.7 | (10.7, 13.0) | 0.92 |
| cc3, median (Q1, Q3) | 13.3 | (12.0, 15.5) | 13.7 | (11.7, 15.8) | 0.53 |
| s2, median (Q1, Q3) | 0.4 | (0.0, 1.0) | 0.7 | (0.3, 1.3) | 0.02 |
| s3, median (Q1, Q3) | 7.0 | (3.0, 14.7) | 6.7 | (4.0, 15.3) | 0.54 |
| tStart CM | 86.57 | (11.62) | 85.91 | (11.51) | 0.70 |
| tSB | 100.14 | (7.29) | 103.71 | (9.31) | 0.03 |
Pronuclei fading (tPNf), t2, t3, t4, t5, t6, t7, t8, t9 = time (h) between ICSI and pronuclei fading, two-, three-, four-, five-, six-, seven-, eight-, and nine-cell stage, respectively; cc2, cc3 = length (h) of the second and third cell cycle, respectively; s2, s3 = synchrony (h) in the division from three to four and five to eight cells, respectively; initiation of compaction (tStart CM) and initiation of blastulation (tSB) is time (h) between ICSI and initiation of compaction and initiation of blastulation, respectively. A p value < 0.05 was considered significant
Fig. 2.
Comparison between the distribution of time-lapse morphokinetic parameters of embryos carrying balanced and unbalanced translocation: t4 = time (h) between ICSI and the four-cell stage; s2 = synchrony (h) in the division from three to four cells; initiation of blastulation (tSB) defined by time (h) between ICSI and initiation of blastulation. The mean (for t4 and tSB) and median (for s2) values are indicated by red dots. *P < 0.05
Table 2.
Blastocyst morphology of embryos with balanced compared with unbalanced translocations
| Blastocyst gradinga | Balanced (n = 61) | Unbalanced (n = 209) | P value | ||
|---|---|---|---|---|---|
| High, n (%) | 24 | (39%) | 41 | (20%) | 0.002 |
| Moderate, n (%) | 29 | (48%) | 115 | (55%) | NS |
| Low, n (%) | 8 | (13%) | 53 | (25%) | 0.046 |
aBlastocyst morphology was evaluated according to Gardner’s scoring system and three subgroups were classified: high (blastocysts or hatched blastocysts with inner cell mass [ICM] and trophectoderm morphology of grade AA/AB/BA), low (delayed embryos at < 9 cells or blastocysts with ICM or trophectoderm morphology of grade C), and moderate (all of the rest). A p value < 0.05 was considered significant
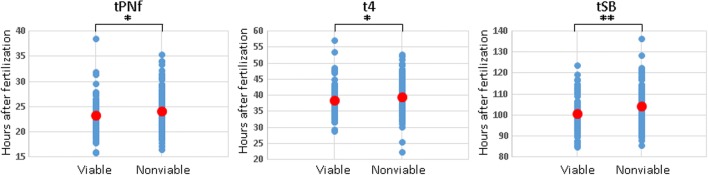
A correlation between morphokinetic parameters and embryo viability has been previously reported in detail [13–18]. We now studied the developmental kinetics of balanced and unbalanced chromosomal translocations which can result in viable fetuses, as well as unbalanced chromosomal translocations which are less likely to result in viable embryos but rather lead more often to implantation failure or miscarriage. The embryos that were labeled as being “potentially viable” were those that carried (1) balanced translocations; (2) unbalanced translocations with translocation derivatives known to be viable; (3) an unbalanced translocation already clinically identified as being viable (baby born in the family); (4) embryos with trisomy of a small sub-telomeric fragment; (5) embryos with mosaicism of balanced/unbalanced translocations. The timing of tSB for the nonviable embryos was significantly delayed compared with the potentially viable ones (104.25 h for nonviable embryos compared to 100.76 h for potentially viable embryos, P = 0.01; Table 3, Fig. 3). This was in addition to a significant delay of t4 in the nonviable embryos compared to the potentially viable embryos (39.58 h vs. 38.57 h, respectively, P < 0.05; Table 3, Fig. 3). Interestingly, the timing of pronuclei fading (tPNf), a very early event during embryo development, was significantly delayed in nonviable embryos compared with the potentially viable ones (24.21 h for nonviable embryos compared to 23.35 h for potentially viable embryos, P = 0.03; Table 3, Fig. 3).
