Abstract
Purpose
This study was conducted in order to investigate the effects of reactive oxygen species (ROS) levels on the seminal plasma (SP) metabolite milieu and sperm dysfunction.
Methods
Semen specimens of 151 normozoospermic men were analyzed for ROS by chemiluminescence and classified according to seminal ROS levels [in relative light units (RLU)/s/106 sperm]: group 1 (n = 39): low (ROS < 20), group 2 (n = 38): mild (20 ≤ ROS < 40), group 3 (n = 31): moderate (40 ≤ ROS < 60), and group 4 (n = 43): high (ROS ≥ 60). A comprehensive analysis of SP and semen parameters, including conventional semen characteristics, measurement of total antioxidant capacity (TAC), sperm DNA fragmentation index (DFI), chromatin maturation index (CMI), H19-Igf2 methylation status, and untargeted seminal metabolic profiling using nuclear magnetic resonance spectroscopy (1H-NMR), was carried out.
Result(s)
The methylation status of H19 and Igf2 was significantly different in specimens with high ROS (P < 0.005). Metabolic fingerprinting of these SP samples showed upregulation of trimethylamine N-oxide (P < 0.001) and downregulations of tryptophan (P < 0.05) and tyrosine/tyrosol (P < 0.01). High ROS significantly reduced total sperm motility (P < 0.05), sperm concentration (P < 0.001), and seminal TAC (P < 0.001) but increased CMI and DFI (P < 0.005). ROS levels have a positive correlation with Igf2 methylation (r = 0.19, P < 0.05), DFI (r = 0.40, P < 0.001), CMI (r = 0.39, P < 0.001), and trimethylamine N-oxide (r = 0.45, P < 0.05) and a negative correlation with H19 methylation (r = − 0.20, P < 0.05), tryptophan (r = − 0.45, P < 0.05), sperm motility (r = − 0.20, P < 0.05), sperm viability (r = − 0.23, P < 0.01), and sperm concentration (r = − 0.30, P < 0.001).
Conclusion(s)
Results showed significant correlation between ROS levels and H19-Igf2 gene methylation as well as semen parameters. These findings are critical to identify idiopathic male infertility and its management through assisted reproduction technology (ART).
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1350-y) contains supplementary material, which is available to authorized users.
Keywords: Sperm DNA fragmentation, Sperm DNA methylation, Oxidative stress, Reactive oxygen species, Total antioxidant capacity
Introduction
Male infertility is a multifactorial condition with complex pathophysiological mechanisms [1, 2]. Despite remarkable advances in diagnosis, a significant number of cases remain unexplained [3]. Current research is focused on the investigation of molecular mechanisms implicated on the etiology of male infertility, including epigenetics and metabolomics [4–6].
Epigenetics include processes like DNA methylation, posttranslational modifications of histones, and chromatin remodeling [7]. The male germ line contains paternally imprinted genes that are primarily silenced through DNA methylation, including the RAS protein-specific guanine nucleotide-releasing factor 1 (RASGRF1), gene trap locus 2 (GTL2), and Igf2-H19 loci [8]. A limited number of imprinted genes have been reported to have a methylation mark in the male germ line that are initiated during germ cell development in embryogenesis, and detailed research has yet been confined to only three of the reported genes, namely IGF2/H19, Rasgrf1, and Gtl2 [9]. However, out of these three, the H19-Igf2 locus is the best-characterized gene with reciprocal paternal Igf2 (insulin-like growth factor 2) and maternal H19 gene expression [10]. Insulator proteins (CCCTC-Binding factor, CTCF) bind to differentially methylated regions of Igf2-H19 to inhibit promoter-enhancer interaction and regulate gene transcription [11].
The expression of H19 gene has an inverse relationship with cellular growth [12]. It has been shown that spermiogenesis is inhibited when expression of H19 gene is high [13]. In contrast, silencing of H19 gene by methylation in the CpG islands of its promoter region results in normal sperm maturation [13–15]. On the contrary, Igf2 gene expression is involved in cellular growth and proliferation. Methylation of the Igf2 gene promoter silences its expression; in male germ cells, factors inducing Igf2 gene methylation inhibit cellular growth [13, 16]. Despite recent studies about the effect of reactive oxygen species (ROS) on methylation of important genes for male fertility [17], no study has yet investigated the impact of ROS-induced changes in the methylation pattern of H19 and Igf2 genes and the possible effects on semen parameters.
Seminal plasma (SP) metabolites also contribute to semen quality by acting as a mixed nutritive-protective medium for sperm maturation and function [18]. ROS may alter the SP metabolite milieu and contribute to sperm dysfunction [19, 20]. In fact, nuclear magnetic resonance (NMR) spectra have displayed significant differences in lactate, citrate, glycerylphosphorylethanolamine, and glycerylphosphorylcholine among semen samples of men with obstructive azoospermia, oligoasthenoteratozoospermia, and spermatogenic failure when compared to healthy men [21]. Furthermore, metabolite imbalance in SP seems to impact assisted reproductive technology (ART) outcomes negatively [19, 22–24]. Nevertheless, the association between ROS-induced alterations in SP metabolites and the effect on semen quality remains unknown.
Despite the existence of studies evaluating the effects of ROS on methylation status of H19-Igf2 [25] and SP metabolites [26, 27], an integrative approach assessing the effect of ROS on both gene methylation and SP metabolite profiling and how these effects translate to semen characteristics have not been carried out yet. Specifically, there is a need to clarify whether the effect of seminal ROS on gene methylation, seminal metabolites, and sperm quality could be level dependent. We therefore designed a study to elucidate whether an association exists between SP ROS levels and alterations of imprinting gene methylation patterns, SP metabolites, and semen parameters. Our objective was to unravel novel pathophysiologic pathways involved in unexplained male infertility that may allow the development of targeted therapies.
Materials and methods
Ethical approval
This study was approved by the Medical Ethics Committee of Avicenna Research Institute (IR.ACECR.Avicenna.REC.94.9).
Study subjects
This prospective study was carried out including fresh semen specimens of 151 normozoospermic male partners of unexplained infertile couples enrolled in infertility treatment at the Avicenna Infertility Clinic affiliated to Avicenna Research Institute, Tehran, Iran, between April and May 2016. Informed consent was obtained from all subjects, which was approved by the Medical Ethics Committee of Avicenna Research Institute. None of the subjects had any history of pelvic and genital infections, chronic diseases (diabetes, cancer, etc.), endocrine abnormalities, chromosomal aberrations, azoospermia, and leukocytospermia. Participants were nonsmokers and had not taken any medication with potential gonado toxic effects within at least 3 months before enrollment.
Semen collection, analysis, and storage
Each patient contributed one specimen which was collected by masturbation into a sterile container in a room located adjacent to the laboratory. Patients were asked to abstain from ejaculation for 3–5 days before collection. After 30-min liquefaction at 37 °C, an aliquot of the sample was used for semen analysis. The sperm concentration, total sperm motility, viability, and sperm morphology were assessed using a CASA according to the WHO guidelines. Sperm parameters were considered normal when sperm concentration was ≥ 15 million/ml, total sperm motility (progressive + nonprogressive) ≥ 40%, viability ≥ 58%, and normal sperm morphology ≥ 4% in compliance with the WHO criteria [28]. Normozoospermia was also confirmed by manual semen analysis. Leukocyte number was assessed using the peroxidase test. None of the specimens had more than one million peroxidase-positive leukocytes per milliliter of semen [28]. The SP and sperm fractions were prepared within 1 h after semen collection. The total semen was centrifuged at 300g for 5 min. SP and sperm were stored at − 20 °C separately until the completion of the measurement of all other parameters. Common semen analysis, ROS, and total antioxidant capacity (TAC) were assessed in fresh samples, and the frozen-thawed specimen was used for DNA fragmentation index (DFI), sperm chromatin maturity index (CMI), DNA extraction, methylation specific PCR (MSP), and NMR assays.