Fig. 3.
Comparison between the distribution of time-lapse morphokinetic parameters of potentially viable and nonviable embryos. Pronuclei fading (tPNf) = time (h) between ICSI and pronuclei fading; t4 = time (h) between ICSI and the four-cell stage; initiation of blastulation (tSB) defined by time (h) between ICSI and initiation of blastulation. The mean values are indicated by red dots. *P < 0.05, **P < 0.01
Infertility risks associated with chromosomal translocations are dependent on the gender of the translocation carrier. While males with balanced translocations are usually infertile, most female carriers are fertile but have a history of repeated spontaneous miscarriages [25]. In view of our results on delayed development of unbalanced embryos and the recurring miscarriages among females carrying chromosomal translocations, we hypothesized that maternal inheritance of translocation may result in an even more pronounced delay of preimplantation embryo development. Seventy-eight percent of embryos carried unbalanced translocation when the females were the translocation carriers, and 77% of embryos carried unbalanced translocation when the males were the translocation carriers. Indeed, analysis of a subgroup of embryos with maternal inheritance of chromosomal translocation (n = 100) demonstrated a significant delay in the timing of almost all of the morphokinetic parameters in embryos carrying unbalanced translocation of maternal origin compared to embryos carrying balanced translocations (tPNf, t2, t3, t4, t6, t7, t8, CC2, S2, and tSB; P < 0.05, Table 4, Supplemental Fig. 1). Likewise, there were significantly fewer high-quality blastocysts and more low-quality blastocysts among unbalanced embryos of maternal origin (P < 0.05; Table 5). In contrast, when performing the same analysis on embryos with unbalanced translocations of paternal origin, only s2 and tSB parameters were significantly delayed (Supplemental Table 2A). Additionally, there were significantly fewer high-quality blastocysts among unbalanced embryos of paternal origin (P < 0.05; Supplemental Table 2B).
Table 4.
The morphokinetic parameters of embryos carrying unbalanced translocations with maternal origin
| Parameter | Balanced (male and female) (n = 61) | Unbalanced (female) (n = 100) | P value | ||
|---|---|---|---|---|---|
| Mean | std | Mean | std | ||
| tPNf | 23.35 | (3.58) | 24.75 | (3.07) | 0.004 |
| t2 | 25.90 | (3.83) | 27.27 | (3.36) | 0.007 |
| t3 | 37.42 | (4.78) | 39.15 | (4.61) | 0.02 |
| t4 | 38.22 | (4.51) | 41.00 | (4.84) | < 0.001 |
| t5 | 50.74 | (7.22) | 52.59 | (7.27) | 0.10 |
| t6 | 53.10 | (6.53) | 55.64 | (6.98) | 0.02 |
| t7, median (Q1, Q3) | 54.9 | (51.3, 58.9) | 58.3 | (53.8, 62.3) | 0.003 |
| t8, median (Q1, Q3) | 58.2 | (52.6, 65.5) | 61.5 | (56.6, 67.0) | 0.03 |
| t9+ | 71.97 | (9.85) | 75.27 | (11.09) | 0.07 |
| cc2, median (Q1, Q3) | 11.7 | (10.7, 12.0) | 12.3 | (11.3, 13.7) | 0.01 |
| cc3, median (Q1, Q3) | 13.3 | (12.0, 15.5) | 13.7 | (12.5, 16.1) | 0.44 |
| s2, median (Q1, Q3) | 0.4 | (0.0, 1.0) | 0.7 | (0.3, 1.5) | 0.01 |
| s3, median (Q1, Q3) | 7.0 | (3.0, 14.7) | 7.0 | (4.3, 15.3) | 0.44 |
| tStart CM | 86.57 | (11.62) | 84.15 | (12.39) | 0.20 |
| tSB | 100.14 | (7.29) | 104.16 | (9.15) | 0.01 |
Pronuclei fading (tPNf),t2, t3, t4, t5, t6, t7, t8, t9 = time (h) between ICSI and pronuclei fading, two-, three-, four-, five-, six-, seven-, eight-, and nine-cell stage, respectively; cc2, cc3 = length (h) of the second and third cell cycle, respectively; s2, s3 = synchrony (h) in the division from three to four and five to eight cells, respectively. Initiation of compaction (tStart CM) and initiation of blastulation (tSB) is time (h) between ICSI and initiation of compaction and initiation of blastulation, respectively. A p value < 0.05 was considered significant
Table 5.