Measurement of ROS in semen
ROS was measured in fresh liquefied semen by chemiluminescence assay using luminol (5-amino-2,3-dihydro-1,4-phthalazinedione). Luminol oxidizes at neutral pH in the presence of ROS resulting in chemiluminescence, which was measured using Cytation™ 3 (BioTek, USA). For the analysis, 1.2 μl of 5 mM luminol (dissolved in dimethyl sulfoxide, Sigma) was added to 40 μl of neat semen sample. ROS levels were determined by measuring chemiluminescence (relative light units (RLU)/s) at 1 min intervals after the addition of luminol, over a total period of 15 min in triplicate and then averaged for each sample. Blank (phosphate-buffered saline solution, PBS), negative control (PBS + luminol), test sample (neat semen sample + luminol), and positive control (H2O2 + luminol) were run in the same plate. To eliminate any variation, the mean control value was subtracted from the mean semen value to give the true value of the test sample. This value was adjusted for sperm concentration and ROS were reported as RLU/s/106 sperm [29], and specimens were classified according to seminal ROS levels into four groups: group 1 (n = 39): low (ROS < 20 RLU/s/106 sperm), group 2 (n = 38): mild (20 RLU/s/106 sperm ≤ ROS < 40 RLU/s/106 sperm), group 3 (n = 31): moderate (40 RLU/s/106 sperm ≤ ROS < 60 RLU/s/106 sperm), and group 4 (n = 43): high (ROS ≥ 60 RLU/s/106 sperm). These four groups were determined based on the previous studies which showed that high ROS level could damage sperm chromatin and decrease ART outcomes [30–32]. For quality control of the assay, intra-assay coefficient of variation (CV) was calculated in three levels of ROS concentrations in 15 replications and all the resultant CVs were less than 10%. The assay sensitivity based on mean ± 2SD signal of zero standard in 15 replications were 1 RLU. The assay relative accuracy was determined using parallelism test and measured to expect the ratio was between 90 and 110%.
Measurement of TAC
TAC of SP was assessed using the TAC test kit (Dain Bioassay Co, Iran), according to the manufacturer’s instructions. The principle of this assay is to measure the ability of aqueous and lipid antioxidants in the SP to inhibit the oxidation of the 2,2′-azino-di-[3-ethyl-benzthiazoline sulfonate] (ABTS) to ABTS+ [33]. The antioxidant capacity of each sample to prevent ABTS oxidation was compared with that of standard Trolox (6-hydroxy-2,5,7,8-tetramethychroman-2-carboxylic acid), a water-soluble tocopherol analog. Samples, as well as Trolox standards, were assayed in duplicate. The Trolox standards and reagents were prepared freshly at the time of assay [33]. Briefly, 180 μl of reagent A, containing ABTS and potassium persulfate, was added to 10 μl of fresh Trolox standard or SP samples in the plate and read (OD0) at 660 nm. Then, 20 μl of the second reagent containing sodium acetate buffer was added to the wells. The plate was incubated for 10 min in the dark and read again (OD1) at 660 nm. Later, the OD1–OD0values were converted to microliter per liter and reported as TAC results.
Assessment of DFI
Sperm DNA fragmentation was assessed by the sperm chromatin dispersion (SCD) test using SDFA kit (Dain Bioassay Co, Iran) [34]. The halosperm assay was based on the sperm characteristics to produce halo following acid denaturation and removal of nuclear proteins [35]. Briefly, 50 μl washed frozen sperm was mixed with 50 μl agarose (6.5%); 20 μl of this mixture was loaded onto a pretreated glass slide and placed on a cold surface (4 °C) for 5 min. Then, the slides were treated with denaturing solution for 7 min and lysing solution for 15 min. Subsequently, the slides were washed for 5 min with distilled water and dehydrated using increasing concentrations of ethanol (70, 90, and 100%), and finally, air-dried slides were stained. Slides were examined with an optical microscope at ×400 magnification and a minimum of 200 spermatozoa were assessed. Sperm with the large halo was classified as normal and those with no or small halo were classified as DNA fragmented sperm. The DFI is expressed as percentage (%) of sperm with fragmented DNA [35].
Assessment of CMI
The degree of sperm chromatin compaction, maturation, and sperm chromatin defects related to nucleoprotein content was assessed using aniline blue staining [36]. An aliquot of washed frozen sperm was diluted (to 1 × 106 sperm/ml concentration) with PBS and centrifuged at 300g for 5 min. The sediment was completely covered and fixed with 3% glutaraldehyde for 5 min at 4 °C. The fixed specimen was used for the preparation of thin smears. Each slide was then stained with aniline blue (Sigma, USA) at room temperature. A minimum of 200 spermatozoa were assessed per specimen at ×1000 magnification using a light microscope. The pink and blue spermatozoa were classified as mature and immature spermatozoa, respectively, and CMI was expressed as the percentage of total sperm count [37].
Sample preparation and NMR spectroscopy
Seminal proteins of the lowest and highest ROS groups (groups 1 and 4) were precipitated by the addition of 500 μl of cold methanol-water (9:1) mixture to 400 μl of the human SP. The mixture was placed at 4 °C for 20 min and then centrifuged at 10,000 rpm for 10 min. The supernatant was then subjected to NMR spectroscopy [38].
1H-NMR acquisition and data processing
The 1H-NMR spectra were acquired using a Bruker DRX500 MHz spectrometer operating at 500.13 MHz, equipped with 5 mm high-quality NMR tubes (Sigma Aldrich, RSA). SP and D2O (10:1 v/v) (deuterium oxide, 99.9% D, Sigma, USA) were mixed and transferred to 5 mm NMR tube. The 1H-NMR spectra of SP samples were acquired at 25 °C using the Carr–Purcell–Meiboom-Gill (CPMG) spin-echo pulse sequence, π/2–tD–π–tD, to attenuate broad signals from high molecular weight components [39]. The acquisition parameters included a 10.5-ms 90̊ pulse, a relaxation delay of 2 s, a spectral width of 8389.26 Hz, an acquisition time of 1.95 s, 32 k data points, 154 scans, and line broadening 0.3 Hz.
The NMR spectra were referenced to solvent within XWIN-NMR. All spectra were manually phased and baseline corrected using the XWIN-NMR (version 3.5, Bruker Spectrospin Ltd., Germany). The regions 0.2–10 ppm were divided into 0.02 ppm wide buckets by the ProMetab software (version prometab_ v3_3) [40] in MATLAB (version 6.5.1, The MathWorks, Cambridge, UK), excluding the region 4.2–5.5 ppm around the water peak. For all spectra, baseline correction, normalization, and alignment were performed using ProMetab software in MATLAB. Then, the data were imported to SIMCA 14.0 (Umetrics, Umea, Sweden) for multivariate statistical analysis.
Each metabolite reference spectrum was identified via single or multiple peaks, characterized by their respective parts per million (ppm) positions as well as their relative intensities. The metabolites were identified according to signal multiplicity and published literature and online databases. The biological databases such as the Human Metabolome Database (HMDB), literature [41–45], Bayesil software [46], Kyoto Encyclopedia of Genes and Genomes (KEGG), and WikiPathways [47] were used to obtain exhaustive information on metabolites.
DNA extraction
DNA was isolated from spermatozoa using TRIzol reagent (Invitrogen, Carlsbad, CA) as previously described [48]. The DNA concentration of each sample was determined by a NanoDrops ND-1000 spectrophotometer (PeqLab, Erlangen, Germany).
Evaluation of gene methylation by MSP
The methylation status in H19 and Igf2 was observed by comparing DNA from semen samples of all four groups. Bisulfite modifications of extracted DNA samples were performed manually. Denaturation of 2 μg of genomic DNA was carried out using NaOH and then modified by sodium bisulfite. DNA samples were purified using Wizard DNA purification resin according to the manufacturer’s instructions (Promega, USA). These DNA samples were again treated with NaOH, precipitated with ethanol, and resuspended in water [44, 49]. Treatment of DNA with bisulfite converts cytosine residues to uracil but leaves 5-methylcytosine residues unaffected. Therefore, DNA that has been treated with bisulfite retains only methylated cytosines. Thus, bisulfite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues, yielding single-nucleotide resolution information about the methylation status of a segment of DNA. The modified DNA was amplified by MSP using specific primers. The primer sequences for methylated (M) and unmethylated (U) PCR are shown in Table 1. Primer pairs were designed to be “M, methylated-specific” by including sequences complementing only unconverted 5-methylcytosines, or, on the converse, “U, unmethylated-specific,” complementing thymines converted from unmethylated cytosines. Unmethylated alleles were differentiated from methylated alleles according to the alterations observed in their sequence via bisulfite treatment of DNA, to change unmethylated cytosines to uracil [50, 51]. Methylation is determined by the ability of the specific primer to achieve amplification. The PCR reactions were carried out using Master Mix PCR buffer (6 μl), primers (50 pmol per reaction), and bisulfite-modified DNA (50 ng) in a final volume, 12 μl. Reactions were carried out at 95 °C for 5 min. Amplification was carried out in Mastercycler Gradient (Eppendorf, Hamburg, Germany) for 35 cycles, and then it was subjected to 4 min extension at 72 °C. Two sets of primers were simultaneously used to determine methylated or unmethylated CpG islands (Table 1). The controls for methylated and unmethylated DNA were human methylated and nonmethylated DNA sets, respectively (Zymo Research, CA, USA). Each PCR reaction product (10 μl) was directly electrophoresed into a 1.5% agarose gel, which was later stained with nucleic acid gel stain (Smobio, Hsinchu City, Taiwan) to allow DNA visualization under UV illumination. The methylation status of the gene was classified as methylated when amplification products were found in the reactions with M primers. The unmethylated status was reported when amplification products were detected in reaction with U primers. A hemimethylated status was reported when amplification products were found in reaction with both U and M primers. When a band is found in the unmethylated sample but not in the control, it was defined as unmethylated. However, when a methylated band was observed in a sample but was absent in the control, the sample was defined as methylated (or hemimethylated in case both unmethylated and methylated bands were found in the sample).
Table 1.
The primer sequences used in MSP analysis
| Genes | Primers sequencing (5′ → 3′) | Primer pair (bp) | |
|---|---|---|---|
| H19(M) | Forward | GGATTTTGTTTTGCGGAAATCAC | 165 |
| Reverse | ATCACGACTCAAACCTCACG | ||
| Igf2(M) | Forward | AGGATTTCGTCGGGAGGCAC | 158 |
| Reverse | CACAAAATCCCGCACCCCG | ||
| H19(U) | Forward | GGGATTTTGTTTTGTGGAAATTAT | 167 |
| Reverse | ATCACAACTCAAACCTCACA | ||
| Igf2(U) | Forward | GTTTAGGATTTTGTTGGGAGGTAT | 162 |
| Reverse | CACAAAATCCCACACCCCA |
M, methylated; U, unmethylated
Statistical analysis
Statistical Package for Social Sciences (SPSS) software (version 16.0, Chicago, IL, USA) was used for statistical analyses. Data was assessed for normality test by Q-Q plots. Descriptive statistics (mean ± 2SE) were also reported for all variables. The analysis of variance (ANOVA) and Kruskal-Wallis were used for comparing the four groups. The post hoc tests in ANOVA/KW were Tukey and Bonferroni. Pearson’s and Spearman correlations were used for parametric and nonparametric variables in the four groups. In all statistical analyses, a P < 0.05was set as significant. The pairwise comparisons in KW were reported by “P value/6.” Cross-tab analysis was carried out to determine the ROS cutoff points associated with an effect on methylation. Statistical software SIMCA 14.0 (Umetrics, Umeå, Sweden) was used for multivariate analysis of the NMR dataset. The metabolic fingerprinting was assessed according to the level of ROS (ROS ≥ 60 RLU/s/106 sperm: high; ROS < 20 RLU/s/106 sperm: low). The obtained matrix including 30 samples and 408 variables was imported into SIMCA 14 and mean centered before multivariate statistical analysis. To show clusters and outliers, principal components analysis (PCA) was performed. Then, orthogonal projections to latent structures discriminant analysis (OPLS-DA) was carried out to find discriminative variables (buckets) between groups 1 and 4 [52]. A supervised pattern recognition approach was used to construct predictive models and identify metabolite fingerprint differences. The quality of the OPLS-DA model was evaluated by R2X, R2Y, and Q2, which were calculated by the default leave-one-out (LOO) procedure. R2 and Q2 are the goodness of fit and goodness of prediction, respectively [53]. A further validation of the model was performed using a receiver-operating characteristic (ROC) analysis generated from 7-fold cross-validation, and the area under the ROC curve (AUC) value was calculated using SPSS 16.0 (SPSS, Inc., Chicago, IL) with 95% confidence interval.
Results
Methylation patterns of H19 and Igf2 genes and ROS levels
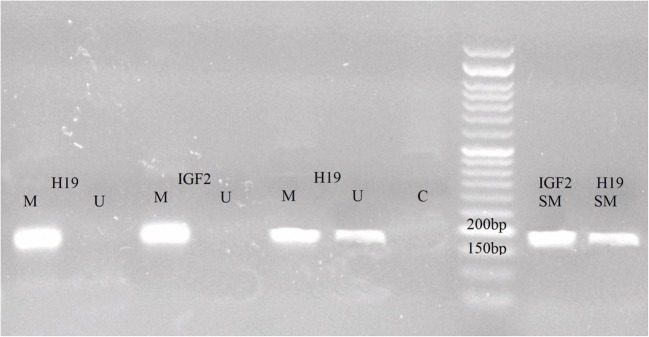
The methylation status in the promoter region of H19 and Igf2 genes was significantly different in specimens with high ROS levels than moderate, mild, and low counterparts (P < 0.005). All specimens had evidence of H19 and Igf2 methylation in the CpG islands of their promoters (Fig. 1). Significant methylation of Igf2, partial and complete, was noted in a large proportion of specimens of groups 2 and 4 (73.6 and 70.7%, respectively) (Supplementary Table 1). In contrast, Igf2 partial and complete methylation was noted in a small proportion of group 1 and 3 specimens (38.5 and 61.3%, respectively) (χ2 = 13.97, df = 3, P < 0.005) (Table 2). Pairwise comparisons were carried out using post hoc tests and the P values are reported in Table 2. H19 also exhibited evidence of significantly increased methylation status, both complete and partial methylation. A large percentage of specimens from groups 1 and 3 were found to have partial or complete H19 methylation (94.9 and 83.9%, respectively) (Supplementary Table 1). In contrast, a small percentage of specimens from groups 2 and 4 exhibited partial or complete H19 methylation (76.3 and 63.4%, respectively) (χ2 = 21.09, df = 3, P < 0.0001). Pairwise comparisons and the P values are shown in Table 2. There were significant changes of sperm H19-Igf2 methylation in specimens with ROS cutoff values of 60 RLU/s/106 sperm or greater. The specificity and sensitivity of ROS in the range of 40–100 RLU/s/106 sperm for H19 and Igf2 methylation are shown in Table 3.
Fig. 1.
Representative image for methylation-specific polymerase chain reaction (MSP) of H19 and Igf2. The primer sets used for amplification were designed as methylated (M) or unmethylated (U). Hemimethylated status was reported when amplification products were detected in reaction with both U and M. SM, standard methylation; C, control
Table 2.
Differential distribution of H19 and Igf2 methylation status according to ROS levels
| Parameters | H19 (%) | Igf2 (%) | |
|---|---|---|---|
| Partial and complete methylation | Group 1 (n = 39) | 94.9 | 38.5 |
| Group 2 (n = 38) | 76.3 | 73.6 | |
| Group 3 (n = 31) | 83.9 | 61.3 | |
| Group 4 (n = 43) | 63.4 | 70.7 | |
| P values | 1 vs 2 | 0.09 | 0.02* |
| 1 vs 3 | 0.16 | 0.14 | |
| 1 vs 4 | 0.000* | 0.000* | |
| 2 vs 3 | 0.14 | 0.09 | |
| 2 vs 4 | 0.000* | 0.09 | |
| 3 vs 4 | 0.000* | 0.000* | |
Group 1: ROS < 20, group 2: 20 ≤ ROS < 40, group 3: 40 ≤ ROS < 60, and group 4: ROS ≥ 60. The Kruskal-Wallis was used for comparison. Pairwise comparisons in KW were reported by “P value/6”
Table 3.