Comparison between the morphology of blastocysts carrying unbalanced translocations originating from females with those of balanced embryos
| Blastocyst gradinga | Balanced (male and female) (n = 61) | Unbalanced (female) (n = 100) | P value | ||
|---|---|---|---|---|---|
| High, n (%) | 24 | (39%) | 16 | (16%) | 0.005 |
| Moderate, n (%) | 29 | (48%) | 55 | (55%) | NS |
| Low, n (%) | 8 | (13%) | 29 | (29%) | 0.044 |
aBlastocyst morphology was evaluated according to Gardner’s scoring system and three subgroups were classified: high (blastocysts or hatched blastocysts with inner cell mass [ICM) and trophectoderm morphology of grade AA/AB/BA), low (delayed embryos at < 9 cells, or blastocysts with ICM or trophectoderm morphology of grade C), and moderate (all of the rest). A p value < 0.05 was considered significant
Discussion
The current study demonstrates, for the first time, delayed preimplantation development of embryos with unbalanced chromosomal translocations compared with balanced ones. Correlating between morphokinetic behavior, embryonic development, and chromosomal abnormality is not a new concept in embryology; however, all previous studies have explored this relationship in terms of numerical (aneuploidy) rather than structural chromosomal abnormalities (translocations). We showed that embryos with unbalanced chromosomal translocation are significantly delayed and that they cleave less synchronously than embryos with balanced translocation. Indeed, t4 and tSB were significantly later and s2 was longer in embryos carrying unbalanced chromosomal translocations compared to balanced ones. Previous studies used time-lapse imaging and demonstrated that embryos with numerical chromosomal abnormalities undergo a significant delay in development compared with euploid embryos. Delayed first and second cleavage divisions and prolonged transition between the two- and four-cell stages were significantly correlated with chromosome aneuploidy [26]. Euploid embryos displayed strict and tightly clustered cell cycle parameters up to the four-cell stage, but only 30% of aneuploidy embryos exhibited values within these normal time ranges [27]. Basile et al. [28] used an algorithm based on the interval for completion of cleavage from two to five cells and the duration of cc3 and observed an increased probability of selecting chromosomally normal embryos. Importantly, the time of division to 5 cells (t5), the duration of the second cell cycle (cc2), and the synchrony of the second and third cell divisions (s2) were found [18] to be significantly correlated with embryo implantation; however, only the median value of s2 was significantly longer in the non-implanting embryos compared with implanting embryos. In contrast, other groups observed no differences between euploid and aneuploid embryos during those early stages of development, although aneuploid embryos had a significant delay in development compared with euploid embryos in the peri-blastulation phase [20, 29, 30] . Several studies reported that the tSB and formation of a full blastocele were delayed in aneuploid embryos compared with euploid embryos [20, 29, 30]. Based on those findings, Campbell et al. [21] established a predictive algorithm which included tSB and tB data to effectively classify the risk for aneuploidy. Their model was then tested on an independent set of transferred blastocysts and demonstrated a correlation between tSB and tB parameters and implantation and live birth rates [19, 21]. The findings of our current study show that delayed preimplantation development is even more pronounced in embryos with structural chromosomal abnormalities, thus providing a new and plausible explanation for infertility, implantation failure, and repeated miscarriages among carriers of balanced translocations.
Our results showed a greater delay in embryos with nonviable unbalanced chromosomal translocation than that observed much earlier during development, i.e., already at pronuclei fading (tPNf). This was in addition to a very significant delay of tSB that was observed in the nonviable compared to the potentially viable embryos. Similarly, the mean tPNf for euploid embryos was reported as being significantly earlier than that of the aneuploid embryos [31]. Other studies had also found that early PNf is a good indicator of embryo quality and viability [32–34]. PNf has been shown to have a significant effect on achieving pregnancy and implantation when implanted embryos had earlier PNf than non-implanted embryos [33, 35, 36]. The findings of the current study support those of earlier works that linked embryonic divisions to blastocyst formation, ploidy, viability, and implantation success [18, 37, 38].