ROS cutoff values in H19 and Igf2 methylation status
| Groups | ROS cutoff RLU/s/106 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40 | 45 | 50 | 55 | 60 | 65 | 70 | 75 | 80 | 85 | 90 | 95 | 100 | ||
| H19 (n = 149) | Specificity (%) | 44.3 | 47 | 49 | 55 | 61.7 | 64.4 | 65.1 | 66.4 | 67.1 | 68.5 | 71.1 | 71.8 | 71.8 |
| Sensitivity (%) | 13.4 | 21.1 | 10.7 | 10.1 | 10.1 | 8.7 | 6.7 | 5.4 | 4.7 | 4.7 | 64 | 3.4 | 2.7 | |
| Kappa | 0.137 | 0.123 | 0.099 | 0.144 | 0.236 | 0.222 | 0.145 | 0.102 | 0.079 | 0.100 | 0.109 | 0.083 | 0.044 | |
| P value | 0.043* | 0.083 | 0.174 | 0.062 | 0.003* | 0.006* | 0.078 | 0.212 | 0.331 | 0.216 | 0.163 | 0.276 | 0.555 | |
| Igf2 (n = 149) | Specificity (%) | 40.3 | 43 | 45 | 51 | 57.7 | 60.4 | 61.1 | 62.4 | 62.4 | 63.8 | 66.4 | 66.4 | 66.4 |
| Sensitivity (%) | 16.1 | 14.8 | 13.4 | 12.8 | 12.8 | 11.4 | 9.4 | 8.1 | 6.7 | 6.7 | 6 | 4.7 | 4 | |
| Kappa | 0.114 | 0.109 | 0.094 | 0.155 | 0.260 | 0.260 | 0.199 | 0.172 | 0.119 | 0.143 | 0.165 | 0.108 | 0.078 | |
| P value | 0.124 | 0.156 | 0.230 | 0.055 | 0.002* | 0.001* | 0.013* | 0.030* | 0.126 | 0.062 | 0.023* | 0.124 | 0.254 | |
| H19/Igf2 (n = 149) | Specificity (%) | 40.3 | 43.0 | 45.0 | 51.0 | 57.7 | 60.4 | 61.1 | 62.4 | 62.4 | 63.8 | 66.4 | 66.4 | 66.4 |
| Sensitivity (%) | 16.1 | 14.8 | 13.4 | 12.8 | 12.8 | 11.4 | 9.4 | 8.1 | 6.7 | 6.7 | 6.0 | 4.7 | 4.0 | |
| Kappa | 0.114 | 0.109 | 0.094 | 0.155 | 0.260 | 0.260 | 0.199 | 0.172 | 0.119 | 0.143 | 0.165 | 0.108 | 0.078 | |
| P value | 0.124 | 0.156 | 0.230 | 0.055 | 0.002* | 0.001* | 0.013* | 0.030* | 0.126 | 0.062 | 0.023* | 0.124 | 0.254 | |
Metabolic fingerprinting and ROS levels
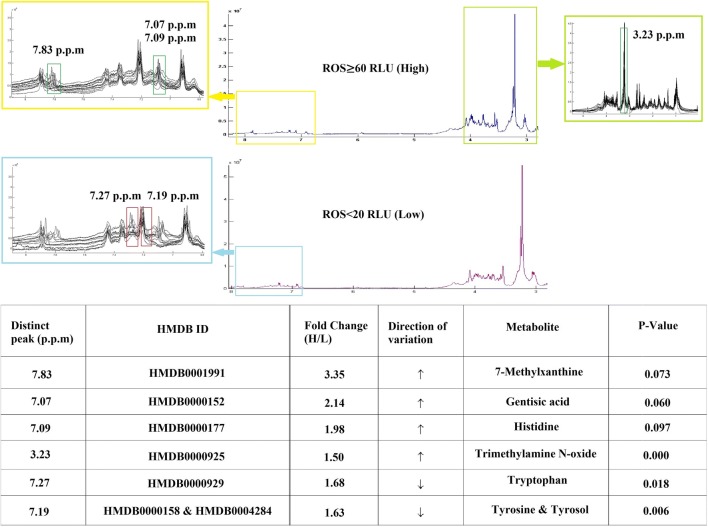
The metabolic fingerprinting was assessed according to the presence of high (ROS ≥ 60 RLU/s/106 sperm) and low (ROS < 20 RLU/s/106 sperm) ROS levels. The obtained matrix having 30 samples (15 samples randomly selected from groups 1 and 4, respectively) and 408 variables was imported into SIMCA 14. The multivariate analysis identified a significant 114 ppm, and then the fold change more than 1.5 was set for these; eventually about 6 ppm was detected. Seven different metabolites were identified according to the literature and online database. Metabolic fingerprinting by 1H-NMR revealed upregulation of trimethylamine N-oxide (P < 0.001) and downregulation of tryptophan (P < 0.05) as well as tyrosine/tyrosol (P < 0.01) in SP samples with high ROS levels (Fig. 2). These metabolites were deemed as major predictors of increased ROS level, verified using 15 samples of ROS ≥ 60 RLU/s/106 sperm and 15 SP controls. The distinct peak (ppm), fold change, and direction of variation of metabolites are summarized in Fig. 2.
Fig. 2.
Metabolic fingerprinting by 1H-NMR. The distinct peak (ppm) fold change and direction of variation of metabolites were identified by 1H-NMR of 15 SP samples with ROS ≥ 60 and 15 with ROS < 20 RLU/s/106 sperm. The metabolites have been exchanged in 3.23, 7.27, and 7.83 ppm with high (≥ 60 RLU/s/106 sperm) and low (< 20 RLU/s/106 sperm) ROS levels. This revealed upregulation of trimethylamine N-oxide and downregulation of tryptophan as well as tyrosine/tyrosol in SP samples with high ROS levels
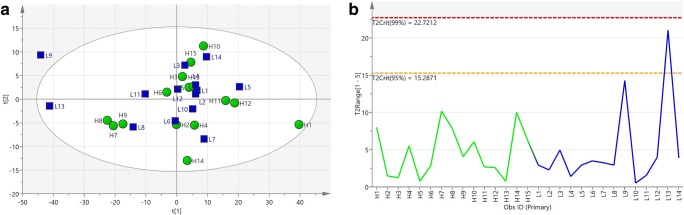
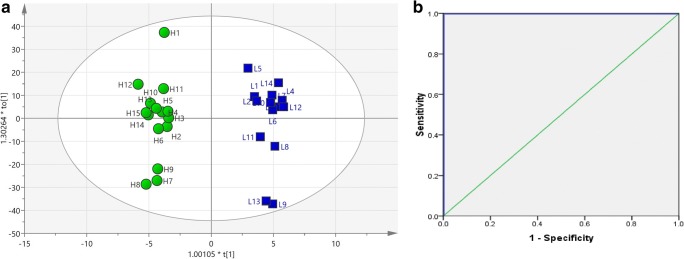
For an overview of the dataset obtained from 1H-NMR spectra of SP samples, pattern recognition, and outlier detection, a multivariate analysis of the two groups was first performed by PCA with R2X = 0.908, Q2 = 0.811. The score plot of PCA showed no separation between the two groups. All samples were in the Hotelling’s T2 99% confidence limit, and no outlier was detected within the PCA score plot (Fig. 3). In order to get a better separation, the supervised OPLS-DA classification model using one orthogonal component and three predictive components was established, and even clearer class discrimination was obtained. The goodness of fit values and predictive ability values (R2X, R2Y, and Q2), respectively, were 0.882, 0.971, and 0.884 (Fig. 4). The predictive model was validated with a P value of 1.69 × 10−6 and AUC = 1.
Fig. 3.
a Principal components analysis of seminal plasma NMR spectra from men being investigated for ROS (n = 20) with R2X = 0.908, Q2 = 0.811. b Hotelling’s T2 line plot of the SP samples. All the samples were in the Hotelling’s T2 99% confidence limit, and no outlier was detected
Fig. 4.
a Score plot of predictive model constructed with supervised orthogonal projections to latent structures discriminant analysis (OPLS-DA). The score plot of the model indicates adequate discrimination between groups (ROS < 20 RLU/s/106 sperm [low] and ROS ≥ 60 RLU/s/106 sperm [high]). The parameters of the model were R2X = 0.882, R2Y = 0.971, Q2 = 0.884. b ROC curve for evaluating the predictive model. Seminal plasma NMR spectra from men being investigated for ROS (n = 20). Area under the ROC curve (AUC) = 1
Semen characteristics and ROS levels
A significant reduction in total sperm motility (P < 0.05) and concentration (P < 0.001) was noted in the specimens with high ROS levels (Table 4), whereas higher CMI and sperm DFI were noted (Fig. 5 and Table 5) in both specimens with moderate (P < 0.05) and high ROS (P < 0.005). In contrast, TAC values were decreased in all specimens when compared to low ROS levels (P < 0.001) (Table 5). Although TAC values decreased in groups 2, 3, and 4, it is interesting to note that the intergroup comparison of TAC showed an increasing trend between group 2 vs 3 (P < 0.05) and group 3 vs 4 (P < 0.01) (Table 5).
Table 4.
Descriptive statistics and comparisons of semen parameters between groups
| Parameters | Mean ± 2SE | P values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 (n = 39) | Group 2 (n = 38) | Group 3 (n = 31) | Group 4 (n = 43) | 1 vs 2 | 1 vs 3 | 1 vs 4 | 2 vs 3 | 2 vs 4 | 3 vs 4 | |
| Age (years) | 35.00 ± 1.64 | 34.52 ± 1.34 | 33.09 ± 3.14 | 35.27 ± 1.62 | NS | NS | NS | NS | NS | NS |
| Volume (ml) | 3.11 ± 0.54 | 3.40 ± 0.60 | 2.91 ± 0.44 | 3.30 ± 0.40 | NS | NS | NS | NS | NS | NS |
| pH | 7.46 ± 0.02 | 7.46 ± 0.02 | 7.48 ± 0.02 | 7.48 ± 0.02 | NS | NS | NS | NS | NS | NS |
| Leukocyte | 14.03 ± 8.88 | 13.97 ± 8.46 | 12.67 ± 6.84 | 8.78 ± 2.80 | NS | NS | NS | NS | NS | NS |
| Motility (%) | 57.82 ± 2.72 | 55.60 ± 2.52 | 55.61 ± 3.00 | 52.13 ± 1.66 | NS | NS | 0.005 | NS | NS | NS |
| Viability (%) | 86.28 ± 1.58 | 87.47 ± 2.28 | 88.71 ± 1.80 | 87.44 ± 2.10 | NS | NS | NS | NS | NS | NS |
| N-morph (%) | 4.38 ± 0.14 | 4.34 ± 0.18 | 4.39 ± 0.40 | 4.07 ± 0.06 | NS | NS | NS | NS | NS | NS |
| SC (× 106/ml) | 50.44 ± 5.90 | 56.32 ± 6.82 | 52.65 ± 7.04 | 32.40 ± 3.14 | NS | NS | 0.0001 | NS | 0.0001 | 0.0001 |
Group 1: ROS < 20, group 2: 20 ≤ ROS < 40, group 3: 40 ≤ ROS < 60, and group 4: ROS ≥ 60. The analysis of variance (ANOVA) was used for comparing these four groups. Data are expressed as means ± SD. P ≤ 0.05, significant; P ≤ 0.001, highly significant
N-morph, normal morphology; SC, sperm concentration; SE, standard error; NS, not significant
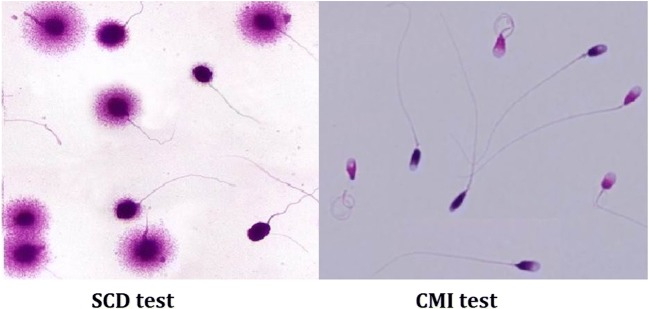
Fig. 5.
Representative image of aniline blue staining (CMI) and sperm chromatin dispersion (SCD) test
Table 5.
Sperm chromatin and seminal antioxidant status in the studied groups
| Parameters | DFI (%) | CMI (%) | TAC (μmol/l) | |
|---|---|---|---|---|
| Mean ± 2SE | Group 1 (n = 39) | 15.18 ± 2.64 | 14.51 ± 2.38 | 1431.04 ± 61.84 |
| Group 2 (n = 38) | 18.58 ± 3.30 | 18.12 ± 4.22 | 739.13 ± 60.82 | |
| Group 3 (n = 31) | 22.59 ± 3.40 | 20.97 ± 3.02 | 943.05 ± 128.98 | |
| Group 4 (n = 43) | 24.26 ± 4.44 | 25.83 ± 4.74 | 1161.07 ± 101.58 | |
| P values | 1 vs 2 | NS | NS | 0.000 |
| 1 vs 3 | 0.03 | 0.021 | 0.000 | |
| 1 vs 4 | 0.002 | 0.0001 | 0.000 | |
| 2 vs 3 | NS | NS | 0.01 | |
| 2 vs 4 | NS | NS | 0.000 | |
| 3 vs 4 | NS | NS | 0.006 | |
Group 1: ROS < 20, group 2: 20 ≤ ROS < 40, group 3: 40 ≤ ROS < 60, and group 4: ROS ≥ 60. The analysis of variance (ANOVA) was used for comparing these four groups. Data are expressed as means ± 2SE. P ≤ 0.05, significant; P ≤ 0.001, highly significant
DFI, DNA fragmentation index; CMI, chromatin maturity index; TAC, total antioxidant capacity; SE, standard error; NS, not significant
Correlation analyses
Pearson’s and Spearman correlations showed that ROS levels were positively correlated with Igf2 methylation (r = 0.19, P = 0.018), DFI (r = 0.40, P = 0.000), CMI (r = 0.39, P = 0.000), and trimethylamine N-oxide (r = 0.45, P = 0.015) and negatively correlated with H19 methylation (r = − 0.20, P = 0.013), tryptophan (r = − 0.45, p = 0.014), sperm motility (r = − 0.20, P = 0.012), sperm viability (r = − 0.23, P = 0.005), and sperm concentration (r = − 0.30, P = 0.000) (Table 6 and Supplementary Table 2).
Table 6.
Correlations between sperm DFI, CMI, TAC, H19/Igf2 methylation level, ROS, metabolites, and conventional semen criteria
| Motility | SC r = 0.49** P = 0.000 |
DFI r = − 0.47* P = 0.032 |
Igf2 r = 0.16* P = 0.048 |
TMN r = − 0.46* P = 0.013 |
Tyr r = 0.41* P = 0.029 |
ROS r = − 0.20* P = 0.012 |
|
| Viability | DFI r = − 0.16* P = 0.043 |
CMI r = − 0.22** P = 0.007 |
ROS r = − 0.23** P = 0.005 |
||||
| N-morph | SC r = 0.17* P = 0.036 |
GA r = − 0.42* P = 0.023 |
|||||
| SC | Motility r = 0.49** P = 0.000 |
N-morph r = 0.17* P = 0.036 |
DFI r = − 0.19* P = 0.018 |
CMI r = − 0.21** P = 0.008 |
TMN r = − 0.61** P = 0.001 |
ROS r = − 0.30** P = 0.000 |
|
| DFI | Motility r = − 0.17* P = 0.032 |
Viability r = − 0.16* P = 0.043 |
SC r = − 0.19* P = 0.018 |
CMI r = 0.67** P = 0.000 |
H19 r = − 0.17* P = 0.037 |
His r = 0.40* P = 0.032 |
ROS r = 0.40** P = 0.000 |
| CMI | Viability r = − 0.22** P = 0.007 |
SC r = − 0.21** P = 0.008 |
DFI r = 0.67** P = 0.000 |
H19 r = − 0.20* P = 0.014 |
Igf2 r = 0.20* P = 0.015 |
ROS r = 0.39** P = 0.000 |
|
| TAC | H19 r = 0.16* P = 0.045 |
TMN r = − 0.48** P = 0.009 |
|||||
| H19 | DFI r = − 0.17* P = 0.037 |
CMI r = − 0.20* P = 0.014 |
TAC r = − 0.16* P = 0.045 |
Igf2 r = − 0.83** P = 0.000 |
Tyr r = 0.40* P = 0.038 |
ROS r = − 0.20* P = 0.013 |
|
| Igf2 | Motility r = 0.16* P = 0.048 |
CMI r = 0.20* P = 0.015 |
H19 r = − 0.83** P = 0.000 |
Trp r = 0.42* P = 0.027 |
ROS r = 0.19* P = 0.018 |
||
| 7-MX (7.83 ppm) | GA) r = 0.905** P = 0.000 |
His r = 0.889** P = 0.000 |
Trp r = 0.61** P = 0.000 |
||||
| GA (7.07 ppm) | N-morph r = − 0.42* P = 0.023 |
7-MX r = 0.905** P = 0.000 |
His r = 0.76** P = 0.000 |
Trp r = 0.46* P = 0.013 |
|||
| His (7.09 ppm) | DFI r = 0.40* P = 0.032 |
7-MX r = 0.889** P = 0.000 |
GA r = 0.76** P = 0.000 |
Trp r = 0.53** P = 0.003 |
|||
| TMN (3.23 ppm) | Motility r = − 0.46* P = 0.013 |
SC r = − 0.61** P = 0.001 |
TAC r = − 0.48** P = 0.009 |
Tyr r = − 0.45* P = 0.016 |
ROS r = 0.45* P = 0.015 |
||
| Tyr (7.27 ppm) | Motility r = 0.41* P = 0.029 |
H19 r = 0.40* P = 0.038 |
TMN r = − 0.45* P = 0.016 |
||||
| Trp (7.19 ppm) | Igf2 r = 0.42* P = 0.027 |
7-MX r = 0.61** P = 0.000 |
GA r = 0.46* P = 0.013 |
His r = 0.53** P = 0.003 |
ROS r = − 0.45* P = 0.014 |
Discussion
Our findings indicate a positive association between ROS levels and alterations in H19-Igf2 methylation as well as metabolite concentration, with an overall detrimental effect on semen quality. The methylation status in the promoter region of H19 and Igf2 genes was markedly different in specimens with high ROS levels than moderate/mild/low counterparts. Metabolic fingerprinting by 1H-NMR revealed upregulation of trimethylamine N-oxide and downregulation of tryptophan as well as tyrosine/tyrosol in SP samples with high ROS levels. Furthermore, reduction in sperm quality was noted in specimens with high ROS levels, including decreased sperm motility, sperm count, and sperm DNA integrity. Based on our novel data, we speculate that deterioration of semen quality and sperm DNA integrity in specimens with moderate/high ROS levels is mediated by alterations in the methylation status of H19-Igf2 genes and SP metabolite concentrations. However, we have not evaluated the potential cause(s) of ROS generation, and data on possible influencing factors such as food consumption trends, environmental exposure, and body mass index (BMI) were not recorded.