Moreover, our results show that when the unbalanced translocation is of maternal origin, the embryos display a significant delay in the timing of almost all of the morphokinetic parameters compared to embryos carrying balanced translocations. Concomitantly, there were significantly fewer high-quality blastocysts among unbalanced embryos of maternal origin. s2 and tSB were the only delayed parameters when the unbalanced translocations were of paternal origin. Reciprocal and Robertsonian translocations are often identified as a cause of male infertility in the presence of a low sperm count due to increasing failure of spermatogenesis and impairment of meiosis [39–42]. Several studies have reported differences in segregation modes in embryos obtained from PGD cycles according to the gender of the reciprocal and Robertsonian translocation carrier [6, 43, 44].
Males and females show very different responses to meiotic disturbances. In spermatogenesis, meiosis is generally arrested and causes male factor infertility when translocated chromosomes are involved. However, when translocated chromosomes are involved in oogenesis, meiosis is typically not halted and thus may continue to form abnormal oocytes that beget abnormal embryos when fertilized. These may implant, but they result in recurrent miscarriage or birth defects [3, 25]. Segregation of homologous chromosomes is hampered in carriers of chromosomal translocations. During meiosis I in reciprocal translocation carriers, quadrivalents are formed between common segments of chromosomes rather than the whole chromosome as in normal cells. An alternative mode of segregation is that of the translocated chromosomes to one pole and the normal homologs to the other pole, thus producing balanced/normal gametes. Chromosomally unbalanced gametes will be produced by adjacent 1 and 2 segregation. In Robertsonian translocation carriers, meiosis pairing of the involved chromosomes results in a trivalent structure, which is formed by association of the translocated chromosome composed of the long arms of two acrocentric chromosomes and the two corresponding normal acrocentric chromosomes. When this trivalent configuration is in the cis-configuration, it promotes an alternative mode of segregation during meiosis I, resulting in two acrocentric chromosomes and a balanced gamete with only the translocated chromosome. The other typical segregation modes are adjacent 1 and 2, which produce unbalanced gametes [43, 45]. Oocytes apparently may be more tolerant to the various segregation modes or to an unbalanced chromosomal content, whereupon the development of the preimplated embryo will be affected accordingly, as described herein.
This study has some limitations. First, all embryos underwent biopsy on day 3. We had recently demonstrated that blastomere biopsy delays compaction and the blastulation, leading to a decrease in their implantation, compared to non-biopsied embryos [46]. However, in the current study, all embryos with balanced translocation (study group) as well as embryos with unbalanced translocation (control group) underwent biopsy on day 3, and the performance of a biopsy similarly affected all the embryos. Second, this study includes cycles performed between 2013 and 2015 in which PGD for translocation carriers was performed by FISH analysis. The accuracy of PGD-FISH analysis on a single-cell biopsy reaches 85%: importantly, our current policy, as described in this study, is to biopsy and analyze two blastomeres, which significantly improves the accuracy of the analysis (see Methods section). With the development of chromosomal microarray analysis (CMA), we implemented this technology on trophectodermal cells biopsied from day 5 blastocysts to this group of patients’ carriers of balanced chromosomal translocation as well since 2015. Another limitation may arise from the fact that previous studies have shown that aneuploidy may also affect embryonic development [19–21, 26–30]. However, in this study, we performed PGD for patients with genetic indications that carry balanced chromosomal translocations rather than the chromosomal screening for aneouplidy done in preimplantation genetic screening/preimplantation genetic testing of aneuploidy. Doing so strengthens our results because translocation has a critical impact on embryo development, whether it is a direct cause or because of its association with aneuploidy. Another point is that the current study did not compare the effects of different types of translocations, i.e., Reciprocal and Robertsonian, on embryonic development due to their unequal numbers, and this important issue warrants further study. Finally, this study has a relatively small sample size, and additional larger studies are needed to confirm our findings.
In conclusion, our data suggest that unbalanced chromosomal translocations, mainly those with nonviable translocations and especially those of maternal origin, will lead to delayed embryo development and asynchronous cleavage, leading to an increased risk of implantation failure or miscarriage.
Electronic supplementary material
(DOCX 21.7 kb)
Comparison between the distribution of time-lapse morphokinetic parameters of embryos carrying balanced translocations with maternal and paternal origin (marked as 1) and embryos carrying unbalanced translocations with maternal origin (marked as 2). Pronuclei fading (tPNf), t2, t3, t4, t6, t7, t8 = time (h) between ICSI and pronuclei fading, two-, three-, four-, six-, seven and eight-cell stage, respectively; cc2 = length (h) of the second cell cycle; s2 = synchrony (h) in the division from three to four cells. Initiation of blastulation (tSB) is defined by time (h) between ICSI and initiation of blastulation. The mean (for tPNf, t2, t3, t4, t6, t7, t8 and tSB) and median (for cc2 and s2) values are indicated by red dots. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 35.4 kb)
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- 1.Liss J, Kiewisz J, Zabielska J, Kulwikowska P, Lukaszuk K. Application of FISH method for preimplantation genetic diagnostics of reciprocal and Robertsonian translocations. Folia Histochem Cytobiol. 2015;53:162–168. doi: 10.5603/FHC.a2015.0017. [DOI] [PubMed] [Google Scholar]
- 2.Fischer J, Colls P, Escudero T, Munne S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94:283–289. doi: 10.1016/j.fertnstert.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 3.Chen CK, Wu D, Yu HT, Lin CY, Wang ML, Yeh HY, Huang HY, Wang HS, Soong YK, Lee CL. Preimplantation genetic diagnosis by fluorescence in situ hybridization of reciprocal and Robertsonian translocations. Taiwan J Obstet Gynecol. 2014;53:48–52. doi: 10.1016/j.tjog.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 4.Loh SF, Wong PC, Jiang B, Yeo GH, Tan AS, Prasath EB, Mathew J, Chan ML, Tan WC, Choolani M, Yap CH, Chong SS. Preimplantation genetic diagnosis of chromosome translocations by analysis of polymorphic short tandem repeats. Singap Med J. 2012;53:648–654. [PubMed] [Google Scholar]
- 5.Stern C, Pertile M, Norris H, Hale L, Baker HW. Chromosome translocations in couples with in-vitro fertilization implantation failure. Hum Reprod. 1999;14:2097–2101. doi: 10.1093/humrep/14.8.2097. [DOI] [PubMed] [Google Scholar]
- 6.Lledo B, Ortiz JA, Morales R, Ten J, de la Fuente PE, Garcia-Ochoa C, et al. The paternal effect of chromosome translocation carriers observed from meiotic segregation in embryos. Hum Reprod. 2010;25:1843–1848. doi: 10.1093/humrep/deq111. [DOI] [PubMed] [Google Scholar]
- 7.Escudero T, Abdelhadi I, Sandalinas M, Munne S. Predictive value of sperm fluorescence in situ hybridization analysis on the outcome of preimplantation genetic diagnosis for translocations. Fertil Steril. 2003;79(Suppl 3):1528–1534. doi: 10.1016/S0015-0282(03)00252-8. [DOI] [PubMed] [Google Scholar]
- 8.Scriven PN, Kirby TL, Ogilvie CM. FISH for pre-implantation genetic diagnosis. J Vis Exp. 2011. 10.3791/2570 [DOI] [PMC free article] [PubMed]
- 9.Colls P, Escudero T, Fischer J, Cekleniak NA, Ben-Ozer S, Meyer B, Damien M, Grifo JA, Hershlag A, Munné S. Validation of array comparative genome hybridization for diagnosis of translocations in preimplantation human embryos. Reprod BioMed Online. 2012;24:621–629. doi: 10.1016/j.rbmo.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Kirkegaard K, Ahlstrom A, Ingerslev HJ, Hardarson T. Choosing the best embryo by time lapse versus standard morphology. Fertil Steril. 2015;103:323–332. doi: 10.1016/j.fertnstert.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Ciray HN, Campbell A, Agerholm IE, Aguilar J, Chamayou S, Esbert M, Sayed S, Time-Lapse User Group Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum Reprod. 2014;29:2650–2660. doi: 10.1093/humrep/deu278. [DOI] [PubMed] [Google Scholar]
- 12.Wong C, Chen AA, Behr B, Shen S. Time-lapse microscopy and image analysis in basic and clinical embryo development research. Reprod BioMed Online. 2013;26:120–129. doi: 10.1016/j.rbmo.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio-Ruiz B, Basile N, Perez Albala S, Bronet F, Remohi J, Meseguer M. Automatic time-lapse instrument is superior to single-point morphology observation for selecting viable embryos: retrospective study in oocyte donation. Fertil Steril. 2016;106:1379–1385. doi: 10.1016/j.fertnstert.2016.07.1117. [DOI] [PubMed] [Google Scholar]