In this study, bisulfite MSP analysis of 151 sperm DNA showed that methylation errors were specific for imprinted genes at higher seminal ROS levels (> 60 RLU/s/106 sperm) (Fig. 1). Our data indicates that the rate of methylation in differentially methylated region (DMR) is reduced in the presence of elevated seminal ROS level, which results in the expression of H19 and inhibition of Igf2 following the binding of CTCF at its promoter. Insulin-like growth factor 2 expressed by Igf2 gene induces growth and division of cells in different tissues, while H19 has a role in the negative regulation (or limiting) of cell proliferation [54, 55]. We speculate that the suppressed expression of Igf2 and induced expression of H19 at high levels of ROS can inhibit spermiogenesis resulting in semen quality deterioration.
The H19-Igf2 locus has been extensively studied over the past years [56–58]. These are located on human chromosome 11p15.5 and expression of H19 is associated with silencing of Igf2. This locus contains the maternally expressed H19 gene and paternally expressed Igf2 gene whose expressions are closely linked. Imprinting at the coding regions of Igf2 and H19 are kept apart from one another via a DMR, which is methylated on the paternal chromosome and remains unmethylated on the maternal chromosome. Expression of Igf2 and H19 genes is regulated by a single 3′-distal enhancer. DMR methylation on the paternal chromosome inhibits CTCF binding to DMR, activating the Igf2 promoter. Hence, Igf2 mRNA is transcribed from the paternal chromosome and H19 is silenced. By contrast, as the maternal chromosome is devoid of methylation on DMR, CTCF binds with it preventing the activation of the Igf2 and transcription of H19 mRNA from the maternal chromosome [59, 60]. Recent investigations corroborate our findings by showing that altered DNA methylation in H19-Igf2 locus of human-imprinted genes was associated with semen quality, disrupted spermatogenesis, and different causes of male infertility, including severe oligozoospermia, Beckwith-Wiedemann syndrome, and Silver-Russell syndrome [13, 14, 16, 37, 61–64].
Our results suggest a link between ROS-induced alterations in seminal metabolites and altered semen quality. Untargeted metabolic profiling by 1H-NMR revealed the upregulation of 7-methylxanthine, gentisic acid, histidine, and trimethylamine N-oxide and the downregulation of tryptophan, tyrosine, and tyrosol in the SP of the group with ROS level ≥ 60 RLU/s/106 sperm. Since these metabolites play significant roles in sperm maturation [65, 66], an imbalance in their SP concentrations might deteriorate semen quality. Metabolites like 7-methylxanthine, gentisic acid, and trimethylamine N-oxide have an essential role in reducing OS and apoptosis and in regulating redox homeostasis [67]. Histidine has antioxidant and anti-inflammatory properties owing to its ROS scavenging action [68, 69]. In our study, elevated concentrations of these metabolites, particularly trimethylamine N-oxide, in specimens with high ROS levels might reflect the activation of antioxidant defense mechanism in the male reproductive tissues, a fact corroborated by the concomitant increase in TAC values. SP levels of tryptophan, tyrosine, and tyrosol were decreased in specimens with high ROS, suggesting an imbalance of antioxidant regulation with detrimental effects on semen quality. Notably, antioxidant levels of tryptophan have been associated with sperm motility, viability, and morphology [70, 71]. Low SP levels of tyrosine, the precursor of thyroid hormones, were found to be associated with sperm maturation, acrosome reaction, and infertility [72–75]. Also, tyrosol has been associated with normal sperm physiology [76, 77] and DNA integrity [78]. In general, our findings are in line with several studies that demonstrated the potential role of SP constituents in male infertility [65, 66, 79–81].
Although many studies have been done on the level of ROS in SP, a definite cutoff value was not determined [29, 82]. In this study, cross-tab analysis showed that ROS cutoff values of 60 (specificity 57.7%, sensitivity 12.8%, P < 0.005), 65 (specificity 60.4%, sensitivity 11.4%, P < 0.005), 70 (specificity 61.1%, sensitivity 9.4%, P < 0.05), 75 (specificity 62.4%, sensitivity 8.1%, P < 0.05), and 90 (specificity 66.4%, sensitivity 6.0%, P < 0.05) RLU/s/106 sperm might be used for categorizing semen specimens based on H19 and Igf2 methylation levels. Although the methylation changes were significantly observed in ROS 60, 65, 70, 75, and 90 RLU/s/106 sperm, lower ROS (60 RLU/s/106 sperm) has better potential for future application in ART. The specificity of 57.7% and the sensitivity of 12.8% for H19-Igf2 methylation levels with a ROS cutoff value of 60 RLU/s/106 sperm indicate the potential of ROS measurement to differentiate normal and abnormal sperm.
Although we studied a population of normozoospermic men, sperm concentration and motility were lower in subjects with ROS ≥ 60 RLU/s/106 sperm than those with low ROS. DNA fragmentation and CMI values were increased in both subjects with moderate (ROS > 40 RLU/s/106 sperm) and high ROS levels (ROS ≥ 60 RLU/s/106 sperm(. Seminal TAC has found to be decreased at ROS levels up to 60 RLU/s/106 sperm. However, TAC levels increased in specimens with ROS ≥ 40 RLU/s/106 sperm, thus suggesting activation of the antioxidant defense mechanism. Overall, the increase in ROS levels was associated with a marked reduction in semen parameters and sperm DNA integrity.
Our study suggests a link between ROS-induced H19-Igf2 methylation and SP metabolites with semen quality. Since H19 and Igf2 are concerned with restriction in cell proliferation and maturation and cellular growth, respectively, our novel data suggest that H19 gene activation and Igf2 gene inhibition by high ROS levels result in impaired spermatogenesis and sperm maturation. The observed ROS-induced changes in concentrations of seminal metabolites and their associations with semen parameters also suggest that metabolites such as trimethylamine N-oxide, tryptophan, tyrosine, and tyrosol are vital for sperm health and maturation. We hypothesize that deterioration of semen quality and sperm DNA integrity in specimens with moderate/high ROS levels is mediated by alterations in the methylation status of H19-Igf2 genes and SP metabolite concentrations. Future studies should focus on the possible beneficiary effects of antioxidant therapy in ameliorating the detrimental molecular effects of ROS on gene methylation and semen quality. Since the use of epigenetic analysis as a marker of fertility is a rapidly expanding area of ROS research, the simultaneous assessment of noncoding and coding RNAs to determine the ROS-induced epigenetic damages warrants further research.
Electronic supplementary material
(DOCX 28 kb)
(DOCX 30 kb)
Acknowledgments
The authors would like to thank all patients and their family members who voluntarily participated in this study. In addition, we thank the director of the Phytochemistry and the Medicinal Chemistry Research Center at Shahid Beheshti University of Medicinal Sciences, SBMU (Iran, Tehran) and the Reproductive Center of Cleveland Clinic (Cleveland, USA) for their assistance.