- 14.Chen AA, Tan L, Suraj V, Reijo Pera R, Shen S. Biomarkers identified with time-lapse imaging: discovery, validation, and practical application. Fertil Steril. 2013;99:1035–1043. doi: 10.1016/j.fertnstert.2013.01.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero J, Meseguer M. Selection of high potential embryos using time-lapse imaging: the era of morphokinetics. Fertil Steril. 2013;99:1030–1034. doi: 10.1016/j.fertnstert.2013.01.089. [DOI] [PubMed] [Google Scholar]
- 16.Herrero J, Tejera A, Albert C, Vidal C, de los Santos MJ, Meseguer M. A time to look back: analysis of morphokinetic characteristics of human embryo development. Fertil Steril. 2013;100:1602–9 e1–4. doi: 10.1016/j.fertnstert.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Kirkegaard K, Agerholm IE, Ingerslev HJ. Time-lapse monitoring as a tool for clinical embryo assessment. Hum Reprod. 2012;27:1277–1285. doi: 10.1093/humrep/des079. [DOI] [PubMed] [Google Scholar]
- 18.Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 19.Campbell A, Fishel S, Laegdsmand M. Aneuploidy is a key causal factor of delays in blastulation: author response to 'A cautionary note against aneuploidy risk assessment using time-lapse imaging'. Reprod BioMed Online. 2014;28:279–283. doi: 10.1016/j.rbmo.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod BioMed Online. 2013;26:477–485. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod BioMed Online. 2013;27:140–146. doi: 10.1016/j.rbmo.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Coonen E, Dumoulin JC, Ramaekers FC, Hopman AH. Optimal preparation of preimplantation embryo interphase nuclei for analysis by fluorescence in-situ hybridization. Hum Reprod. 1994;9:533–537. doi: 10.1093/oxfordjournals.humrep.a138540. [DOI] [PubMed] [Google Scholar]
- 23.Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- 24.Kim HY. Statistical notes for clinical researchers: assessing normal distribution (2) using skewness and kurtosis. Restor Dent Endod. 2013;38:52–54. doi: 10.5395/rde.2013.38.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta UR, Rajitha P, Pidugu VK, Dalal AB. Cytogenetic abnormalities in 1162 couples with recurrent miscarriages in southern region of India: report and review. J Assist Reprod Genet. 2011;28:145–149. doi: 10.1007/s10815-010-9492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies S, Christopikou D, Tsorva E, Karagianni A, Handyside AH, Mastrominas M. Delayed cleavage division and prolonged transition between 2 and 4 cell stages in embryos identified as aneuploidy at 8 cell stage in array CGH. Hum Reprod. 2012;27:ii84–iii6. doi: 10.1093/humrep/des027. [DOI] [Google Scholar]
- 27.Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, Reijo Pera RA. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101:699–704. doi: 10.1016/j.fertnstert.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Patel DV, Shah PB, Kotdawala AP, Herrero J, Rubio I, Banker MR. Morphokinetic behavior of euploid and aneuploid embryos analyzed by time-lapse in embryoscope. J Hum Reprod Sci. 2016;9:112–118. doi: 10.4103/0974-1208.183511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, Liu J. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med Genet. 2014;7:38. doi: 10.1186/1755-8794-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chawla M, Fakih M, Shunnar A, Bayram A, Hellani A, Perumal V, Divakaran J, Budak E. Morphokinetic analysis of cleavage stage embryos and its relationship to aneuploidy in a retrospective time-lapse imaging study. J Assist Reprod Genet. 2015;32:69–75. doi: 10.1007/s10815-014-0372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wharf E, Dimitrakopoulos A, Khalaf Y, Pickering S. Early embryo development is an indicator of implantation potential. Reprod BioMed Online. 2004;8:212–218. doi: 10.1016/S1472-6483(10)60518-4. [DOI] [PubMed] [Google Scholar]
- 33.Fancsovits P, Toth L, Takacs ZF, Murber A, Papp Z, Urbancsek J. Early pronuclear breakdown is a good indicator of embryo quality and viability. Fertil Steril. 2005;84:881–887. doi: 10.1016/j.fertnstert.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 34.Neuber E, Rinaudo P, Trimarchi JR, Sakkas D. Sequential assessment of individually cultured human embryos as an indicator of subsequent good quality blastocyst development. Hum Reprod. 2003;18:1307–1312. doi: 10.1093/humrep/deg269. [DOI] [PubMed] [Google Scholar]
- 35.Aguilar J, Motato Y, Escriba MJ, Ojeda M, Munoz E, Meseguer M. The human first cell cycle: impact on implantation. Reprod BioMed Online. 2014;28:475–484. doi: 10.1016/j.rbmo.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Lemmen JG, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod BioMed Online. 2008;17:385–391. doi: 10.1016/S1472-6483(10)60222-2. [DOI] [PubMed] [Google Scholar]
- 37.Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto S, Kato N, Saeki K, Morimoto Y. Selection of high-potential embryos by culture in poly (dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril. 2012;97:332–337. doi: 10.1016/j.fertnstert.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 39.Vozdova M, Oracova E, Kasikova K, Prinosilova P, Rybar R, Horinova V, Gaillyova R, Rubes J. Balanced chromosomal translocations in men: relationships among semen parameters, chromatin integrity, sperm meiotic segregation and aneuploidy. J Assist Reprod Genet. 2013;30:391–405. doi: 10.1007/s10815-012-9921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Y, Du RC, Jiang YT, Wu J, Li LL, Liu RZ. Impact of chromosomal translocations on male infertility, semen quality, testicular volume and reproductive hormone levels. J Int Med Res. 2012;40:2274–2283. doi: 10.1177/030006051204000625. [DOI] [PubMed] [Google Scholar]
- 41.Zhang HG, Wang RX, Li LL, Sun WT, Zhang HY, Liu RZ. Male carriers of balanced reciprocal translocations in Northeast China: sperm count, reproductive performance, and genetic counseling. Genet Mol Res. 2015;14:18792–18798. doi: 10.4238/2015.December.28.28. [DOI] [PubMed] [Google Scholar]
- 42.Sobotka V, Vozdova M, Heracek J, Rubes J. A rare Robertsonian translocation rob(14;22) carrier with azoospermia, meiotic defects, and testicular sperm aneuploidy. Syst Biol Reprod Med. 2015;61:245–250. doi: 10.3109/19396368.2015.1045089. [DOI] [PubMed] [Google Scholar]
- 43.Nishikawa N, Sato T, Suzumori N, Sonta S, Suzumori K. Meiotic segregation analysis in male translocation carriers by using fluorescent in situ hybridization. Int J Androl. 2008;31:60–66. doi: 10.1111/j.1365-2605.2007.00759.x. [DOI] [PubMed] [Google Scholar]
- 44.Ko DS, Cho JW, Lee HS, Kim JY, Kang IS, Yang KM, Lim CK. Preimplantation genetic diagnosis outcomes and meiotic segregation analysis of robertsonian translocation carriers. Fertil Steril. 2013;99:1369–1376. doi: 10.1016/j.fertnstert.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Oliver-Bonet M, Benet J, Sun F, Navarro J, Abad C, Liehr T, Starke H, Greene C, Ko E, Martin RH. Meiotic studies in two human reciprocal translocations and their association with spermatogenic failure. Hum Reprod. 2005;20:683–688. doi: 10.1093/humrep/deh654. [DOI] [PubMed] [Google Scholar]
- 46.Bar-El, et al. Blastomere biopsy for PGD delays embryo compaction and blastulation: a time-lapse microscopic analysis. JARG. 2016;33:1449–1457. doi: 10.1007/s10815-016-0813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 21.7 kb)
Comparison between the distribution of time-lapse morphokinetic parameters of embryos carrying balanced translocations with maternal and paternal origin (marked as 1) and embryos carrying unbalanced translocations with maternal origin (marked as 2). Pronuclei fading (tPNf), t2, t3, t4, t6, t7, t8 = time (h) between ICSI and pronuclei fading, two-, three-, four-, six-, seven and eight-cell stage, respectively; cc2 = length (h) of the second cell cycle; s2 = synchrony (h) in the division from three to four cells. Initiation of blastulation (tSB) is defined by time (h) between ICSI and initiation of blastulation. The mean (for tPNf, t2, t3, t4, t6, t7, t8 and tSB) and median (for cc2 and s2) values are indicated by red dots. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 35.4 kb)