Author’s roles
Mahsa Darbandi (data interpretation, study design, execution, analysis, manuscript drafting and revision), Sara Darbandi (data interpretation, study design, execution, analysis, manuscript drafting and revision), Ashok Agarwal (data interpretation, study design, analysis, manuscript review, revision and critical discussion), Saradha Baskaran (data interpretation, manuscript preparation, review and revision), Sulagna Dutta (data interpretation, manuscript preparation, review and revision), Pallav Sengupta (data interpretation, manuscript preparation, review and revision), Hamid Reza Khorram Khorshid (data interpretation, study design, manuscript drafting), Sandro Esteves (data interpretation, manuscript review and revision), Kambiz Gilany (acquisition of data, analysis), Mehdi Hedayati (data interpretation, study design, execution), Fatemeh Nobakht (acquisition of data, execution, analysis), Mohammad Mehdi Akhondi (data interpretation, study design), Niknam Lakpour (acquisition of data, study design, execution), and Mohammad Reza Sadeghi (data interpretation, study design, manuscript drafting and critical discussion). All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants
All applicable international, national, and/or institutional guidelines for the use of human tissues were followed. All procedures performed in studies involving human subjects were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
The authors consider that the first two authors should be regarded as joint first authors.
References
- 1.Jarow JP, Sharlip ID, Belker AM, Lipshultz LI, Sigman M, Thomas AJ, Schlegel PN, Howards SS, Nehra A, Damewood MD, Overstreet JW, Sadovsky R, Male Infertility Best Practice Policy Committee of the American Urological Association Inc. Best practice policies for male infertility. J Urol. 2002;167:2138–2144. doi: 10.1016/S0022-5347(05)65109-9. [DOI] [PubMed] [Google Scholar]
- 2.Darbandi S, Darbandi M. Lifestyle modifications on further reproductive problems. Cresco J Reprod Sci. 2016;1:1–2. [Google Scholar]
- 3.Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. 2012;38:576–594. doi: 10.1590/S1677-55382012000500002. [DOI] [PubMed] [Google Scholar]
- 4.Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res-Rev Mutat. 2011;727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Esteves SC. A clinical appraisal of the genetic basis in unexplained male infertility. J Hum Reprod Sci. 2013;6:176–182. doi: 10.4103/0974-1208.121419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovac JR, Pastuszak AW, Lamb DJ. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril. 2013;99:998–1007. doi: 10.1016/j.fertnstert.2013.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. New York: Springer; 2014. [DOI] [PubMed] [Google Scholar]
- 8.Li J-Y, Lees-Murdock DJ, Xu G-L, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Gomes MVM, Pelosi GG. Epigenetic vulnerability and the environmental influence on health. Exp Biol Med. 2013;238:859–865. doi: 10.1177/1535370213490630. [DOI] [PubMed] [Google Scholar]
- 10.Biermann K, Steger K. Epigenetics in male germ cells. J Androl. 2007;28:466–480. doi: 10.2164/jandrol.106.002048. [DOI] [PubMed] [Google Scholar]
- 11.Klenova EM, Morse HC, Ohlsson R, Lobanenkov VV. The novel BORIS+CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/S1044-579X(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 12.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Dev. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 13.Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 14.Olszewska M, Barciszewska MZ, Fraczek M, Huleyuk N, Chernykh VB, Zastavna D, et al. Global methylation status of sperm DNA in carriers of chromosome structural aberrations. Asian J Androl. 2016;19:117–124. doi: 10.4103/1008-682X.168684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 16.Boissonnas CC, El Abdalaoui H, Haelewyn V, Fauque P, Dupont JM, Gut I, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunes S, Agarwal A, Henkel R, Mahmutoglu A, Sharma R, Esteves S, et al. Association between promoter methylation of MLH1 and MSH2 and reactive oxygen species in oligozoospermic men—a pilot study. Andrologia. 2017;50(3):1–6. doi: 10.1111/and.12903. [DOI] [PubMed] [Google Scholar]
- 18.Juyena NS, Stelletta C. Seminal plasma: an essential attribute to spermatozoa. J Androl. 2012;33:536–551. doi: 10.2164/jandrol.110.012583. [DOI] [PubMed] [Google Scholar]
- 19.Minai-Tehrani A, Jafarzadeh N, Gilany K. Metabolomics: a state-of-the-art technology for better understanding of male infertility. Andrologia. 2016;48(6):609–616. doi: 10.1111/and.12496. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0—the human metabolome database in 2013. Biochem Biophys Res Commun. 2012:D801–7. [DOI] [PMC free article] [PubMed]
- 21.Deepinder F, Chowdary HT, Agarwal A. Role of metabolomic analysis of biomarkers in the management of male infertility. Expert Rev Mol Diagn. 2007;7:351–358. doi: 10.1586/14737159.7.4.351. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, Suzuki R, Suzuki F, Hayashi C, Utsunomiya T, Yaegashi N, Arima T. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–1591. doi: 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagami M, Nagai T, Fukami M, Yamazawa K, Ogata T. Silver-Russell syndrome in a girl born after in vitro fertilization: partial hypermethylation at the differentially methylated region of PEG1/MEST. J Assist Reprod Gen. 2007;24:131–136. doi: 10.1007/s10815-006-9096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrell DT, Aston KI, Oliva R, Emery B, De Jonge C. The “omics” of human male infertility: integrating big data in a systems biology approach. Cell Tissue Res. 2016;363:295–312. doi: 10.1007/s00441-015-2320-7. [DOI] [PubMed] [Google Scholar]
- 25.Bhusari SS, Dobosy JR, Fu V, Almassi N, Oberley T, Jarrard DF. Superoxide dismutase 1 knockdown induces oxidative stress and DNA methylation loss in the prostate. Epigenet. 2010;5:402–409. doi: 10.4161/epi.5.5.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasperczyk A, Dobrakowski M, Czuba ZP, Horak S, Kasperczyk S. Environmental exposure to lead induces oxidative stress and modulates the function of the antioxidant defense system and the immune system in the semen of males with normal semen profile. Toxicol Appl Pharmacol. 2015;284:339–344. doi: 10.1016/j.taap.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Moazamian R, Polhemus A, Connaughton H, Fraser B, Whiting S, Gharagozloo P, et al. Oxidative stress and human spermatozoa: diagnostic and functional significance of aldehydes generated as a result of lipid peroxidation. MHR: Basic Sci Reprod Med. 2015;21:502–515. doi: 10.1093/molehr/gav014. [DOI] [PubMed] [Google Scholar]
- 28.WHO . WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010. [Google Scholar]
- 29.Homa ST, Vessey W, Perez-Miranda A, Riyait T, Agarwal A. Reactive oxygen species (ROS) in human semen: determination of a reference range. J Assist Reprod Genet. 2015;32:757–764. doi: 10.1007/s10815-015-0454-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shamsi MB, Venkatesh S, Pathak D, Deka D, Dada R. Sperm DNA damage & oxidative stress in recurrent spontaneous abortion (RSA) Indian J Med Res. 2011;133:550–551. [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, Zheng T, Huang J, Mo J, Zhou H, Liu M, Gao X, Yu B. Association of semen cytokines with reactive oxygen species and histone transition abnormalities. J Assist Reprod Gen. 2016;33:1239–1246. doi: 10.1007/s10815-016-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal A, Sharma R, Gupta S, Harlev A, Ahmad G, du Plessis SS, et al. Oxidative stress in human reproduction: shedding light on a complicated phenomenon. Cham: Springer International Publishing; 2017. [Google Scholar]
- 33.Mahfouz R, Sharma R, Sharma D, Sabanegh E, Agarwal A. Diagnostic value of the total antioxidant capacity (TAC) in human seminal plasma. Fertil Steril. 2009;91:805–811. doi: 10.1016/j.fertnstert.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Fernández JL, Muriel L, Goyanes V, Segrelles E, Gosálvez J, Enciso M, et al. Halosperm® is an easy, available, and cost-effective alternative for determining sperm DNA fragmentation. Fertil Steril. 2005;84:860. doi: 10.1016/j.fertnstert.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Esteves SC, Sharma RK, Gosálvez J, Agarwal A. A translational medicine appraisal of specialized andrology testing in unexplained male infertility. Int Urol Nephrol. 2014;46:1037–1052. doi: 10.1007/s11255-014-0715-0. [DOI] [PubMed] [Google Scholar]
- 36.Barnard L, Aston KI. Spermatogenesis: methods and protocols. New York: Humana; 2012. [Google Scholar]
- 37.Zini A, Agarwal A. Sperm chromatin: biological and clinical applications in male infertility and assisted reproduction. New York: Springer; 2011. [Google Scholar]
- 38.Paiva C, Amaral A, Rodriguez M, Canyellas N, Correig X, Ballescà J, et al. Identification of endogenous metabolites in human sperm cells using proton nuclear magnetic resonance (1H-NMR) spectroscopy and gas chromatography-mass spectrometry (GC-MS) Andrology. 2015;3:496–505. doi: 10.1111/andr.12027. [DOI] [PubMed] [Google Scholar]
- 39.Brown FF, Campbell ID, Kuchel PW. Human erythrocyte metabolism studies by 1H spin echo NMR. FEBS Lett. 1977;82:12–16. doi: 10.1016/0014-5793(77)80875-2. [DOI] [PubMed] [Google Scholar]
- 40.Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Commun. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 41.Niederberger C. The “omics” of human male infertility: integrating big data in a systems biology approach. J Urol. 2016;196:295–312. doi: 10.1016/j.juro.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Kostidis S, Addie RD, Morreau H, Mayboroda OA, Giera M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: a tutorial. Anal Chim Acta. 2017;980:1–24. doi: 10.1016/j.aca.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Pechlivanis A, Kostidis S, Saraslanidis P, Petridou A, Tsalis G, Mougios V, Gika HG, Mikros E, Theodoridis GA. 1H NMR-based metabonomic investigation of the effect of two different exercise sessions on the metabolic fingerprint of human urine. J Proteome Res. 2010;9:6405–6416. doi: 10.1021/pr100684t. [DOI] [PubMed] [Google Scholar]
- 44.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 45.Höcker M, Rosenberg I, Xavier R, Henihan RJ, Wiedenmann B, Rosewicz S, Podolsky DK, Wang TC. Oxidative stress activates the human histidine decarboxylase promoter in AGS gastric cancer cells. J Biol Chem. 1998;273:23046–23054. doi: 10.1074/jbc.273.36.23046. [DOI] [PubMed] [Google Scholar]
- 46.Ravanbakhsh S, Liu P, Bjordahl TC, Mandal R, Grant JR, Wilson M, et al. Seminal plasma metabolomics approach for the diagnosis of unexplained male infertility. PLoS One. 2015;10:1–13. [Google Scholar]
- 47.Alonso A, Marsal S, Julià A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:1–20. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darbandi M, Darbandi S, Khorshid HRK, Akhondi MM, Mokarram P, Sadeghi MR. A simple, rapid and economic manual method for human sperm DNA extraction in genetic and epigenetic studies. Middle East Fertil Soc J, 2017.
- 49.Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Epigenet Protoc. 2011;79:11–21. doi: 10.1007/978-1-61779-316-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi J-M, Chan TA, van Neste L, van Criekinge W, van den Bosch S, van Engeland M, Ting AH, Jair K, Yu W, Toyota M, Imai K, Ahuja N, Herman JG, Baylin SB. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:1–9. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metabolomics. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Anal Chem. 2008;80:7562–7570. doi: 10.1021/ac800954c. [DOI] [PubMed] [Google Scholar]
- 54.Moore GE, Ishida M, Demetriou C, Al-Olabi L, Leon LJ, Thomas AC, et al. The role and interaction of imprinted genes in human fetal growth. Philos Trans R Soc B. 2015;370:1–12. doi: 10.1098/rstb.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nordin M, Bergman D, Halje M, Engström W, Ward A. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014;47:189–199. doi: 10.1111/cpr.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:1–17. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanduri C. Long noncoding RNAs: lessons from genomic imprinting. BBA-Gene Regul Mech. 1859;2016:102–111. doi: 10.1016/j.bbagrm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Kingsley SL, Deyssenroth MA, Kelsey KT, Awad YA, Kloog I, Schwartz JD, Lambertini L, Chen J, Marsit CJ, Wellenius GA. Maternal residential air pollution and placental imprinted gene expression. Environ Int. 2017;108:204–211. doi: 10.1016/j.envint.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabory A, Ripoche M-A, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 60.Arney KL. H19 and Igf2—enhancing the confusion? Trends Genet. 2003;19:17–23. doi: 10.1016/S0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 61.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 64.Stouder C, Somm E, Paoloni-Giacobino A. Prenatal exposure to ethanol: a specific effect on the H19 gene in sperm. Reprod Toxicol. 2011;31:507–512. doi: 10.1016/j.reprotox.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Maher AD, Patki P, Lindon JC, Want EJ, Holmes E, Craggs M, Nicholson JK. Seminal oligouridinosis: low uridine secretion as a biomarker for infertility in spinal neurotrauma. Clin Chem. 2008;54:2063–2066. doi: 10.1373/clinchem.2008.112219. [DOI] [PubMed] [Google Scholar]
- 66.Gupta A, Mahdi AA, Ahmad MK, Shukla KK, Jaiswer SP, Shankhwar SN. 1H NMR spectroscopic studies on human seminal plasma: a probative discriminant function analysis classification model. J Pharm Biomed Anal. 2011;54:106–113. doi: 10.1016/j.jpba.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 67.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Son DO, Satsu H, Shimizu M. Histidine inhibits oxidative stress- and TNF-α-induced interleukin-8 secretion in intestinal epithelial cells. FEBS Lett. 2005;579:4671–4677. doi: 10.1016/j.febslet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 69.Peterson JW, Boldogh I, Popov VL, Saini SS, Chopra AK. Anti-inflammatory and antisecretory potential of histidine in Salmonella-challenged mouse small intestine. Lab Investig. 1998;78:523–534. [PubMed] [Google Scholar]
- 70.Jiménez-Trejo F, Tapia-Rodríguez M, Cerbón M, Kuhn DM, Manjarrez-Gutiérrez G, Mendoza-Rodríguez CA, Picazo O. Evidence of 5-HT components in human sperm: implications for protein tyrosine phosphorylation and the physiology of motility. Reproduction. 2012;144:677–685. doi: 10.1530/REP-12-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Sheshtawy RI, El-Nattat WS, Sabra HA. Effect of addition of catalase with or without L-tryptophan on cryopreservation of bull extended semen and conception rate. Glob Vet. 2013;11:280–284. [Google Scholar]
- 72.Naz RK, Rajesh PB. Role of tyrosine phosphorylation in sperm capacitation/acrosome reaction. Reprod Biol Endocrin. 2004;2:1–12. doi: 10.1186/1477-7827-2-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pukazhenthi BS, Long JA, Wildt DE, Ottinger MA, Armstrong DL, Howard J. Regulation of sperm function by protein tyrosine phosphorylation in diverse wild felid species. J Androl. 1998;19:675–685. [PubMed] [Google Scholar]
- 74.Sati L, Cayli S, Delpiano E, Sakkas D, Huszar G. The pattern of tyrosine phosphorylation in human sperm in response to binding to zona pellucida or hyaluronic acid. Reprod Sci. 2014;21:573–581. doi: 10.1177/1933719113504467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta GS. Proteomics of spermatogenesis. New York: Springer; 2005. [Google Scholar]
- 76.Banihani SA. Semen quality as affected by olive oil. Int J Food Prop. 2017;20:1901–1906. [Google Scholar]
- 77.Gomes VPM, Torres C, Rodriguez-Borges JE, Paiva-Martins F. A convenient synthesis of hydroxytyrosol monosulfate metabolites. J Agric Food Chem. 2015;63:9565–9571. doi: 10.1021/acs.jafc.5b04307. [DOI] [PubMed] [Google Scholar]
- 78.Negri L, Benaglia R, Monti E, Morenghi E, Pizzocaro A, Setti PEL. Effect of superoxide dismutase supplementation on sperm DNA fragmentation. Arch Ital Urol Androl. 2017;89:212–218. doi: 10.4081/aiua.2017.3.212. [DOI] [PubMed] [Google Scholar]
- 79.Bieniek JM, Drabovich AP, Lo KC. Seminal biomarkers for the evaluation of male infertility. Asian J Androl. 2016;18:426–433. doi: 10.4103/1008-682X.175781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiao S, Wu W, Chen M, Tang Q, Xia Y, Jia W, et al. Seminal plasma metabolomics approach for the diagnosis of unexplained male infertility. PLoS One. 2017;12:1–13. doi: 10.1371/journal.pone.0181115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X, Hu C, Dai J, Chen L. Metabolomics analysis of seminal plasma in infertile males with kidney-yang deficiency: a preliminary study. Evid-Based Compl Alt. 2015;2015:1–8. doi: 10.1155/2015/892930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agarwal A, Ahmad G, Sharma R. Reference values of reactive oxygen species in seminal ejaculates using chemiluminescence assay. J Assist Reprod Gene. 2015;32:1721–1729. doi: 10.1007/s10815-015-0584-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 28 kb)
(DOCX 30 kb)